Introduction
Colon cancer is one of the most common (1). In recent years, improvements in early
screening techniques and treatment strategies has largely decreased
the morbidity and mortality of colon cancer in the United States
(1). However, these regions do not
include developing countries such as China. Furthermore, the
incidence is increased in younger patients (2). Previous studies have focussed on early
diagnosis and clinical treatment of colon cancer (3,4),
however, the mechanism of action remains unknown. Therefore,
investigating the mechanism of colon cancer has important practical
significance.
microRNAs (miRNAs) are a class of small non-coding
RNAs that are 19–25 nucleotides long (5). They regulate gene expression by binding
to their target mRNAs, resulting in the degradation of mRNA or
repression of mRNA translation (6).
Previous studies have reported that miRNAs are involved in the
development of cancer (7) and serve
a role in the development of chemotherapy resistance in different
tumours (8,9). Therefore, targeting miRNAs may be
developed as a novel method of treating cancer and improve the
responses of patients to chemotherapy.
miR-15 is a miRNA that has been extensively studied.
miR-15 is aberrantly regulated in various types of cancer and is
associated with cell proliferation, angiogenesis and metastasis
(10). However, the role of miR-15
in cancer remains unclear. Some studies have suggested that miR-15
functions as an oncogene, whereas others regard it as a tumour
suppressor (11,12). The effects of miR-15 on chemotherapy
are also inconsistent (13,14) and to the best of our knowledge, there
have been no studies investigating the potential effect of miR-15
on colon cancer chemotherapy.
In the current study, miR-15 was detected in
matching colon cancer and para-carcinoma tissues, the effect of
miR-15 on colon cancer chemotherapy was analysed and its associated
mechanism of action was investigated. These results may improve
understanding of the process of chemotherapy resistance and the
search for potential intervention targets.
Materials and methods
Patients
In the present study, miR-15 was detected in 62
paired colon cancer and para-cancerous colon tissues collected from
patients who underwent surgical resection to treat colon cancer at
Linyi People's Hospital (Shandong, China)between May 2011 and
December 2014. The mean range of the enrolled patients was
50.8±16.4 years old (range, 34–65 years) and the male-female ratio
was 42:20. The final diagnosis for each patient was confirmed by
two double-blinded pathologists. Following resection, the tissues
were rapidly frozen and stored in liquid nitrogen. All procedures
performed in the present study that involved human participants
were in accordance with the ethical standards of the institutional
and/or national research committee, and with the 1964 Helsinki
Declaration and its later amendments or comparable ethical
standards. The current study was approved by the Ethics Committee
of Linyi People's Hospital (Linyi, China) and informed consent was
obtained from all participants.
Cell culture and transfection
Human colorectal carcinoma HCT116 cells (Cell Center
of Institute of Basic Medical Science, Chinese Academy of Medical
Science, Shanghai, China) were cultured in Dulbecco's Modified
Eagle's Medium (Hyclone; GE Healthcare, Logan, UT, USA)
supplemented with 10% fetal bovine serum (GIBCO; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), 100 U/ml penicillin and 100
µg/ml streptomycin at 37°C in a humidified atmosphere containing 5%
CO2 and 95% air.
HCT116 cells were plated in 6-well plates
(1×105/per well) and the chemosynthetic miR-15 mimics
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) were
transfected into cells using Lipofectamine™ 2000
(Invitrogen; Thermo Fisher Scientific Inc.) following the
manufacturer's protocol. The scrambled sequence was used as
negative control (Invitrogen; Thermo Fisher Scientific Inc.). The
concentration of miR-15/NC was 50 nmol/l in each well.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from frozen tissues or cells
using TRIzol® (Invitrogen; Thermo Fisher Scientific
Inc.) and reverse-transcribed into cDNA. In brief, 5× RT buffer (4
µl), primer (0.1 µg), 10 mM dNTPs (1 µl), RNA sample (1 µg),
transcriptase (1 µl), and RNAase-free water (20 µl) were mixed. The
mixture was incubated at 25°C for 5 min, 42°C for 30 min, 85°C for
5 min and then stored at 4°C. All above reagents were supplied by
Takara Biotechnology Co. Ltd, (Dalian, China) SYBR Green PCR
Mastermix (Takara Biotechnology Co. Ltd) was used to perform qPCR.
PCR conditions were as follows: denaturation at 96°C for 3 min, 35
cycles of amplification at 94°C for 10 sec/cycle and then 58°C for
30 sec. miR-15 expression was normalized to that of U6, whereas
levels of nuclear factor-κB (NF-κB), B cell lymphoma-2 (BCL-2) and
B cell lymphoma-extra large (BCL-XL) mRNA were normalized to that
of GAPDH. Expression level was analyzed using the 2−∆∆Cq
method (15). The primers (Takara
Biotechnology Co. Ltd) for PCR are presented in Tables I and II.
 | Table I.Primers for reverse transcription. |
Table I.
Primers for reverse transcription.
| Primer names | Primer sequence
(5′-3′) |
|---|
| U6 |
AAAATATGGAACGCTTCACGAATTTG |
| miR-15 |
GTCGTATCCAGTGCAGGGTCCGAGGT |
|
|
ATTCGCACTGGATACGACCACAAA |
 | Table II.Primers for qPCR. |
Table II.
Primers for qPCR.
| Primer names | Primer sequence
(5′-3′) |
|---|
| miR-15 | F:
TAGCAGCACATAATGGTTTGTG |
|
| R:
GTCGTATCCAGTGCAGGGTCCGAGGT |
| U6 | F:
CTCGCTTCGGCAGCACATATACT |
|
| R:
ACGCTTCACGAATTTGCGTGTC |
| GAPDH | F:
ATGGGGAAGGTGAAGGTCG |
|
| R:
GGGGTCATTGATGGCAACAATA |
| BCL2 | F:
GGTGGGGTCATGTGTGTGG |
|
| R:
CGGTTCAGGTACTCAGTCATCC |
| BCL-XL | F:
GAGCTGGTGGTTGACTTTCTC |
|
| R:
TCCATCTCCGATTCAGTCCCT |
Luminescent cell viability assay
The cells transfected with miR-15/NC
(1×104/per well) were seeded into a 96-well plate and
treated with 5, 10, 20, 40 and 80 µg/ml 5-Fluorouracil (5-Fu,
Tjkingyork company, Hangzhou, China) or 2.5, 5, 10, 20, 40 µg/ml
Oxaliplatin (OX, Zhejian Hisun Pharmaceutical Co., Ltd, Zhejiang,
China) for 48 h at 37°C. Cellular viability was measured using Cell
Titer-Glo™ (Promega Corporation, Madison, WI, USA)
following the manufacturer's protocol.
Measurement of apoptosis by flow
cytometry
Flow cytometry (BD Biosciences, Franklin Lakes, NJ,
USA) was used to detect the apoptosis of cells transfected with
miR-15 or-NC and treated with 5-Fu or OX. The apoptosis kit was
obtained from BD Biosciences. Propidium iodide and Annexin
V-fluorescein-isothiocyonate stains were used to measure late and
early apoptosis, respectively. The stains were incubated for 15 min
at room temperature in the dark.
Plasmid construction and
dual-luciferase assay
To generate the luciferase reporter with NF-κB
activity, the canonical NF-κB recognising sequence (GGG RNN YYC
CGG GRN NYY CC) was synthesized and inserted into the promoter
region of the pGL3 basic vector (Promega Corporation) lined with
MluI and XhoI. To measure the interaction between
miR-15 and its target mRNAs, the sequence containing predicted
target sites within the p50 or p65 mRNAs was synthesized and cloned
downstream of the firefly luciferase coding region in the pGL3
control vector (Promega Corporation) lined with SacI and
HindIII. The plasmids and miR-15 or NC were co-transfected
into HCT116 cells using Lipofectamine™ 2000 (Invitrogen;
Thermo Fisher Scientific Inc.) following the manufacturer's
protocol. At 24 h post-transfection, cells were lysed and
luciferase activity was measured using a dual-luciferase reporter
test kit (Promega Corporation, Madison, WI, USA). The normalization
was completed by comparison with Renilla luciferase
activity. The primers (Takara Biotechnology Co. Ltd, Dalian, China)
were shown in Table III.
 | Table III.Primers for reporter genes
sequence. |
Table III.
Primers for reporter genes
sequence.
| Primer names | Primer sequence
(5′-3′) |
|---|
|
pMIR-reporter-P50 | F:
CCATGTTTGCTGCTGCTGGTGA |
|
| R:
AGCTTCACCAGCAGCAGCAAACATGGAGCT |
|
pMIR-reporter-P65 | F:
CCGAGTTTCAGCAGCTGCTGAA |
|
| R:
AGCTTTCAGCAGCTGCTGAAACTCGGAGCT |
| NF-κB activity
reporter | F:
CGCGTGGGACTTTCCGGGGAACCCCC |
|
| R:
TCGAGGGGGTTCCCCGGAAAGTCCCA |
Western blot analysis
Protein was extracted from cells transfected with
miR-15 or NC using radioimmunoprecipitation assay lysis buffer
(Beyotime Institute of Biotechnology, Haimen, China) and protein
concentration was determined using the BCA assay kit (Beyotime
Institute of Biotechnology). Equal amounts of protein (20 µg/per
lane) were loaded onto 12% SDS-PAGE and transferred onto PVDF
membranes. The membranes were blocked with 5% skimmed milk powder
at 37°C for 2 h. Then, membranes were incubated with primary
antibodies (all Santa Cruz Biotechnology, Inc., Dallas, TX, USA),
including mouse anti-human p50 (cat no. sc-53744, 1:200), mouse
anti-human BCL-2 (cat no. sc-509, 1:200), rabbit anti-human BCL-XL
(cat no. sc-7195, 1:200) and mouse anti-human β-actin (cat no.
sc-130300, 1:500) at 4°C overnight and they were subsequently
incubated with horse radish peroxide-conjugated bovine
anti-mouse/rabbit secondary antibody(Santa Cruz Biotechnology,
Inc., 1:2,000; cat no. sc-2371/2370) at 37°C for 2 h. An immunoblot
was generated using the ECL western blotting detection system
(Thermo Fisher Scientific Inc.).
Statistical analysis
All data were presented as the mean ± standard
deviation. The gene expression level, apoptotic cells percent, NFKB
and luminescence activity between two groups were compared using
Student's t test. The chemosensitivity was compared by two-way
analysis of variance. Multiple comparisons between the groups was
completed using the Student-Newman-Keul's method. SPSS software was
used to analyse data (version 10.0; SPSS, Inc., Chicago, IL, USA).
P<0.05 was regarded as indicating a statistically significant
difference.
Results
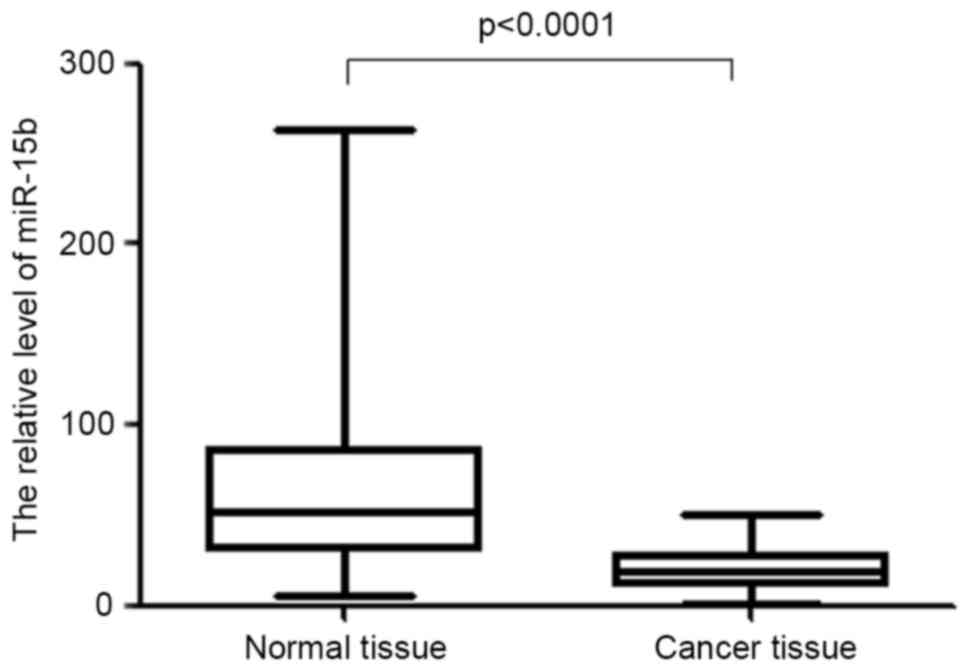
miR-15 is significantly downregulated
in colon cancer
To determine the role of miR-15 in colon cancer
chemotherapy, the expression of miR-15 in cancerous and
para-cancerous tissues from 62 patients with colon cancer was
measured. The results demonstrated that in the majority of cases
(56/62), miR-15 was significantly decreased in cancerous tissue
compared with matched para-cancerous tissue (P<0.0001, Fig. 1). The decrease in miR-15 expression
in colon cancer tissue may affect the sensitivity of colon cancer
tissue to chemotherapy.
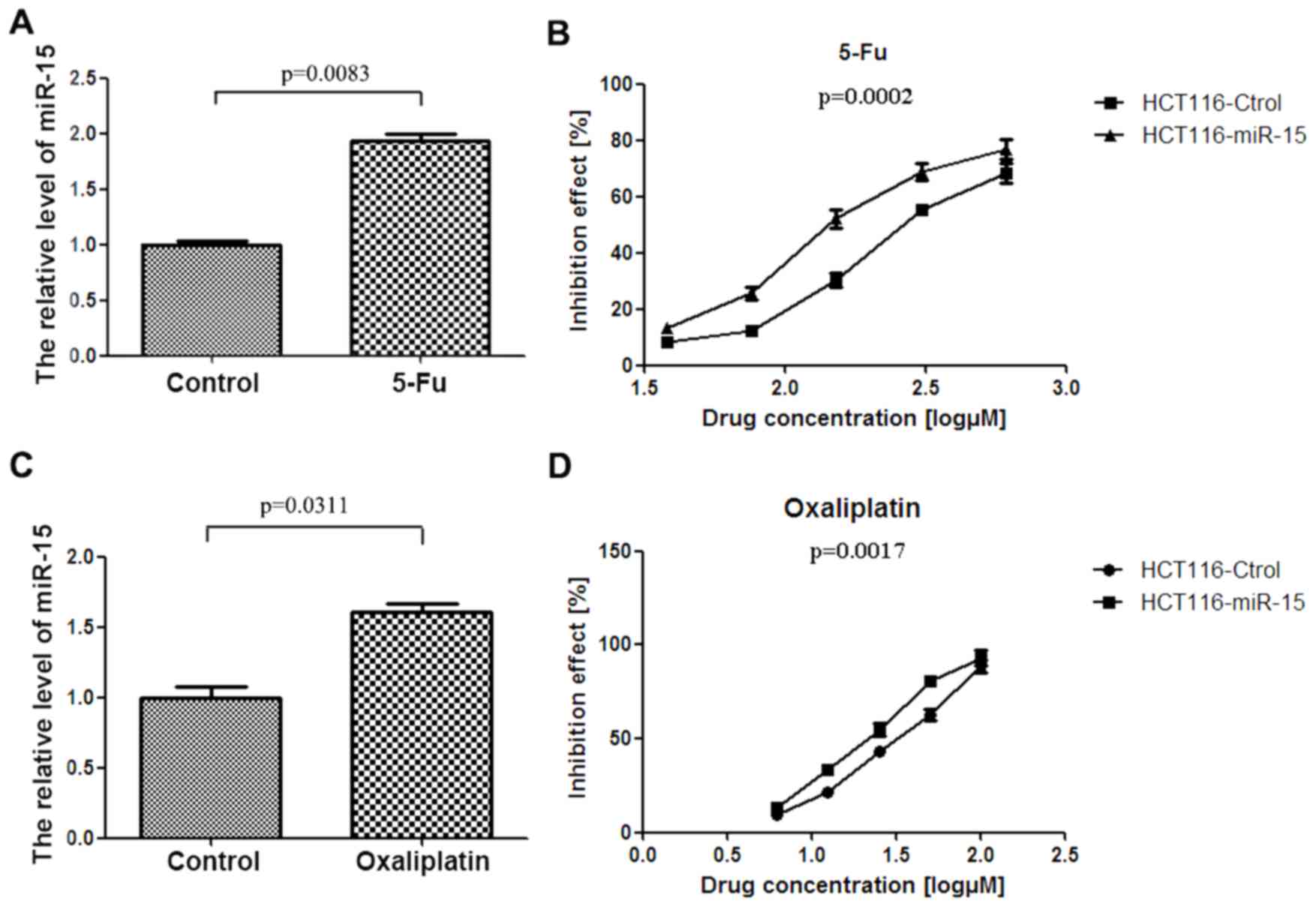
miR-15 improves the chemosensitivity
of colon cancer cells
To determine the association between
chemotherapeutics and miR-15 expression, HCT116 cells were treated
with 5-Fu and OX, two reagents commonly used to treat colon cancer,
and the change in expression of miR-15 was assessed (Fig. 2). The results demonstrated that
miR-15 expression was significantly upregulated in HCT116 cells
treated with 5-Fu and OX (P<0.01; Fig. 2A and P<0.05; Fig. 2C).
The effect of miR-15 on the response of HCT116 cells
to 5-Fu and OX was detected using a luminescent cell viability
assay. The results indicated that miR-15 significantly increased
the inhibitory effect of 5-Fu or OX on HCT116 cell viability (both
P<0.01; Fig. 2B and D). The half
maximal inhibitory concentrations (IC50) for 5-Fu in NC
and miR-15 transfected cells were 289.6 µM and 93.99 µM
respectively, whereas the IC50 for OX in NC and miR-15
transfected cells were 50.27 µM and 20.07 µM, respectively. These
results suggest that miR-15 increases the sensitivity of colon
cancer cells to 5-Fu and OX chemotherapy.
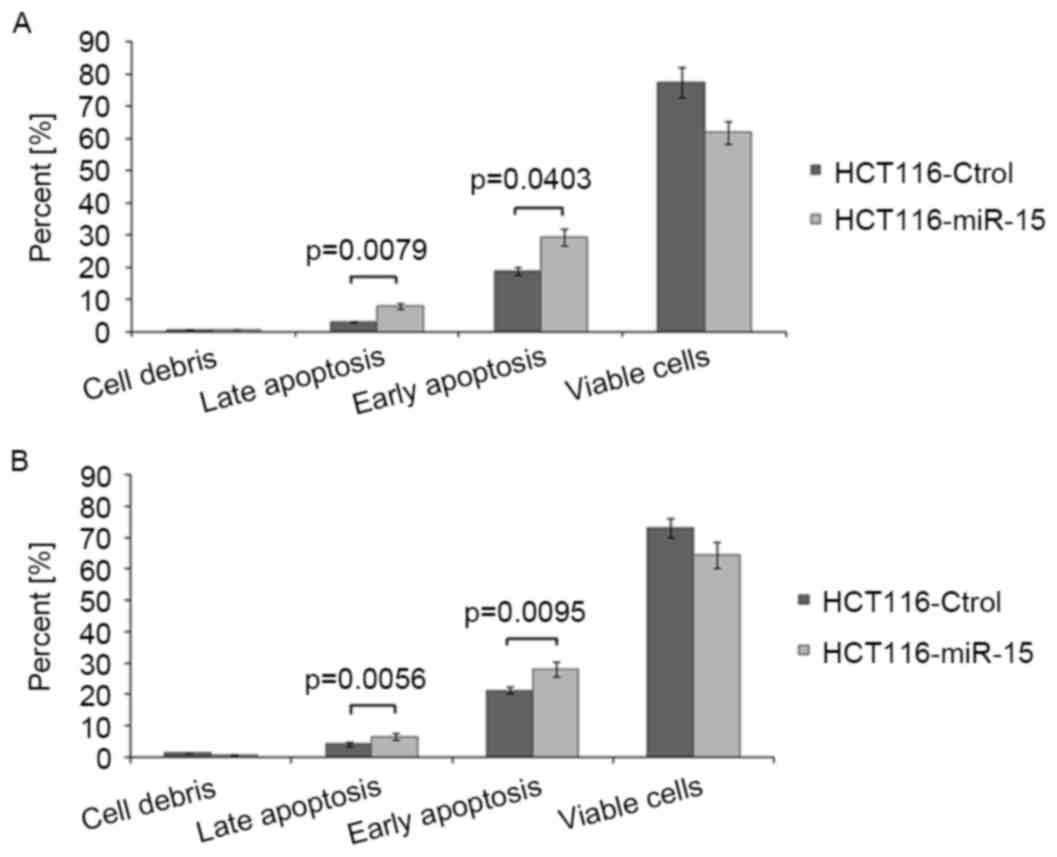
miR-15 promotes 5-Fu- or OX-induced
apoptosis in colon cancer cells
To determine the possible mechanism of miR-15
increasing chemosensitivity, the level of apoptosis induced by 5-Fu
or OX in HCT116 cells transfected with miR-15 was measured. The
results demonstrated that the rate of apoptosis was significantly
increased in miR-15 transfected cells compared with control cells
(P<0.05; Fig. 3) following
treatment with OX or 5-Fu. This indicates that miR-15 promotes
apoptosis induced by chemotherapeutic agents in HCT116 cells.
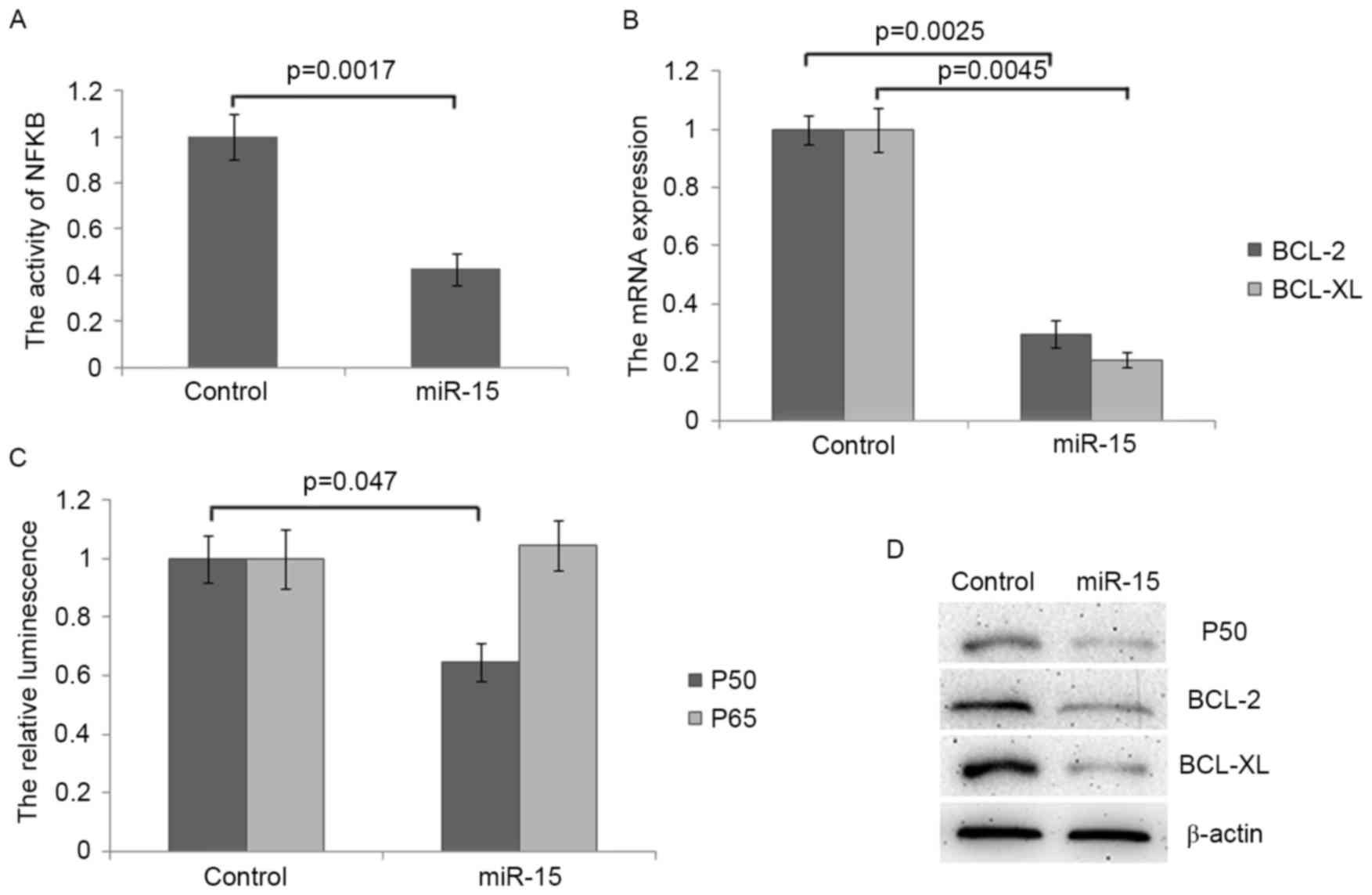
miR-15 targets NF-κB and inhibits the
expression of its target gene
The activation of NF-κB promotes the transcription
of several anti-apoptotic factors including BCL-2 and BCL-XL
(16) and it has been demonstrated
that miR-15 inhibits the expression of BCL-2 (17). Thus, the current study investigated
whether miR-15 inhibited the activation of NF-κB in colon cancer
cells. It was demonstrated that the transcriptional activity of
NF-κB was significantly decreased in miR-15 transfected cells
compared with control cells (P<0.01; Fig. 4A). Following transfection with
miR-15, levels of BCL-2 and BCL-XL mRNA, two direct targets of
NF-κB exhibiting anti-apoptotic functions, were also significantly
decreased (both P<0.01; Fig. 4B).
These results suggest that miR-15 inhibits the activation of NF-κB
in colon cancer. The potential binding sites for miR-15 within p50
and p65, two subunits of the transcription factor NF-κB, were
screened and a potential binding site was identified within the
open reading frame of either p50 or p65, which is located between
2,470 and 2,476 nucleotides (nt) and between 1,231 and 1,236 nt in
the mRNA of p50 and p65. The results of the dual-luciferase assay
demonstrated that miR-15 significantly inhibits the expression of
p50 but not of p65 (Fig. 4C). The
decrease in the expression of p50 protein following miR-15
transfection was confirmed by western blot analysis (Fig. 4D). Furthermore, the expression of
BCL-2 and BCL-XL protein were also decreased in miR-15 transfected
cells (Fig. 4D). Taken together,
these results indicate that miR-15 increased the chemosensitivity
of colon cancer cells to 5-Fu and OX by repressing the NF-κB
signalling pathway and promoting apoptosis.
Discussion
Chemotherapy is an important method of treating
cancer. A large number of patients with cancer have benefitted from
tumour regression and increased survival following the development
of various chemotherapeutic agents (18). However, the development of
chemoresistance, in which cancer cells exhibit little or no
response to chemotherapy drugs, poses a challenge to the treatment
of cancer (19).
Previous studies have identified some of the
mechanisms of chemoresistance, which include increased drug efflux
(20), decreased drug activation
(21) and increased DNA repair
activity (22). Recently, several
studies have indicated the regulatory role of miRNAs in response to
chemotherapy (23–25). For example, miR-214 induces cisplatin
resistance in certain types of ovarian cancer by targeting the
phosphatase and tensin homolog/phosphoinositide 3-kinase/Akt
pathway (26). In addition, high
levels of miR-378 may reverse the chemoresistance of lung
adenocarcinoma cells to cisplatin by inhibiting the expression of
secreted clusterin (27). miRNA is
therefore an appealing target to increase effectiveness of various
drugs.
It has been demonstrated that miR-15 serves an
important role in the tumourigenesis of colorectal cancer (10). However, its expression and function
remain unclear. Xi et al (28) determined that miR-15 was
significantly overexpressed in colorectal cancer tissues and thus
may function as an oncogene. By contrast, Wang et al
(29) identified that miR-15 was
downregulated in colorectal cancer tissues and may act as a tumour
suppressor. In the present study, the expression of miR-15 was
decreased in the majority of colon cancer tissues compared with
para-cancerous tissues, supporting the role of miR-15 as a tumour
suppressor in colon cancer.
miR-15 is able to regulate chemoresistance in
addition to regulating cell proliferation, apoptosis, angiogenesis
and metastasis (30–33). Xia et al (34) identified that miR-15 may enhance the
sensitivity of gastric cancer cells to anticancer drugs by
targeting BCL2 and promoting apoptosis. Reduced levels of miR-15
are also associated with chemotherapeutic resistance in human
tongue cancer cells by targeting BMI1 (14). However, it has also been identified
that increased expression of miR-15 is associated with decreased
sensitivity to cisplatin and the poor prognosis of patients with
lung adenocarcinoma by suppressing the expression of
phosphatidylethanolamine-binding protein 4 (13). The results of the present study
indicate that the upregulation of miR-15 may increase the
sensitivity of colon cancer cells to 5-Fu and OX by promoting
apoptosis.
To determine the action of miRNA and the role of
apoptosis in the development of colon cancer, the target mRNAs of
miR-15 associated with apoptosis were detected and analysed. The
results demonstrated that NF-κB was a target of miR-15 and that
miR-15 inhibits the activation of NF-κB by repressing the
expression of p50. Previous studies have demonstrated that NF-κB
inhibits apoptosis by transcriptionally activating anti-apoptotic
proteins (35,36). This includes BCL-2 and BCL-XL,
members of the BCL-2 family of apoptosis regulators (37,38). In
the present study, it was demonstrated that the downregulation of
NF-κB decreased the expression of BCL-2 and BCL-XL, which may
contribute to the induction of apoptosis in colon cancer cells by
miR-15.
In conclusion, the current study demonstrated that
miR-15 was downregulated in colon cancer tissues and may act as a
tumour suppressor. Exogenous overexpression of miR-15 may increase
the response of colon cancer cells to 5-Fu and OX by inhibiting the
NF-κB/BCL-2/BCL-XL signalling pathway and inducing apoptosis.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li L and Ma BB: Colorectal cancer in
Chinese patients: Current and emerging treatment options. Onco
Targets Ther. 7:1817–1828. 2014.PubMed/NCBI
|
|
3
|
Loree JM and Kopetz S: Recent developments
in the treatment of metastatic colorectal cancer. Ther Adv Med
Oncol. 9:551–564. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Boland PM and Ma WW: Immunotherapy for
colorectal cancer. Cancers. 9:pii: E502017. View Article : Google Scholar
|
|
5
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
King TD, Suto MJ and Li Y: The
Wnt/β-catenin signaling pathway: A potential therapeutic target in
the treatment of triple negative breast cancer. J Cell Biochem.
113:13–18. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Magee P, Shi L and Garofalo M: Role of
microRNAs in chemoresistance. Ann Transl Med. 3:3322015.PubMed/NCBI
|
|
10
|
Zhao C, Wang G, Zhu Y, Li X, Yan F, Zhang
C, Huang X and Zhang Y: Aberrant regulation of miR-15b in human
malignant tumors and its effects on the hallmarks of cancer. Tumour
Biol. 37:177–183. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang WL, Zhang JH, Wu XZ, Yan T and Lv W:
miR-15b promotes epithelial-mesenchymal transition by inhibiting
SMURF2 in pancreatic cancer. Int J Oncol. 47:1043–1053. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lovat F, Fassan M, Gasparini P, Rizzotto
L, Cascione L, Pizzi M, Vicentini C, Balatti V, Palmieri D,
Costinean S and Croce CM: miR-15b/16-2 deletion promotes B-cell
malignancies. Proc Natl Acad Sci USA. 112:pp. 11636–11641. 2015;
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao Z, Zhang L, Yao Q and Tao Z: miR-15b
regulates cisplatin resistance and metastasis by targeting PEBP4 in
human lung adenocarcinoma cells. Cancer Gene Ther. 22:108–114.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sun L, Yao Y, Liu B, Lin Z, Lin L, Yang M,
Zhang W, Chen W, Pan C, Liu Q, et al: miR-200b and miR-15b regulate
chemotherapy-induced epithelial-mesenchymal transition in human
tongue cancer cells by targeting BMI1. Oncogene. 31:432–445. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Konishi T, Sasaki S, Watanabe T, Kitayama
J and Nagawa H: Overexpression of hRFI inhibits
5-fluorouracil-induced apoptosis in colorectal cancer cells via
activation of NF-kappaB and upregulation of BCL-2 and BCL-XL.
Oncogene. 25:3160–3169. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cimmino A, Calin GA, Fabbri M, Iorio MV,
Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et
al: miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl
Acad Sci USA. 102:pp. 13944–13949. 2005; View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cunningham D, Allum WH, Stenning SP,
Thompson JN, Van de Velde CJ, Nicolson M, Scarffe JH, Lofts FJ,
Falk SJ, Iveson TJ, et al: Perioperative chemotherapy versus
surgery alone for resectable gastroesophageal cancer. N Engl J Med.
355:11–20. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wilson TR, Longley DB and Johnston PG:
Chemoresistance in solid tumours. Ann Oncol. 17 Suppl 10:x315–x324.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gottesman MM, Fojo T and Bates SE:
Multidrug resistance in cancer: Role of ATP-dependent transporters.
Nat Rev Cancer. 2:48–58. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Barnes MJ, Estlin EJ, Taylor GA, Aherne
GW, Hardcastle A, McGuire JJ, Calvete JA, Lunec J, Pearson AD and
Newell DR: Impact of polyglutamation on sensitivity to raltitrexed
and methotrexate in relation to drug-induced inhibition of de novo
thymidylate and purine biosynthesis in CCRF-CEM cell lines. Clin
Cancer Res. 5:2548–2558. 1999.PubMed/NCBI
|
|
22
|
Dabholkar M, Vionnet J, Bostick-Bruton F,
Yu JJ and Reed E: Messenger RNA levels of XPAC and ERCC1 in ovarian
cancer tissue correlate with response to platinum-based
chemotherapy. J Clin Invest. 94:703–708. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Amirkhah R, Farazmand A, Irfan-Maqsood M,
Wolkenhauer O and Schmitz U: The role of microRNAs in the
resistance to colorectal cancer treatments. Cell Mol Biol
(Noisy-le-grand). 61:17–23. 2015.PubMed/NCBI
|
|
24
|
Dehghanzadeh R, Jadidi-Niaragh F, Gharibi
T and Yousefi M: MicroRNA-induced drug resistance in gastric
cancer. Biomed Pharmacother. 74:191–199. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang G, Zhao R, Zhao X, Chen XI, Wang D,
Jin Y, Liu XI, Zhao CI, Zhu Y, Ren C, et al: MicroRNA-181a enhances
the chemotherapeutic sensitivity of chronic myeloid leukemia to
imatinib. Oncol Lett. 10:2835–2841. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang H, Kong W, He L, Zhao JJ, O'Donnell
JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV and Cheng JQ:
MicroRNA expression profiling in human ovarian cancer: miR-214
induces cell survival and cisplatin resistance by targeting PTEN.
Cancer Res. 68:425–433. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen X, Jiang Y, Huang Z, Li D, Chen X,
Cao M, Meng Q, Pang H, Sun L, Zhao Y and Cai L: miRNA-378 reverses
chemoresistance to cisplatin in lung adenocarcinoma cells by
targeting secreted clusterin. Sci Rep. 6:194552016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xi Y, Formentini A, Chien M, Weir DB,
Russo JJ and Ju J, Kornmann M and Ju J: Prognostic values of
microRNAs in colorectal cancer. Biomarker Insights. 2:113–121.
2006.PubMed/NCBI
|
|
29
|
Wang X, Wang J, Ma H, Zhang J and Zhou X:
Downregulation of miR-195 correlates with lymph node metastasis and
poor prognosis in colorectal cancer. Med Oncol. 29:919–927. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wu CS, Yen CJ, Chou RH, Chen JN, Huang WC,
Wu CY and Yu YL: Downregulation of microRNA-15b by hepatitis B
virus X enhances hepatocellular carcinoma proliferation via
fucosyltransferase 2-induced Globo H expression. Int J Cancer.
134:1638–1647. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kedmi M, Ben-Chetrit N, Körner C, Mancini
M, Ben-Moshe NB, Lauriola M, Lavi S, Biagioni F, Carvalho S,
Cohen-Dvashi H, et al: EGF induces microRNAs that target
suppressors of cell migration: miR-15b targets MTSS1 in breast
cancer. Sci Signal. 8:ra292015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zheng X, Chopp M, Lu Y, Buller B and Jiang
F: miR-15b and miR-152 reduce glioma cell invasion and angiogenesis
via NRP-2 and MMP-3. Cancer Lett. 329:146–154. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li J, Chen Y, Guo X, Zhou L, Jia Z, Tang
Y, Lin L, Liu W and Ren C: Inhibition of miR-15b decreases cell
migration and metastasis in colorectal cancer. Tumour Biol.
37:8765–8773. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun
S, Hong L, Liu J and Fan D: miR-15b and miR-16 modulate multidrug
resistance by targeting BCL2 in human gastric cancer cells. Int J
Cancer. 123:372–379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rayet B and Gelinas C: Aberrant rel/nfkb
genes and activity in human cancer. Oncogene. 18:6938–6947. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Barkett M and Gilmore TD: Control of
apoptosis by Rel/NF-kappaB transcription factors. Oncogene.
18:6910–6924. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cheng Q, Lee HH, Li Y, Parks TP and Cheng
G: Upregulation of Bcl-x and Bfl-1 as a potential mechanism of
chemoresistance, which can be overcome by NF-kappaB inhibition.
Oncogene. 19:4936–4940. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xia Y, Shen S and Verma IM: NF-κB, an
active player in human cancers. Cancer Immunol Res. 2:823–830.
2014. View Article : Google Scholar : PubMed/NCBI
|