Introduction
Hepatocellular carcinoma (HCC) is one of the most
harmful types of cancer in humans; it is highly malignant and the
prognosis of patients is poor (1).
Recently, the incidence and mortality rates of HCC have been
increasing, particularly across Eastern Asia, including in China.
According to data taken from the National Central Cancer Registry
in 2015, there were 466,100 new HCC cases and 422,100
HCC-associated mortalities in China in 2015, making HCC one of the
top five malignant tumors in the incidence and mortality rates
(2). The median survival time
following HCC diagnosis is very short (usually <6 months) as the
tumors are difficult to resect, due to high recurrence rates
following surgery, and due to poor responses to chemotherapy and
radiotherapy (3). Currently the
molecular mechanisms that lead to the onset, development and
metastasis of HCC remain to be elucidated.
It has been suggested that microRNAs (miRNAs) may be
used as diagnostic and prognostic biomarkers for various types of
tumors. miRNAs are endogenous, evolutionarily conserved, serum
stable, non-coding RNAs (4); these
characteristics mean that they are well suited to be used as
biomarkers. miRNAs are also key regulators of gene transcription.
Therefore, determining the regulatory mechanisms of miRNAs may
improve understanding of cancer pathogenesis. Genome-wide miRNA
profiling studies have revealed that there are numerous
differentially expressed miRNAs in HCC, including miR-223 (5,6).
Furthermore, it has been demonstrated that the expression of
miR-223 is reduced in HCC liver biopsy specimens compared with
normal adjacent liver tissues (6,7).
The increased expression of miR-223 is usually
observed in cells with a myeloid lineage, as miR-223 serves an
important role in granulopoiesis (8). The aberrant expression of miR-223 has
been identified in leukemia and lymphoma (9). In addition, miR-223 is involved in the
tumorigenesis of solid types of cancer; it was demonstrated that
the overexpression of miR-223 inhibits HeLa cell proliferation and
tumor growth in nude mice (10).
This inhibitory role of miR-223 has also been observed in other
tumors, including osteosarcoma (11). In HCC, the downregulation of miR-223
is correlated with the upregulation of stathmin1, a potential
target of miR-223 that is frequently overexpressed in cancer
(6). Insulin-like growth factor-1
receptor (IGF-1R) is another potential target of miR-223;
eosinophil production is increased in miR-223-/- mice, potentially
due to the effects of miR-223 on IGF-1R (12).
IGF-1R is a key protein involved in the IGF-1
signaling pathway and is a highly conserved regulatory module
involved in cell proliferation, differentiation and metabolism
(13). IGF-1 is primarily secreted
by the liver following stimulation by the growth hormone (14). It binds to its receptor IGF-1R, which
is a transmembrane receptor tyrosine kinase and is activated by
phosphorylation. Activation of IGF-1R leads to the downstream
activation of the protein kinase B (Akt) and extracellular
signal-regulated kinase (ERK) signaling pathways, two pathways that
are linked to the development of numerous types of cancer including
prostate cancer, gastric cancer, non-small cell lung cancer,
ovarian cancer and HCC (15). The
dysregulation of IGF-1R has been identified in multiple malignant
tumors, including HCC, prostate and pancreatic cancer (16).
Based on these previous reports, it was hypothesized
that the IGF-1 signaling pathway may mediate the regulatory role of
miR-223 in HCC. In the present study, the role of miR-223 in cell
proliferation, as well as its possible influence on IGF-1R, Akt and
ERK, was investigated.
Materials and methods
Cell culture
The HCC cell line Hep3B [hepatitis B virus
(HBV)-positive and hepatitis C virus (HCV)-negative; ATCC,
Manassas, VA, USA] was used in the current study. Cells were
cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10%
(v/v) fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.).
The cells were incubated were at 37°C in a humidified hood with 5%
CO2.
Cell transfections
The predicted binding sites between miR-223 and
IGF-1R were obtained using the TargetScanHuman online software 7.1
(http://www.targetscan.org/vert_71/).
All transfections were performed using Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Cells were grown
to ~70% confluence and divided into four groups: The blank (BL)
control group in which cells were treated with culture media; the
miR-223 mimic group in which cells were transfected with 100 pmol
miR-223 mimic (5′-UGUCAGUUUGUCAAAUACCCC-3′); the miR-223 inhibitor
group in which cells were transfected with 100 pmol miR-223
inhibitor (5′-UGGGGUAUUUGACAAACUGACA-3′); the NC group in which
cells were transfected with 100 pmol non-specific control RNA
(5′-CAGUACUUUUGUGUAGUACAA-3′). Nucleotides were designed and
synthesized by Shanghai GenePharma Co. Ltd. (Shanghai, China). The
cells were harvested 48 h after transfection for RT-qPCR, western
blot and cell apoptosis analyses.
Tumor samples
A total of 30 HCC tumor tissue samples were used to
analyze the levels of miR-223 and IGF-1R in HCC. Samples were
randomly selected from the pathology department of the First
Affiliated Hospital to Zhejiang University (Hangzhou, China).
Tissue samples were collected from 30 patients (49±11 years old; 24
males and 6 females) that underwent curative surgery for HCC at the
hospital. The sample collection dates were from August 2012 to June
2015. Samples were stored in liquid nitrogen (−196°C) until further
experiments. The study protocol was approved by the Ethics
Committee of the First Affiliated Hospital, College of Medicine,
Zhejiang University and informed consent was obtained from each
patient or their family prior to surgery.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells using
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.).
Total RNA from tissue samples was extracted using the RNAprep Pure
Tissue kit (Tiangen Biotech Co., Ltd., Beijing, China).
Complementary DNA was synthesized from RNA using the PrimeScript RT
Master Mix reagent kit (Takara Biotechnology, Co., Ltd., Dalian,
China). The primer sequences used for reverse transcription were as
follows: miR-223,
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTGGGGT-3′; U6,
5′-AAAATATGGAACGCT-3′ and IGF-1R, 5′-TGCATACAGAATTATTTTATTT-3′. The
oligo dT primer was used for the other genes. qPCR was performed
using the SYBR premix EX Taq kit (Takara Biotechnology, Co., Ltd.).
The thermocycling conditions were as follows: 95°C for 30 sec, and
40 cycles of 95°C for 5 sec and 60°C for 34 sec. U6 was used as
control for miR-223. GAPDH was used as control for the other genes.
The primers used for qPCR were as follows: miR-223, forward,
5′-GTGCAGGGTCCGAGGT-3′ and reverse, 5′-CGGGCTGTCAGTTTGTCA-3′; U6,
forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′; IGF-1R, forward,
5′-GGACAGGTCAGAGGGTTTC-3′ and reverse, 5′-CTCGTAACTCTTCTCTGTGCC-3′;
Akt, forward, 5′-ATCGTCGCCAAGGATGAGGT-3′ and reverse,
5′-TCTCGTGGTCCTGGTTGTAG-3′; ERK, forward,
5′-CCTAAGGAAAAG-CTCAAAGA-3′ and reverse,
5′-AAAGTGGATAA-GCCAAGAC-3′; GAPDH, forward,
5′-ATGGGTGTGAACCATGAGAAGTATG-3′ and reverse,
5′-GGTGCAGGAGGCATTGCT-3′. The relative expressions of the target
genes were normalized to GAPDH expression using the
2−ΔΔCq method (17).
Western blot analysis
Cells were lysed in sodium dodecyl sulfate (SDS)
lysis buffer (Beyotime Institute of Biotechnology, Shanghai,
China). Protein concentrations were determined using the Bradford
Protein Assay kit (Beyotime Institute of Biotechnology). For each
sample, 30 µg/lane of total proteins were separated using 12%
SDS-PAGE. Proteins were then transferred onto nitrocellulose
membranes (EMD Millipore, Billerica, MA, USA), blocked with 5%
bovine serum albumin (Gibco; Thermo Fisher Scientific, Inc.) at
room temperature for 2 h. The membranes were probed with the
relevant primary antibodies overnight at 4°C. The primary
antibodies used were as follows: Rabbit polyclonal to IGF-1R
(G1155; 1:1,000; cat. no. BS1182; Biocompare, South San Francisco,
CA, USA); rabbit polyclonal to ERK 1/2 (1:1,000; cat. no.
ab196883); rabbit polyclonal to phosphorylated (p)-ERK 1/2
(1:1,000; cat. no. ab214362); rabbit polyclonal to Akt (1:1,000;
cat. no. ab8805); rabbit polyclonal to p-Akt (1:1,000; cat. no.
ab38449); and rabbit polyclonal to GAPDH (1:1,000; cat. no. ab9485;
all Abcam, Cambridge, MA, USA). Membranes were then incubated with
horseradish peroxidase-conjugated goat-anti rabbit immunoglobulin G
secondary antibody (cat. no. SC-2030, 1:2,000; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA). Signals were detected using
an enhanced chemiluminescence kit (Pierce; Thermo Fisher
Scientific, Inc.).
Cell proliferation assay
Cell proliferation was measured using a Cell
Counting Kit-8 (CCK-8) assay. A total of 24 h after transfection,
3,000 cells/well were seeded into a 96-well plate. CCK-8 reagent
(Sangon Biotech, Co., Ltd., Shanghai, China) was added 72 h
following seeding. Optical density was measured at 450 nm using a
microplate reader.
Cell apoptosis assay
Cell apoptosis was measured using Annexin V
apoptosis assay kit and 7-amino-actinomycin D staining kit (both
Sangon Biotech, Co., Ltd.) and measured on BD LSRFortessa Cell
Analyzer and analyzed by BD FACSDIVA software version 8.0.1 (BD
Biosciences, Franklin Lakes, NJ, USA).
Statistical analysis
All statistical analyses were performed using SPSS
software version 16.0 (SPSS, Inc., Chicago, IL, USA). Continuous
variables were expressed as the mean ± standard deviation.
Comparison of the relative mRNA expression and cell proliferation
rate among multiple groups was assessed by one-way analysis of
variance followed by Fisher's Least Significant Difference test.
Correlations between miR-223 and IGF-1R mRNA levels in tumor
tissues were analyzed using the linear regression method. P<0.05
was considered to indicate a statistically significant
difference.
Results
miR-223 regulates IGF-1R expression in
HCC
It was determined whether the expression of IGF-1R
is regulated by miR-223. The position 224–231 within the
3′-untranslated coding region of IGF-1R was predicted to be the
binding region of miR-223 (Fig. 1A).
Following transfection of miR-223 mimic, the relative expression of
IGF-1R mRNA was reduced by 38.8% compared with the blank control
(P<0.01; Fig. 1B). The expression
of IGF-1R protein was also reduced in cells transfected with
miR-223 mimic (Fig. 1C). However,
transfection with the miR-223 inhibitor did not alter the levels of
IGF-1R mRNA or protein. These results indicate that IGF-1R
expression is regulated by miR-223.
 | Figure 1.miR-223 regulates IGF-1R expression in
the Hep3B cell line. (A) The 224–231 of the IGF-1R 3′-untranslated
region is the putative target site of miR-223. (B) Relative mRNA
expression of IGF-1R in cells transfected with miR-223 mimic,
miR-223 inhibitor and NC, compared with the BL. The expression of
IGF-1R mRNA was normalized to GAPDH. (C) Expression of IGF-1R
protein detected by western blotting in cells transfected with
miR-223 mimic, miR-223 inhibitor or NC, compared with the BL group.
GAPDH was used as a loading control. Quantified data are presented
as the mean ± standard deviation. **P<0.01 vs. BL. BL, blank
control; NC, negative control RNA; mimic, miR-223 mimic; inhibitor,
miR-223 inhibitor; IGF-1R, insulin-like growth factor-1 receptor;
miR-223, microRNA-223. |
The effect of miR-223 on HCC cell
proliferation and apoptosis
Subsequently, the role of miR-223 on Hep3B cell
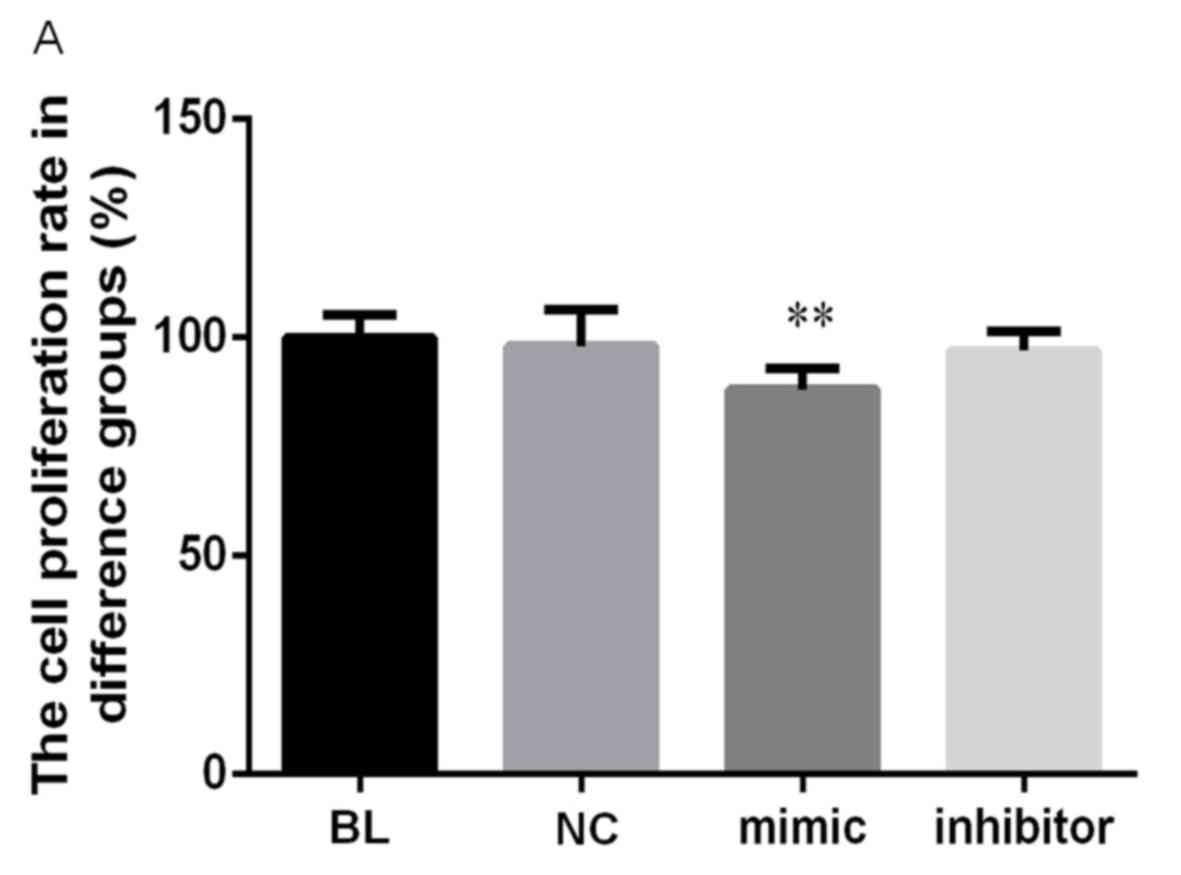
proliferation and apoptosis was investigated. The results of the
CCK-8 assay demonstrated that cell proliferation was significantly
suppressed following transfection of miR-223 mimic (P<0.01;
Fig. 2A). The relative expression of
IGF-1R mRNA was reduced by 11.3% compared with controls (P<0.01;
Fig. 2A). Furthermore, the rate of
cell apoptosis was significantly increased following transfection
with miR-223 mimic compared with the control, from 8.16 to 28.16%
(P<0.01; Fig. 2B and C).
Transfection with NC or miR-223 inhibitor did not significantly
affect cell proliferation or apoptosis compared with the control.
These results indicate that upregulation of miR-223 may reduce the
tumorigenicity of HCC cells in vitro.
miR-223 reduces p-Akt and p-ERK
expression
Since activation of Akt and ERK are essential for
tumor progression, it was investigated whether miR-223
overexpression affected the expression and phosphorylation of these
molecules. The results of western blotting indicated that levels of
p-Akt and p-ERK were reduced following transfection with miR-223
mimic (Fig. 3A). Notably, levels of
total Akt and ERK were also reduced. Levels of Akt and ERK mRNA
were also measured and it was demonstrated that levels of Akt and
ERK mRNA were significantly decreased following transfection
(P<0.01; Fig. 3B). These results
indicate that miR-223 affects downstream signaling of the IGF-1
pathway at the mRNA and protein levels.
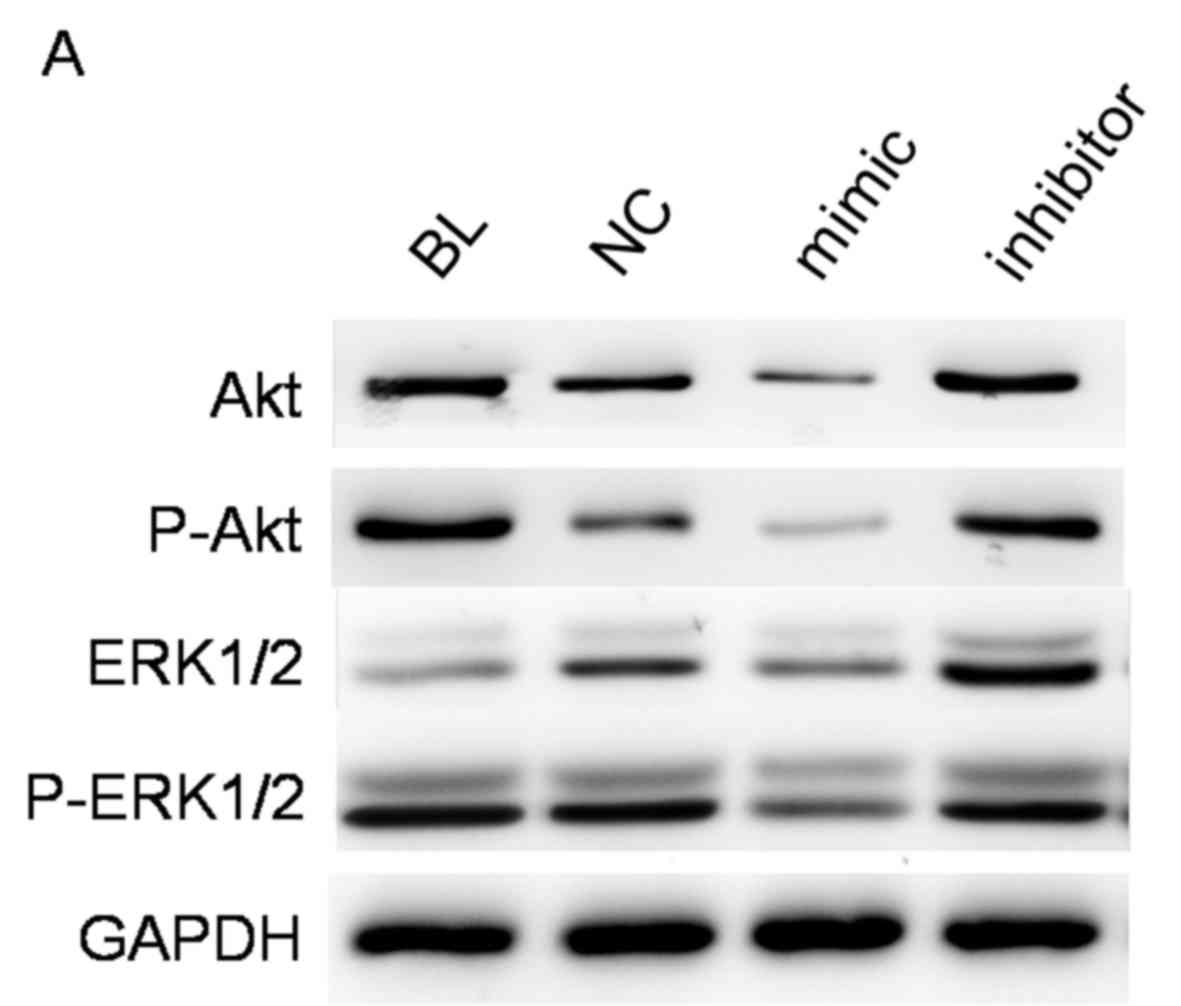 | Figure 3.miR-223 regulates downstream signaling
of the IGF-1 pathway. The expression of Akt and ERK were measured
in cells transfected with miR-223 mimic, miR-223 inhibitor or NC
and compared with the BL group. (A) The expression of total Akt,
total ERK, and their active forms p-Akt and p-ERK were measured by
western blot analysis. (B) Relative mRNA level of Akt and ERK
normalized to GAPDH. Quantified data are presented as the mean ±
standard deviation. **P<0.01 vs. BL. BL, blank control; NC,
negative control RNA; mimic, miR-223 mimic; inhibitor, miR-223
inhibitor; IGF-1R, insulin-like growth factor-1 receptor; miR-223,
microRNA-223; p-, phosphorylated; Akt, protein kinase B; ERK,
extracellular-signal regulated kinase. |
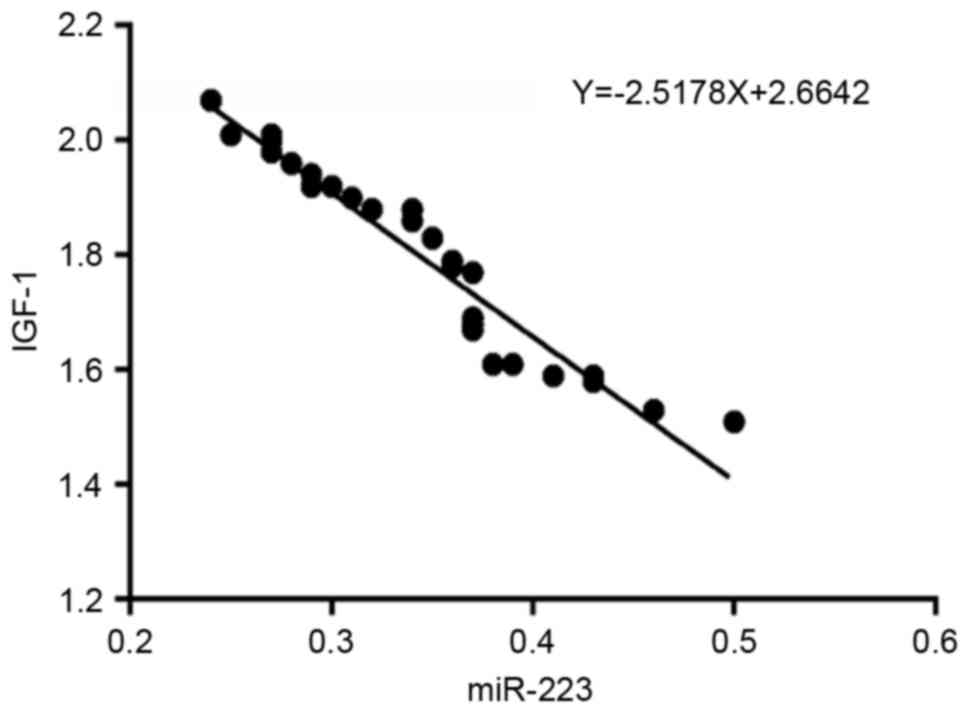
miR-223 expression is negatively
correlated with IGF-1R expression
To confirm the correlation between miR-223 and
IGF-1R, their levels were measured in 30 HCC tissues. As presented
in Fig. 4, a significant inverse
correlation was identified between miR-223 and IGF-1R mRNA
expression (regression equation: Y=−2.5178X+2.6642, r=−0.965).
Therefore, taken together, the results of the present study
indicate that IGF-1R is directly regulated by miR-223.
Discussion
It has been determined that miR-223 expression is
downregulated in HCC tumor tissue compared with matching adjacent
normal liver tissue (6). Therefore,
the aim of the present study was to assess whether miR-223 may act
as a tumor suppressor in HCC by inhibiting cell proliferation and
promoting apoptosis. Furthermore, the current study aimed to
identify the method by which miR-223 acts as a tumor suppressor.
One potential mechanism by which it may act may be via the direct
modulation of IGF-1R and the downstream regulation of Akt and ERK.
Jung and Suh (13) indicated that
the Akt and ERK pathways are involved in cell proliferation and
apoptosis by regulating the expression of Bcl-2, p27, forkhead box
protein (FOXO) transcription factors and BAD.
The most common risk factors for HCC are chronic
infection with viral hepatitis B and C, and dietary exposure to
aflatoxin B1 (2). Since
miR-223 may be a negative regulator of inflammation (8), the chronic inflammatory response in the
liver may be explained by miR-223 repression. Although miR-223 is
commonly downregulated in HCC tumor tissue, its levels of
expression in the sera of patients with HCC are variable. Xu et
al (18) reported elevated
miR-223 levels, whereas Bhattacharya et al (7) reported reduced miR-223 levels in the
sera of patients with HCC. These contrasting results suggest that
serum miR-223 may not be suitable as a standard biomarker for HCC;
further studies are required to evaluate its diagnostic value.
The role of miR-223 in cell proliferation has been
observed in multiple HCC cell lines. Wong et al (6) identified that miR-223 expression was
reduced in 11 HBV-related cell lines, 4 HCV-related cell lines and
3 non-B or non-C-related cell lines, including the Hep3B cell line
used in the current study. The results of a previous study also
indicated that the IGF-1 signaling pathway is a crucial regulatory
route of miR-223 in HeLa cells (10). IGF-1-mediated regulation by miR-223
has also been reported in Lewis lung carcinoma cells (19). Furthermore, it has been demonstrated
that the transcription factor FOXO1 mediates the influence of
miR-223 on the proliferation of HCT116 colorectal cancer cells,
HeLa cervical cancer cells and HuH-7 hepatoma cells (20). Activation of the Akt pathway results
in the downstream activation FOXO1; therefore, the involvement of
miR-223 in the IGF-1 pathway does not merely occur via IGF-1R,
other factors are also involved. However, regulation of the IGF-1
pathway using miRNAs or miRNA-targeting oligonucleotides is an
attractive strategy for the treatment of cancer. Several ongoing
clinical trials have already tested the efficiency and safety of
drugs targeting IGF-1R, including the IGF-1R antibodies MK-0646 and
the IGF-1R tyrosine kinase inhibitor OSI-906 (21,22).
The regulation of miR-223 expression may, at least
partially, be induced by the p53 R175H gain-of function mutation.
This mutation inhibits the miR-223 promoter in colon and breast
cancer cell lines and is associated with chemoresistance in tumor
cells (23). Inhibition of oncogenic
Notch signaling may induce miR-223 expression in T-lineage acute
lymphoblastic leukemia cells, implicating that Notch may be another
regulator of miR-223 expression in cancer cells (24).
Although miR-223 has proven to be a tumor suppressor
in HCC, miR-223 may serve opposing roles in the migration and
invasion of different types of cancer cells. In esophageal cancer
cells, miR-223 seems to suppress cell migration and invasion
(25), whereas in metastatic gastric
cancer cells and recurrent ovarian cancer, miR-223 is overexpressed
and promotes metastasis (26,27).
These contrasting results indicate that further studies are
required to determine the roles miR-223 serves in different types
of cancer.
Theoretically, repression of miR-223 is supposed to
promote IGF-1R production. However, in the present study,
transfection with miR-223 inhibitor did not significantly increase
IGF-1R production compared with the control. This may be due to the
fact that miR-223 is already downregulated in HCC tumor cells and
further inhibition of miR-223 may not lead to the marked promotion
of IGF-1R expression. Another possible explanation is that IGF-1R
is under the regulation of many factors and the effect of
inhibiting miR-223 expression may be restricted by those other
factors.
In conclusion, it has been demonstrated that miR-223
serves a role as a tumor suppressor in HCC cells by regulating
IGF-1R expression. There were a few limitations of the current
study. Only one HCC cell line was tested; experiments in other HCC
cell lines should be performed to verify if the regulation of
IGF-1R by miR-223 is universal in HCC cells. Furthermore, the
current study was an in vitro study; in vivo studies,
for example, studies investigating the effects of miR-223 on tumor
formation in nude mice, are required to confirm the regulating role
of miR-223 via the IGF-1 pathway. miR-223 may have other targets
besides IGF-1R, therefore future studies should investigate whether
miR-223 also regulates other target genes in HCC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JZ designed and supervised the study. CZ performed
the experiments and analyzed the data. CZ and JZ discussed the
results and wrote the study together.
Ethics approval and consent to
participate
The study protocol was approved by the Ethics
Committee of the First Affiliated Hospital, College of Medicine,
Zhejiang University and informed consent was obtained from each
patient or their family prior to surgery.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidmeiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kew MC: Hepatocellular carcinoma:
Epidemiology and risk factors. J Hepatocellular Carcinoma.
1:115–125. 2014. View Article : Google Scholar
|
|
4
|
Chen K and Rajewsky N: The evolution of
gene regulation by transcription factors and microRNAs. Nat Rev
Genet. 8:93–103. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li LM, Hu ZB, Zhou ZX, Chen X, Liu FY,
Zhang JF, Shen HB, Zhang CY and Zen K: Serum microRNA profiles
serve as novel biomarkers for HBV infection and diagnosis of
HBV-positive hepatocarcinoma. Cancer Res. 70:9798–9807. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wong QW, Lung RW, Law PT, Lai PB, Chan KY,
To KF and Wong N: MicroRNA-223 is commonly repressed in
hepatocellular carcinoma and potentiates expression of stathmin1.
Gastroenterology. 135:257–269. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bhattacharya S, Steele R, Shrivastava S,
Chakraborty S, Di Bisceglie AM and Ray RB: Serum miR-30e and
miR-223 as novel noninvasive biomarkers for hepatocellular
carcinoma. Am J Pathol. 186:242–247. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ramkissoon SH, Mainwaring LA, Ogasawara Y,
Keyvanfar K, McCoy JP Jr, Sloand EM, Kajigaya S and Young NS:
Hematopoietic-specific microRNA expression in human cells. Leuk
Res. 30:643–647. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fazi F, Racanicchi S, Zardo G, Starnes LM,
Mancini M, Travaglini L, Diverio D, Ammatuna E, Cimino G, Lo-Coco
F, et al: Epigenetic silencing of the myelopoiesis regulator
microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell. 12:457–466.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jia CY, Li HH, Zhu XC, Dong YW, Fu D, Zhao
QL, Wu W and Wu XZ: miR-223 suppresses cell proliferation by
targeting IGF-1R. PLoS One. 6:e270082011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li G, Cai M, Fu D, Chen K, Sun M, Cai Z
and Chen B: Heat shock protein 90B1 plays an oncogenic role and is
a target of microRNA-223 in human osteosarcoma. Cell Physiol
Biochem. 30:1481–1490. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lu TX, Lim E, Besse JA, Itskovich S,
Plassard AJ, Fulkerson PC, Aronow BJ and Rothenberg ME: miR-223
deficiency increases eosinophil progenitor proliferation. J
Immunol. 190:1576–1582. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jung HJ and Suh Y: Regulation of IGF-1
signaling by microRNAs. Front Genet. 5:4722015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fürstenberger G and Senn H: Insulin-like
growth factors and cancer. Lancet Oncol. 3:298–302. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Martini M, De Santis M, Braccini L,
Gulluni F and Hirsch E: PI3K/AKT signaling pathway and cancer: An
updated review. Ann Med. 46:372–383. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Samani A, Yakar S, LeRoith D and Brodt P:
The role of the IGF system in cancer growth and metastasis:
Overview and recent insights. Endocr Rev. 28:20–47. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu J, Wu C, Che X, Wang L, Yu D, Zhang T,
Huang L, Li H, Tan W, Wang C and Lin D: Circulating MicroRNAs,
miR-21, miR-122, and miR-223, in patients with hepatocellular
carcinoma or chronic hepatitis. Mol Carcinog. 50:136–142. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nian W, Ao X, Wu Y, Huang Y, Shao J, Wang
Y, Chen Z, Chen F and Wang D: miR-223 functions as a potent tumor
suppressor of the Lewis lung carcinoma cell line by targeting
insulin-like growth factor-1 receptor and cyclin-dependent kinase
2. Oncol Lett. 6:359–366. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu L, Li H, Jia CY, Cheng W, Yu M, Peng M,
Zhu Y, Zhao Q, Dong YW, Shao K, et al: MicroRNA-223 regulates FOXO1
expression and cell proliferation. FEBS Lett. 586:1038–1043. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Reidy-Lagunes DL, Vakiani E, Segal MF,
Hollywood EM, Tang LH, Solit DB, Pietanza MC, Capanu M and Saltz
LB: A phase 2 study of the insulin-like growth factor-1 receptor
inhibitor MK-0646 in patients with metastatic, well-differentiated
neuroendocrine tumors. Cancer. 118:4795–4800. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pitts TM, Tan AC, Kulikowski GN, Tentler
JJ, Brown AM, Flanigan SA, Leong S, Coldren CD, Hirsch FR,
Varella-Garcia M, et al: Development of an integrated genomic
classifier for a novel agent in colorectal cancer: Approach to
individualized therapy in early development. Clin Cancer Res.
16:3193–3204. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Masciarelli S, Fontemaggi G, Di Agostino
S, Donzelli S, Carcarino E, Strano S and Blandino G:
Gain-of-function mutant p53 downregulates miR-223 contributing to
chemoresistance of cultured tumor cells. Oncogene. 33:1601–1608.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gusscott S, Kuchenbauer F, Humphries RK
and Weng AP: Notch-mediated repression of miR-223 contributes to
IGF1R regulation in T-ALL. Leuk Res. 36:905–911. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li S, Li Z, Guo F, Qin X, Liu B, Lei Z,
Song Z, Sun L, Zhang HT, You J and Zhou Q: miR-223 regulates
migration and invasion by targeting Artemin in human esophageal
carcinoma. J Biomed Sci. 18:242011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li X, Zhang Y, Zhang H, Liu X, Gong T, Li
M, Sun L, Ji G, Shi Y, Han Z, et al: miRNA-223 promotes gastric
cancer invasion and metastasis by targeting tumor suppressor
EPB41L3. Mol Cancer Res. 9:824–833. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Laios A, O'Toole S, Flavin R, Martin C,
Kelly L, Ring M, Finn SP, Barrett C, Loda M, Gleeson N, et al:
Potential role of miR-9 and miR-223 in recurrent ovarian cancer.
Mol Can. 7:352008. View Article : Google Scholar
|