Introduction
Osteosarcoma is the most common histological form of
primary bone cancer in children and adolescents (1). The incidence rates of osteosarcoma in
Americans under the age of 20 are estimated to be about 5.0 per
million per year for the general population (1). It occurs more frequently in the
metaphyseal region of tubular long bones, with 42% occurring in the
femur, 19% in the tibia, and 10% in the humerus (1).
Due to complex nuclear and highly unstable genomes,
osteosarcoma usually shows both numerical and structural
chromosomal changes (2).
Osteosarcoma often presents in Paget's disease of the bone, which
may be caused by a mutation in the TNFRSF11A gene on chromosome
18q22 (3). Osteosarcoma is a
characteristic of Li-Fraumeni syndrome-1 (LFS1), which is caused by
a mutation of the TP53 gene, and of Li-Fraumeni syndrome-2 (LFS2),
which is caused by a mutation of the CHEK2 gene. Sadikovic et
al (4) used 10 pediatric
osteosarcoma tissue samples for integrative whole-genome analysis
of promoter methylation, gene expression, and DNA copy number.
Consequently, hypomethylation, overexpression, and copy number gain
were identified for the histone cluster 2 genes on chromosomes
1q21.1-q21.3. They also discovered i) the loss of chromosome
8p21.3-p21.2, ii) lower expression of TNFRSF10A, DOCK5, and
TNFRSF10D genes, iii) copy number gain of chromosome 6p21.1-p12.3,
and iv) amplification-related overexpression of RUNX2.
Currently, multidrug resistance (MDR) is an urgent
problem to be solved in osteosarcoma treatment. The human MDR gene
1 (MDR1) reportedly plays an important role in the drug resistance
process in osteosarcoma (5). A
recent study shows that the expression of trichorhinophalangeal
syndrome type 1 (Trps1) is directly related to MDR1/P-gp, and Trps1
can promote MDR1/P-gp gene expression in osteosarcoma cell lines.
Therefore, Trps1 should be a promising molecular target for
reversing drug resistance in osteosarcoma (6).
Although there has been some improvement in the
pathogenesis, diagnosis, and treatment of osteosarcoma in recent
years, the mechanism underlying the development of osteosarcoma
remains obscure. Furthermore, efforts have rarely been intended at
implementing system biology-based analyses to elaborate the
underpinning pathological molecular mechanisms of osteosarcoma.
Comprehensive analysis of latent causal genes within a pathway
and/or a network framework could provide more holistic insights
than classical single-gene analyses. In the present study, we first
identified an osteosarcoma-related gene module (OSM) by combining
the genome-wide association studies (GWAS) dataset of osteosarcoma
with whole protein-protein interaction (PPI) network. Functions of
genes contained in the OSM were also explored through WebGestalt
and ToppGene online tools. Potential osteosarcoma diagnosis and
treatment targets were further screened through topological and
survival analyses of the OSM. This study should help understand the
molecular mechanisms of osteosarcoma and provide a valuable
pipeline for other types of disease.
Materials and methods
Data sources
Results from MutSigCV (broadinstitute.org/cancer/cga/mutsig) analysis on
somatic mutations in osteosarcoma were obtained from dbGaP
(ncbi.nlm.nih.gov/gap) (NCBI dbGaP study
accession: phs000699.v1.p1 and dbGaP analysis accession:
pha003862). The datasets consisted of 58 pairs of osteosarcoma and
normal adjacent tissues. Forty-nine percent of the patients were
male and 47% had metastases at diagnosis. Five-year overall
survival was 49% with 33% for patients with metastatic disease and
64% for patients with localized disease. Illumina HiSeq 2000 was
used for the whole genome sequencing of all samples. Genes with a
P-value <1 were selected as seeds to identify the disease module
underlying osteosarcoma.
The human PPI data containing 16,022 nodes and
228,122 edges were obtained from Hu's study (7), which collected and integrated two PPI
datasets, i.e., Protein Interaction Network Analysis (PINA)
platform (May 21, 2014) (8) and a
human interactome compiled by a recent study (9).
Disease module identification
The OSM was identified using dmGWAS version 3.0
(10), an R package that implements
heuristic local search algorithms to recognize candidate
subnetworks or genes associated with complex diseases by
incorporating the association signal from GWAS datasets into the
PPI network, which has been efficiently applied in identifying
disease modules. Osteosarcoma-related seed genes with their
corresponding tested P-values and PPI networks were used as input,
and other parameters were set as per the dmGWAS recommendations.
Ultimately, the candidate disease modules were ranked in accordance
with their normalized module scores and empirical P-values.
To test the non-randomness of the identified module,
we first generated 1,000 random networks with the same number of
nodes and edges as the identified OSM by utilizing the ‘networkx’
module in python (https://www.python.org/). Then, we computed the
average values of clustering coefficients and the shortest-path
distance for each random network. By counting the number of random
networks with the average clustering coefficient (NCC)
greater than that of the OSM as well as the number of random
networks with the average shortest-path distance (NSD)
less than that of the OSM, we could evaluate the significance level
of non-randomness by calculating the empirical P-value
(NCC/1000 and NSD/1000).
Functional enrichment analysis of the
OSM
The functional features of the OSM were examined
through WebGestalt (11) and
ToppGene (12). WebGestalt
incorporates the information from multiple sources to detect the
biological themes out of the given gene lists, including
identifying the significantly enriched gene ontology (GO) terms. GO
terms with a P<0.05 were considered significantly enriched.
ToppGene was chosen to analyze the Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathways enriched in the OSM. The pathways with
false discovery rate (FDR) <0.05 were considered significantly
enriched.
Pathway crosstalk analysis of the
significantly enriched pathways
Further, we carried out pathway crosstalk analysis
to investigate the interactions among significantly enriched
pathways (13). To demonstrate the
overlap between any pair of pathways, we adopted two measurements,
i.e., the Overlap Coefficient
(OC)=|A∩B|min(|A|,|B|)
and the Jaccard Coefficient
(JC)=|A∩BA∪B|
where A and B denote the lists of genes contained
in the two tested pathways. We carried out the following
procedure to establish the pathway crosstalk: i) We chose a set of
pathways with FDR <0.05 for crosstalk analysis; ii) we
calculated the number of shared OS genes between any pair of
pathways. Pathway pairs with <2 overlapped OS genes were
discarded; iii) we computed the overlap of all pairs of pathways
and ranked them in accordance with their OC and JC values; and iv)
we used the Cytoscape (14) software
to visualize the chosen pathway crosstalk.
Inferring essential genes in the
OSM
Previous studies reported that hub genes (high
degree) and bottleneck genes (high betweenness) in gene networks
are more important for cell survival (15–17). In
addition, another study reported that minimum dominating sets of
proteins (MDSets) play a topologically and biologically important
role in controllability in protein interaction networks (18). Briefly, MDSet is an optimized subset
of nodes from where each remaining node can be immediately reached
by a single interaction. To identify the essential genes in the
OSM, we ranked all module genes on the basis of their degree and
betweenness centrality and obtained the MDSets within the OSM.
Overlapping genes among hub genes, bottleneck genes, and genes
contained in the MDSets were considered valuable for osteosarcoma
progression.
Statistical analysis
All of the statistical analyses were conducted in R
3.4.1 (r-project.org/) and Python 2.7.11
(python.org/downloads/release/python-2711/).
Networkx module of Python was used for the non-randomness test of
identified gene module. P<0.05 was used as the criteria for the
identification of significantly enriched functions.
Results
Identification of disease module
associated with osteosarcoma
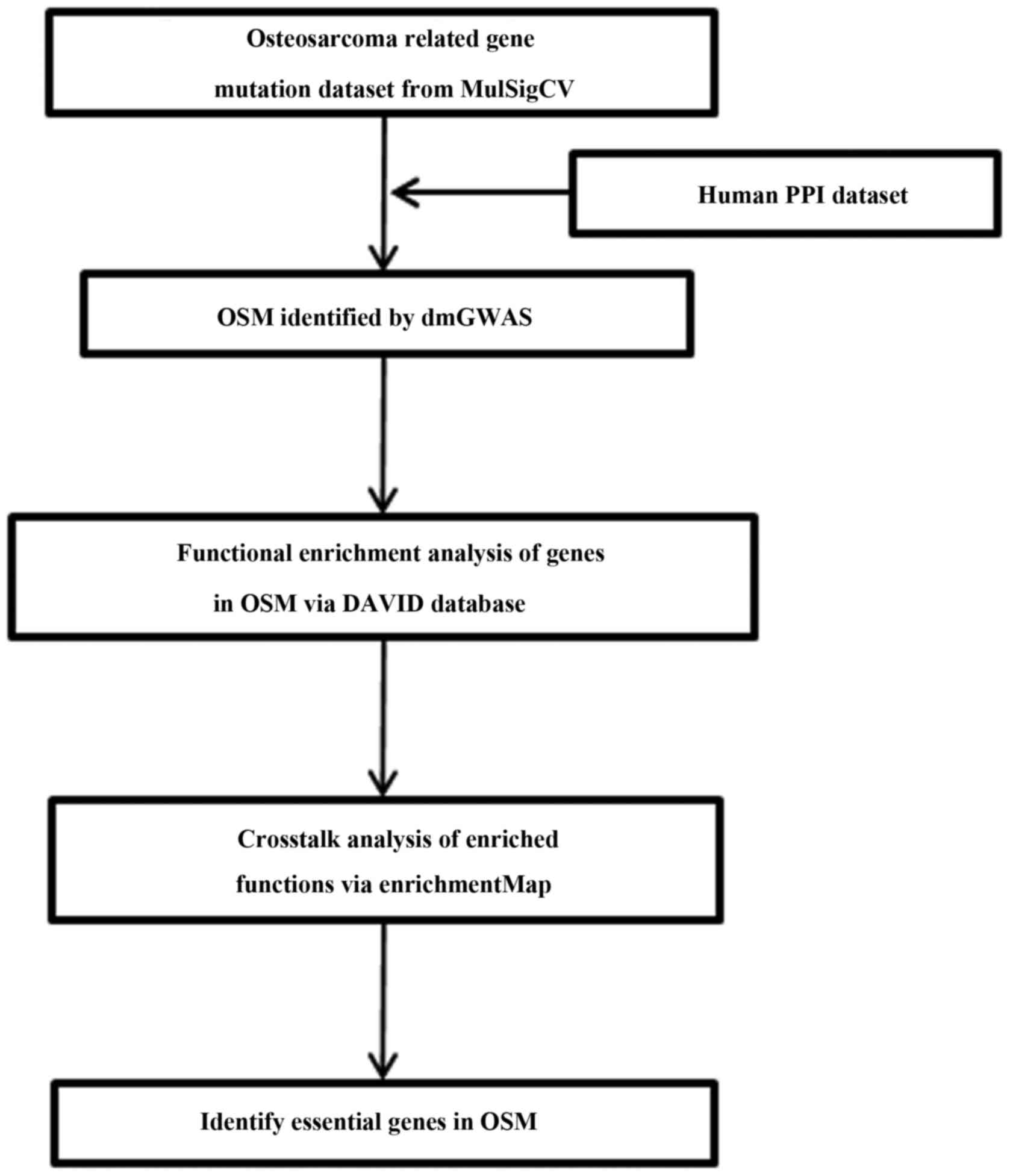
Fig. 1 illustrates
the flow chart of this study. By taking advantage of dmGWAS
(10), candidate disease modules
related to osteosarcoma were deduced by incorporating the
association signal from GWAS into the PPI network. The top 150
candidate modules with the highest module scores and empirical
P<0.001 were selected. The OSM was obtained by merging these
modules and excluding the redundant entries, which resulted in a
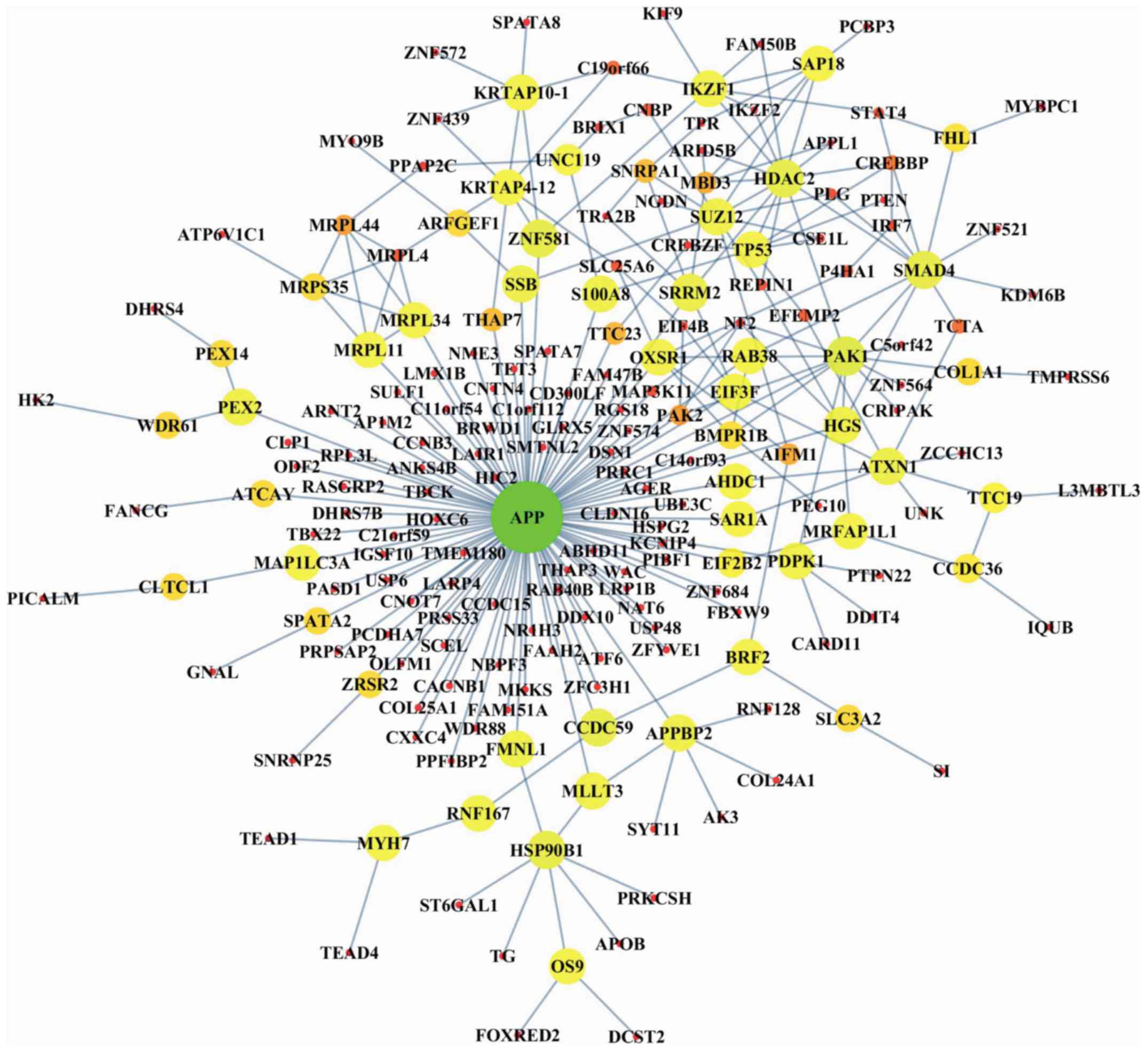
subnetwork containing 201 nodes and 268 edges (Fig. 2). To test for the non-randomness of
the identified disease module, 1,000 random subnetworks were
generated, and their corresponding average clustering coefficient
and the mean shortest-path distance were compared with the
corresponding values of the OSM. For these random subnetworks, the
average clustering coefficient was 0.01, which is statistically
significantly smaller than that of the OSM (clustering coefficient,
0.07; empirical P<0.001). The average shortest-path distance for
the 1000 random subnetwork was 4.75, which was statistically
significantly higher than that of the OSM (shortest-path distance,
3.37; empirical P<0.001). This indicated that our identified OSM
was a non-random network. Notably, some of the genes within the
OSM, such as small nuclear ribonucleoprotein U11/U12 subunit 25
(SNRNP25) (19,20), cyclin B3 (CCNB3) (21), tumor protein p53 (TP53) (22,23),
OS9, endoplasmic reticulum lectin (OS9) (24), and phosphatase and tensin homolog
(PTEN) (25), have been reportedly
associated with osteosarcoma in previous studies.
Significantly enriched functions of
the OSM
In this study, significantly enriched GO terms and
KEGG pathways of the OSM were identified through WebGestalt and
ToppGene online tools, respectively. Consequently, a total of 30 GO
terms (Table I) and 32 KEGG pathways
(Table II) were significantly
associated with the OSM. GO terms related to response to stimulus
(e.g., response to estradiol and to epidermal growth factor)
(26,27) and intracellular signal transduction
(e.g., hippo signaling) (28) were
obtained in this study; these were consistent with the previous
findings about osteosarcoma. Terms directly related to protein or
nucleic acid binding (e.g., RNA polymerase II transcription factor
binding, protein N-terminus binding, lipoprotein particle receptor
binding, single-stranded RNA binding, and damaged DNA binding) and
catalytic activity (e.g., deacetylase activity, phosphotransferase
activity, phosphate group as a acceptor, and kinase activity of
nucleobase-containing compounds) were also included. GO terms
related to translation (e.g., cytoplasmic translation) were also
enriched in OS genes. In line with previous studies, several
pathways, such as mTOR signaling pathway (ranked 5th) (29,30),
Hippo signaling pathway (ranked 9th) (31), PI3K-Akt signaling pathway (ranked
10th) (30,32), MAPK signaling pathway (ranked 11th)
(33), Wnt signaling pathway (ranked
22nd) (34,35), p53 signaling pathway (ranked 24th)
(36–38), and TGF-β signaling pathway (ranked
27th) (38,39), were enriched in OS genes. In
addition, several cancer-related pathways were identified, such as
Pathways in cancer, Proteoglycans in cancer, central carbon
metabolism in cancer, and transcriptional misregulation in cancer.
Moreover, cell proliferation, survival, and apoptosis-associated
biological processes comprising cell cycle, FoxO signaling pathway,
protein processing in the endoplasmic reticulum, RNA transport, and
focal adhesion were also obtained.
 | Table I.Gene ontology terms enriched in the
osteosarcoma-related gene module. |
Table I.
Gene ontology terms enriched in the
osteosarcoma-related gene module.
| Category | GO terms | Cell process | P-value | Genes included in
the GO term |
|---|
| Biological
process | GO:0070482 | Response to oxygen
levels |
1.43×10−4 | COL1A1; CREBBP;
AGER; HDAC2; SMAD4; PAK1; DDIT4; PTEN; TP53; HSP90B1; AIFM1;
ARNT2 |
| Biological
process | GO:0048511 | Rhythmic
process |
2.15×10−3 | NR1H3; CREBBP;
PASD1; FANCG; HDAC2; SMAD4; PTEN; BMPR1B; TP53; EIF2B2 |
| Biological
process | GO:0032355 | Response to
estradiol |
2.19×10−3 | COL1A1; APOB; MBD3;
PTEN; AIFM1; ARNT2 |
| Biological
process | GO:0002181 | Cytoplasmic
translation |
3.02×10−3 | EIF4B; SSB; UNK;
EIF3F |
| Biological
process | GO:0031349 | Positive regulation
of defense response |
4.05×10−3 | NR1H3; CREBBP;
PTPN22; SLC25A6; IRF7; PAK1; PAK2; PDPK1; S100A8; HSP90B1;
CARD11 |
| Biological
process | GO:0071826 | Ribonucleoprotein
complex subunit organization |
6.05×10−3 | CLP1; EIF4B; CNOT7;
BRIX1; RPL3L; MRPL11; ZRSR2 |
| Biological
process | GO:0035329 | Hippo
signaling |
6.46×10−3 | NF2; TEAD1;
TEAD4 |
| Biological
process | GO:0055076 | Transition metal
ion homeostasis |
7.64×10−3 | TMPRSS6; APP;
SMAD4; S100A8; PICALM |
| Biological
process | GO:0016570 | Histone
modification |
7.68×10−3 | SAP18; CREBBP;
KDM6B; SUZ12; HDAC2; SMAD4; WAC; MBD3; TP53; WDR61; ARID5B |
| Biological
process | GO:0070849 | Response to
epidermal growth factor |
8.13×10−3 | COL1A1; PDPK1;
TPR |
| Molecular
function | GO:0001085 | RNA polymerase II
transcription factor binding |
1.13×10−4 | CREBBP; HDAC2;
SMAD4; TEAD1; TEAD4; TP53; MKKS |
| Molecular
function | GO:0019205 |
Nucleobase-containing compound kinase
activity |
1.31×10−3 | CLP1; NME3; AK3;
CARD11 |
| Molecular
function | GO:0005057 | Signal transducer
activity, downstream of receptor |
3.19×10−3 | SMAD4; MAP3K11;
PAK1; PAK2; BMPR1B; OXSR1 |
| Molecular
function | GO:0003727 | Single-stranded RNA
binding |
5.23×10−3 | EIF4B; ATXN1;
TRA2B; CNBP |
| Molecular
function | GO:0016776 | Phosphotransferase
activity, phosphate group as acceptor |
7.33×10−3 | NME3; AK3;
CARD11 |
| Molecular
function | GO:0047485 | Protein N-terminus
binding |
1.61×10−2 | SRRM2; PEX14; TP53;
THAP7 |
| Molecular
function | GO:0019213 | Deacetylase
activity |
1.67×10−2 | SAP18; HDAC2;
MBD3 |
| Molecular
function | GO:0070325 | Lipoprotein
particle receptor binding |
1.84×10−2 | APOB; HSP90B1 |
| Molecular
function | GO:0003735 | Structural
constituent of ribosome |
2.10×10−2 | MRPL4; MRPS35;
RPL3L; MRPL34; MRPL11 |
| Molecular
function | GO:0003684 | Damaged DNA
binding |
2.30×10−2 | CREBBP; FANCG;
TP53 |
| Cellular
component | GO:0005788 | Endoplasmic
reticulum lumen |
1.07×10−3 | OS9; COL1A1;
COL24A1; APOB; P4HA1; PRKCSH; HSP90B1; FOXRED2; COL25A1 |
| Cellular
component | GO:0000118 | Histone deacetylase
complex |
7.72×10−3 | SAP18; APPL1;
HDAC2; MBD3 |
| Cellular
component | GO:0070603 | SWI/SNF
superfamily-type complex |
1.43×10−2 | SUZ12; APPL1;
HDAC2; MBD3 |
| Cellular
component | GO:0017053 | Transcriptional
repressor complex |
2.46×10−2 | PASD1; APPL1;
HDAC2; MBD3 |
| Cellular
component | GO:0005840 | Ribosome |
2.86×10−2 | REPIN1; MRPL4;
MRPS35; RPL3L; MRPL34; MRPL11; MRPL44 |
| Cellular
component | GO:0008023 | Transcription
elongation factor complex |
2.90×10−2 | MLLT3; PEX2;
WDR61 |
| Cellular
component | GO:0044454 | Nuclear chromosome
part |
4.17×10−2 | NR1H3; IKZF1;
CREBBP; SUZ12; REPIN1; HDAC2; SMAD4; MBD3; SSB; TP53; DSN1 |
| Cellular
component | GO:0031983 | Vesicle lumen |
4.48×10−2 | APOB; APP; PLG;
HSP90B1 |
| Cellular
component | GO:0030139 | Endocytic
vesicle |
4.52×10−2 | AP1M2; SYT11;
RAB38; APOB; HSP90B1; FMNL1; PICALM |
| Cellular
component | GO:0031984 | Organelle
subcompartment |
4.77×10−2 | AP1M2; ARFGEF1;
SULF1; RAB38; APP; ZFYVE1; ST6GAL1; CLTCL1 |
 | Table II.Kyoto Encyclopedia of Genes and
Genomes pathways enriched in the osteosarcoma-related gene
module. |
Table II.
Kyoto Encyclopedia of Genes and
Genomes pathways enriched in the osteosarcoma-related gene
module.
| Pathways | P-value | False discovery
rate | Genes included in
the pathway |
|---|
| Hepatitis B |
1.01×10−6 |
2.47×10−4 | STAT4, TP53, SMAD4,
HSPG2, PTEN, IRF7, CREBBP |
| Pathways in
cancer |
1.67×10−6 |
2.15×10−3 | RASGRP2, CREBBP,
HDAC2, TP53, APPL1, SMAD4, HSP90B1, PTEN, TPR, ARNT2 |
| Prostate
cancer |
1.80×10−5 |
1.78×10−3 | PTEN, TP53, CREBBP,
HSP90B1, PDPK1 |
| Hepatitis C |
1.23×10−4 |
4.72×10−3 | CLDN16, PDPK1,
IRF7, TP53, NR1H3 |
| mTOR signaling
pathway |
6.68×10−5 |
4.72×10−3 | PTEN, PDPK1, DDIT4,
EIF4B |
| Cell cycle |
8.84×10−5 |
4.72×10−3 | SMAD4, CCNB3,
HDAC2, TP53, CREBBP |
| Renal cell
carcinoma |
9.72×10−5 |
4.72×10−3 | CREBBP, ARNT2,
PAK2, PAK1 |
| FoxO signaling
pathway |
1.28×10−4 |
4.72×10−3 | SMAD4, PTEN, CCNB3,
CREBBP, PDPK1 |
| Hippo signaling
pathway |
2.44×10−4 |
7.23×10−3 | SMAD4, BMPR1B, NF2,
TEAD1, TEAD4 |
| PI3K-Akt signaling
pathway |
2.38×10−4 |
7.23×10−3 | DDIT4, HSP90B1,
PTEN, PDPK1, COL1A1, TP53, EIF4B |
| MAPK signaling
pathway |
3.29×10−4 |
8.85×10−3 | RASGRP2, TP53,
CACNB1, MAP3K11, PAK1, PAK2 |
| Protein processing
in endoplasmic reticulum |
3.75×10−4 |
9.24×10−3 | OS9, SAR1A, PRKCSH,
HSP90B1, ATF6 |
| RNA transport |
4.06×10−4 |
9.25×10−3 | EIF3F, EIF2B2, TPR,
SAP18, EIF4B |
| T cell receptor
signaling pathway |
5.59×10−4 |
1.18×10−2 | PDPK1, CARD11,
PAK1, PAK2 |
| Huntington's
disease |
6.85×10−4 |
1.35×10−2 | CLTCL1, HDAC2,
TP53, SLC25A6, CREBBP |
| Thyroid hormone
signaling pathway |
8.98×10−4 |
1.40×10−2 | PDPK1, HDAC2, TP53,
CREBBP |
| Endometrial
cancer |
8.92×10−4 |
1.40×10−2 | PTEN, TP53,
PDPK1 |
| Focal adhesion |
8.41×10−4 |
1.40×10−2 | PTEN, PDPK1, PAK1,
PAK2, COL1A1 |
| Proteoglycans in
cancer |
8.60×10−4 |
1.40×10−2 | PDPK1, HSPG2, TP53,
PAK1, EIF4B |
| Colorectal
cancer |
1.49×10−3 |
2.20×10−2 | SMAD4, APPL1,
TP53 |
| Ribosome |
1.56×10−3 |
2.20×10−2 | MRPL11, MRPL34,
MRPL4, RPL3L |
| Wnt signaling
pathway |
1.83×10−3 |
2.39×10−2 | SMAD4, TP53,
CREBBP, CXXC4 |
| Central carbon
metabolism in cancer |
1.86×10−3 |
2.39×10−2 | HK2, PTEN,
TP53 |
| p53 signaling
pathway |
2.02×10−3 |
2.50×10−2 | CCNB3, PTEN,
TP53 |
| Chronic myeloid
leukemia |
2.38×10−3 |
2.82×10−2 | SMAD4, HDAC2,
TP53 |
| Peroxisome |
3.42×10−3 |
3.89×10−2 | DHRS4, PEX2,
PEX14 |
| TGF-β signaling
pathway |
3.54×10−3 |
3.89×10−2 | SMAD4, BMPR1B,
CREBBP |
| Influenza A |
3.78×10−3 |
3.99×10−2 | PLG, IRF7, SLC25A6,
CREBBP |
| Protein digestion
and absorption |
4.30×10−3 |
4.24×10−2 | SLC3A2, COL24A1,
COL1A1 |
| Transcriptional
misregulation in cancer |
4.18×10−3 |
4.24×10−2 | HDAC2, TP53, ARNT2,
MLLT3 |
| Thyroid cancer |
4.95×10−3 |
4.73×10−2 | TP53, TPR |
| Galactose
metabolism |
5.29×10−3 |
4.89×10−2 | HK2, SI |
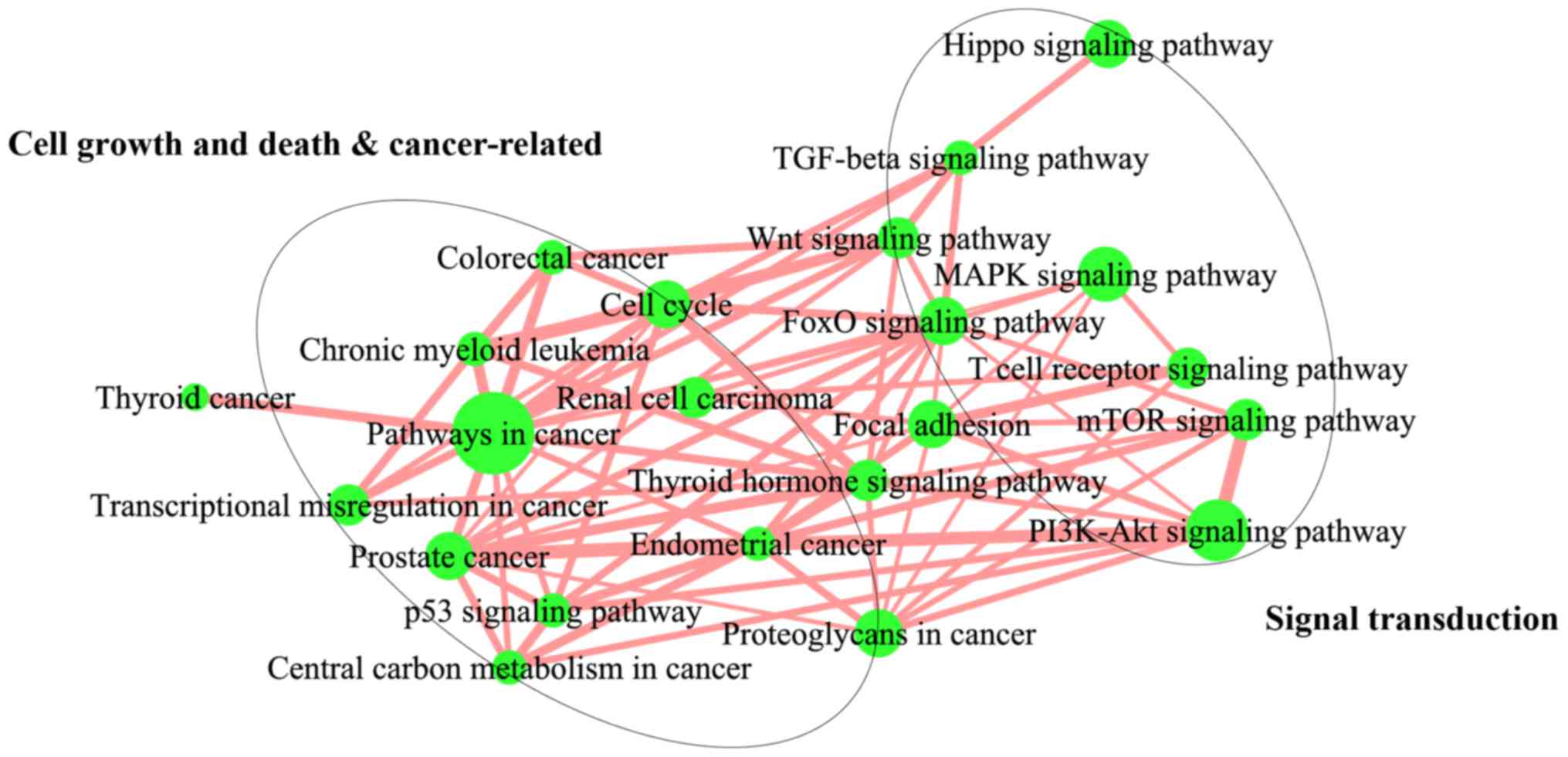
Crosstalk among significantly enriched
pathway
To understand how significantly enriched pathways
interact with each other, we carried out a pathway crosstalk
analysis among the 32 significantly enriched pathways. The approach
was based on the assumption that two pathways were considered to
interact with each other if they shared a proportion of the OS
genes. With this criterion for pathway crosstalk analysis, 22
pathways were selected. All the pathway pairs were used to build
the pathway crosstalk, and the overlapping level between two
pathways was measured on the basis of the average scores of the
coefficients OC and JC. Based on their crosstalk, these pathways
could be roughly divided into two main modules, each of which
included pathways that shared more interactions compared with other
pathways and might involve the same or similar biological processes
(Fig. 3). One module primarily
comprised cell growth, death, and cancer-related pathways, such as
those associated with the cell cycle, p53 signaling pathway,
central carbon metabolism in cancer, transcriptional misregulation
in cancer, and chronic myeloid leukemia. Conversely, the second
module was mainly dominated by signal transduction-related
pathways, including mTOR signaling pathway, FoxO signaling pathway,
Hippo signaling pathway, PI3K-Akt signaling pathway, Wnt signaling
pathway, and MAPK signaling pathway. Furthermore, these two modules
were linked by a couple of pathway interactions.
Essential genes in the OSM
To identify the essential genes within the OSM, we
first ranked the OS genes based on their degree as well as
betweenness centrality and obtained the MDSets simultaneously. The
genes intersecting among these three gene lists were considered as
essential OS genes, which included APP, APPBP2, ATXN1, HSP90B1,
IKZF1, KRTAP10-1, PAK1, PDPK1, SMAD4, SUZ12, and TP53. Some of
these genes, such as HSP90B1 (30,39),
PAK1 (40), SMAD4 (31,41), and
TP53 (22,23), have already been reported to be
associated with osteosarcoma. Besides, PDPK1 plays an important
role in the mTOR signaling pathway and PI3K-Akt signaling pathway,
which are reportedly involved in the pathogenesis of osteosarcoma.
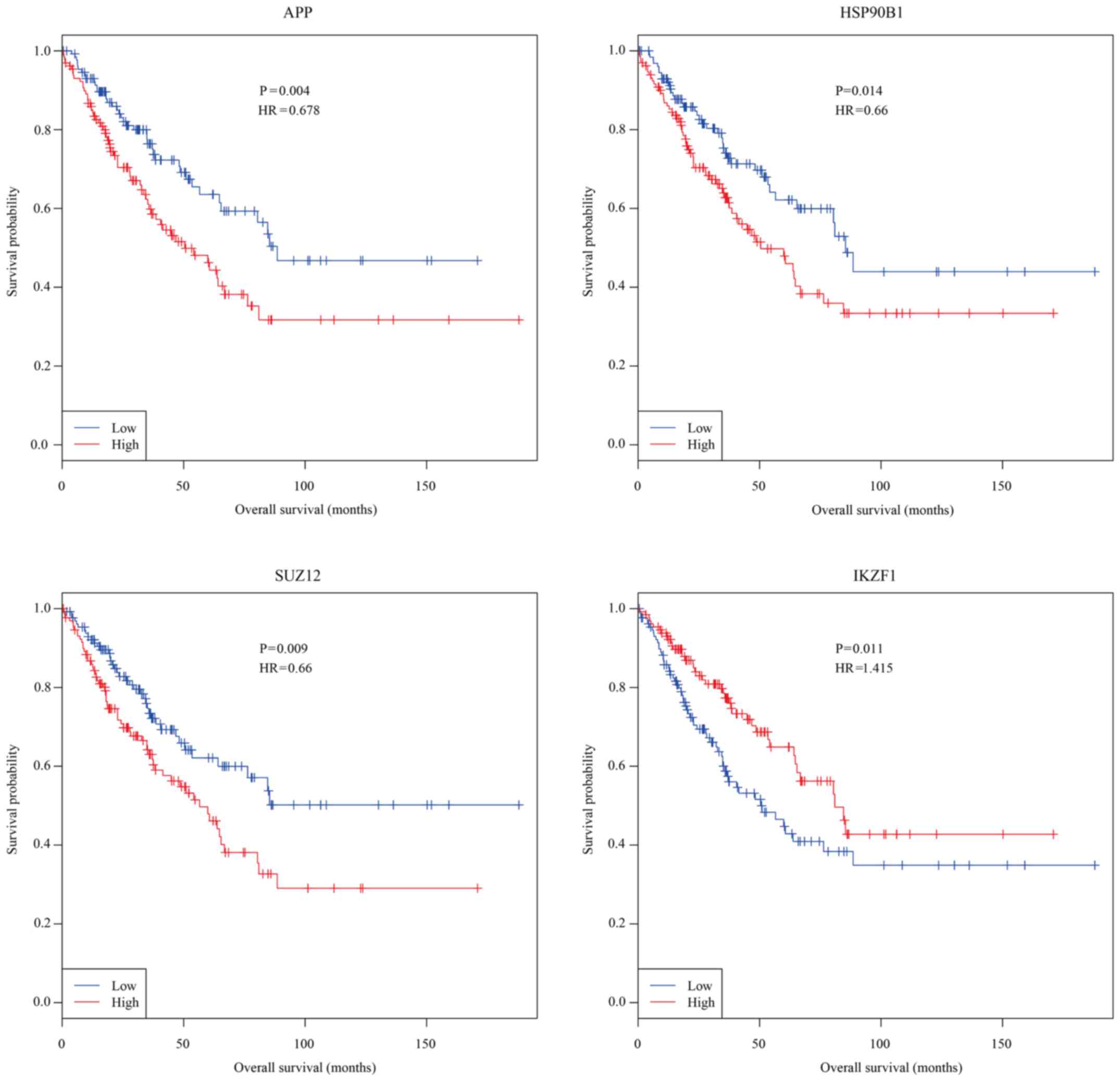
Kaplan-Meier analysis for overall survival with osteosarcoma and
the expression values of essential genes from the Cancer Genome
Atlas (TCGA) identified APP, HSP90B1, SUZ12, and IKZF1 as
prognosis-related genes (Fig. 4).
High expressions of APP (median survival for low and high APP
expression patients is 95 and 64 months), HSP90B1 (median survival
for low and high HSP90B1 expression patients is 90 and 60 months),
and SUZ12 (median survival for low and high SUZ12 expression
patients is 97 and 64 months) were significantly associated with
shorter overall survival with osteosarcoma, whereas high expression
of IKZF1 (median survival for low and high APP expression patients
is 57 and 81 months) was significantly associated with longer
overall survival with osteosarcoma.
Discussion
With the development of high-throughput technology,
more and more genes/proteins have been identified as associated
with osteosarcoma. Over the past ten years, much has been learnt
from studies on animals, human subjects, or cell models about the
molecular mechanisms underlying osteosarcoma. However, a
comprehensive understanding of the biological process associated
with osteosarcoma pathogenesis at the molecular level remains
largely unclear. It is imperative to decipher the latent
pathogenesis of osteosarcoma at the systems biology level. In this
work, we first identified an OSM by combining the GWAS dataset of
osteosarcoma with whole PPI network. A relatively comprehensive
human physical interactome that integrated PPI data from various
sources has been used in this study to reduce the limitations due
to incomplete and noisy human interactome. Functions of genes
contained in the OSM and potential osteosarcoma diagnosis and
treatment targets were further explored.
The OSM we identified was a non-random network and
11 essential OS genes have been identified as essential in the
module. Some of the essential in the module, such as HSP90B1
(29,38), PAK1 (39), SMAD4 (30,40), and
TP53 (21,22), have already been reported to be
related to osteosarcoma, suggesting the reliability of the methods
we used in this study. In addition, although several OS genes have
been found as not directly involved in the pathogenesis of
osteosarcoma, they are a part of biological pathways that play
important roles in osteosarcoma development. PDPK1 plays an
important role in the mTOR signaling pathway and PI3K-Akt signaling
pathway, which are reportedly involved in the pathogenesis of
osteosarcoma. DDIT4 and EIF4B were included in the OSM; both of
them are involved in mTOR signaling pathway and PI3K-Akt signaling
pathway, which are both closely associated with osteosarcoma. Some
of the genes, such as STAT4 and LRP1B, have not been demonstrated
to be related to osteosarcoma. Some of the members of the same
family with those unproved genes, such as STAT3 (42,43) and
LRP1 (19), have shown their
relevance in osteosarcoma in recent studies.
Kaplan-Meier curve analysis was performed to explore
the associations between overall survival and these 11 essential OS
genes in patients with osteosarcoma. Consequently, four essential
OS genes, namely APP, HSP90B1, SUZ12, and IKZF1, were found to be
significantly associated with overall survival with osteosarcoma
(P<0.05). Importantly, SUZ12 polycomb repressive complex 2
subunit (SUZ12) has been identified at the breakpoints of a
recurrent chromosomal translocation reported in endometrial stromal
sarcoma. Thus, the OS genes identified as essential are a list of
potential candidates and are reliable for researchers for further
exploration.
In conclusion, our study provides a pipeline for
systematically analyzing genome-wide datasets of osteosarcoma.
Several valuable biomarkers which should be helpful for
understanding the underlying mechanisms of osteosarcoma were
identified.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and material
The datasets generated and/or analyzed during the
current study are available in the dbGaP (ncbi.nlm.nih.gov/gap) (NCBI dbGaP study accession:
phs000699.v1.p1 and dbGaP analysis accession: pha003862).
Authors' contributions
YZ and FY made substantial contributions to the
conception and design, and interpretation of the data. YZ and FY
revised the manuscript critically for important intellectual
content and gave final approval of the version to be published. YZ
and FY agreed to be accountable for all aspects of the work in
ensuring that questions related to the accuracy or integrity of any
part of the study are appropriately investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ottaviani G and Jaffe N: The epidemiology
of osteosarcoma. Cancer Treat Res. 152:3–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yang J, Cogdell D, Yang D, Hu L, Li H,
Zheng H, Du X, Pang Y, Trent J, Chen K and Zhang W: Deletion of the
WWOX gene and frequent loss of its protein expression in human
osteosarcoma. Cancer Lett. 291:31–38. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sparks AB, Peterson SN, Bell C, Loftus BJ,
Hocking L, Cahill DP, Frassica FJ, Streeten EA, Levine MA, Fraser
CM, et al: Mutation screening of the TNFRSF11A gene encoding
receptor activator of NF kappa B (RANK) in familial and sporadic
Paget's disease of bone and osteosarcoma. Calcif Tissue Int.
68:151–155. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sadikovic B, Yoshimoto M, Chilton-MacNeill
S, Thorner P, Squire JA and Zielenska M: Identification of
interactive networks of gene expression associated with
osteosarcoma oncogenesis by integrated molecular profiling. Hum Mol
Genet. 18:1962–1975. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ueda K, Cardarelli C, Gottesman MM and
Pastan I: Expression of a full-length cDNA for the human ‘MDR1’
gene confers resistance to colchicine, doxorubicin, and
vinblastine. Proc Natl Acad Sci USA. 84:3004–3008. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jia M, Hu J, Li W, Su P, Zhang H, Zhang X
and Zhou G: Trps1 is associated with the multidrug resistance of
osteosarcoma by regulating MDR1 gene expression. FEBS Lett.
588:801–810. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hu YS, Xin J, Hu Y, Zhang L and Wang J:
Analyzing the genes related to Alzheimer's disease via a network
and pathway-based approach. Alzheimers Res Ther. 9:292017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cowley MJ, Pinese M, Kassahn KS, Waddell
N, Pearson JV, Grimmond SM, Biankin AV, Hautaniemi S and Wu J: PINA
v2.0: Mining interactome modules. Nucleic Acids Res. 40:(Database
Issue). D862–D865. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Menche J, Sharma A, Kitsak M, Ghiassian
SD, Vidal M, Loscalzo J and Barabási AL: Disease networks.
Uncovering disease-disease relationships through the incomplete
interactome. Science. 347:12576012015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jia P, Zheng S, Long J, Zheng W and Zhao
Z: dmGWAS: Dense module searching for genome-wide association
studies in protein-protein interaction networks. Bioinformatics.
27:95–102. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang J, Duncan D, Shi Z and Zhang B:
WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): Update 2013.
Nucleic Acids Res. 41:(Web Server Issue). W77–W83. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen J, Bardes EE, Aronow BJ and Jegga AG:
ToppGene Suite for gene list enrichment analysis and candidate gene
prioritization. Nucleic Acids Res. 37:(Web Server Issue).
W305–W311. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hu Y, Pan Z, Hu Y, Zhang L and Wang J:
Network and pathway-based analyses of genes associated with
Parkinson's disease. Mol Neurobiol. 54:4452–4465. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
He X and Zhang J: Why do hubs tend to be
essential in protein networks? PLoS Genet. 2:e882006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jeong H, Mason SP, Barabasi AL and Oltvai
ZN: Lethality and centrality in protein networks. Nature.
411:41–42. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yu H, Kim PM, Sprecher E, Trifonov V and
Gerstein M: The importance of bottlenecks in protein networks:
Correlation with gene essentiality and expression dynamics. PLoS
Comput Biol. 3:e592007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wuchty S: Controllability in protein
interaction networks. Proc Natl Acad Sci USA. 111:7156–7160. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xing P, Liao Z, Ren Z, Zhao J, Song F,
Wang G, Chen K and Yang J: Roles of low-density lipoprotein
receptor-related protein 1 in tumors. Chin J Cancer. 35:62016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang J, Annala M, Ji P, Wang G, Zheng H,
Codgell D, Du X, Fang Z, Sun B, Nykter M, et al: Recurrent
LRP1-SNRNP25 and KCNMB4-CCND3 fusion genes promote tumor cell
motility in human osteosarcoma. J Hematol Oncol. 7:762014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shibayama T, Okamoto T, Nakashima Y, Kato
T, Sakurai T, Minamiguchi S, Kataoka TR, Shibuya S, Yoshizawa A,
Toguchida J and Haga H: Screening of BCOR-CCNB3 sarcoma using
immunohistochemistry for CCNB3: A clinicopathological report of
three pediatric cases. Pathol Int. 65:410–414. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bousquet M, Noirot C, Accadbled F, de
Gauzy Sales J, Castex MP, Brousset P and Gomez-Brouchet A:
Whole-exome sequencing in osteosarcoma reveals important
heterogeneity of genetic alterations. Ann Oncol. 27:738–744. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Saalfrank A, Janssen KP, Ravon M,
Flisikowski K, Eser S, Steiger K, Flisikowska T, Müller-Fliedner P,
Schulze É, Brönner C, et al: A porcine model of osteosarcoma.
Oncogenesis. 5:e2102016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yotov WV, Hamel H, Rivard GE, Champagne
MA, Russo PA, Leclerc JM, Bernstein ML and Levy E: Amplifications
of DNA primase 1 (PRIM1) in human osteosarcoma. Genes Chromosomes
Cancer. 26:62–69. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xi Y and Chen Y: PTEN plays dual roles as
a tumor suppressor in osteosarcoma cells. J Cell Biochem.
118:2684–2692. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kersting C, Gebert C, Agelopoulos K,
Schmidt H, van Diest PJ, Juergens H, Winkelmann W, Kevric M,
Gosheger G, Brandt B, et al: Epidermal growth factor receptor
expression in high-grade osteosarcomas is associated with a good
clinical outcome. Clin Cancer Res. 13:2998–3005. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mantovani FB, Morrison JA and Mutsaers AJ:
Effects of epidermal growth factor receptor kinase inhibition on
radiation response in canine osteosarcoma cells. BMC Vet Res.
12:822016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Deel MD, Li JJ, Crose LE and Linardic CM:
A review: Molecular aberrations within hippo signaling in bone and
soft-tissue sarcomas. Front Oncol. 5:1902015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Niu NK, Wang ZL, Pan ST, Ding HQ, Au GH,
He ZX, Zhou ZW, Xiao G, Yang YX, Zhang X, et al: Pro-apoptotic and
pro-autophagic effects of the Aurora kinase A inhibitor alisertib
(MLN8237) on human osteosarcoma U-2 OS and MG-63 cells through the
activation of mitochondria-mediated pathway and inhibition of p38
MAPK/PI3K/Akt/mTOR signaling pathway. Drug Des Devel Ther.
9:1555–1584. 2015.PubMed/NCBI
|
|
30
|
Li G, Cai M, Fu D, Chen K, Sun M, Cai Z
and Cheng B: Heat shock protein 90B1 plays an oncogenic role and is
a target of microRNA-223 in human osteosarcoma. Cell Physiol
Biochem. 30:1481–1490. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ma J, Huang K, Ma Y, Zhou M and Fan S: The
TAZ-miR-224-SMAD4 axis promotes tumorigenesis in osteosarcoma. Cell
Death Dis. 8:e25392017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Meng ZJ, Wu N, Liu Y, Shu KJ, Zou X, Zhang
RX, Pi CJ, He BC, Ke ZY, Chen L, et al: Evodiamine inhibits the
proliferation of human osteosarcoma cells by blocking PI3K/Akt
signaling. Oncol Rep. 34:1388–1396. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tingting R, Wei G, Changliang P, Xinchang
L and Yi Y: Arsenic trioxide inhibits osteosarcoma cell
invasiveness via MAPK signaling pathway. Cancer Biol Ther.
10:251–257. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cai Y, Cai T and Chen Y: Wnt pathway in
osteosarcoma, from oncogenic to therapeutic. J Cell Biochem.
115:625–631. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Du X, Yang J, Yang D, Tian W and Zhu Z:
The genetic basis for inactivation of Wnt pathway in human
osteosarcoma. BMC Cancer. 14:4502014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Alkhalaf M and Jaffal S: Potent
antiproliferative effects of resveratrol on human osteosarcoma
SJSA1 cells: Novel cellular mechanisms involving the ERKs/p53
cascade. Free Radic Biol Med. 41:318–325. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Deng Z, Liu X, Jin J, Xu H, Gao Q, Wang Y
and Zhao J: Histone deacetylase inhibitor trichostatin a promotes
the apoptosis of osteosarcoma cells through p53 signaling pathway
activation. Int J Biol Sci. 12:1298–1308. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sun Y, Xia P, Zhang H, Liu B and Shi Y:
P53 is required for Doxorubicin-induced apoptosis via the TGF-beta
signaling pathway in osteosarcoma-derived cells. Am J Cancer Res.
6:114–125. 2015.PubMed/NCBI
|
|
39
|
Suzuki S and Kulkarni AB: Extracellular
heat shock protein HSP90beta secreted by MG63 osteosarcoma cells
inhibits activation of latent TGF-beta1. Biochem Biophys Res
Commun. 398:525–531. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhou Y, Zhao RH, Tseng KF, Li KP, Lu ZG,
Liu Y, Han K, Gan ZH, Lin SC, Hu HY and Min DL: Sirolimus induces
apoptosis and reverses multidrug resistance in human osteosarcoma
cells in vitro via increasing microRNA-34b expression. Acta
Pharmacol Sin. 37:519–529. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yu D, An F, He X and Cao X: Curcumin
inhibits the proliferation and invasion of human osteosarcoma cell
line MG-63 by regulating miR-138. Int J Clin Exp Pathol.
8:14946–14952. 2015.PubMed/NCBI
|
|
42
|
Jiang R, Zhang C, Liu G, Gu R and Wu H:
MicroRNA-126 inhibits proliferation, migration, invasion and EMT in
osteosarcoma by targeting ZEB1. J Cell Biochem. 118:3765–3774.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yun HM, Park KR, Quang TH, Oh H, Hong JT,
Kim YC and Kim EC: 4-parvifuran inhibits metastatic and invasive
actions through the JAK2/STAT3 pathway in osteosarcoma cells. Arch
Pharm Res. 40:601–609. 2017. View Article : Google Scholar : PubMed/NCBI
|