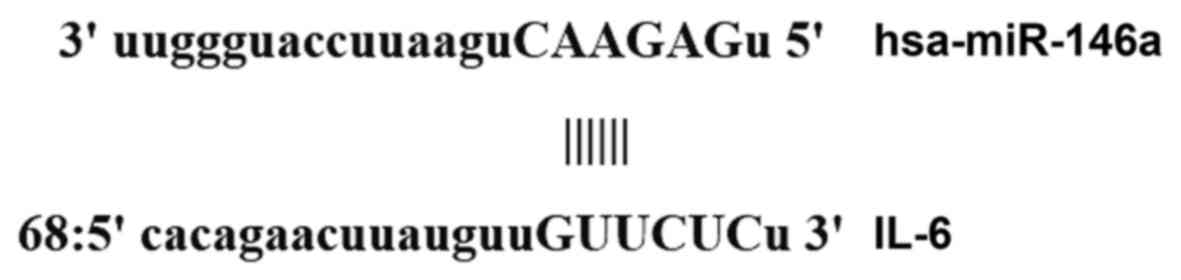Introduction
Paraquat is a type of bipyridine compound that
damages the majority of internal organs following ingestion,
particularly the heart, liver, lungs and kidney (1–3). Due to its
wide range of usage, it has become the herbicide with the highest
acute poisoning mortality rate (4).
The high mortality rates of paraquat caused by incorrect intake or
suicidal oral ingestion have been a huge challenge for health care
workers.
In organs damaged by paraquat poisoning, the lungs
present the most evident inflammatory characteristics at an early
stage, including impaired alveolar epithelial cells, intra-alveolar
hemorrhage and edema, inflammatory cell infiltration, and
irreversible fibrosis in the alveoli and pulmonary interstitium
(5). Anti-inflammatory treatments
are involved in the majority of early treatment regimens for
paraquat poisoning. In addition, certain key genes with large
differences in expression during the occurrence and development of
inflammation have become candidate targets for gene therapy.
Interleukin (IL)-6, a type of lymphokine produced by
activated monocytes and tissue macrophages, is an important factor
in immune response. IL-6 transforms B-cell precursors into cells
that produce antibodies, promote the growth and differentiation of
primary bone marrow-derived cells, and enhance the lysis function
of natural killer cells (6–8). To date, studies investigating the
role of mRNA and microRNA (miRNA or miR) molecules in the
regulation mechanism of IL-6 have achieved promising results. For
instance, miR-365 was observed to negatively regulate IL-6
expression in 293 cells and HeLa cells (9). However, to the best of our knowledge,
studies on the regulation and upstream miRNA of IL-6 in lung injury
due to paraquat poisoning have not been conducted. miR-146a is a
member of the miR-146 family, which is primarily involved in the
regulation of inflammation and the innate immune system (10). To the best of our knowledge miR-146a
has not been previously investigated in association with paraquat
poisoning and its association with IL-6 has not been examined
(11).
In the present study, the expression levels of IL-6
mRNA and protein in the macrophages, peripheral blood mononuclear
cells (PBMCs) and serum of patients with paraquat poisoning-induced
lung injury were determined, and the association between IL-6 and
miR-146a was investigated.
Materials and methods
Patients
A total of 26 patients with lung injury caused by
paraquat poisoning who received treatments at the Affiliated
Hospital of Jining Medical University (Jining, China) between
August 2013 and February 2017 were included in the present study.
In addition, 33 healthy subjects who undertook physical
examinations at this hospital in the same period were included into
the control group. Blood and alveolar lavage fluid were collected
from all patients and healthy subjects. Among the 26 patients with
lung injury caused by paraquat poisoning, 16 were males and 10 were
females (age range, 18–56 years; median age, 39 years). In the
control group, 18 individuals were males and 15 were females (age
range, 20–58; median age, 41 years). Patients with lung injury
caused by paraquat poisoning did not present complications or
infection in the heart, liver or kidney, and did not suffer from
immune or immune-associated diseases, such as diabetes and tumors.
Subjects in the control group requested fiber bronchoscopy due to
symptoms including foreign body sensation at pharynx or chest
tightness. None of the healthy subjects in the control group
exhibited abnormal results in the pulmonary function, chest X-ray
and fiber bronchoscopy, or were smokers. All procedures were
approved by the Ethics Committee of Jining Medical University
(Jining, China). Written informed consents were obtained from all
patients or their families.
Samples
To collect pulmonary macrophages, alveolar lavage
was initially performed 4–6 times (20 ml each time) at the
bronchial opening. A total of 40–60% fluid was recollected,
filtered (100 µm pore size) and centrifuged at 1,000 × g for 10 min
at 4°C. Next, the precipitates were subjected to cytological
classification according to a previously published method (12) and the cell density was adjusted to
1–3×106 cells/ml in PBS. Phthalocyanine blue (Guangzhou
Chemical Reagent Factory, Guangzhou, China) staining was conducted
at room temperature for 3 min, indicating that up to 90% cells were
viable. All cells were seeded onto clean glass slides and cultured
in Dulbecco's modified Eagle's medium (Thermo Fisher Scientific,
Inc., Waltham, MA, USA) at 37°C for 60 min. Subsequent to washing
off any floating cells using 0.5 mol/l PBS, the remaining adherent
cells were macrophages, which were stored at −20°C. To examine
whether miR-146a expression altered the expression of IL-6,
pulmonary macrophages were transfected with either 100 nM
agomiR-negative control (NC; forward, 5′-UAAACGGGUGACAGGUUUUAUC-3′
and reverse, 5′-GAUAAAUCCUGUCACCCGUUUA-3′) or 100 nM agomiR-146a
(forward, 5′-UGAGAACUGAAUUCCAUGGGUU-3′ and reverse,
5′-AACCCAUGGAAUUCAGUUCUCA-3′; Guangzhou RiboBio Co., Ltd.,
Guangzhou, China) using Lipofectamine® 2000 (Life
Sciences; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. Following 48 h the transfected cells were
used for subsequent experiments.
Peripheral blood (10–15 ml) was collected from all
participants and stored at 4°C for 1–2 h. Next, the serum was
separated by centrifugation at 400 × g at 4°C for 10 min, and
aliquots were added into Eppendorf tubes (100 µl in each tube)
prior to storage at −70°C.
In order to collect PBMCs, anticoagulant venous
blood was mixed with an equal volume of serum-free Iscove's
modified Dulbecco's medium (Thermo Fisher Scientific, Inc.) and the
mixture was added onto the surface of human lymphocyte separation
solution (5 ml; Cedarlane, Burlington, ON, Canada). Following
centrifugation at 400 × g for 30 min, the middle mist layer was
gently aspirated into tubes, and mixed with 5 times of volumes of
Hank's solution (Beijing Solarbio Science & Technology Co.,
Ltd., Beijing, China), followed by centrifugation at 300 × g for 10
min. Subsequent to washing twice with PBS, the cells were diluted
into a density of 1×106/ml, and a total of
3×106 cells were seeded into culture dishes (bottom
area, 9 cm2). After cultivation at 37°C and under 5%
CO2 for 2 h, the cells that adhered at the bottom of the
dishes were identified as PBMCs by microscopic evaluation.
Non-adhered cells were washed off with PBS and PBMCs were
trypsinized and collected for further use.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from 100 µl liquid samples
or 3×106 cells using TRIeasy™ reagent following the
manufacturer's protocol (cat. no. 10606ES60; Yeasen Biotechnology
Co., Ltd., Shanghai, China). The concentration and quality of RNA
was measured using ultraviolet spectrophotometry (Nanodrop ND2000;
Thermo Fisher Scientific, Inc., Waltham, MA, USA). Subsequently,
cDNA was obtained by RT from 1 µg RNA and stored at −20°C. The RT
of mRNA was performed using the TIANScript II cDNA First Strand
Synthesis kit (KR107; Tiangen Biotech Co., Ltd., Beijing, China),
and RT of miRNA was conducted using the miRcute miRNA cDNA First
Strand Synthesis kit (KR201; Tiangen Biotech Co., Ltd.).
In order to detect the mRNA expression of IL-6, the
SuperReal PreMix (SYBR Green) RT-qPCR kit (FP204; Tiangen Biotech
Co., Ltd.) was used, with GAPDH serving as an internal reference.
The primer sequences were as follows: IL-6,
5′-GGCACTGGCAGAAAACAACC-3′ (forward) and
5′-GCAAGTCTCCTCATTGAATCC-3′ (reverse); GAPDH,
5′-GGGAAACTGCGGCGTGAT-3′ (forward) and 5′-AAAGGTGGAGGAGTGGGT−3′
(reverse). The reaction system (20 µl) was composed of 10 µl
RT-qPCR Mix, 0.5 µl upstream primer, 0.5 µl downstream primer, 2 µl
cDNA and 7 µl ddH2O. The qPCR conditions involved an
initial denaturation at 95°C for 30 sec, followed by 45 cycles of
denaturation at 95°C for 5 sec and elongation at 57°C for 30 sec
(iQ5; Bio-Rad Laboratories, Inc., Hercules, CA, USA). The
2−ΔΔCq method (13) was
used to calculate the relative expression of IL-6 mRNA against
GAPDH. Each sample was tested in triplicate.
The expression of miR-146a was determined by miRcute
miRNA RT-PCR Kit (FP401; Tiangen Biotech Co., Ltd.), using U6 as an
internal reference. The primer sequences used were as follows:
miR-146a, 5′-CGGCGGTGAGAACTGAATTCCA-3′ (forward) and
5′-GTGCAGGGTCCGAGGT-3′ (reverse); U6, 5′-CTCGCTTCGGCAGCACA-3′
(forward) and 5′-AACGCTTCACGAATTTGCGT-3′ (reverse). The reaction
system (20 µl) consisted of 10 µl RT-qPCR Mix, 0.5 µl upstream
primer, 0.5 µl downstream universal primer, 2 µl cDNA and 7 µl
ddH2O. The reaction protocol involved an initial
denaturation at 95°C for 5 min, followed by 40 cycles of
denaturation at 95°C for 10 sec, annealing at 60°C for 20 sec and
elongation at 72°C for 10 sec (iQ5; Bio-Rad Laboratories, Inc.).
The 2−ΔΔCq method was used to calculate the relative
expression of miR-146a against U6. Each sample was tested in
triplicate.
Western blotting
According to a previously published study (14), precooled radioimmunoprecipitation
assay lysis buffer [600 µl; containing 50 mM Tris, 1 mM EDTA, 150
mM NaCl, 0.1% sodium dodecyl sulfate (SDS), 1% Triton X-100 and 1%
sodium deoxycholate; Beyotime Institute of Biotechnology, Shanghai,
China] was used to lyse PBMCs for 50 min on ice. Next, the mixture
was centrifuged at 12,000 × g at 4°C for 5 min. The protein
concentration of the obtained supernatant was determined by a
bicinchoninic acid protein concentration determination kit
(RTP7102; Real-Times Biotechnology Co., Ltd., Beijing, China).
Protein samples (20 µg) were then mixed with SDS loading buffer,
followed by denaturation in boiling water bath for 5 min, and were
then subjected to 10% SDS-polyacrylamide gel electrophoresis. The
resolved proteins were transferred to polyvinylidene difluoride
membranes on ice for 2 h at 100 V and blocked with 5% skimmed milk
for 1 h at room temperature. Subsequently, the membranes were
incubated overnight at 4°C with the following primary antibodies:
Rabbit anti-human polyclonal IL-6 antibody (1:1,000; ab6672; Abcam,
Cambridge, UK) and rabbit anti-human β-actin antibody (1:5,000;
ab129348; Abcam). Following extensive washing with PBS with Tween
20 three times for 15 min each, the membranes were incubated with
goat anti-rabbit horseradish peroxidase-conjugated secondary
antibody (1:3,000; ab6721; Abcam) at room temperature for 1 h. The
samples were further washed with PBS with Tween 20 three times for
15 min each, and then the membrane was developed with an enhanced
chemiluminescence detection kit (ab65623; Abcam). Image Lab version
3.0 software (Bio-Rad Laboratories, Inc.) was used to acquire and
analyze imaging signals. The relative content of IL-6 protein was
expressed in terms of the IL-6/β-actin ratio.
Enzyme-linked immunosorbent assay
(ELISA)
According to a previously published study (12), the IL-6 level in the serum obtained
from patients and healthy controls was tested using the human IL-6
ELISA kit (ab178013; Abcam, Cambridge, UK). Briefly, standards (50
µl), samples (10 µl sample liquid and 40 µl diluent) and blank were
added to predefined wells in 96-well microplates. In the standard
and sample wells, horseradish peroxidase-labelled conjugates (100
µl) were added prior to sealing the plates for incubation at 37°C
for 1 h. Subsequent to washing the plates five times with wash
solution provided in the kit, substrates A (50 µl) and B (50 µl)
were added into each well. After incubation at 37°C for 15 min,
stop solution (50 µl) was added into each well, and the absorbance
of the wells was measured at 450 nm within 15 min using a
Multiskan™ FC microplate reader (Thermo Fisher Scientific,
Inc.).
Bioinformatics analysis and
dual-luciferase reporter assay
Bioinformatics prediction is a powerful tool for the
investigation of the functions of miRNAs. Thus, to understand the
regulatory mechanism of IL-6, various databases were used to
predict miRNA molecules that may regulate IL-6, including miRanda
(www.microrna.org/microrna/home.do), TargetScan
(www.targetscan.org), PiTa (genie.weizmann.ac.il/pubs/mir07/mir07_data.html),
RNAhybrid (bibiserv.techfak.uni-bielefeld.de/rnahybrid) and
PicTar (pictar.mdc-berlin.de/). It was
identified that miR-146a was able to potentially regulate IL-6
(Fig. 1).
According to the bioinformatics results, wild-type
(WT) and mutant (MT) seed regions of miR-146a in the
3′-untranslated region (UTR) of IL-6 gene were chemically
synthesized in vitro, the Spe-1 and Hind III restriction
sites were added, and then the regions were cloned into pMIR-REPORT
luciferase reporter plasmids. According to a previously published
study (13), plasmids (0.8 µg) with
WT or MT 3′-UTR DNA sequences were co-transfected with 100 nM
agomiR-NC (forward, 5′-UAAACGGGUGACAGGUUUUAUC-3′ and reverse,
5′-GAUAAAUCCUGUCACCCGUUUA-3′) or 100 nM agomiR-146a mimics
(forward, 5′-UGAGAACUGAAUUCCAUGGGUU-3′ and reverse,
5′-AACCCAUGGAAUUCAGUUCUCA-3′; Guangzhou RiboBio Co., Ltd.) into
293T cells (The Cell Bank of Type Culture Collection of Chinese
Academy of Sciences, Shanghai, China) using Lipofectamine
2000® (Thermo Fisher Scientific, Inc.). After
cultivation for 48 h the cells were lysed using a dual-luciferase
reporter assay kit (Promega Corp., Fitchburg, WI, USA) according to
the manufacturer's protocol, and the fluorescence intensity was
measured using GloMax 20/20 luminometer (Promega Corp.). Using the
Renilla fluorescence activity as an internal reference, the
fluorescence values of each group of cells were measured.
Statistical analysis
The results were analyzed using SPSS version 18.0
statistical software (IBM Corp., Armonk, NY, USA). The data are
expressed as the means ± standard deviation. Data were tested for
normality, and multigroup measurement data were analyzed using
one-way analysis of variance. In the case of homogeneity of
variance, the least significant difference and Student-Newman-Keuls
methods were used. In the case of heterogeneity of variance,
Tamhane's T2 or Dunnett's T3 method was used. P<0.05 was
considered to indicate a statistically significant difference.
Results
Patients with paraquat
poisoning-induced lung injury exhibit higher IL-6 mRNA levels
compared with healthy subjects
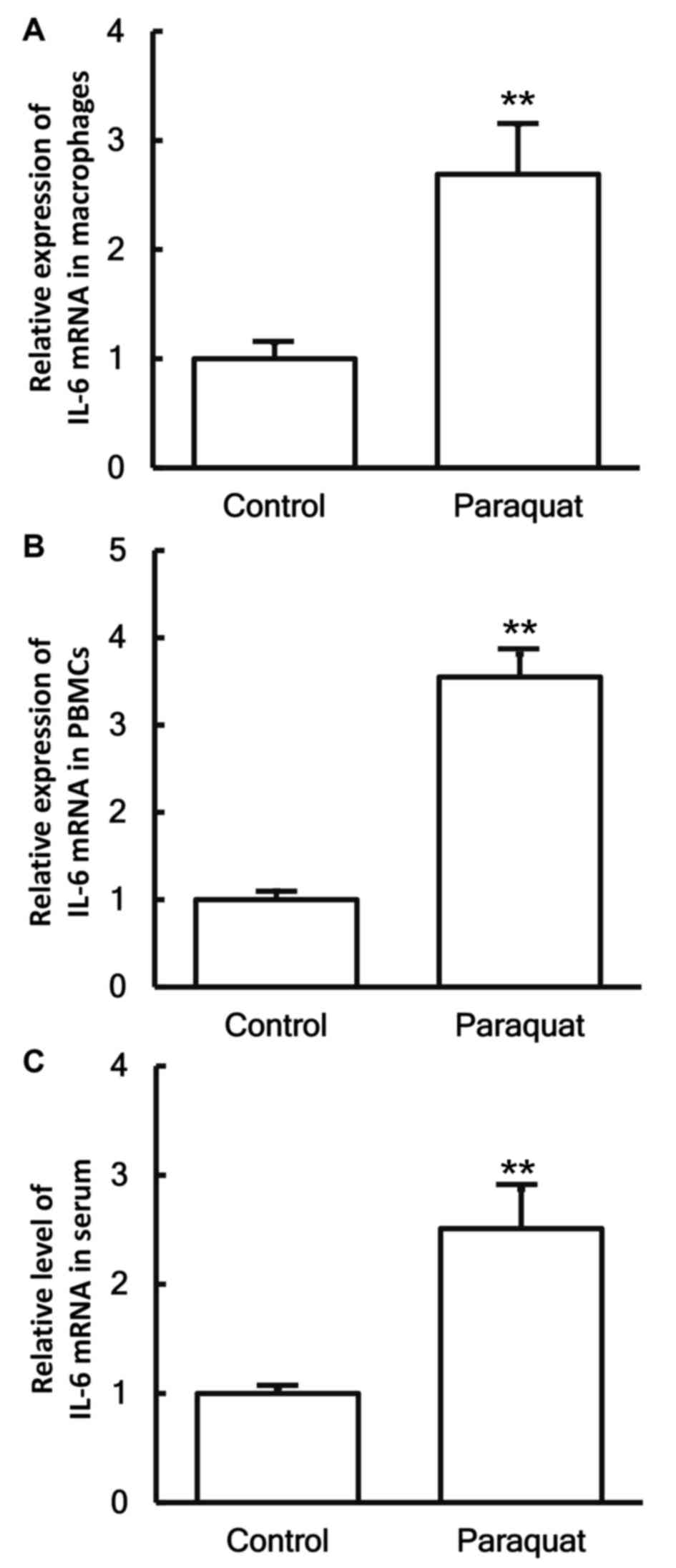
To measure the expression of IL-6 mRNA in different
samples, RT-qPCR was conducted. The data revealed that the levels
of IL-6 mRNA in the pulmonary macrophages, PBMCs and serum of
patients with lung injury caused by paraquat poisoning were all
significantly higher when compared with those in the control group
(P<0.05; Fig. 2A-C).
IL-6 protein expression in macrophages
and PBMCs is upregulated in patients with lung injury caused by
paraquat poisoning
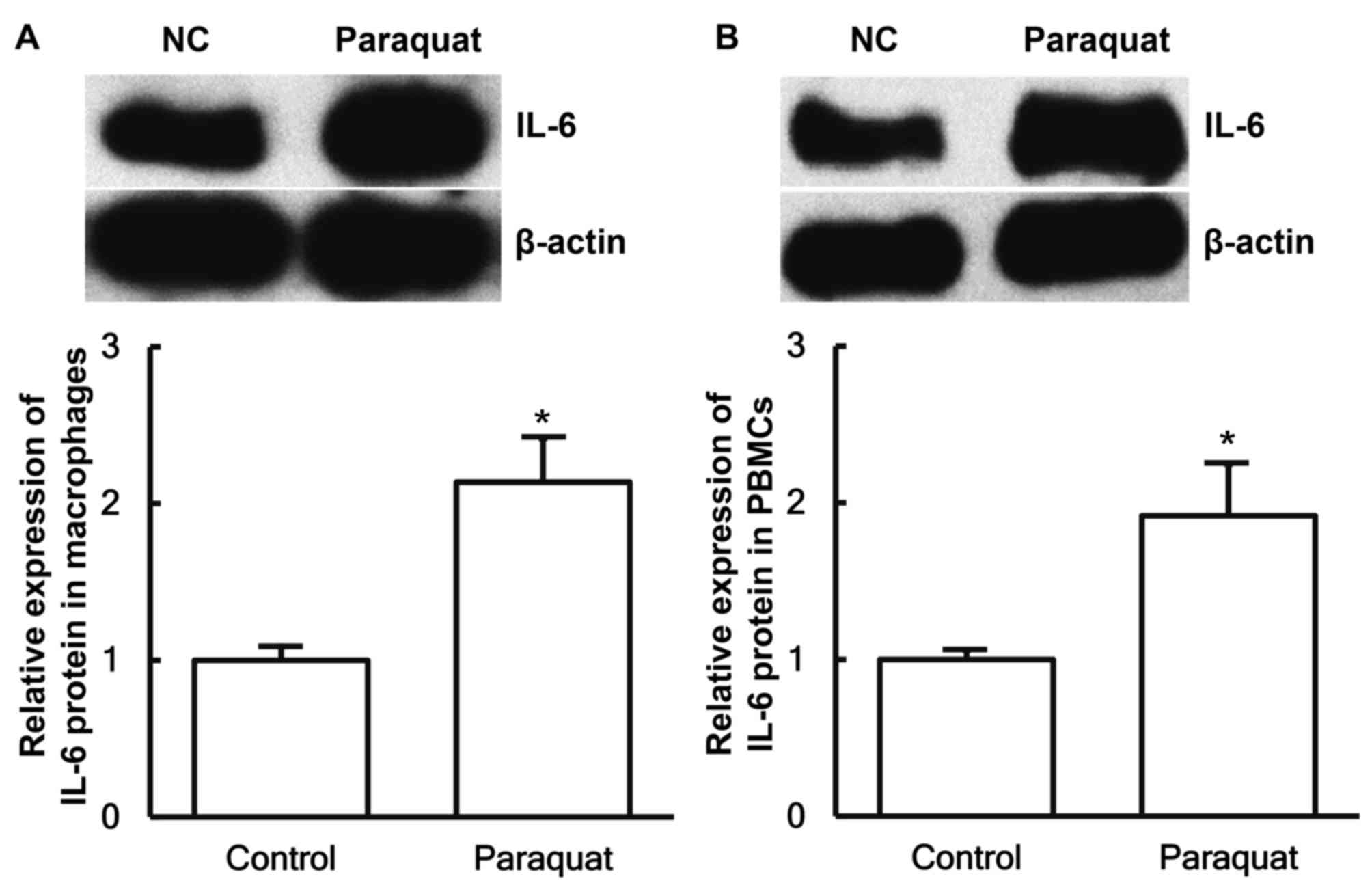
To determine the expression of IL-6 protein in
macrophages and PBMCs, western blotting was performed. The data
demonstrated that the IL-6 protein expression levels in macrophages
and PBMCs from patients with lung injury caused by paraquat
poisoning were significantly elevated compared with those obtained
from healthy subjects (P<0.05; Fig.
3A and B).
Higher serum IL-6 content in patients
with lung injury caused by paraquat poisoning
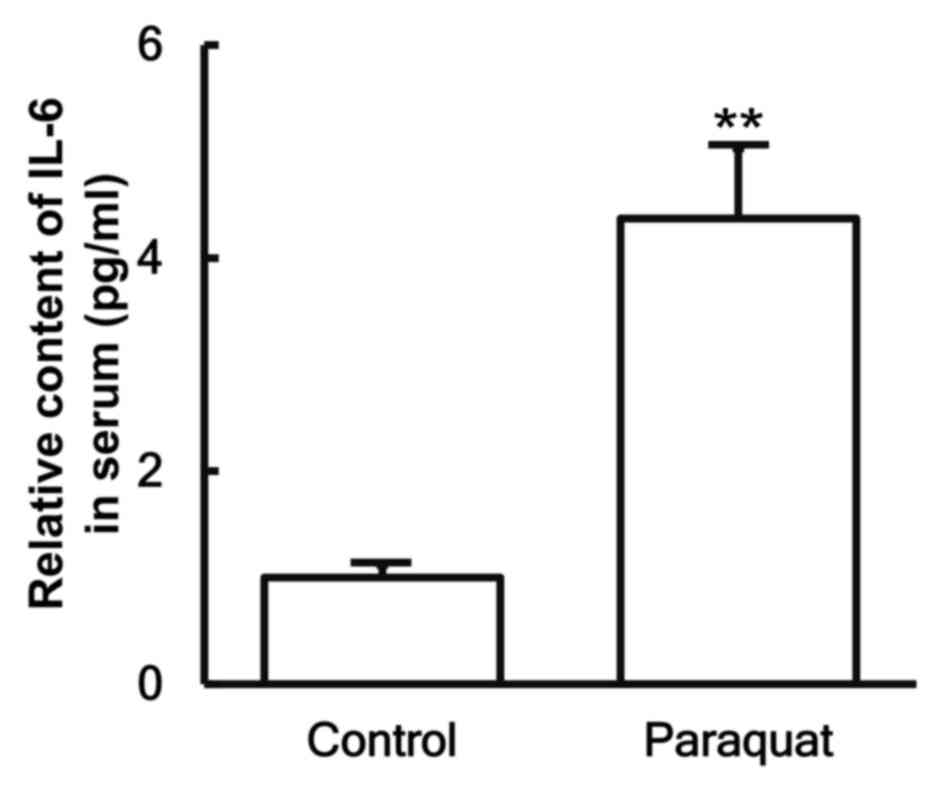
To examine the secretion of IL-6 in the blood, ELISA
was performed. The data indicated that the IL-6 content in the
serum of patients with lung injury caused by paraquat poisoning was
significantly increased as compared with that in healthy subjects
(P<0.05; Fig. 4).
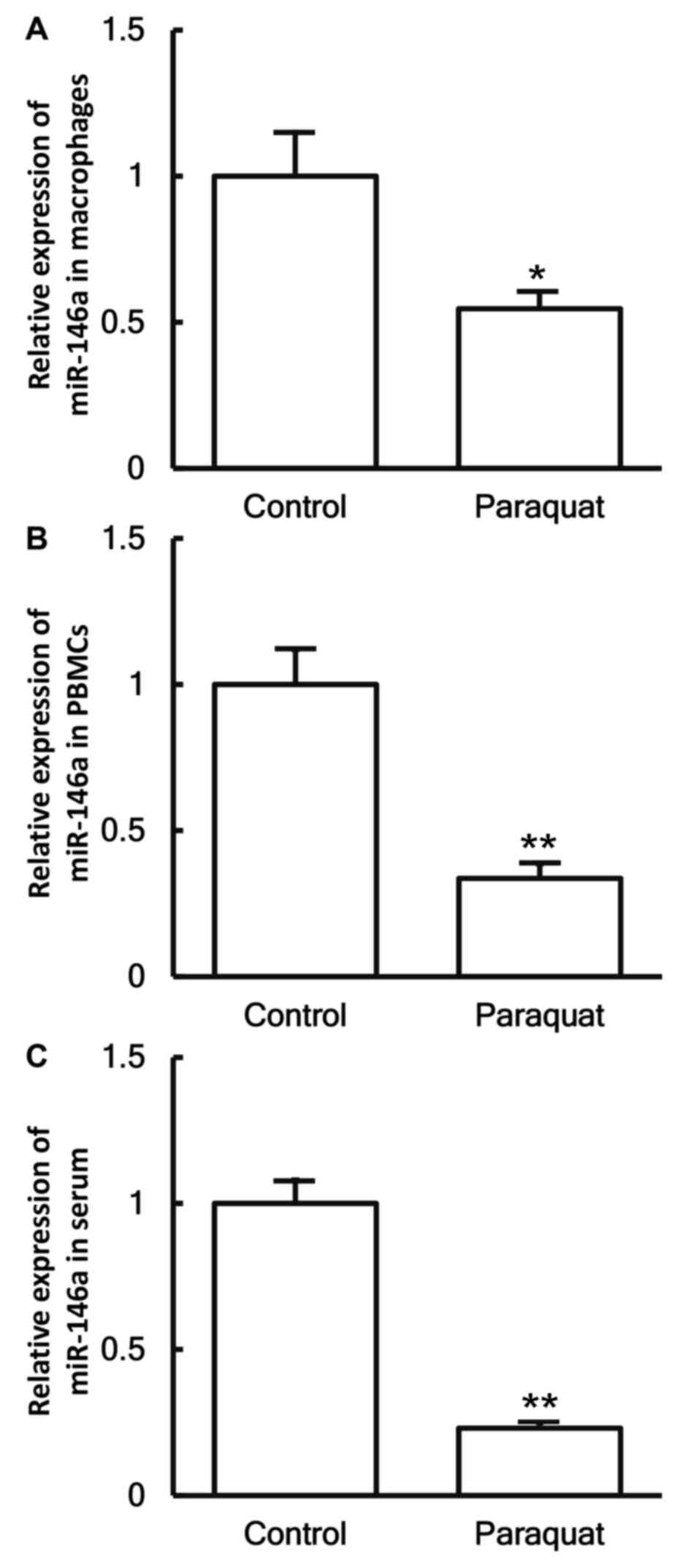
Reduced miR-146a levels in patients
with lung injury caused by paraquat poisoning
To investigate the levels of miR-146a in the cells
and serum, RT-qPCR was employed. The data demonstrated that the
expression levels of miR-146a in the macrophages, PBMCs and serum
of patients with lung injury caused by paraquat poisoning were
significantly decreased in comparison with those in healthy
subjects (P<0.05; Fig. 5A-C).
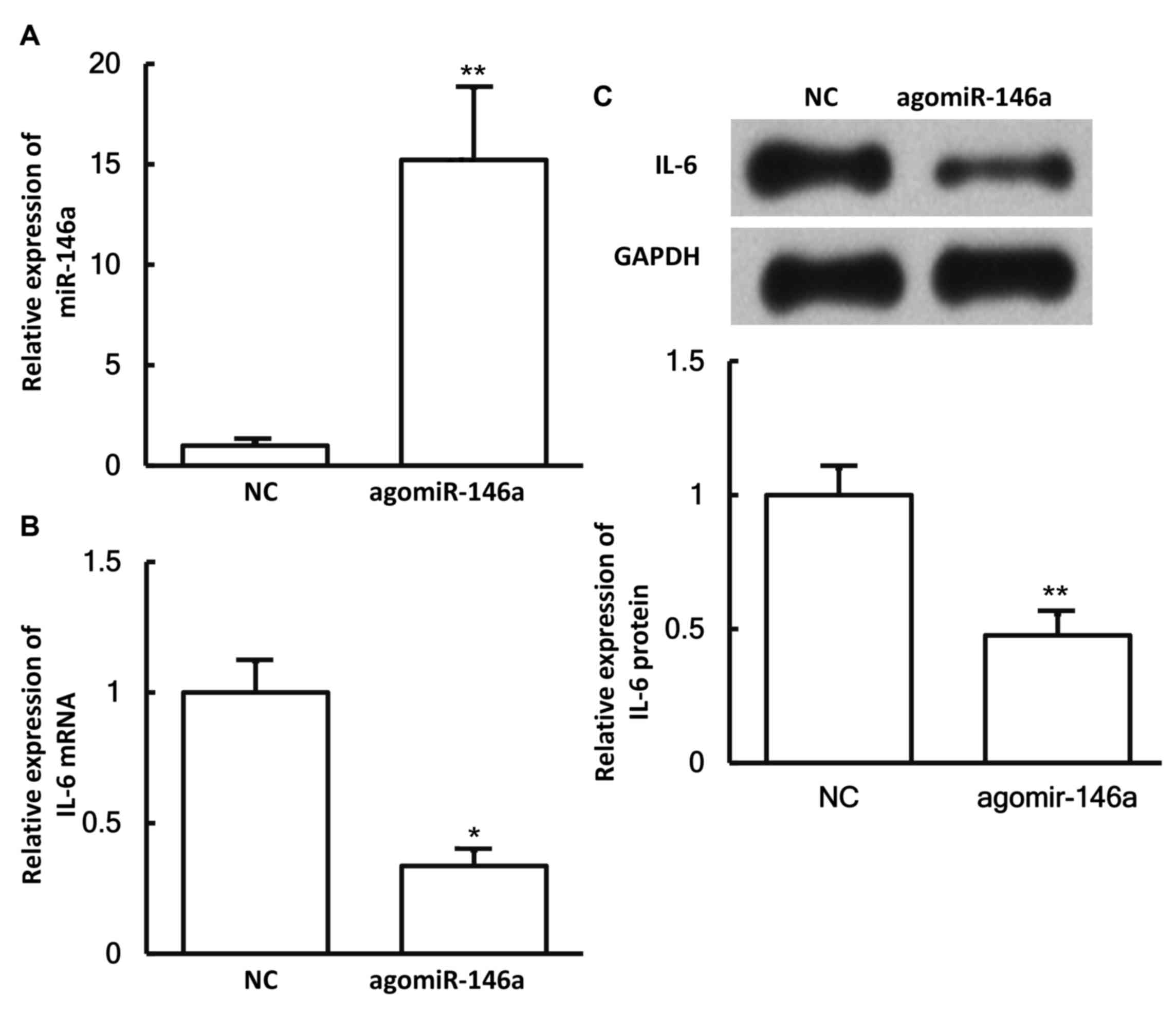
miR-146a expression downregulates the
expression of IL-6 in pulmonary macrophages
To examine whether miR-146a expression affects the
expression of IL-6, pulmonary macrophages were transfected with
agomiR-146a and then subjected to RT-qPCR and western blotting. The
results revealed that the levels of miR-146a in cells transfected
with agomiR-146a were significantly higher in comparison with those
in the negative control group (P<0.05; Fig. 6A). In addition, the expression levels
of IL-6 mRNA and protein in cells transfected with agomiR-146a were
significantly lower as compared with those in the negative control
group (both P<0.05; Fig. 6B and
C). These results suggest that miR-146a expression
downregulates the expression of IL-6 in pulmonary macrophages.
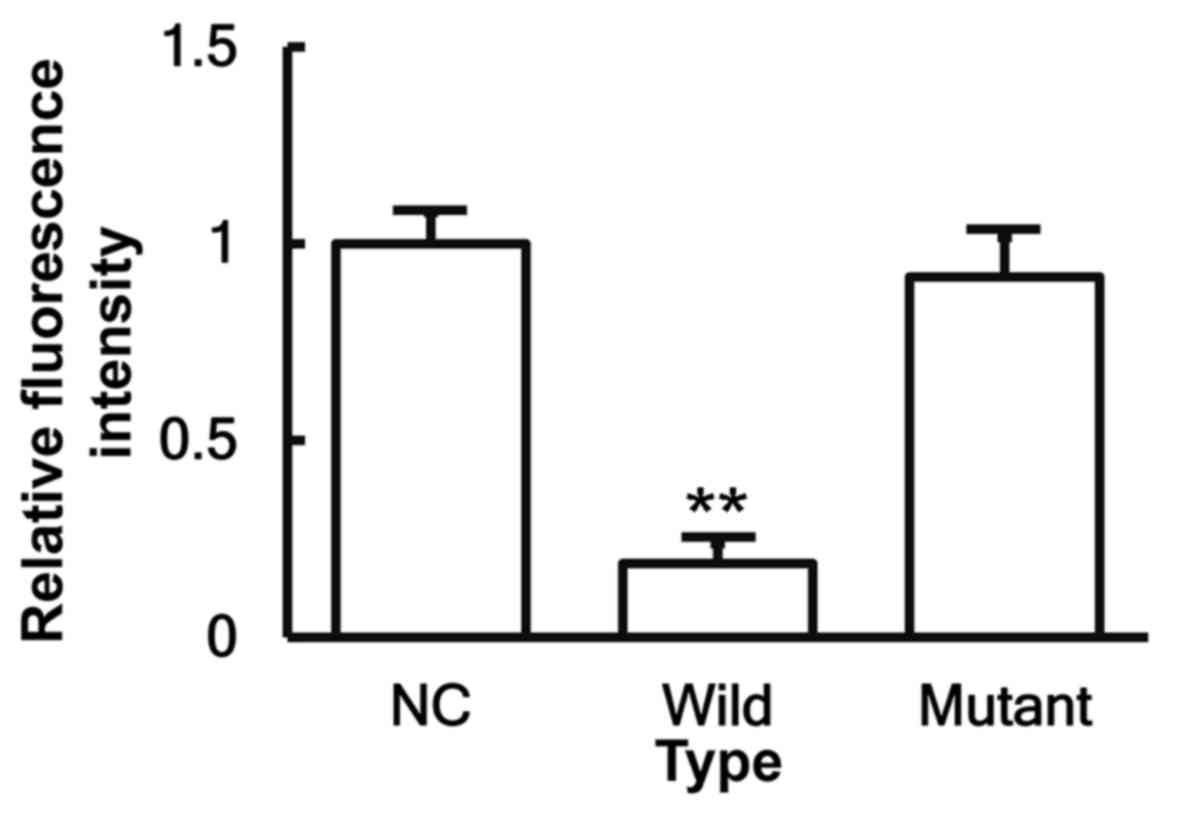
miR-146a binds to the 3′-UTR seed
region of IL-6 mRNA to regulate its expression
To identify the interaction between miR-146a and the
3′-UTR of IL-6 mRNA, a dual-luciferase reporter assay was
performed. The fluorescence value of cells co-transfected with
miR-146a mimics and pMIR-REPORT-WT luciferase reporter plasmids was
significantly reduced when compared with that of cells transfected
with miR-NC (P<0.05). By contrast, the fluorescence value of
cells co-transfected with miR-146a mimics and pMIR-REPORT-MT
luciferase reporter plasmids was not significantly different from
that of cells transfected with miR-NC (P>0.05; Fig. 7). These results indicate that
miR-146a is able to bind to the 3′-UTR seed region of IL-6 mRNA in
order to regulate its expression.
Discussion
Paraquat has been registered as a type of herbicide
since 1962, and is widely used to date (15), and it is the second most used
compound among all herbicides (16).
However, paraquat poisoning has been reported to result in a high
fatality rate (4), with no specific
antidote currently known for this poisoning. One of the
characteristics of early paraquat poisoning is a strong
inflammatory reaction. IL-6 is an inflammatory factor that has been
extensively studied. It induces the production of C-reactive
proteins and fibrinogen in inflammation, and promotes thrombosis
(17). Increased levels of IL-6 in
the body may cause inflammatory diseases, including rheumatoid
arthritis and Crohn's disease, due to binding to the IL-6 receptors
(18). In rheumatoid arthritis, IL-6
stimulates the secretion of inflammation mediators by T lymphocytes
and B lymphocytes, facilitates the maturation and differentiation
of B lymphocytes, and increases the effects of IL-1β and tumor
necrosis factor α (TNF-α). In inflammation, IL-6 presents
chemotaxis to other inflammatory cells, such as neutrophilic
lymphocytes and mononuclear macrophages (19). These observations suggest that IL-6
serves important roles in inflammation responses. In the present
study, it was demonstrated that the expression levels of IL-6 mRNA
and protein were upregulated in the pulmonary macrophages, PBMCs
and serum of patients with lung injury caused by paraquat
poisoning, consistent with the inflammatory characteristics of
early lung injury.
miRNA widely participates in various
pathophysiological processes, including the proliferation, invasion
and migration of tumor cells, hypertension, diabetes mellitus and
atherosclerosis (20,21). It has been reported that expression
of miR-146a is abnormal in autoimmune diseases, such as rheumatoid
arthritis (22,23). Clinical and animal models of
osteoarthritis revealed that miR-146a is associated with pain in
osteoarthritis (24–26). Furthermore, multiple nuclear factor-κB
(NF-κB) binding sites exist in the promoter region of the miR-146a
gene, while lipopolysaccharide, IL-1 and TNF-α promote the
expression of miR-146a in an NF-κB-dependent manner (27–30). In the
present study, bioinformatics analysis demonstrated that miR-146a
and IL-6, two genes that are closely involved in inflammation, may
have a regulatory association, and that IL-6 may be a direct target
gene of miR-146a. The results also demonstrated that miR-146a was
downregulated and IL-6 was upregulated in pulmonary macrophages and
PBMCs, suggesting that the immune system of the body negatively
regulated the cleavage of IL-6 by miR-146a and promoted immune
responses by enhancing the expression of IL-6. In addition, reduced
expression of miR-146a and enhanced expression of IL-6 were
detected in the serum, indicating that these serum levels may
reflect the inflammation responses and tissue damages in lung
injury caused by paraquat poisoning. Finally, a dual-luciferase
reporter assay demonstrated that IL-6 was a direct target gene of
miR-146a, since overexpression of miR-146a reduced the fluorescence
intensity of the IL-6 luciferase reporter plasmid.
In conclusion, the present study demonstrated that
the increased expression of IL-6 in patients with lung injury
caused by paraquat poisoning was associated with a decreased
expression of miR-146a. A limitation of the present study is the
small sample size, which should be increased in future studies. In
addition, additional experiments should be designed to test the
effect of paraquat on other cell types or in an animal model. The
present study provides a basis for the development of novel
treatments for lung injury caused by paraquat poisoning.
Acknowledgements
The authors would like to thank Dr Luning Jiang from
the Department of Respiratory Medicine, Affiliated Hospital of
Jining Medical University (Jining, China).
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YL designed the study and analyzed the data. WW
performed the experiments and analyzed the data.
Ethics approval and consent to
participate
All procedures were approved by the Ethics Committee
of Jining Medical University (Jining, China). Written informed
consents were obtained from all patients or their families.
Consent for publication
All patients provided written informed consent for
the publication of their data.
Competing interests
The authors confirm that they have no competing
interests.
References
|
1
|
Mullick FG, Ishak KG, Mahabir R and
Stromeyer FW: Hepatic injury associated with paraquat toxicity in
humans. Liver. 1:209–221. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nagao M, Takatori T, Inoue K, Shimizu M,
Terazawa K and Akabane H: Immunohistochemical localization and
dynamics of paraquat in small intestine, liver and kidney.
Toxicology. 63:167–182. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dinis-Oliveira RJ, Remião F, Duarte JA,
Ferreira R, Navarro Sánchez A, Bastos ML and Carvalho F:
P-glycoprotein induction: An antidotal pathway for paraquat-induced
lung toxicity. Free Radic Biol Med. 41:1213–1224. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hwang KY, Lee EY and Hong SY: Paraquat
intoxication in Korea. Arch Environ Health. 57:162–166. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yamashita M, Yamashita M and Ando Y: A
long-term follow-up of lung function in survivors of paraquat
poisoning. Hum Exp Toxicol. 19:99–103. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Anderson AE, Pratt AG, Sedhom MA, Doran
JP, Routledge C, Hargreaves B, Brown PM, Cao Lê KA, Isaacs JD and
Thomas R: IL-6-driven STAT signalling in circulating CD4+
lymphocytes is a marker for early anticitrullinated peptide
antibody-negative rheumatoid arthritis. Ann Rheum Dis. 75:466–473.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tezono K, Sarker KP, Kikuchi H, Nasu M,
Kitajima I and Maruyama I: Bioactivity of the vascular endothelial
growth factor trapped in fibrin clots: Production of IL-6 and IL-8
in monocytes by fibrin clots. Haemostasis. 31:71–79.
2001.PubMed/NCBI
|
|
8
|
Vila N, Reverter JC, Yagüe J and Chamorro
A: Interaction between interleukin-6 and the natural anticoagulant
system in acute stroke. J Interferon Cytokine Res. 20:325–329.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xu Z, Xiao SB, Xu P, Xie Q, Cao L, Wang D,
Luo R, Zhong Y, Chen HC and Fang LR: miR-365, a novel negative
regulator of interleukin-6 gene expression, is cooperatively
regulated by Sp1 and NF-kappaB. J Biol Chem. 286:21401–21412. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sonkoly E, Ståhle M and Pivarcsi A:
MicroRNAs and immunity: Novel players in the regulation of normal
immune function and inflammation. Semin Cancer Biol. 18:131–140.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ye EA and Steinle JJ: miR-146a suppresses
STAT3/VEGF pathways and reduces apoptosis through IL-6 signaling in
primary human retinal microvascular endothelial cells in high
glucose conditions. Vision Res. 139:15–22. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Aspelund Stjärne A, Hammarström H,
Inghammar M, Larsson H, Hansson L, Christensson B and Påhlman LI:
Heparin-binding protein, lysozyme, and inflammatory cytokines in
bronchoalveolar lavage fluid as diagnostic tools for pulmonary
infection in lung transplanted patients. Am J Transplant.
18:444–452. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
He X, Ping J and Wen D: MicroRNA-186
regulates the invasion and metastasis of bladder cancer via
vascular endothelial growth factor C. Exp Ther Med. 14:3253–3258.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu H, Song J and Ma J: Paraquat is
approved for continued use in the European Union. Pest Sci
Administ. 25:36–37. 2004.
|
|
16
|
Yamamoto M, Toda M, Tanaka K, Sugita T,
Sasaki S, Uneyama C and Morikawa K: Study on usage of pesticides in
various countries. Kokuritsu Iyakuhin Shokuhin Eisei Kenkyusho
Hokoku. 92–100. 2007.PubMed/NCBI
|
|
17
|
Tang YH, Vital S, Russell J, Seifert H and
Granger DN: Interleukin-6 mediates enhanced thrombus development in
cerebral arterioles following a brief period of focal brain
ischemia. Exp Neurol. 271:351–357. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Suzuki Y, Matsui T, Ito H, Ashida T,
Nakamura S, Motoya S, Matsumoto T, Sato N, Ozaki K, Watanabe M and
Hibi T: Circulating interleukin 6 and albumin, and infliximab
levels are good predictors of recovering efficacy after dose
escalation infliximab therapy in patients with loss of response to
treatment for crohn's disease: A prospective clinical trial.
Inflamm Bowel Dis. 21:2114–2122. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schaper F and Rose-John S: Interleukin-6:
Biology, signaling and strategies of blockade. Cytokine Growth
Factor Rev. 26:475–487. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liz J and Esteller M: lncRNAs and
microRNAs with a role in cancer development. Biochim Biophys Acta.
1859:169–176. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Varshney J and Subramanian S: MicroRNAs as
potential target in human bone and soft tissue sarcoma
therapeutics. Front Mol Biosci. 2:312015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Abou-Zeid A, Saad M and Soliman E:
MicroRNA 146a expression in rheumatoid arthritis: Association with
tumor necrosis factor-alpha and disease activity. Genet Test Mol
Biomarkers. 15:807–812. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pauley KM, Satoh M, Chan AL, Bubb MR,
Reeves WH and Chan EK: Upregulated miR-146a expression in
peripheral blood mononuclear cells from rheumatoid arthritis
patients. Arthritis Res Ther. 10:R1012008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li X, Kroin JS, Kc R, Gibson G, Chen D,
Corbett GT, Pahan K, Fayyaz S, Kim JS, van Wijnen AJ, et al:
Altered spinal microRNA-146a and the microRNA-183 cluster
contribute to osteoarthritic pain in knee joints. J Bone Miner Res.
28:2512–2522. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li X, Gibson G, Kim JS, Kroin J, Xu S, van
Wijnen AJ and Im HJ: MicroRNA-146a is linked to pain-related
pathophysiology of osteoarthritis. Gene. 480:34–41. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yamasaki K, Nakasa T, Miyaki S, Ishikawa
M, Deie M, Adachi N, Yasunaga Y, Asahara H and Ochi M: Expression
of MicroRNA-146a in osteoarthritis cartilage. Arthritis Rheum.
60:1035–1041. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Larner-Svensson HM, Williams AE, Tsitsiou
E, Perry MM, Jiang X, Chung KF and Lindsay MA: Pharmacological
studies of the mechanism and function of interleukin-1beta-induced
miRNA-146a expression in primary human airway smooth muscle. Respir
Res. 11:682010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cameron JE, Yin Q, Fewell C, Lacey M,
McBride J, Wang X, Lin Z, Schaefer BC and Flemington EK:
Epstein-Barr virus latent membrane protein 1 induces cellular
MicroRNA miR-146a, a modulator of lymphocyte signaling pathways. J
Virol. 82:1946–1958. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Taganov KD, Boldin MP, Chang KJ and
Baltimore D: NF-kappaB-dependent induction of microRNA miR-146, an
inhibitor targeted to signaling proteins of innate immune
responses. Proc Natl Acad Sci USA. 103:12481–12486. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Curtale G, Citarella F, Carissimi C,
Goldoni M, Carucci N, Fulci V, Franceschini D, Meloni F, Barnaba V
and Macino G: An emerging player in the adaptive immune response:
microRNA-146a is a modulator of IL-2 expression and
activation-induced cell death in T lymphocytes. Blood. 115:265–273.
2010. View Article : Google Scholar : PubMed/NCBI
|