Introduction
Arrhythmogenic cardiomyopathy (AC) is a clinically
and genetically heterogeneous heart muscle disease estimated to
affect ~1/5,000 individuals (1). The
pathological hallmark of AC consists of progressive myocardial
atrophy followed by fibro-fatty replacement (2). Palpitations, syncope, ventricular
tachycardia, right heart failure and even sudden cardiac death
(SCD) are typical clinical manifestations of AC (2).
In recent years, great advances have been made in
the understanding of the pathogenesis of AC (3). Mutations in genes that encode
desmosomal proteins (desmoglein-2, desmocollin-2, plakoglobin,
plakophilin and desmoplakin) are thought to cause AC. Approximately
50% of patients with AC have one or more mutations in their
desmosomal genes; thus, AC is referred to as ‘a disease of the
desmosome’ (4). In addition to
desmosomal genes, mutations in non-desmosomal genes such as TGFβ3,
TMEM43, RYR2, DES, and TTN are also involved in AC (5–9).
Mutations in disease-causing genes have been
identified in AC patients with a variable frequency (10,11), and
there is limited knowledge regarding the molecular disease
mechanisms leading to the condition in individuals with pathogenic
gene mutations. Since it is difficult to obtain myocardial tissue
for protein studies, the majority of previous studies investigated
the molecular disease mechanisms by culturing the keratinocytes of
patients with AC (12,13). These studies revealed two disease
mechanisms induced by the pathogenic mutations: Haploinsufficiency
and dominant-negative effects.
In the present study, a systematic sequence analysis
was performed for five selected desmosomal genes obtained from
patients with AC who underwent heart transplantation. The aim of
the study was to identify novel AC-associated variations or known
mutations of the five desmosomal genes and to observe their effects
on desmosomal protein expression, in order to further our
understanding of the mechanism underlying the effects of desmosomal
gene mutations on AC pathogenesis.
Materials and methods
Study patients
Patients enrolled in the present study included 23
individuals, 70% of whom were male, aged 41.4±13.8 years (range,
18–68 years) with AC who underwent heart transplantation at Fuwai
Hospital (Beijing, China) between 2005 and 2012. Subjects were
diagnosed from the pathological results of a biopsy. All patients
gave written informed consent for this investigation, which was
approved by the Ethical Committee of Fuwai Hospital. Routine
medical history, physical examinations and laboratory tests were
collected.
Tissue samples
Cardiac tissue samples were collected from the
patients with AC who underwent heart transplantation. Healthy
control hearts were obtained from autopsies or deceased donors with
no history of heart disease.
Electrocardiography
Twelve-lead electrocardiogram was carried out during
echocardiographic examination. Data analyses (Epsilon wave, T wave
inversion and ventricular tachycardia with left bundle branch
block) were performed.
Echocardiographic studies
Two-dimensional echocardiographic studies were
performed and data were analyzed with EchoPAC version 112 (GE
Healthcare, Chicago, IL, USA). From 2-dimensional echocardiography,
we assessed proximal right ventricular outflow tract (RVOT)
diameter in the parasternal short-axis view and right ventricular
basal diameter (RV diameter). Ventricular wall structure was
assessed through RVOT, RV diameter, left ventricular (LV)
end-diastolic volume, LV end-systolic volume, and left ventricular
ejection fraction.
Genetic analysis
Genomic DNA from the cohort of 23 patients with AC
was extracted from the myocardial tissue samples using a QIAamp DNA
Mini kit (Qiagen China Co., Ltd., Shanghai, China). The DNA
sequence of these patients was screened by target capture
sequencing, then verified by Sanger sequencing (14,15). All
coding exons of the desmosomal genes (DSG2, NM_001943.3, DSC2,
NM_024422.3, JUP, NM_021991.2, PKP2, NM_004572.3 and DSP,
NM_004415.2) were enriched using custom-made SureSelectXT Target
Enrichment arrays (Agilent Technologies, Inc., Santa Clara, CA,
USA). HiSeq 2000 was used to perform next-generation sequencing as
previously described (16).
Common polymorphisms were excluded based on their
presence in the 1,000 genome databases (ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/), yhSNP
(http://yh.genomics.org) and Hapmap (http://www.ncbi.nlm.nih.gov/snp/). Variants were
considered to be a possible AC-associated variation if they
exhibited the following (3): i)
Absence from the 1,000 genome database; ii) the variation was
expected to have a drastic effect on the protein (nonsense
mutation, frame shift mutation or the mutated residue was highly
conserved among species); iii) the variation was predicted to be
damaging by polymorphism phenotyping and sorting intolerant from
tolerant.
Sanger sequencing
Sanger sequencing were performed according to method
described previously (17). A 600
base pair fragment containing the candidate variant was polymerase
chain reaction-amplified from genomic DNA of the index case. Base
sequences of the primer pairs and annealing temperature are listed
in Table I. Each amplicon was
bi-directionally sequenced, followed by electrophoresis on an ABI
3730XL Genetic Analyzer (Perkin-Elmer; Applied Biosystems Inc.,
Foster City, CA, USA).
 | Table I.Base sequence of the primer pairs. |
Table I.
Base sequence of the primer pairs.
| Location | Forward primer
(5′-3′) | Reverse primer
(5′-3′) | TM (°C) |
|---|
| DSG2 |
|
|
|
|
c.2390T>A |
CAGTTGTGTCGTAGGATCTC |
ACTGCCAGAGGAGTAGGTAT | 55.4 |
|
c.593A>G |
TCTAATGCCAGATACTCTTGTG |
GAGAGTTTAAAGATGGGCAAC | 57.0 |
| PKP2 |
|
|
|
|
c.746G>C |
AAGTGCCAGCTCATGCTGTC |
CTACACGCACAGCGATTACC | 57.4 |
|
c.2422delC |
TAGCGATTTCTTCCCAGGGTC |
AGCAGTTGAGGAGCGAAGAG | 57.5 |
Immunohistochemistry
When available, dependent on the patients
willingness to donate cardiac tissue, the formalin-fixed cardiac
tissue samples obtained from the patients with AC were examined for
the distribution of intercellular junction proteins. Antigens were
exposed in the paraffin-embedded tissue sections using high
pressure antigen-recovery techniques. Following washing three times
with PBS and blocking with goat serum at room temperature for 20
min, the tissue sections were subsequently treated with
commercially-available rabbit antibodies targeting connexin-43
(Cx43; 3512; 1:100; Cell Signaling Technology, Inc., Danvers, MA,
USA), N-cadherins (ab18203; 1:1,000; Abcam, Cambridge, UK),
junction plakoglobin (JUP; 2309; 1:50; Cell Signaling Technology,
Inc.), plakophilin 2 (PKP2; ab151402; 1:200; Abcam) and desmoglein
2 (DSG2; ab85632; 1:200; Abcam) overnight at 4°C. Following washing
with PBS three times, tissue sections were treated with anti-rabbit
(pv6001) and anti-mouse (pv6002; both Zhongshan Golden Bridge
Biotechnology Co., Ltd., Beijing, China) secondary antibodies and
visualized with 3,3′-diaminobenzidine (ZLI-9017; Zhongshan Golden
Bridge Biotechnology Co., Ltd.) via light microscopy (DMI-600B;
Leica Microsystems, Inc., Buffalo Grove, IL, USA). In addition,
myocardial tissue sections obtained from two age-matched controls
with no clinical or autopsy evidence of heart disease were also
examined by immunohistochemistry. Only two age-matched controls
were used to the valuable nature of the tissues. Although there
were only two controls, repeated independent experiments were
performed to ensure the reliability of the data.
Western blotting
Snap frozen tissue samples (80 µg) were homogenized
with a buffer (8 M Urea, 50 mM Tris-HCL, 100 mM NaCl, 1 mM DTT, 1%
Triton-X-100, pH 7.5) supplemented with protease inhibitors (1:100)
as previously described (12).
Following centrifugation for 15 min at 4°C (3,000 × g), the
supernatant was extracted and the protein concentration was
determined via bicinchoninic acid assay. Protein (30 µg) was loaded
onto 4–12% NuPAGE Bis-Tris Protein Gels (NP0321BOX; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) and electrotransferred to a
polyvinylidene difluoride membrane. The membrane was subsequently
blocked by 5% bovine serum albumin for 1 h at room temperature.
Assessment of the cytoskeletal proteins was performed using the
following primary antibodies: Rabbit anti-DSG2 (ab85632; 1:1,000),
anti-JUP (2309; 1:50), anti-Cx43 (3512; 1:100) and mouse anti-PKP2
(ab151402; 1:200), and the loading control was β-tubulin (2146;
1:1,000; Cell Signaling Technology, Inc.) was used as the internal
reference. The membrane was incubated for 2 h at room temperature
and was subsequently washed three times (5 min each) with 10 ml
TBST. Following this, the membrane was incubated with horseradish
peroxidase-conjugated anti-rabbit (A0208; 1:1,000) and anti-mouse
secondary antibodies (A0216; 1:1,000; both Beyotime Institute of
Biotechnology, Beijing, China). Following washing three times with
TBST, the blots were visualized with a chemiluminescence imaging
instrument (ProteinSimple, Santa, Clara, CA, USA).
Statistical analyses
Each independent experiment was repeated at least
three times, and at least three independent experiments were
performed. Expression levels were quantified using ImageJ software
(V2.1.4.7; National Institutes of Health, Bethesda, MA, USA). Data
were expressed as the mean ± standard deviation. GraphPad Prism
software (version 5; GraphPad Software, Inc., La Jolla, CA, USA)
was used for the statistical analyses. P<0.05 was considered to
indicate a statistically significant difference.
Results
Genetic analysis
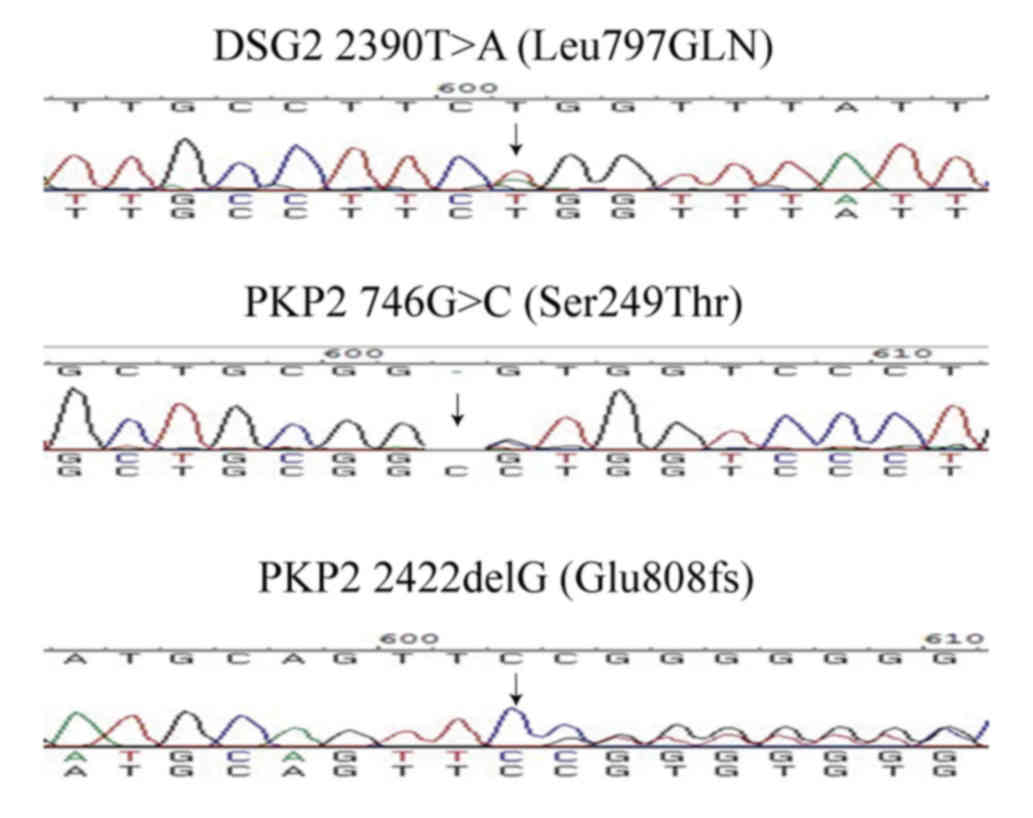
Six variations (four in DSG2, two in PKP2) were
identified in the desmosome genes (Table II). Two of the variations were
single nucleotide polymorphisms, and one of the variations was not
verified by Sanger sequencing; thus, it was a false positive from
next generation sequencing. The three variations, DSG2 L797Q, PKP2
S249T and E808fsX30 with possible functional alteration were absent
in the 1,000 genome database. Their position was highly conserved
and function prediction indicated that the variations were
damaging. Therefore, the three variants were considered to be
unique to the patients with AC and determined to be AC-associated
variants (Table III). The DNA
sequencing results of the three variations are shown in Fig. 1.
 | Table II.Variations in the desmosome genes of
the 23 patients with AC. |
Table II.
Variations in the desmosome genes of
the 23 patients with AC.
| Gene | Variation type | Nucleotide
change | Amino acid
change |
PolyPhena | SIFTb |
|---|
| DSG2 | SNP | c.618T>A | p.Ala206Ala | − | 0.60 |
| DSG2 | Missense | c.2390T>A | p.Leu797Gln | 0.998 (++) | 0.95 |
| DSG2 | Missense | c.593A>G | p.Tyr198Cys | 0.995 | 0.95 |
| DSG2 | SNP | c.161C>T | p.Ala54Val | Benign | 0.55 |
| PKP2 | Missense | c.746G>C | p.Ser249Thr | 0.97 (++) | 1 |
| PKP2 | Frameshift | c.2422delC | p.Glu808fsX30 |
|
|
 | Table III.Potential AC-associated variations
identified in the 23 patients. |
Table III.
Potential AC-associated variations
identified in the 23 patients.
| Gene | Type of
variation | Nucleotide
change | Amino acid
change | Patient index
no. |
PolyPhena | SIFTb |
|---|
| DSG2 | Missense | c.2390T>A | p.Leu797Gln | 1 | 0.998 (++) | 0.95 |
| PKP2 | Missense | c.746G>C | p.Ser249Thr | 1 | 0.97 (++) | 1 |
| PKP2 | Frameshift | c.2422delC | p.Glu808fsX30 | 1 | N/Ac | N/Ac |
Pathological and clinical
characteristics
Among the 23 patients diagnosed with AC who received
heart transplantation, two patients carried the three desmosomal
gene variations. The clinical characteristics of the patients are
summarized in Table IV. The first
symptoms of the two patients were dyspnea and arrhythmia,
respectively. Patient 1 had a family history of SCD, with the
patient's mother having suffered from SCD at 52 years of age, and
also had atrial fibrillation and regional RV dyskinesia. Patient 2
also had a family history of SCD (the patient's brother suffered
SCD when 14 years old), along with ventricular tachycardia. The two
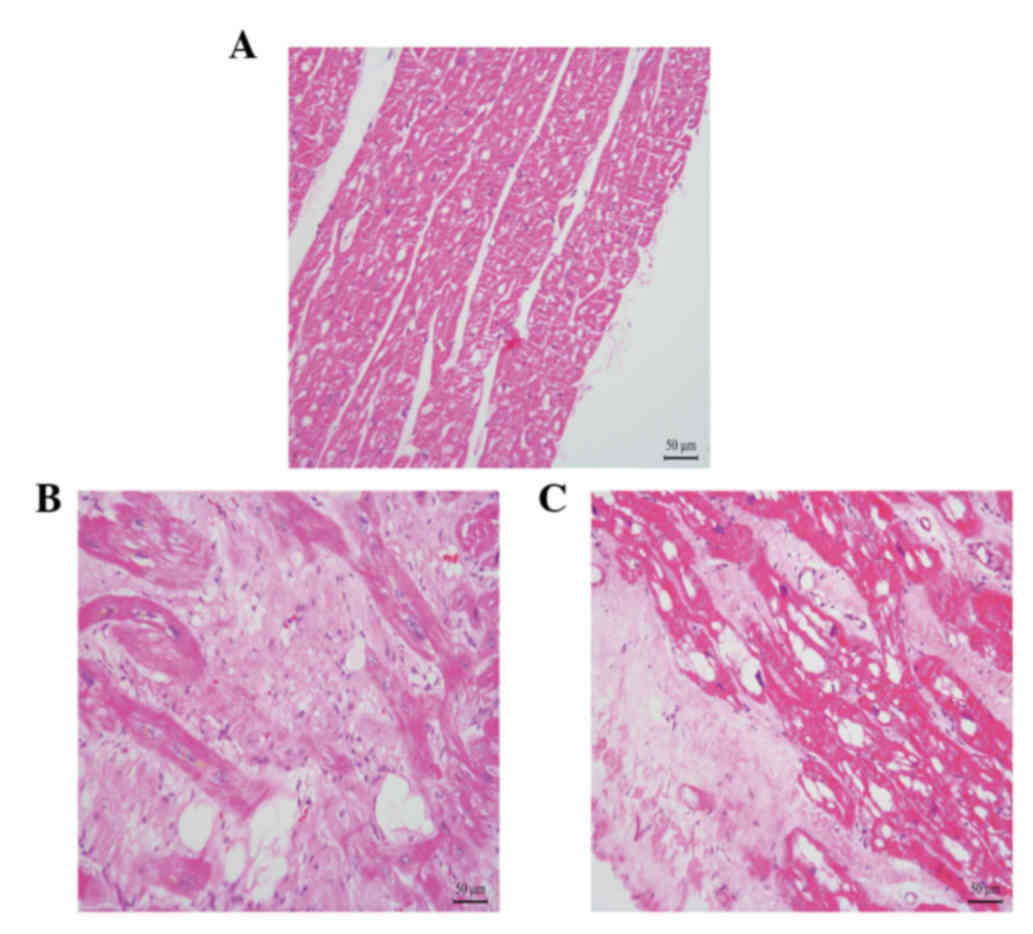
patients were both in end-stage heart failure (NYHA III–IV). The
pathological characteristic of the right ventricular myocardium in
the two patients is shown in Fig. 2.
Compared with the control, the number of myocardiocytes in the
myocardium of the patients with AC had markedly decreased, and
numerous fibro-fatty had infiltrated into the myocardium.
 | Table IV.Clinical characteristics of the two
patients with arrhythmogenic cardiomyopathy-associated
variations. |
Table IV.
Clinical characteristics of the two
patients with arrhythmogenic cardiomyopathy-associated
variations.
| No. | Age (yr)/sex | Presentation | FH | Epsilon wave | T wave
inversion | VT with LBBB | IMG |
|---|
| 1 | 38/M | Dyspnea | + | − | − | − | + |
| 2 | 30/M | Arrhythmia | + | − | − | + | + |
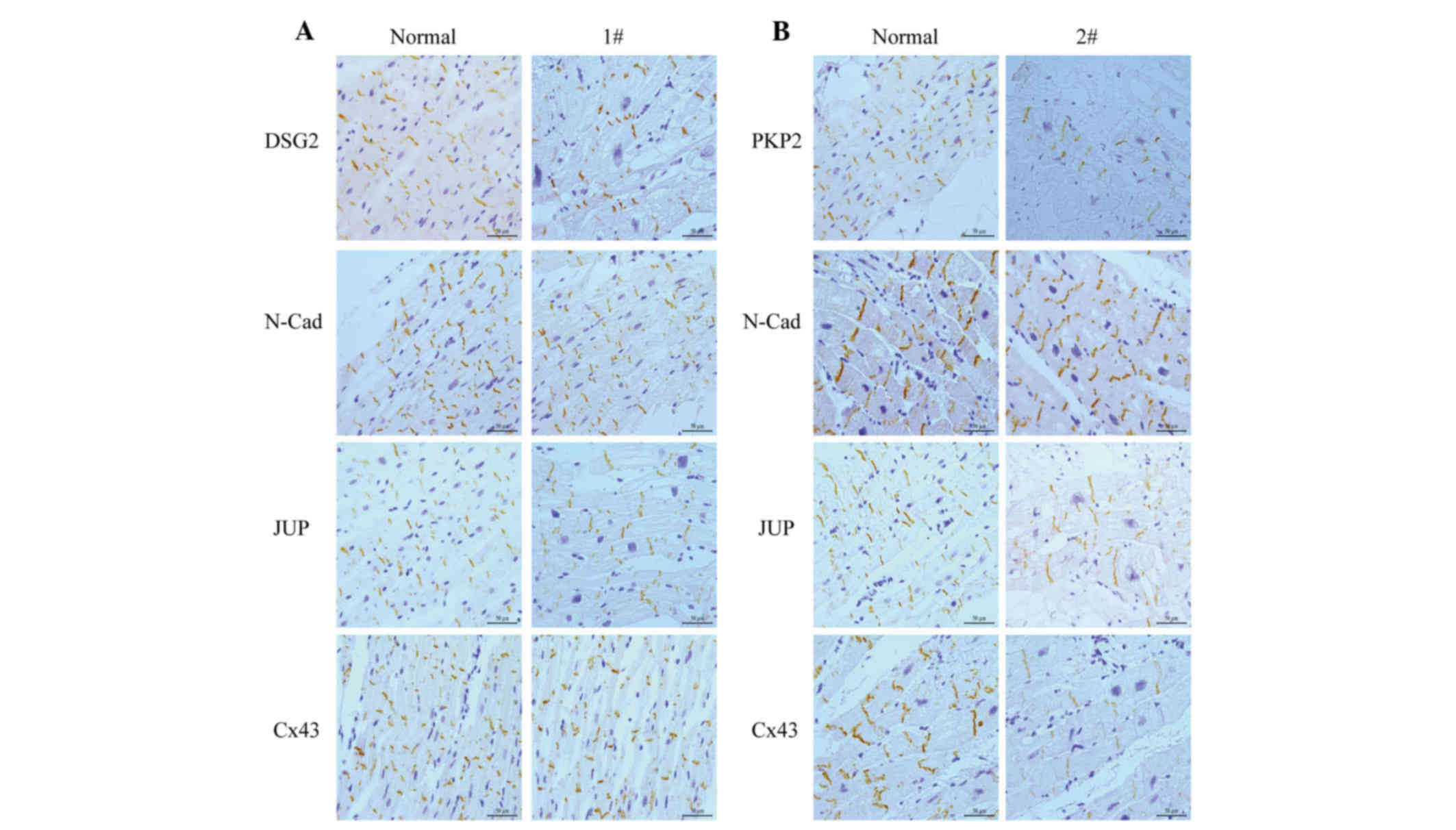
Change in location of the intercalated
disc proteins in the two patients
Myocardial samples were obtained from the two
individuals. These myocardial tissue samples were sectioned and
stained for various junctional proteins, including N-cadherin,
DSG2, PKP2, and JUP, as well as Cx43.
The primary component of the adherens junction
within the intercalated disc, which was N-cadherin, was present and
comparable with the controls in all samples (Fig. 3). In the myocardial tissue of patient
1 (Fig. 3A), who carried DSG2 L797Q,
the staining of DSG2 produced no change compared with the control.
Staining of samples from this patient was comparable to the
controls for other antibodies, including Cx43 and JUP.
Immunohistochemical staining performed in the
myocardial tissue of patient 2 (Fig.
3B), who carried the PKP2 point mutation and deletion in exon
12 demonstrated that the positive expression of PKP2 is reduced
compared with the control. The other intercalated disc proteins,
JUP and Cx43, also showed a marked reduction in positive expression
at the intercalated disc.
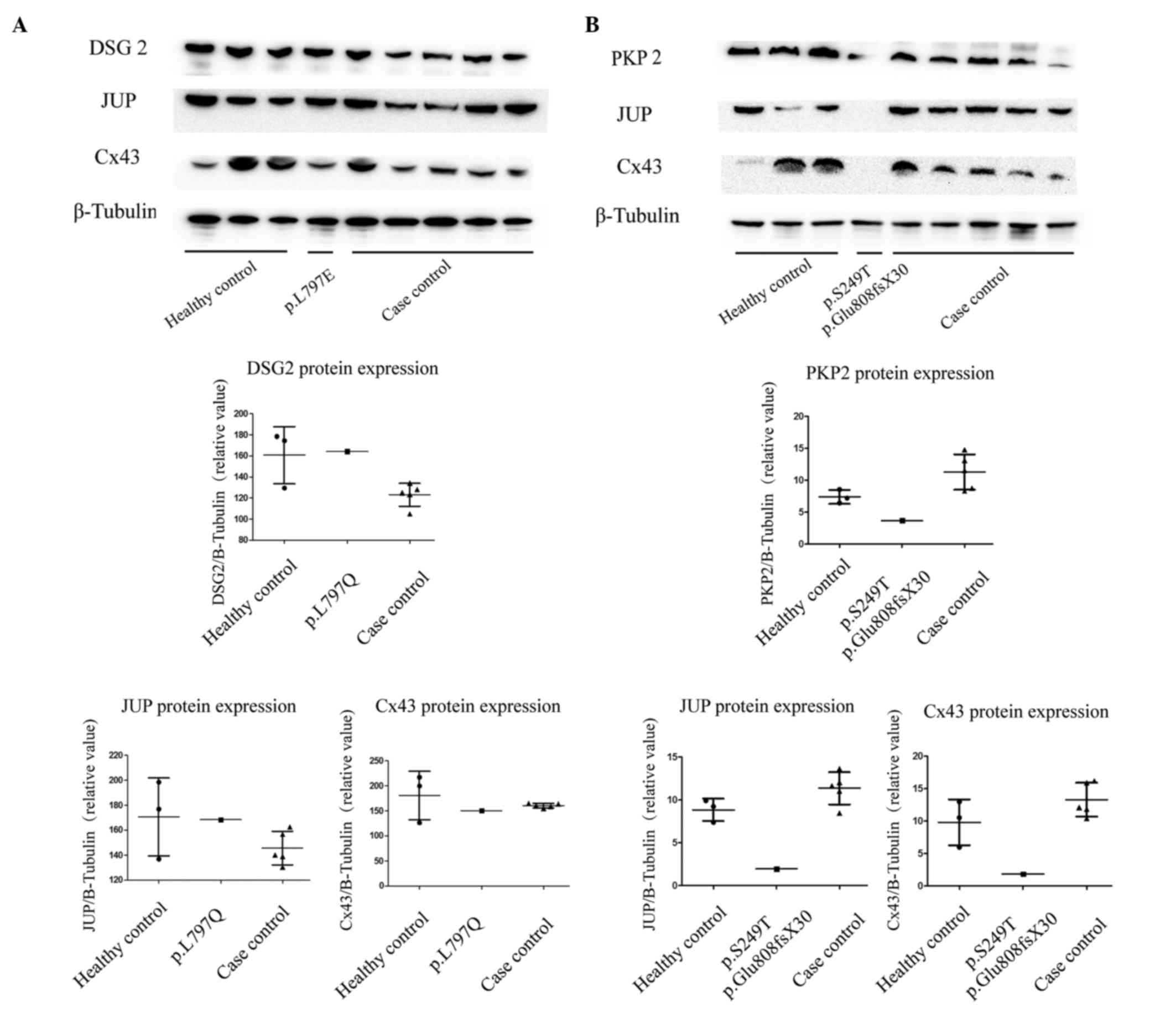
Change in expression levels of
intercalated disc protein in the two patients
Immunoblotting was performed on the right ventricle
to identify potential AC-specific cardiac protein expression
patterns.
As shown in Fig. 4A,
the total expression levels of DSG2 in the right ventricle remained
unchanged in the heterozygous patient, compared with the healthy
controls and case controls without this variation. The expression
levels of the other proteins in the intercalated disc (JUP, Cx43)
were similar in the myocardium of the three groups. In the carrier
with the PKP2 variation, the expression levels of PKP2 were
markedly decreased compared with the healthy controls and
non-carriers. Notably, the expression levels of JUP and Cx43 were
also decreased in patient 2 compared with the healthy controls and
case controls.
Discussion
AC is an inheritable structural heart disease first
described by Frank et al (18) in 1978. Since 50% of the diagnosed
patients carried the desmosome gene mutation (19), AC became known as a ‘desmosome
disease’. To date, 12 genes have been identified as AC-causing
genes (20), and ~300 genetic
variants of these genes have been classified as damaging (21). The majority of pathogenic mutations
have been identified in genes encoding desmosomal proteins, with
the PKP2 gene being responsible for ~35–40% of cases (3,22).
Mutations in the genes DSP, DSG2, and DSC2 are responsible for
~15–20% of AC cases (20). Thus,
genetic counseling is important to identify patients who are at
high risk of the disease.
The discovery of novel genes has been limited by the
reduced penetrance and variable expressivity associated with AC
(23). A previous study reported
that in individuals with a desmosomal mutation, only ~30–50% will
fulfill the diagnostic criteria for AC (24). Modifier genes are hypothesized to
account for much of the variation between individuals, even within
the same family. In addition, numerous environmental factors have a
role in disease expression, including age, gender, strenuous
exercise, drugs, hormones, infection/inflammation and emotional
stress (25).
In the present study, we identified three variations
and amongst these variations, a heterozygous variant of DSG2,
L797Q, was newly found. In the myocardium of patients with the
L797Q variant, the protein expression levels of DSG2 are similar in
both the variation carrier and the controls. The expression levels
of other proteins in intercalated discs remained unchanged. These
results, combined with the results of the location of DSG2 observed
in the myocardial tissue, indicated that the intracellular
localization and expression level of proteins in intercalated disc
were normal. Therefore, we speculated that this variant may be
non-pathogenic, fir example it may be a disease modifier gene, and
other pathogenic genes remain unrecognized. Furthermore, these
findings indicated that even this position is a highly conserved
intracellular anchor domain, and the predicted functions indicated
that the variation is damaging. It is unreasonable to consider it
as a disease-causing mutation.
Since family members with identical genotypes and
even monozygotic twins show significant differences in symptoms,
presence and distribution of structural changes, and rate of
disease progression, modifier genes are likely responsible for
phenotypic heterogeneity (26).
Therefore, detection of modifier genes is helpful to further
understand the disease mechanism underlying AC.
Two variants pertaining to PKP2, S249T and E808fsX30
were also observed, which were all carried by one patient.
PKP2 is an important desmosomal protein. Dominant
mutations in PKP2 were reported in ~25–50% of patients with AC,
suggesting that PKP2 is the major disease gene (27). The mutation rates of PKP2 in Chinese
subjects were reported to be 18–39% (28,29) and
58% (30). Unexpectedly, in the
present study, the mutation rate of PKP2 was observed to be 4%,
which may be due to the small sample size.
The PKP2 S249T variant has been previously
identified (17,30). Variants were considered to be a
possible AC-related variation if they were: i) Absent in 1000
genome databases; ii) a drastic effect was expected in the protein
(nonsense mutation, frame shift mutation, or the mutated residue
was highly conserved among species); or iii) predicted to be
damaging by PolyPhen. As SIFT.PKP2 S249T fuifilled the
above-mentioned conditions via a change in amino acid sequence, its
location in a highly conserved domain and the prediction of its
mutational function, this variant is considered to be an
AC-associated mutation.
There is a 10% chance of missense mutation of PKP2,
resulting in truncated proteins (20,27). In
the present study, the heterozygous point mutation S249T and
frameshift mutation E808fsX30 were demonstrated to be associated
with decreased PKP2 protein expression. Although E808fsX30 resulted
in a truncated PKP2 protein, the protein expression levels
decreased >50%. Therefore, the two mutations may result in
protein instability and degradation, similarly to other PKP2
mutations (Q59L, Q62K, R79X, C796R), which have exhibit a similar
mode of action (31–33). In addition to the decreased
expression levels of PKP2, the expression levels of JUP and Cx43
also decreased in the mutation carrier. This suggests that since
PKP2, JUP, and Cx43 are structural proteins in the intercalated
disc, the degradation of PKP2 could influence the expression of JUP
and Cx43. The possible mechanism underlying this process requires
further investigation.
A previous study reported that loss of plakophilin-2
expression leads to decreased sodium current and slower conduction
velocity in cultured cardiac myocytes (34); thus, decreased sodium current due to
the loss of PKP2 expression may result in arrhythmia. Future
experiments will be necessary to address this possibility.
Further research in to the disease-associated
mutations and protein expression pattern associated with desmosomal
gene mutations will provide a better understanding of the molecular
mechanisms underlying AC. To date, the effects of desmosomal gene
mutations have most commonly been studied in artificial expression
systems using transfected cell cultures and transgenic mice that
overexpress the mutant proteins (12). The present study examined both the
location and expression levels of these proteins in the myocardial
tissue of patients with mutations, and revealed the potential
molecular mechanism underlying these mutations in AC.
As the most common variants, desmosomal gene
mutations are closely related to several clinical characteristics,
including T wave inversion, epsilon wave, age at AC onset and
ventricular arrhythmias. A definite genotype-phenotype relationship
in patients with AC caused by desmosomal gene mutations is helpful
in the diagnosis of AC. Although considerable progress has been
made in the research of genotype-phenotype relationship in patients
with AC, numerous challenges remain due to the genetic and clinical
heterogeneity in these patients. The current study did not include
genotype-phenotype correlation analysis due to the small sample
size; as a result, it could not describe the genetic background of
the AC index of patients who received heart transplantation
comprehensively. In addition, the use of in vitro functional
tests to reveal the pathogenic mechanism underlying these mutations
is required.
This report described variations of five desmosomal
genes in the AC index of patients who underwent heart
transplantation. The results enabled the identification of three
potential AC-associated gene variations, including DSG2 L797Q, PKP2
S249T, and E808fsX30; two of them [PKP2 S249T (Ser249Thr) and
E808fsX30 (Glu808fsX30)] may reduce protein stability and promote
degradation. Conversely, DSG2 L797Q exhibited modifying effects in
AC. The data uncovered one desmosomal gene variation in AC which
has modifying effect and two potential AC-related mutations, this
provides clues for early diagnosis of AC patients in the future.
Furthermore, we detected the expression pattern of related protein
in intercalated disc and provided potential disease mechanism of
the mutations.
Acknowledgements
The authors of the present study are grateful to the
patients and their families for their support and collaboration, to
Dr Zhigang Wang for her advice on data analyses, and to Dr Bi Huang
for his valuable suggestions in the preparation of this manuscript.
The present study was supported by research grants from the CAMS
Innovation Fund for the Medical Sciences (grant no. 2016-I2M-1-015)
and the National Natural Science Foundation of China (grant no.
81470424).
Glossary
Abbreviations
Abbreviations:
|
AC
|
arrhythmogenic cardiomyopathy
|
|
SCD
|
sudden cardiac death
|
|
DSG2
|
desmoglein2
|
|
DSC2
|
desmocollin2
|
|
PKP2
|
plakophilin2
|
|
DSP
|
desmoplakin
|
References
|
1
|
Awad MM, Calkins H and Judge DP:
Mechanisms of disease: Molecular genetics of arrhythmogenic right
ventricular dysplasia/cardiomyopathy. Nat Clin Pract Cardiovasc
Med. 5:258–267. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dalal D, Nasir K, Bomma C, Prakasa K,
Tandri H, Piccini J, Roguin A, Tichnell C, James C, Russell SD, et
al: Arrhythmogenic right ventricular dysplasia: A United States
experience. Circulation. 112:3823–3832. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fressart V, Duthoit G, Donal E, Probst V,
Deharo JC, Chevalier P, Klug D, Dubourg O, Delacretaz E, Cosnay P,
et al: Desmosomal gene analysis in arrhythmogenic right ventricular
dysplasia/cardiomyopathy: Spectrum of mutations and clinical impact
in practice. Europace. 12:861–868. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Saffitz JE: Arrhythmogenic cardiomyopathy
and abnormalities of cell-to-cell coupling. Heart Rhythm. 6(8
Suppl): S62–S65. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Beffagna G, Occhi G, Nava A, Vitiello L,
Ditadi A, Basso C, Bauce B, Carraro G, Thiene G, Towbin JA, et al:
Regulatory mutations in transforming growth factor-beta3 gene cause
arrhythmogenic right ventricular cardiomyopathy type 1. Cardiovasc
Res. 65:366–373. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Merner ND, Hodgkinson KA, Haywood AF,
Connors S, French VM, Drenckhahn JD, Kupprion C, Ramadanova K,
Thierfelder L, McKenna W, et al: Arrhythmogenic right ventricular
cardiomyopathy type 5 is a fully penetrant, lethal arrhythmic
disorder caused by a missense mutation in the TMEM43 gene. Am J Hum
Genet. 82:809–821. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tiso N, Stephan DA, Nava A, Bagattin A,
Devaney JM, Stanchi F, Larderet G, Brahmbhatt B, Brown K, Bauce B,
et al: Identification of mutations in the cardiac ryanodine
receptor gene in families affected with arrhythmogenic right
ventricular cardiomyopathy type 2 (arvd2). Hum Mol Genet.
10:189–194. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
van Tintelen JP, Van Gelder IC, Asimaki A,
Suurmeijer AJ, Wiesfeld AC, Jongbloed JD, van den Wijngaard A, Kuks
JB, van Spaendonck-Zwarts KY, Notermans N, et al: Severe cardiac
phenotype with right ventricular predominance in a large cohort of
patients with a single missense mutation in the des gene. Heart
Rhythm. 6:1574–1583. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Taylor M, Graw S, Sinagra G, Barnes C,
Slavov D, Brun F, Pinamonti B, Salcedo EE, Sauer W, Pyxaras S, et
al: Genetic variation in titin in arrhythmogenic right ventricular
cardiomyopathy-overlap syndromes. Circulation. 124:876–885. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bauce B, Nava A, Beffagna G, Basso C,
Lorenzon A, Smaniotto G, De Bortoli M, Rigato I, Mazzotti E,
Steriotis A, et al: Multiple mutations in desmosomal proteins
encoding genes in arrhythmogenic right ventricular
cardiomyopathy/dysplasia. Heart Rhythm. 7:22–29. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lorenzon A, Beffagna G, Bauce B, De
Bortoli M, Li Mura IE, Calore M, Dazzo E, Basso C, Nava A, Thiene G
and Rampazzo A: Desmin mutations and arrhythmogenic right
ventricular cardiomyopathy. Am J Cardiol. 111:400–405. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rasmussen TB, Hansen J, Nissen PH,
Palmfeldt J, Dalager S, Jensen UB, Kim WY, Heickendorff L, Mølgaard
H, Jensen HK, et al: Protein expression studies of desmoplakin
mutations in cardiomyopathy patients reveal different molecular
disease mechanisms. Clin Genet. 84:20–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rasmussen TB, Palmfeldt J, Nissen PH,
Magnoni R, Dalager S, Jensen UB, Kim WY, Heickendorff L, Mølgaard
H, Jensen HK, et al: Mutated desmoglein-2 proteins are incorporated
into desmosomes and exhibit dominant-negative effects in
arrhythmogenic right ventricular cardiomyopathy. Hum Mutat.
34:697–705. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liang WC, Tian X, Yuo CY, Chen WZ, Kan TM,
Su YN, Nishino I, Wong LC and Jong YJ: Comprehensive target
capture/next-generation sequencing as a second-tier diagnostic
approach for congenital muscular dystrophy in Taiwan. PLoS One.
12:e01705172017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jiang B, Chen Y, Xu B, Hong N, Liu R, Qi M
and Shen L: Identification of a novel missense mutation of MIP in a
Chinese family with congenital cataracts by target region capture
sequencing. Sci Rep. 7:401292017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Miyanaga A, Masuda M, Tsuta K, Kawasaki K,
Nakamura Y, Sakuma T, Asamura H, Gemma A and Yamada T: Hippo
pathway gene mutations in malignant mesothelioma: Revealed by RNA
and targeted exon sequencing. J Thorac Oncol. 10:844–851. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kinnear C, Glanzmann B, Banda E,
Schlechter N, Durrheim G, Neethling A, Nel E, Schoeman M, Johnson G
and van Helden PD: Exome sequencing identifies a novel TTC37
mutation in the first reported case of Trichohepatoenteric syndrome
(THE-S) in South Africa. BMC Med Genet. 18:262017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Frank R, Fontaine G, Vedel J, Mialet G,
Sol C, Guiraudon G and Grosgogeat Y: Electrocardiology of 4 cases
of right ventricular dysplasia inducing arrhythmia. Arch Mal Coeur
Vaiss. 71:963–972. 1978.(In French). PubMed/NCBI
|
|
19
|
Basso C, Corrado D, Marcus FI, Nava A and
Thiene G: Arrhythmogenic right ventricular cardiomyopathy. Lancet.
373:1289–1300. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Campuzano O, Alcalde M, Allegue C,
Iglesias A, García-Pavía P, Partemi S, Oliva A, Pascali VL, Berne
P, Sarquella-Brugada G, et al: Genetics of arrhythmogenic right
ventricular cardiomyopathy. J Med Genet. 50:280–289. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
van der Zwaag PA, Jongbloed JD, van den
Berg MP, van der Smagt JJ, Jongbloed R, Bikker H, Hofstra RM and
van Tintelen JP: A genetic variants database for arrhythmogenic
right ventricular dysplasia/cardiomyopathy. Hum Mutat.
30:1278–1283. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kapplinger JD, Landstrom AP, Salisbury BA,
Callis TE, Pollevick GD, Tester DJ, Cox MG, Bhuiyan Z, Bikker H,
Wiesfeld AC, et al: Distinguishing arrhythmogenic right ventricular
cardiomyopathy/dysplasia-associated mutations from background
genetic noise. J Am Coll Cardiol. 57:2317–2327. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Murray B: Arrhythmogenic right ventricular
dysplasia/cardiomyopathy (ARVD/C): A review of molecular and
clinical literature. J Genet Couns. 21:494–504. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Towbin JA: Arrhythmogenic right
ventricular cardiomyopathy: A paradigm of overlapping disorders.
Ann Noninvasive Electrocardiol. 13:325–326. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sen-Chowdhry S, Syrris P, Pantazis A,
Quarta G, McKenna WJ and Chambers JC: Mutational heterogeneity,
modifier genes and environmental influences contribute to
phenotypic diversity of arrhythmogenic cardiomyopathy. Circ
Cardiovasc Genet. 3:323–330. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Calkins H: Arrhythmogenic right
ventricular dysplasia. Curr Probl Cardiol. 38:103–123. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gerull B, Heuser A, Wichter T, Paul M,
Basson CT, McDermott DA, Lerman BB, Markowitz SM, Ellinor PT,
MacRae CA, et al: Mutations in the desmosomal protein plakophilin-2
are common in arrhythmogenic right ventricular cardiomyopathy. Nat
Genet. 36:1162–1164. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Qiu X, Liu W, Hu D, Zhu T, Li C, Li L, Guo
C, Liu X, Wang L, Zheng H, et al: Mutations of plakophilin-2 in
Chinese with arrhythmogenic right ventricular
dysplasia/cardiomyopathy. Am J Cardiol. 103:1439–1444. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wu SL, Wang PN, Hou YS, Zhang XC, Shan ZX,
Yu XY and Deng M: Mutation of plakophilin-2 gene in arrhythmogenic
right ventricular cardiomyopathy. Chin Med J (Engl). 122:403–407.
2009.PubMed/NCBI
|
|
30
|
Bao J, Wang J, Yao Y, Wang Y, Fan X, Sun
K, He DS, Marcus FI, Zhang S, Hui R, et al: Correlation of
ventricular arrhythmias with genotype in arrhythmogenic right
ventricular cardiomyopathy. Circ Cardiovasc Genet. 6:552–556. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Joshi-Mukherjee R, Coombs W, Musa H,
Oxford E, Taffet S and Delmar M: Characterization of the molecular
phenotype of two arrhythmogenic right ventricular cardiomyopathy
(arvc)-related plakophilin-2 (pkp2) mutations. Heart Rhythm.
5:1715–1723. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hall C, Li S, Li H, Creason V and Wahl JK
III: Arrhythmogenic right ventricular cardiomyopathy plakophilin-2
mutations disrupt desmosome assembly and stability. Cell Commun
Adhes. 16:15–27. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kirchner F, Schuetz A, Boldt LH, Martens
K, Dittmar G, Haverkamp W, Thierfelder L, Heinemann U and Gerull B:
Molecular insights into arrhythmogenic right ventricular
cardiomyopathy caused by plakophilin-2 missense mutations. Circ
Cardiovasc Genet. 5:400–411. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sato PY, Musa H, Coombs W, Guerrero-Serna
G, Patino GA, Taffet SM, Isom LL and Delmar M: Loss of
plakophilin-2 expression leads to decreased sodium current and
slower conduction velocity in cultured cardiac myocytes. Circ Res.
105:523–526. 2009. View Article : Google Scholar : PubMed/NCBI
|