Introduction
Multiple myeloma (MM) is an age-related monoclonal
plasma cell cancer (1). Although
recent drug discoveries have greatly improved survival rates, MM
currently remains an incurable B-cell malignancy. The initiation
and progression of the disease like bone destruction might be
caused by the abnormal secretion of related cytokines, activation
of oncogenes and molecular genetic abnormalities. Although recent
studies on therapies [such as immunomodulatory drugs (IMiDs) and
proteasome inhibitors] have greatly improved clinical outcomes
(such as survival, the median survival has increased to over 5
years), MM continues to be an incurable disease (2).
The desensitization mechanism of G protein-coupled
receptors (GPCRs) terminates signaling initiated by G
protein-coupled receptor kinases (GRKs). GRKs are a family of
serine/threonine protein kinases; this family of proteins
phosphorylates agonist-activated GPCRs specifically. The GRK family
is composed of seven members, named GRK1-7, among which GRK2, 3 and
6 are highly expressed in the immune system (3,4). GRKs
specifically phosphorylate GPCRs, leading to GPCR uncoupling and
the binding of regulatory proteins, such as arrestins; the
phosphorylated receptors thus block further activation by G
proteins and initiate receptor internalization (5,6). Thus,
GRKs effectively reduce the level of functional receptors on the
cell membrane, which results in decreased receptor-mediated
signaling. Recent studies have reported that, compared to normal
tissues, G protein-coupled receptor kinase 6 (GRK6) is highly
expressed in tumor tissues, such as colonic carcinoma (7), liver cancer and medulloblastoma
(8,9). Mutations in GRK6 were also detected in
gastric cancer (10) and breast
cancer. These studies demonstrate that GRK6 might have a certain
relationship with tumor occurrence and development, and
prognosis.
As previously mentioned, recent studies have shown
that GRK6 is involved in many kinds of cell signaling pathways and,
to a certain extent, has an impact on many other related cell
biology activities, including the regulation of cell apoptosis and
the cell cycle. Our previous study showed that down-regulating GRK6
expression can significantly inhibit the proliferation of MM MM1R
cells, and stop the cell cycle in the G0/G1 phase by decreasing the
expression levels of Cyclin D1 and CDK4. Therefore, we aimed to
further investigate the possible role of GRK6 during tumorigenesis
in MM cell lines. In this study, the expression of the GRK6 gene
was observed in MM1R cells and cell samples isolated from MM
patients. Furthermore, this study demonstrated that GRK6 can
modulate apoptosis in MM cells.
Materials and methods
Cell culture
The MM cell lines (NCI-H929, RPMI8226, U266, OPM2,
MM1S and MM1R) were obtained from the American Type Culture
Collection (ATCC, Manassas, VA, USA), and cultured in RPMI 1640
medium supplemented with 10% fetal bovine serum (both Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA). Cell lines were
incubated at 37°C in a 5% CO2 atmosphere. GRK6
expression levels were detected in all cell lines (NCI-H929,
RPMI8226, U266, OPM2, MM1S and MM1R), and cell functional
experiments were performed in MM1R cell subsequently.
MM samples
MM patients and healthy individuals provided
informed consent, and the study complied with the Declaration of
Helsinki and its amendments. 28 individuals who were admitted to
the Affiliated Hospital of Xuzhou Medical College were enrolled in
this study, which included 16 patients newly diagnosed with MM in
the Department of Hematology from January 2015 to June 2016 and 12
healthy individuals. All subjects met the diagnostic criteria of MM
according to the International Myeloma Working Group (IMWG). Bone
marrow samples were collected from all 16 MM patients before
treatment. Healthy individuals were predominantly bone marrow
transplant donors and were not exposed to any known cytotoxic
treatment.
Samples from the 16 patients with MM were first
treated with Ficoll-Hypaque (GE Healthcare, Shanghai, China) and
underwent gradient centrifugation to obtain mononuclear cells.
Myeloma cells were isolated from bone marrow by magnetic cell
separation (MACS; Miltenyi Biotec GmbH, Bergisch Gladbach, Germany)
according to the manufacturer's protocol. The purity of the
CD138+MM cells was analyzed by flow cytometry (FACScan cytometer;
BD Biosciences, Franklin Lakes, NJ, USA) and determined to be ≥90%.
The study was approved by the local ethics committee.
Lentiviral vectors and infection
GRK6 specific shRNA sequences were cloned into GV246
lentiviral vectors. Restructured plasmids were named shRNA-1,
shRNA-2, shRNA-3 and shRNA-4 (Table
I). An empty vector was used as the control plasmid. pSPXA2 and
pMD2.G plasmids recombined the restructured lentiviral vectors, and
then they were cotransfected into 293FT cells with Lipofectamine
2000 (both Invitrogen; Thermo Fisher Scientific, Inc.). The viral
particles were harvested and concentrated by ultracentrifugation.
MM1R cells were treated 8 µg/ml polybrene (Sigma-Aldrich; Merck
KGaA, Germany) after recombinant virus infection. GFP-positive
cells were isolated using puromycin (0.5 µg/ml) after the cells
were cultured for 4 days. shRNA-4 had the highest inhibition rate
of GRK6 and was used in the further experiments.
 | Table I.GRK6 specific shRNA
sequences. |
Table I.
GRK6 specific shRNA
sequences.
| ID | 5′ | Stem | Loop | Stem | 3′ |
|---|
|
GRK6−1-RNAi-a | Ccgg |
ctGAATGTCTTTGGGCTGGAT | CTCGAG |
ATCCAGCCCAAAGACATTCAG | TTTTTg |
|
GRK6−1-RNAi-b | aattcaaaaa |
ctGAATGTCTTTGGGCTGGAT | CTCGAG |
ATCCAGCCCAAAGACATTCAG |
|
|
GRK6−2-RNAi-a | Ccgg |
caGTAGGTTTGTAGTGAGCTT | CTCGAG |
AAGCTCACTACAAACCTACTG | TTTTTg |
|
GRK6−2-RNAi-b | aattcaaaaa |
caGTAGGTTTGTAGTGAGCTT | CTCGAG |
AAGCTCACTACAAACCTACTG |
|
|
GRK6−3-RNAi-a | Ccgg |
caGCATCTACTTCAACCGTTT | CTCGAG |
AAACGGTTGAAGTAGATGCTG | TTTTTg |
|
GRK6−3-RNAi-b | aattcaaaaa |
caGCATCTACTTCAACCGTTT | CTCGAG |
AAACGGTTGAAGTAGATGCTG |
|
|
GRK6−4-RNAi-a | Ccgg |
ccTCGACAGCATCTACTTCAA | CTCGAG |
TTGAAGTAGATGCTGTCGAGG | TTTTTg |
|
GRK6−4-RNAi-b | aattcaaaaa |
ccTCGACAGCATCTACTTCAA | CTCGAG |
TTGAAGTAGATGCTGTCGAGG |
|
Apoptosis analysis
To assess the level of cell apoptosis, Annexin V PE
and 7AAD (both from eBioscience; Thermo Fisher Scientific, Inc.)
were used to stain cells according to the manufacturer's
instructions. The percentages of early apoptotic cells (Annexin V
PE+/7AAD-) and late apoptotic cells (Annexin V PE+/7AAD+) were
analysed in this study using CellQuest software in a FACScan
cytometer (BD Biosciences).
Western blotting
Protein was isolated using ProteoJET Mammalian Cell
Lysis Reagent (Beyotime Institute of Biotechnology, Haimen, China)
according to the manufacturer's instructions. Centrifugation
(12,000 × g, 15 min, 4°C) was performed to isolate the proteins.
They were then denatured and subjected to sodium dodecyl sulfate
polyacrylamide gel electrophoresis (SDS-PAGE) and western blotting.
Rabbit monoclonal anti-GAPDH (Sigma-Aldrich; Merck KGaA), anti-GRK6
(dilution, 1:1,000, cat. no. 5878), anti-STAT3 (dilution, 1:2,000;
cat. no. 4904), anti-pSTAT3 (dilution, 1:1,000; cat. no. 4093),
anti-Bax (dilution, 1:1.000; cat. no. 2772) and anti-Bcl-2
(dilution, 1:1,000; cat. no. 2872; all from Cell Signaling
Technology, Inc., Danvers, MA, USA) antibodies were used in the
present study. We also used horseradish peroxidase-conjugated
anti-rabbit secondary antibodies (dilution, 1:1,000; Cell Signaling
Technology, Inc.). The resulting bands were visualized using ECL
(Thermo Fisher Scientific, Inc.) and densitometric analysis
conducted using ImageJ software (v.1.4.3.67; National Institutes of
Health, Bethesda, MD, USA).
Reverse transcription-quantitative PCR
RT-(qPCR) analysis
Total RNA was extracted from cells and clinical
samples using TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). Complementary DNA was reverse transcribed using 2.5 µg of
RNA as a template. A Roche LC480 real-time PCR machine and a SYBR
Green kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) were
used to perform the qPCR analysis. 10 µl of the SYBR Green PCR
master mix, 50 ng cDNA and 250 nM of each primer were mixed to make
up a total volume of 20 µl, which was used for all PCR experiments.
Each sample was analysed in triplicate and the analysis was
repeated three times. The thermal cycling conditions were as
follows: 10 min at 95°C, followed by 40 cycles of 95°C for 15 sec
and 60°C for 60 sec. A dissociation curve was used to monitor
amplification. The relative expression of the target genes was
calculated using the 2−ΔΔCq method (11). The primer sequences were as follows:
GRK6, forward: 5′-CAGCCCATGGAGCTCGAGAAC-3′ and reverse:
5′-GGTGCAAAACTGTTAAACGGCGC-3′; GAPDH, forward:
5′-AGAAGGCTGGGGCTCATTTG-3′ and reverse: 5′-AGGGGCCATCCACAGTCTTC-3′.
GAPDH was used as the endogenous control.
Statistical analysis
The data was analyzed by SPSS statistical software
(v.24.0; SPSS, Inc., Chicago, IL, USA), which was expressed as mean
± standard deviation. Significant differences between groups were
determined by Student's t-test. For analyses between more than two
groups, Test was performed after the one-way ANOVA and Tukey's
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Expression of GRK6 in MM cell
lines
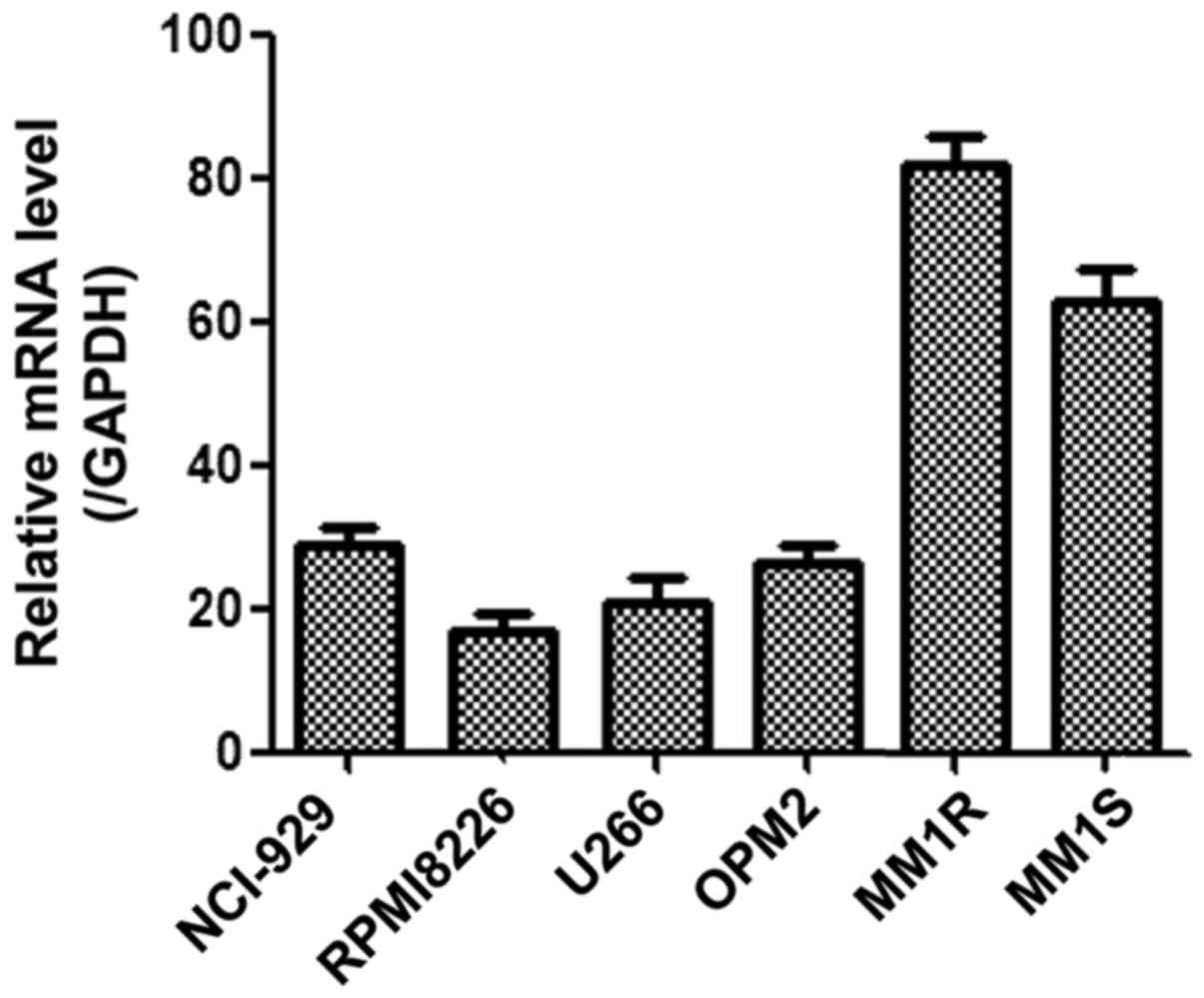
The expression levels of GRK6 mRNA in MM cells lines
NCI-H929, RPMI8226, U266, OPM2, MM1S and MM1R were measured using
qPCR analysis. The result showed that the GRK6 expression levels
varied between the different cell lines and that GRK6 expression
was significantly higher in MM1R cells than in the other cells
(Fig. 1).
GRK6 gene expression in patients with
MM
We analyzed the expression of GRK6 in 16 MM patients
before treatment (Table II). A
statistically significant 2–4-fold increase in GRK6 expression was
found in the MM patients compared with the healthy individuals
(P<0.05; Fig. 2A). Further
analyses indicated that GRK6 expression in the MM patients was
unrelated to clinical features, such as bone destruction (Fig. 2B), or ISS stage (Fig. 2C) or patient sex (Fig. 2D). The non-significant results may be
due to an insufficient sample size.
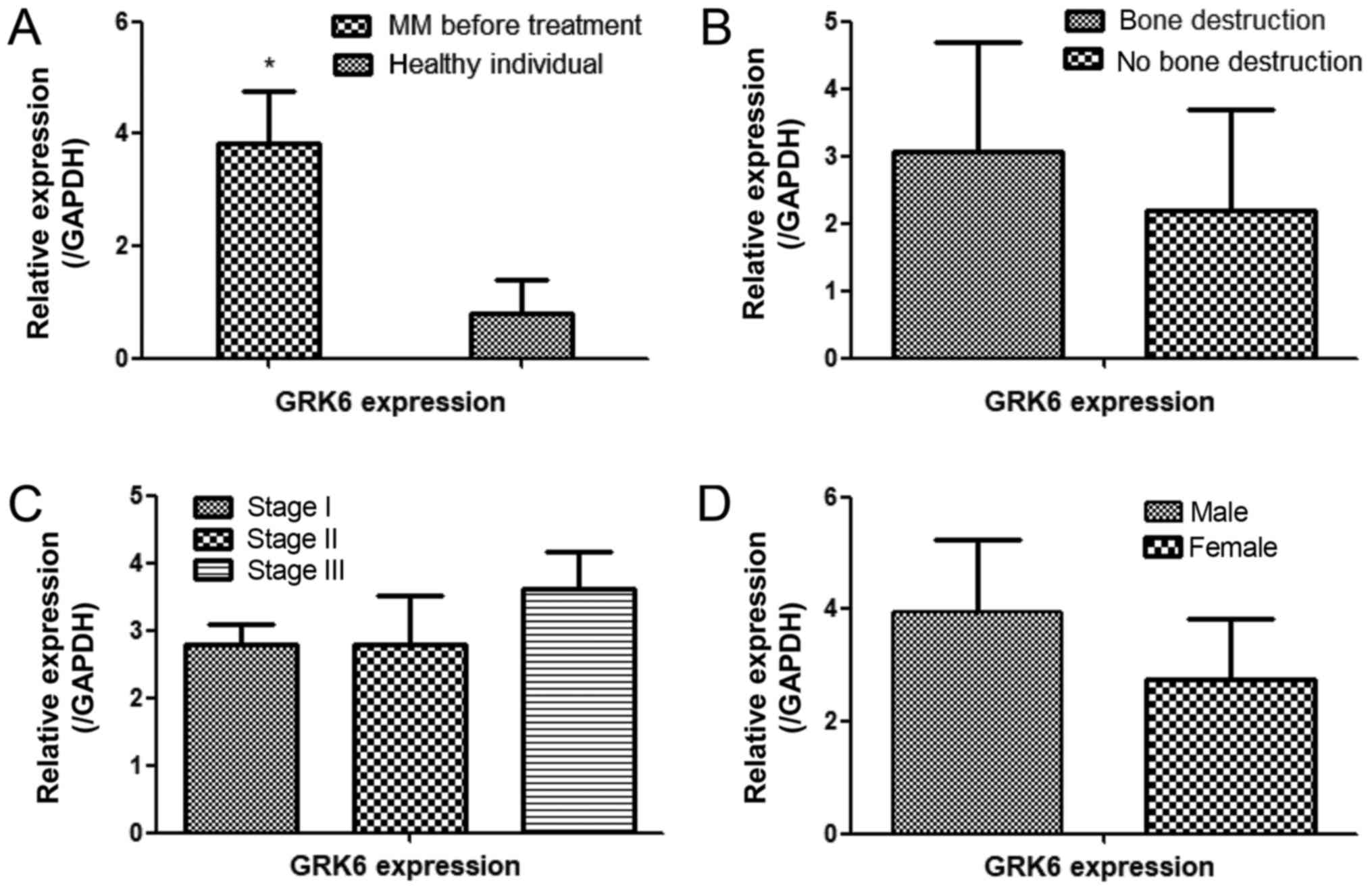 | Figure 2.qPCR analysis of the relative
expression of GRK6 in MM patient bone marrow. (A) The expression of
GRK6 in MM patients before treatment and in healthy controls *.
GRK6 expression before treatment was not significantly associated
with clinical features when MM patients were stratified according
to their (B) level of bone destruction (bone destruction, n=12; no
bone destruction, n=4), (C) ISS stage (stage I, n=5; stage II, n=4;
stage III, n=7) and (D) sex (female, n=4; male, n=12) (*P<0.05
vs. the healthy individuals). GRK6, G protein-coupled receptor
kinase 6; MM, multiple myeloma. |
 | Table II.Characteristics of patients before
treatment. |
Table II.
Characteristics of patients before
treatment.
| Clinical
characteristics | N (%) (n=16) |
|---|
| Age, years |
|
| ≤65 | 68 (11) |
| ≥65 | 32 (5) |
| Sex |
|
| Male | 75 (12) |
|
Female | 25 (4) |
| ISS stage |
|
| I | 31 (5) |
| II | 25 (4) |
| III | 44 (7) |
| Bone destruction |
|
| Yes | 75 (12) |
| No | 25 (4) |
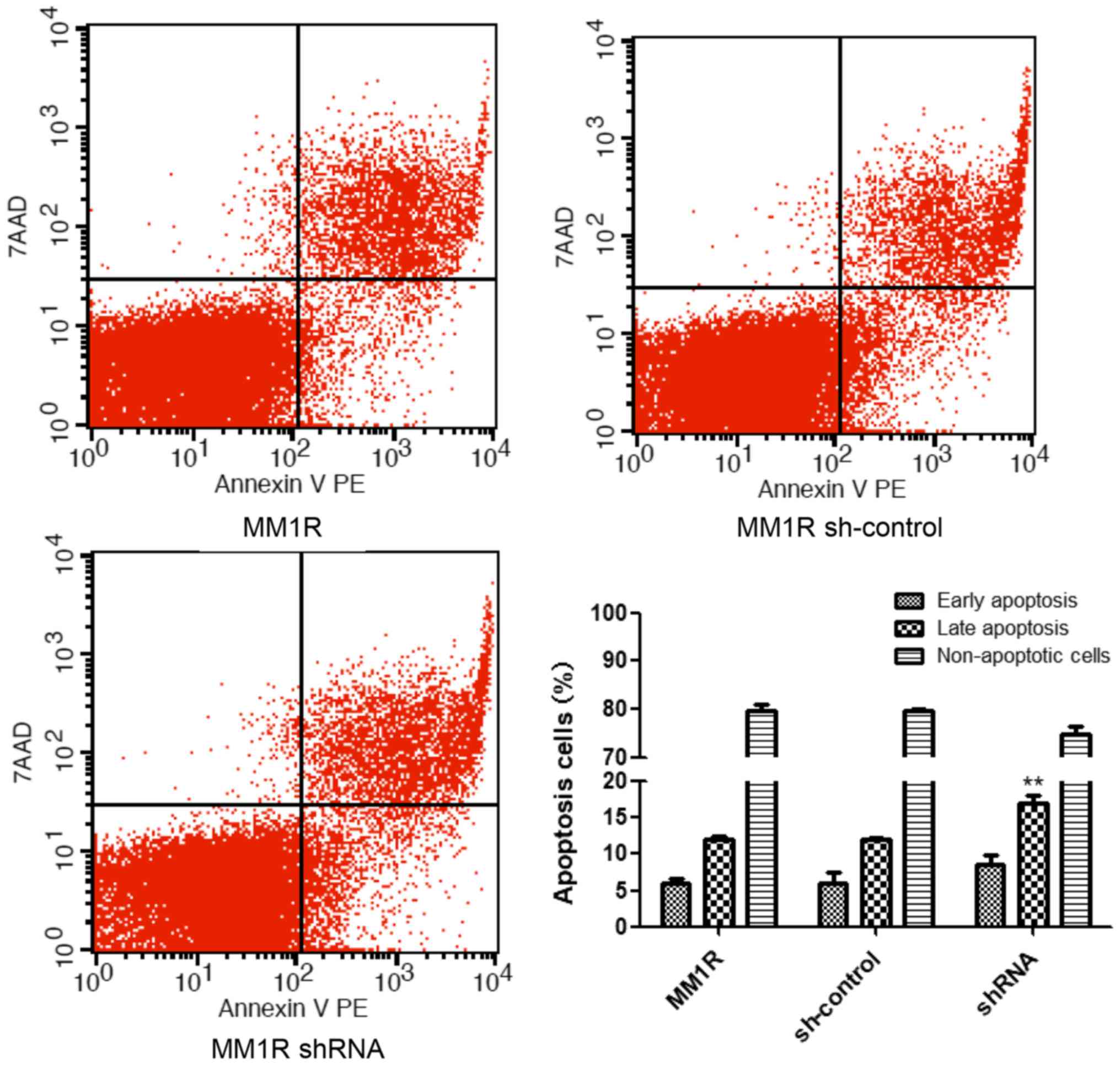
GRK6 regulates apoptosis in MM1R
cells
Decreasing GRK6 expression could significantly lead
to the apoptosis of MM1R cells. The MM1R cells stably expressing
shRNAs were analyzed by flow cytometry; analyses mainly focused on
early apoptotic cells (Annexin V PE+/7AAD-) and late apoptotic
cells (Annexin V PE+/7AAD+). A higher rate of late apoptosis was
observed in cells treated with the shRNA against GRK6. As
demonstrated, GRK6 inhibition is lethal to MM1R cells, especially
cells in late apoptosis (P<0.01; Fig.
3).
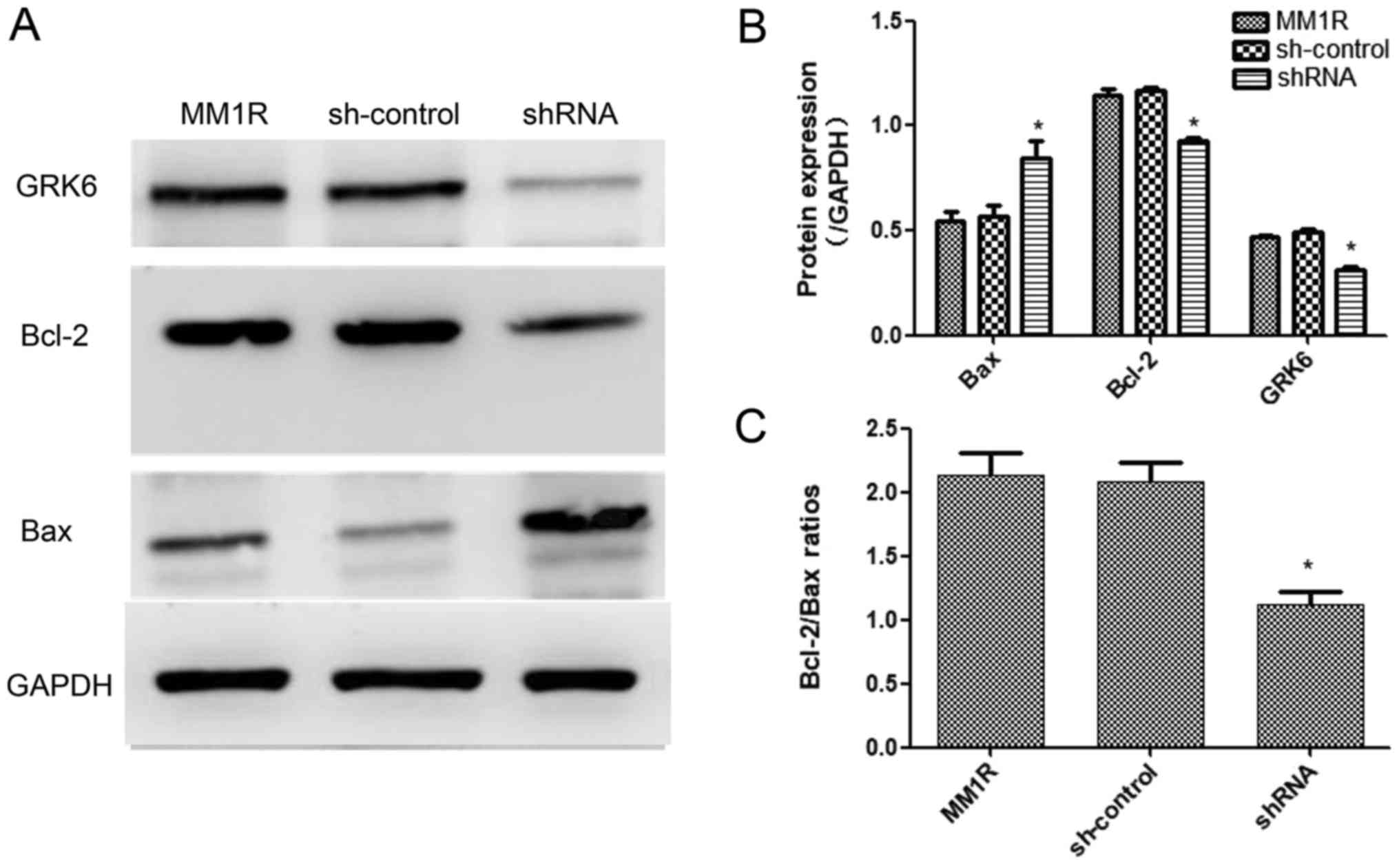
GRK6 altered the expression of
apoptosis-associated proteins
As shown in Fig. 4,
the expression of the pro-apoptotic protein, Bax, was increased and
an apoptosis-inhibiting protein, Bcl-2, was decreased (P<0.05;
Fig. 4A and B). Bcl-2/Bax ratios
indicated that down-regulating GRK6 influenced the apoptosis rate
of MM1R cells (P<0.05; Fig. 4C).
These results might explain how GRK6 affects the apoptosis
signaling pathway in MM1R cells.
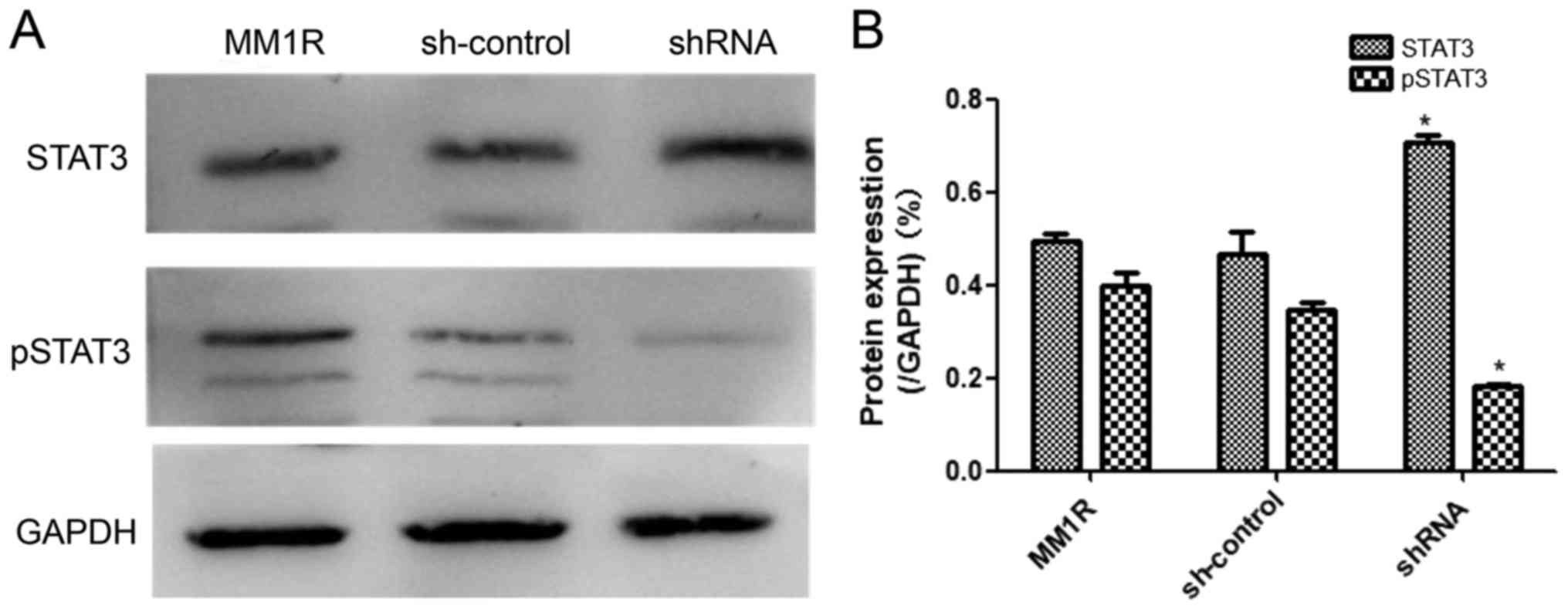
GRK6 phosphorylates STAT3 signaling
pathway
GRKs serve an important role in the apoptosis
signaling pathway in cells by phosphorylating, and thus regulating
the activity of, agonist-occupied GPCRs. Here, we attempted to
further elucidate these processes by inhibiting the expression of
GRK6. GRK6 inhibits apoptosis through a STAT3-dependent mechanism.
Phosphorylation of STAT3 plays a crucial role in mediating cell
apoptosis and proliferation. Therefore, the effect of GRK6
inhibition on the STAT3 signaling pathway was examined in MM1R
cells. As displayed in Fig. 5, the
inhibition of GRK6 in MM1R cells using the shRNA resulted in a
decrease in STAT3 phosphorylation (P<0.05). Quantified western
blotting results showed that pSTAT3 was significantly decreased,
but contradictorily, the concentration of STAT3 increased.
Discussion
GRK6 is specifically expressed in lymphoid tissues
and myeloma, and is absent from or weakly expressed in most primary
human somatic tissues; it is similarly expressed in mice (12). The SDF-1/CXCR4 axis (13), and the HSP90 and STAT3 signaling
pathway (14–16) play significant roles in MM. Recent
studies have shown that GRK6 is involved in many kinds of cell
signaling (17). Therefore, multiple
signaling pathways might regulate and promote the pathogenesis of
MM via GRK6.
Previous studies have shown that GRK6 is highly
expressed in colorectal adenocarcinoma, and that gastric carcinoma
(18), lung cancer (19) and breast carcinoma (10) might be caused by mutations in GRK6.
In our study, compared with healthy individuals, GRK6 was
differently expressed in newly diagnosed MM patients, which
supports the hypothesis that GRK6 might act as an endogenous
promoter of oncogenes. The high expression of GRK6 might inhibit
tumor cell apoptosis and promote cell invasion during MM
progression.
In a previous study, we found that GRK6 plays an
important role in cell proliferation and cell cycle arrest in MM1R
cells (data not shown). In this study, we verified that GRK6 is
extensively expressed in MM cell lines; among which MM1R cells have
the highest levels of GRK6 expression. Using the lentivirus
containing shRNAs against GRK6, we induced GRK6 knockdown in MM1R
cells. As assumed, down-regulating GRK6 induced apoptosis in a
large number of MM1R cells. By contrast, the non-silencing
lentivirus was nonlethal, demonstrating that inhibiting GRK6 might
be lethal to myeloma cells. GRK6 is involved in multiple networks
relevant in myeloma, such as EDNRB (20) and CXCR4 (21). STAT3, which is an oncogene (22,23),
mediates various cytokines, such as IL6. STAT3 has been shown to
play a role in activating myeloma cell lines (24), primary myeloma cells (25) and many other cancer cells (23). Inhibition of the STAT3 signaling
pathway in myeloma might lead to both cellular apoptosis and
sensitization to chemotherapeutics (23,26,27). In
this study, the expression level of phosphorylated STAT3 was
significantly decreased. It should be noted that the expression of
STAT3 increased after transfection with the shRNA. To elucidate
this mechanism, future studies should focus on detecting the level
of STAT3 in the cytoplasm and nucleus of cells after transfections
with GRK6 shRNA or inhibitors. Notably, a significant decrease in
the expression of CDK4 and CyclinD1, which could suppress cell
growth, was detected in MM1R cells in which GRK6 was inhibited
(data not shown). In addition, we detected the obvious
up-regulation of Bax and down-regulation of Bcl-2, which play key
roles in the progression of apoptosis in GRK6-deficient myeloma
cells. Our results confirmed that the STAT3 signaling pathway is
involved in the induction of apoptosis in GRK6-down-regulated MM
cells.
Overall, this study postulated that GRK6 might be
involved in inhibiting tumor cell apoptosis in MM1R cells. The
results indicate that GRK6 may be an important target in the
treatment of MM and that GRK6 inhibitors may ameliorate the
disease.
Acknowledgements
We would like to thank all patients for their
collaboration in this study. We appreciate the help given by staffs
at the Xuzhou Medical College/Laboratory of Transplantation and
Immunology for their support during the present study. We also
thank all the help given by department of Hematology, Beijing
Chao-Yang Hospital.
Funding
Not applicable.
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
ZZ and WC analyzed and interpreted the patient data,
performed the experiments related to the plasma cells, and were
major contributors in writing the manuscript. LZ designed the study
and was a major contributor in sample collection. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
Patients and healthy individuals provided informed
consent, and the study complied with the Declaration of Helsinki
and its amendments. The present study was approved by the Clinical
Research Ethics Committee of Xuzhou Medical University Affiliated
Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
MM
|
multiple myeloma
|
|
GRK6
|
G protein-coupled receptor kinase
6
|
|
GPCRs
|
G protein-coupled receptors
|
References
|
1
|
Kuehl WM and Bergsagel PL: Multiple
myeloma: Evolving genetic events and host interactions. Nat Rev
Cancer. 2:175–187. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rajkumar SV and Kumar S: Multiple myeloma:
Diagnosis and treatment. Mayo Clin Proc. 91:101–119. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chuang TT, Sallese M, Ambrosini G, Parruti
G and De Blasi A: High expression of beta-adrenergic receptor
kinase in human peripheral blood leukocytes. Isoproterenol and
platelet activating factor can induce kinase translocation. J Biol
Chem. 267:6886–6892. 1992.PubMed/NCBI
|
|
4
|
Loudon RP, Perussia B and Benovic JL:
Differentially regulated expression of the G-protein-coupled
receptor kinases, betaARK and GRK6, during myelomonocytic cell
development in vitro. Blood. 88:4547–4557. 1996.PubMed/NCBI
|
|
5
|
DeWire SM, Ahn S, Lefkowitz RJ and Shenoy
SK: Beta-arrestins and cell signaling. Annu Rev Physiol.
69:483–510. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gurevich EV and Gurevich VV: Arrestins:
Ubiquitous regulators of cellular signaling pathways. Genome Biol.
7:2362006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ek IJ and Otterstad HK: Health care for
the aged is better than its start! A status report from communities
in Vestfold in 1988–89. Fag Tidsskr Sykepleien. 78:26–28. 1990.(In
Norwegian). PubMed/NCBI
|
|
8
|
Kasiske BL: Relationship between vascular
disease and age-associated changes in the human kidney. Kidney Int.
31:1153–1159. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kumar SK, Rajkumar SV, Dispenzieri A, Lacy
MQ, Hayman SR, Buadi FK, Zeldenrust SR, Dingli D, Russell SJ, Lust
JA, et al: Improved survival in multiple myeloma and the impact of
novel therapies. Blood. 111:2516–2520. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Stephens P, Edkins S, Davies H, Greenman
C, Cox C, Hunter C, Bignell G, Teague J, Smith R, Stevens C, et al:
A screen of the complete protein kinase gene family identifies
diverse patterns of somatic mutations in human breast cancer. Nat
Genet. 37:590–592. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fong AM, Premont RT, Richardson RM, Yu YR,
Lefkowitz RJ and Patel DD: Defective lymphocyte chemotaxis in
beta-arrestin2- and GRK6-deficient mice. Proc Natl Acad Sci USA.
99:7478–7483. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chudziak D, Spohn G, Karpova D, Dauber K,
Wiercinska E, Miettinen JA, Papayannopoulou T and Bönig H:
Functional consequences of perturbed CXCL12 signal processing:
Analyses of immature hematopoiesis in GRK6-deficient mice. Stem
Cells Dev. 24:737–746. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nieto-Lluis C, Garcia-Pérez M and
Cabrera-Santos A: Influence of sympathetic withdrawal on early
postinfarction arrhythmias. Acta Physiol Hung. 70:351–356.
1987.PubMed/NCBI
|
|
15
|
Li YP: GRK6 expression in patients with
hepatocellular carcinoma. Asian Pac J Trop Med. 6:220–223. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Taipale M, Krykbaeva I, Koeva M, Kayatekin
C, Westover KD, Karras GI and Lindquist S: Quantitative analysis of
HSP90-client interactions reveals principles of substrate
recognition. Cell. 150:987–1001. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nakaya M, Tajima M, Kosako H, Nakaya T,
Hashimoto A, Watari K, Nishihara H, Ohba M, Komiya S, Tani N, et
al: GRK6 deficiency in mice causes autoimmune disease due to
impaired apoptotic cell clearance. Nat Commun. 4:15322013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Forbes S, Clements J, Dawson E, Bamford S,
Webb T, Dogan A, Flanagan A, Teague J, Wooster R, Futreal PA and
Stratton MR: COSMIC 2005. Br J Cancer. 94:318–322. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yao S, Zhong L, Liu J, Feng J, Bian T,
Zhang Q, Chen J, Lv X, Chen J and Liu Y: Prognostic value of
decreased GRK6 expression in lung adenocarcinoma. J Cancer Res Clin
Oncol. 142:2541–2549. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Freedman NJ, Ament AS, Oppermann M,
Stoffel RH, Exum ST and Lefkowitz RJ: Phosphorylation and
desensitization of human endothelin A and B receptors. Evidence for
G protein-coupled receptor kinase specificity. J Biol Chem.
272:17734–17743. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vroon A, Heijnen CJ, Raatgever R, Touw IP,
Ploemacher RE, Premont RT and Kavelaars A: GRK6 deficiency is
associated with enhanced CXCR4-mediated neutrophil chemotaxis in
vitro and impaired responsiveness to G-CSF in vivo. J Leukoc Biol.
75:698–704. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bromberg JF, Wrzeszczynska MH, Devgan G,
Zhao Y, Pestell RG, Albanese C and Darnell JE Jr: Stat3 as an
oncogene. Cell. 98:295–303. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Frank DA: STAT3 as a central mediator of
neoplastic cellular transformation. Cancer Lett. 251:199–210. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Brocke-Heidrich K, Kretzschmar AK, Pfeifer
G, Henze C, Löffler D, Koczan D, Thiesen HJ, Burger R, Gramatzki M
and Horn F: Interleukin-6-dependent gene expression profiles in
multiple myeloma INA-6 cells reveal a Bcl-2 family-independent
survival pathway closely associated with Stat3 activation. Blood.
103:242–251. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bharti AC, Shishodia S, Reuben JM, Weber
D, Alexanian R, Raj-Vadhan S, Estrov Z, Talpaz M and Aggarwal BB:
Nuclear factor-kappaB and STAT3 are constitutively active in CD138+
cells derived from multiple myeloma patients, and suppression of
these transcription factors leads to apoptosis. Blood.
103:3175–3184. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nelson EA, Walker SR, Kepich A, Gashin LB,
Hideshima T, Ikeda H, Chauhan D, Anderson KC and Frank DA:
Nifuroxazide inhibits survival of multiple myeloma cells by
directly inhibiting STAT3. Blood. 112:5095–5102. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pathak AK, Bhutani M, Nair AS, Ahn KS,
Chakraborty A, Kadara H, Guha S, Sethi G and Aggarwal BB: Ursolic
acid inhibits STAT3 activation pathway leading to suppression of
proliferation and chemosensitization of human multiple myeloma
cells. Mol Cancer Res. 5:943–955. 2007. View Article : Google Scholar : PubMed/NCBI
|