Introduction
Cerebral small vessel disease (SVD), an intrinsic
disorder of the brain's perforating arterioles, which could induce
vascular dementia, cognitive decline or disability (1,2). SVD
refers to a set of pathological processes the two most frequent
types of which were cerebral amyloid angiopathy and
hypertension-associated small vessel diseases (3–5). The
damages of SVD on brain are usually lacunar infarcts, microbleeds
and white matter lesions. Common SVD can be detected via
neuroimaging, whereas small vessels (including small arteries and
venules) cannot be diagnosed by neuroimaging. In this condition,
postmortem plays an important role in detecting microvascular
pathology (6,7). Owing to the development of
high-throughput sequencing technology, long non-coding RNAs
(lncRNAs) have been discovered in a wide range of biological
processes. The functions of lncRNAs in SVD are not well
characterized, and the identification of lncRNA biomarkers is
challenging. lncRNAs compete with competitive endogenous RNAs
(ceRNAs) by acting as miRNA sponges to regulate expression level of
other transcripts (8). Several ceRNA
related databases have also been developed to facilitate
interference of lncRNA function. Collectively, this data
underscores the importance of lncRNA interactions with ceRNAs, and
indicates that integration of expression profiles and network
analysis could enable identification of risk lncRNAs and the
underlying tumor pathology of SVD. Up to now, the pathogenesis of
SVD is basically unknown. The pathogenesis of SVD is still not
clear, so we need to find molecular markers to prevent.
It is reported that human gene transcripts which
encode proteins account for approximatly 2% of all transcripts, and
the remaining transcripts are non-coding RNAs (ncRNAs), which based
on length could divide into two subclasses lncRNA and small ncRNA
(9,10). lncRNAs, which often longer than 200
nucleotides, lack open reading frames compared with mRNAs and
through chromatin structural alternation and transcriptional
modulation regulate gene transcriptions (11–14).
Recent studies have identified that lncRNAs played a pivotal role
in tumorigenesis and tumor metastasis (15). Prostate cancer (PC) associated loci
were mostly located at lncRNA regions. Jin et al reported
that 52 loci were enriched in lncRNA regions and SNP rs3787016
which was located at lncRNA regions was risk-associated with PC
(16). In human ovarian cancer
SKOV3.ip 1 cell line, lncRNA 578 was downregulated while lncRNA 583
was upregulated, compared with parental SKOV3 cell line
respectively (17).
ceRNAs modulate gene expression in trans and account
for major parts of gene regulators (18). MicroRNA recognition elements (MREs),
a common sequences of ceRNAs, which recognized by microRNAs through
sequence complementarity and usually induce gene repression
(19). Complicated interaction
network between gene transcripts have MREs which regulate each
other are called ceRNA crosstalk (20). Zhou et al constructed a breast
cancer-related ceRNA network by integrating miRNA expression data
and mRNA expression data (21). In
human ovarian cancer, 10 lncRNAs which related to ceRNA network
were determined as potential biomarkers (22).
According to the criteria reported by Cao et
al (23), we constructed a SVD
associated ceRNA network by integrating mRNA and lncRNA profiling,
which contained 236,952 miRNA-mRNA interactions and 359
lncRNA-miRNA intersections. We used a hypergeometric test to
evaluate enriched significance of miRNAs related with lncRNAs and
mRNAs. A significant co-expressed lncRNA-mediated ceRNA network
(LMCN) was constructed which contained 21 lncRNAs, 129 mRNA and 141
ceRNA interaction pairs. In the prediction of lncRNA functions, 4
significant gene oncology terms contained 7 genes were identified.
In adition, we identified 29 mRNAs enriched pathways which were
significantly related with lncRNAs. These results demonstrated that
LMCNs were useful for identifying potential biomarkers associated
with SVD.
Materials and methods
Data processing
miRNA-mRNA interactions and lncRNA-miRNA
intersections which associated with SVD were downloaded from
starBase 2.0 database (http://starbase.sysu.edu.cn/), whilie mRNA and lncRNA
expression profiling were downloaded from ArrayExpress database
(https://www.ebi.ac.uk/arrayexpress/).
The mRNA and lncRNA expression profiling comprised of 30 pairs of
human SVD and normal control samples from EMBL-EBI dataset whose
dataset number was E-MTAB-3408, which employed mapping between
probes and genes to perform.
Genes of expression profiling were intersected with
mRNAs of miRNA-mRNA intersections and lncRNAs of lncRNA-miRNA
intersections respectively. A new expression profiling which
contained 7,649 mRNAs and 27 lncRNAs was identified.
Identification of potential ceRNA
interactions
Hypergeometric test was used to evaluate enriched
significance of miRNAs which were interacted both with lncRNAs and
mRNAs. The P-values were subjected to false discovery rate (FDR)
method, which were calculated as follows:
P=1-∑t=0x(Kt)(N-KM-t)(NM)
A FDR <0.01 was considered as the threshold.
Identification of sub-LMCN
Co-expression analysis of lncRNA-mRNA interactions
was carried out to identify lncRNA-mRNA pairs. In the control and
the disease groups, Pearson correlation coefficients of interaction
pairs were calculated. The Pearson correlation coefficient was
calculated as follows:
ρX,Y=cov(X,Y)σXσY
cov (X,Y) represents the covariance of variables X
and Y. σX and σY are the
standard deviations of X and Y, respectively.
lncRNA-mRNA interactions with Pearson correlation coefficients
>0.8 were considered as significant co-expression ceRNA
interactions.
Gene Ontology (GO) prediction of
mRNAs
Functional enrichment analysis of mRNAs, which
utilized Gene Ontology Consortium (http://www.geneontology.org), significantly correlated
with lncRNAs, which predicted the function of lncRNAs using the
function of mature mRNAs. lncRNA functions were predicted by mature
mRNA functions. P-value =0.05 was used as the threshold to perform
functional enrichment analysis for mRNAs of sub-LMCN.
Pathway prediction of lncRNAs
The pathway database selected for this analysis is
the Reactome (https://reactome.org), which uses
Fisher's test to identify lncRNA enriched pathways. Pathways with
P-values <0.05 were considered as mRNA enriched pathways which
maybe mediated by lncRNAs. These pathways were putative
lncRNA-mediated pathways.
Results
Identification of miRNA-mRNA
interactions and lncRNA-miRNA intersections
SVD associated mRNA and lncRNA expression profiling
data which were downloaded from ArrayExpress database contained 30
pairs of disease and control samples and we obtained a profile data
of 12,628 genes were identified via mapping between probes and
genes.
A new gene expression profile of 7,676 genes
including 7,649 mRNA and 27 lncRNAs was obtained by crossing the
genes in the expression profile data with the mRNAs and lncRNAs in
the above miRNA-mRNA interactions and lncRNA-miRNA interactions,
respectively. Furthermore, the interactions containing
new-expressed gene (mRNA or lncRNA) were screen out from miRNA-mRNA
interactions and lncRNA-miRNA interactions, and we obtained 236,952
pairs of miRNA-mRNA interactions and 359 pairs separately.
Identification of potential ceRNA
interactions
We used hypergeometric test to evaluate enrichment
significance of miRNAs which were interacted with lncRNAs and
mRNAs. The P-values were corrected by false discovery rate method
and the LMCN was filtrated from the lncRNA-mRNA interactions with
P-values <0.01. As a result, the LMCN contained 27 lncRNAs,
7,229 mRNA, and 28,871 lncRNAs-mRNA interrelationship pairs.
Identification of a highly competitive
sub-LMCN
The partial sub-networks revealed a more detailed
picture of how the lncRNAs synergized with competing mRNAs, while
the LMCN could provide a global view of all possible competing
ceRNA interactions. Therefore, we derived a high-competing
lncRNA-related sub-network (sub-LMCN) from the LMCN by co-expressed
analysis the lncRNA-mRNA interactions screened above. Pearson
correlation coefficient threshold of lncRNA-mRNA interactions in
the control and disease groups were calculated respectively.
lncRNA-mRNAs interactions with Pearson correlation coefficient
threshold >0.8 were considered as significant co-expressed ceRNA
interactions. These ceRNAs constructed a network which contained 21
lncRNA, 129 mRNA and 141 interaction pairs (Fig. 1).
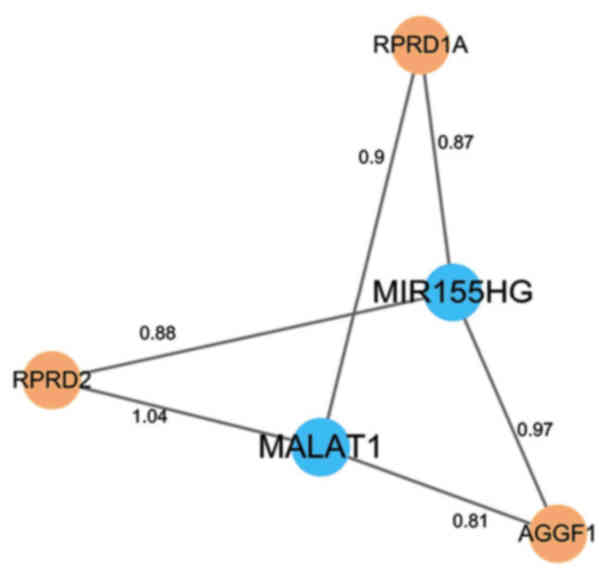
What's more, a cooperative with competition network
was identified from the sub-LMCN. SVD associated lncRNAs such as
MIR155 host gene (MIR155HG) and metastasis-associated lung
adenocarcinoma transcript 1 (MALAT1) competed with other mRNAs in
the sub-LMCN (Fig. 2).
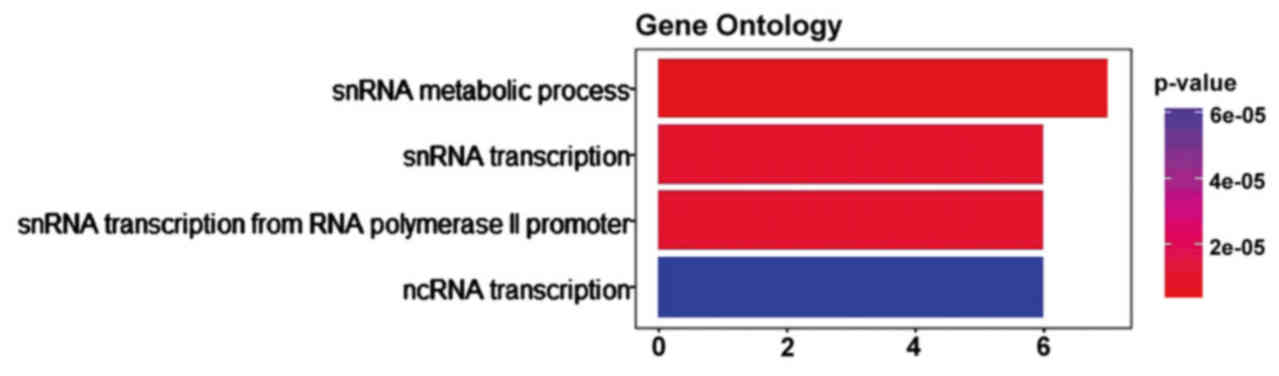
Prediction of lncRNA functions
The functions of mature mRNAs were used to predict
lncRNA functions. mRNAs were enriched via GO analysis and we
identified 4 GO terms, including snRNA metabolic process
(P=0.0037), snRNA transcription (P=0.0058), snRNA transcription
from RNA polymerase II promoter (P=0.0058) and ncRNA transcription
(P=0.0280) (Fig. 3). The snRNA
metabolic process including 7 genes (RPRD2, RPRD1A, NOP10, POLR2A,
SNAPC3, POLR2H, TAF13) and each of the remaining three terms
contain 6 genes (RPRD2, RPRD1A, POLR2A, SNAPC3, POLR2H, TAF13).
Fisher test was used to identify lncRNA enrichment
pathways, which were selected from Reactome database. As results,
29 enriched pathways of mRNA enrichemt significantly correlated
with lncRNA were identified, which maybe mediated by lncRNAs
(Table I).
 | Table I.Twenty-nine lncRNA-mediated pathways
were identified by Fisher test. |
Table I.
Twenty-nine lncRNA-mediated pathways
were identified by Fisher test.
| Pathway names | Corrected
P-values |
|---|
| Processing of capped
intron-containing pre-mRNA | 0.024321 |
| Elongation arrest and
recovery | 0.033775 |
| Gene expression | 0.033775 |
| Nuclear import of Rev
protein | 0.033775 |
| Pausing and recovery
of HIV elongation | 0.033775 |
| Pausing and recovery
of Tat-mediated HIV elongation | 0.033775 |
| RNA polymerase II
pre-transcription events | 0.033775 |
| Transcriptional
regulation by small RNAs | 0.033775 |
| Transport of
ribonucleoproteins into the host nucleus | 0.033775 |
| Transport of the
SLBP dependant mature mRNA | 0.033775 |
| Transport of the
SLBP independent mature mRNA | 0.033775 |
| Viral messenger RNA
synthesis | 0.033775 |
| Formation of RNA
Pol II elongation complex | 0.034444 |
| Influenza life
cycle | 0.034444 |
| Interactions of Rev
with host cellular proteins | 0.034444 |
| mRNA splicing -
minor pathway | 0.034444 |
| RNA polymerase II
promoter escape | 0.034444 |
| RNA polymerase II
transcription elongation | 0.034444 |
| RNA polymerase II
transcription initiation | 0.034444 |
| RNA polymerase II
transcription initiation and promoter clearance | 0.034444 |
| RNA polymerase II
transcription pre-initiation and promoter opening | 0.034444 |
| RNA polymerase III
abortive and retractive initiation | 0.034444 |
| RNA polymerase III
transcription | 0.034444 |
| Transcription | 0.034444 |
| Transport of mature
mRNA derived from an intronless transcript | 0.034444 |
| Transport of mature
mRNAs derived from intronless transcripts | 0.034444 |
| Antigen processing:
ubiquitination and proteasome degradation | 0.034552 |
| Influenza
infection | 0.036016 |
| Regulatory RNA
pathways | 0.049813 |
Discussion
ceRNAs mutually regulate miRNAs expression through
competing mechanisms, which are important for physiological and
pathological processes of a variety of cancers. It has developed
several valuable ceRNA resources to promote the lncRNAs functional
analysis such as DIANA-LncBase (24), starBase (25) and LncACTdb (26). Here, we have used a more
comprehensive approach to construct a functional LMCN across 30
pairs of samples. In our study, we constructed a ceRNAs network by
integrating lncRNA and mRNA co-expression profiling, which
contained 27 lncRNAs, 7,649 mRNA and an expression data of 7,676
genes. And then, we obtained 236,952 pairs of miRNA-mRNA
interactions and 359 pairs of lncRNA-miRNAs intersections through
extracting from miRNA-mRNA interactions and lncRNA-miRNA
interactions separately. Hypergeometric test was used to evaluate
enrichment significance of miRNAs, which has interaction with both
lncRNA and mRNA. The P-values were corrected by FDR method, and
lncRNA-mRNA intersections (LMCN) with corrected P-values <0.01
were screened, which was containing 27 lncRNAs, 7,229 mRNAs and
28,871 pairs of LMCNs. In addition, co-expression analysis of
lncRNA-mRNA interactions screened above, which was to calculate the
Pearson correlation coefficients of these interactions in the
control and disease groups. The interactions between Pearson
correlation coefficients larger than 0.8 were screened for ceRNA
interactions that were significantly co-expressed and network maps
were constructed, which contained 21 lncRNAs, 129 mRNAs and 141
pairs of interactions. However, the lncRNA and miRNA with most
nodes in the interaction network and the correlation with patents
prognosis should be further analyzed, we will add the analysis in
our next study.
By exploring the highly competitive sub-network of
the LMCN, we identified a co-competing module from sub-LMCN that
MALAT1 and MIR155HG comprised a ceRNA regulating module that
competed with RPRD1A, RPRD2, and AGGF1. lncRNA MALAT1, a well-known
lncRNA, is associated with pathogenesis and progress of variety of
tumors (27). MIR155HG, the primary
microRNA of miR-155, which plays an essential role in
hematopoiesis, inflammation, and tumorigenesis (28). Blood-based biomarkers are critical
for prediction of SVD patient survival. We observed MALAT1
expression in white blood cells in a tissue-specific RNA analysis
(29) and in several
blood-associated RNASeq profiles. These data indicated that MALAT1
can be secreted into the circulation and is therefore a potential
blood-based biomarker for SVD.
Functional enrichment analysis of mRNAs
significantly correlated with lncRNAs, which predicted the function
of lncRNAs using the function of mature mRNAs. Seven genes,
including RPRD2, RPRD1A, NOP10, POLR2A, SNAPC3, POLR2H, TAF13, were
obtained through GO enrichment. In addition, 29 mRNA enriched
pathways significantly associated with lncRNAs was obtained via
Fisher test. The results demonstrated that the cancer-associated
LMCN can be used to accelerate biomarker discovery and SVD
therapeutic development. However, lack of KEGG analysis is the
limitation of our study, we will offer KEGG analysis results in our
future manuscript. What's more, we will evaluate our predictions
using clinical examples in future study.
In conclusion, we identified 7 potential lncRNAs and
29 possible lncRNA-mediated pathways associated with SVD through
enrichment. These findings may contribute to deepen understanding
of SVD mechanisms.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HW contributed to the conception of the study. HY
contributed significantly to perform the experiments and wrote the
manuscript; XY contributed significantly to analysis and helped in
the writing of the manuscript; XZ performed the data analyses. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ceRNAs
|
competitive endogenous RNAs
|
|
SVD
|
cerebral small vessel disease
|
|
ncRNAs
|
non-coding RNAs
|
|
lncRNA
|
long non-coding RNA
|
|
LMCN
|
lncRNA-mediated ceRNA network
|
|
MALAT1
|
metastasis-associated lung
adenocarcinoma transcript 1
|
|
MIR155HG
|
MIR155 host gene
|
References
|
1
|
Pantoni L: Cerebral small vessel disease:
From pathogenesis and clinical characteristics to therapeutic
challenges. Lancet Neurol. 9:689–701. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wardlaw JM, Smith C and Dichgans M:
Mechanisms of sporadic cerebral small vessel disease: Insights from
neuroimaging. Lancet Neurol. 12:483–497. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
van der Flier WM, van Straaten EC, Barkhof
F, Verdelho A, Madureira S, Pantoni L, Inzitari D, Erkinjuntti T,
Crisby M, Waldemar G, et al: Small vessel disease and general
cognitive function in nondisabled elderly: The LADIS study. Stroke.
36:2116–2120. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pantoni L, Sarti C, Alafuzoff I, Jellinger
K, Munoz DG, Ogata J and Palumbo V: Postmortem examination of
vascular lesions in cognitive impairment: A survey among
neuropathological services. Stroke. 37:1005–1009. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hu X, De Silva TM, Chen J and Faraci FM:
Cerebral vascular disease and neurovascular injury in ischemic
stroke. Circ Res. 120:449–471. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lammie GA, Brannan F, Slattery J and
Warlow C: Nonhypertensive cerebral small-vessel disease. An autopsy
study. Stroke. 28:2222–2229. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dalkara T and Alarcon-Martinez L: Cerebral
microvascular pericytes and neurogliovascular signaling in health
and disease. Brain Res. 1623:3–17. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Das S, Ghosal S, Sen R and Chakrabarti J:
lnCeDB: Database of human long noncoding RNA acting as competing
endogenous RNA. PLoS One. 9:e989652014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Birney E, Stamatoyannopoulos JA, Dutta A,
Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis
ET, Thurman RE, et al: Children's Hospital Oakland Research
Institute: Identification and analysis of functional elements in 1%
of the human genome by the ENCODE pilot project. Nature.
447:799–816. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kapranov P, Cheng J, Dike S, Nix DA,
Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J,
Hofacker IL, et al: RNA maps reveal new RNA classes and a possible
function for pervasive transcription. Science. 316:1484–1488. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guo X, Gao L, Wang Y, Chiu DK, Wang T and
Deng Y: Advances in long noncoding RNAs: Identification, structure
prediction and function annotation. Brief Funct Genomics. 15:38–46.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Martianov I, Ramadass A, Barros Serra A,
Chow N and Akoulitchev A: Repression of the human dihydrofolate
reductase gene by a non-coding interfering transcript. Nature.
445:666–670. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et
al: Functional demarcation of active and silent chromatin domains
in human HOX loci by noncoding RNAs. Cell. 129:1311–1323. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Y, Tao Y and Liao Q: Long noncoding
RNA: A crosslink in biological regulatory network. Brief Bioinform.
Apr 24–2017.(Epub ahead of print).
|
|
16
|
Jin G, Sun J, Isaacs SD, Wiley KE, Kim ST,
Chu LW, Zhang Z, Zhao H, Zheng SL, Isaacs WB, et al: Human
polymorphisms at long non-coding RNAs (lncRNAs) and association
with prostate cancer risk. Carcinogenesis. 32:1655–1659. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu SP, Yang JX, Cao DY and Shen K:
Identification of differentially expressed long non-coding RNAs in
human ovarian cancer cells with different metastatic potentials.
Cancer Biol Med. 10:138–141. 2013.PubMed/NCBI
|
|
18
|
Kartha RV and Subramanian S: Competing
endogenous RNAs (ceRNAs): New entrants to the intricacies of gene
regulation. Front Genet. 5:82014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhou X, Liu J and Wang W: Construction and
investigation of breast-cancer-specific ceRNA network based on the
mRNA and miRNA expression data. IET Syst Biol. 8:96–103. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhou M, Wang X, Shi H, Cheng L, Wang Z,
Zhao H, Yang L and Sun J: Characterization of long non-coding
RNA-associated ceRNA network to reveal potential prognostic lncRNA
biomarkers in human ovarian cancer. Oncotarget. 7:12598–12611.
2016.PubMed/NCBI
|
|
23
|
Cao Y, Wang P, Ning S, Xiao W, Xiao B and
Li X: Identification of prognostic biomarkers in glioblastoma using
a long non-coding RNA-mediated, competitive endogenous RNA network.
Oncotarget. 7:41737–41747. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Paraskevopoulou MD, Georgakilas G,
Kostoulas N, Reczko M, Maragkakis M, Dalamagas TM and Hatzigeorgiou
AG: DIANA-LncBase: Experimentally verified and computationally
predicted microRNA targets on long non-coding RNAs. Nucleic Acids
Res. 41(D1): D239–D245. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42(D1): D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang P, Ning S, Zhang Y, Li R, Ye J, Zhao
Z, Zhi H, Wang T, Guo Z and Li X: Identification of
lncRNA-associated competing triplets reveals global patterns and
prognostic markers for cancer. Nucleic Acids Res. 43:3478–3489.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li Y, Bao C, Gu S, Ye D, Jing F, Fan C,
Jin M and Chen K: Associations between novel genetic variants in
the promoter region of MALAT1and risk of colorectal cancer.
Oncotarget. 8:92604–92614. 2017.PubMed/NCBI
|
|
28
|
Wu X, Wang Y, Yu T, Nie E, Hu Q, Wu W, Zhi
T, Jiang K, Wang X, Lu X, et al: Blocking MIR155HG/miR-155 axis
inhibits mesenchymal transition in glioma. Neuro-oncol.
19:1195–1205. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cabili MN, Trapnell C, Goff L, Koziol M,
Tazon-Vega B, Regev A and Rinn JL: Integrative annotation of human
large intergenic noncoding RNAs reveals global properties and
specific subclasses. Genes Dev. 25:1915–1927. 2011. View Article : Google Scholar : PubMed/NCBI
|