Introduction
With the growth of aging population in China,
osteoporosis (OP) has become one of the most common chronic
diseases (1). OP is a metabolic
orthopedic disease characterized by decreased bone mass, reduced
bone density, and degraded bone structure. OP is mostly present in
elderly and postmenopausal women, and the incidence is higher in
females than in males (2,3). In patients with OP, the balance of bone
metabolism in the body is broken and bone absorption is much
greater than bone formation. Patient experience bone pain,
kyphosis, shortened body, decreased respiratory function, and
fractures, which seriously harm the whole body (4). At present, calcium, physical activity
and bone resorption inhibitors are widely used in the treatment of
OP, but these measures can only alleviate the patient's symptoms,
and cannot prevent the occurrence of the disease (5,6). Studies
have shown that bone marrow mesenchymal stem cells (BMSCs), as the
main source of osteoblasts in humans, have multi-directional
differentiation potential and can differentiate into osteoblasts,
chondrocytes and adipocytes under certain conditions. Therefore,
BMSCs play a very important role in the treatment of OP (7,8).
MicroRNAs are a class of non-coding RNAs of ~22
nucleotides in length that are widely conserved across species.
MicroRNAs degrade mRNA and inhibit translation by specifically
binding to the 3′-untranslated region (3′-UTR) of messenger RNA of
the target gene (9,10). MicroRNAs are involved in the
transcriptional regulation of many cells or other basic
physiological activities such as cell survival, cell proliferation,
and apoptosis (11). MicroRNA-21 is
differentially expressed in cardiovascular diseases, cancer, renal
and digestive system diseases, and thus participates in their
pathophysiological process (12).
However, whether microRNA-21 is differentially expressed in
patients with OP and whether it can regulate the differentiation of
BMSCs into osteoblasts, thereby affecting the balance of bone
metabolism, is still unknown. Therefore, this study investigated
the expression of microRNA-21 in OP patients and its influence on
the osteogenic differentiation of BMSCs.
Materials and methods
Materials
Fetal bovine serum and carbon dioxide incubator was
obtained from Thermo Fisher Scientific, Inc. (Waltham, MA, USA),
and inverted optical microscope from Olympus Corp. (Tokyo, Japan).
SD rats were purchased from the Animal Experimental Center of the
Second Affiliated Hospital of Harbin Medical University (Harbin,
China), and microRNA-21 mimics, inhibitors and negative controls
(NCs) from Invitrogen (Invitrogen; Thermo Fisher Scientific).
DMEM/F12 culture medium was provided by Wisent Biotechnology
(Nanjing, China). Trypsin digestion solution was obtained from
Beyotime Institute of Biotechnology (Haimen, China). Complete
culture medium (F12 medium + 10% fetal bovine serum + 1%
cyanine-streptomycin), osteogenic medium (F12 medium + 10% fetal
bovine serum + 10 mM β-sodium glycerophosphate + 0.05 mM vitamin C
+ 10 mM dexamethasone + 1% cyanine-streptomycin), alkaline
phosphatase (ALP) staining solution (10 ml distilled water + 5 ml
3% β-sodium glycerophosphate + 5 ml 2% pentobarbital + 10 ml 2%
calcium chloride + 1 ml 2% magnesium sulfate, pH 9.4) were prepared
by the authors of this study. Alizarin red staining medium (Cyagen
Biosciences, Inc., Santa Clara, CA, USA), TRIzol (Sigma-Aldrich;
Merck KGaA, St. Louis, MO, USA), reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) primers (Invitrogen; Thermo
Fisher Scientific, Inc.), reverse transcription kit and reverse
transcription-polymerase chain reaction (both from Applied
Biosystems; Thermo Fisher Scientific, Inc., Foster City, CA, USA),
cyan-streptomyces (Beyotime Institute of Biotechnology), RT-qPCR
instrument and SYBR-Green (both from Roche Diagnostics GmbH,
Mannheim, Germany), pipette (Thermo Fisher Scientific, Inc.) and
RNase-free tips (Qiagen Sciences, Inc., Germantown, MD, USA) were
used in our experiments. All other experimental reagents were
purchased from Sigma-Aldrich (Sigma-Aldrich; Merck KGaA).
Collection of clinical samples and
microRNA detection
During the period from June 2016 to December 2017,
bone tissue was collected from patients with OP and normal subjects
at the Department of Orthopaedic Surgery of the Second Affiliated
Hospital of Qiqihar Medical University (Qiqihar, China). The study
was approved by the Ethics Committee of the Second Affiliated
Hospital of Qiqihar Medical University. Signed informed consents
were obtained from the patients or the guardians.
Patients with diabetes mellitus,
hyperprolactinemia, ovariectomy, liver cancer, and malabsorption
syndrome affect bone metabolic disease were excluded
Bone tissues of 48 OP patients were set as
experimental group, and bone tissues of 48 normal subjects were set
as control group. The expression of microRNA-17, microRNA-20a,
microRNA-21, microRNA-26a, microRNA-29b, and microRNA-106b was
detected. Briefly, bone tissues were ground in liquid nitrogen,
followed by addition of TRIzol to extract total RNA. Reverse
transcription was performed to synthesize DNA according to the
manufacturer's instructions. cDNA samples were stored at −20°C
before use. qPCR reaction system included 10 µl SYBR-Green, 7 µl
ddH2O, 1 µl upstream primers, 1 µl downstream primers,
and 1 µl cDNA. qPCR reaction conditions were: 95°C for 30 sec,
followed by 40 cycles of 95°C for 5 sec, 60°C for 30 sec and 72°C
for 30 sec. The experiment was performed in triplicate, and the
expression of each microRNA was normalized to U6 endogenous control
using 2−ΔΔCq method (Table
I) (13).
 | Table I.qPCR primer sequences for microRNA
detection. |
Table I.
qPCR primer sequences for microRNA
detection.
| Gene | Forward | Reverse |
|---|
| microRNA-21 |
CGCCGTAGCTTATCAGACTG |
CAGCCACAAAAGAGCACAAT |
| U6 |
GCTTCGGCAGCACATATACTAAAAT |
CGCTTCACGAATTTGCGTGTCAT |
Primary culture and subculture of rat
BMSCs
A total of 24 4-week-old SD female rats (70–90 g)
were maintained under specific pathogen-free conditions (22°C, 12-h
light/12-h dark cycles, and 50–55% humidity) and were used to
isolate tibias and femurs under aseptic conditions. Bone tissues
were cleaned and sterilized with 75% alcohol, and the epiphyses
were cut off. A syringe (1 ml) was used to inject complete culture
fluid into the cavity to flush bone marrow, and this procedure was
performed 3 times until the bone became transparent. The flushed
bone marrow was mixed with 4 ml of culture medium, inoculated into
a 25-cm2 perforated cell culture flask and cultured in
an incubator (37°C, 5% CO2). Medium was replaced every
48 h. When cell confluence reached 80%, digestion with trypsin and
subculture were performed. Subculture was performed 3 times before
subsequent experiments.
Cell transfection
Cell transfection was performed when confluence of
rat BMSCs reached 50–60%. Cells were starved for 2 h in serum-free
Opti media. Lipofectamine 2000 was used to transfect 50 nM
microRNA-21 mimics/NC or 100 nM microRNA-21 inhibitor/NC into
cells. Cells were observed 6 h after transfection and the medium
was replaced.
Osteogenic differentiation of rat
BMSCs
Subculture was performed 3 times and BMSCs were
collected. Osteogenic induction medium was added 24 h after
transfection with microRNA-21 mimic, inhibitor and NC. Induction
was performed for 10 days, medium was replaced with fresh
osteogenic induction medium every 3 days, and each experiment was
repeated 3 times.
Identification of osteogenic
differentiation of BMSCs and comparison between groups
After transfection and osteogenic induction, cells
were washed 3 times with PBS, followed by fixation in 4%
paraformaldehyde at room temperature for 20 min. After that,
incubation with ALP staining solution for 4 h and alizarin red
staining solution for 30 min was performed. Cells were washed with
PBS and observed under an inverted light microscope.
Differentiation ability was determined according to the area and
color of mineralized nodule.
Detection of the expression of
osteogenesis-related genes
After transfection and osteogenic induction, RT-qPCR
was used to detect the expression of intracellular osteogenic genes
including ALP, osteocalcin (OCN), bone morphogenetic protein 2
(BMP2) and runt-related transcription factor 2 (Runx2) using
methods given in ‘Collection of clinical samples and microRNA
detection’ (Table II).
 | Table II.qPCR primer sequences for the
detection of the expression of osteogenesis-related genes. |
Table II.
qPCR primer sequences for the
detection of the expression of osteogenesis-related genes.
| Gene | Forward | Reverse |
|---|
| ALP |
ACAACCTGACTGACCCTTCG |
TCATGATGTCCGTGGTCAAT |
| OCN |
GGACCATCTTTCTGCTCACTC |
CTGCTTGGACATGAAGGCTT |
| BMP2 |
CCTTGCTGACCACCTGAACT |
AACATGGAGATTGCGCTGA |
| Runx2 |
AGAAGGCACAGACAGAAGCTT-GA |
AGGAATGCGCCCTAAATCACT |
| GAPDH |
CATCACTGCCACCCAGAAGAC |
CCAGTGAGCTTCCCGTTCAG |
Statistical analysis
All data were expressed as mean ± SEM and were
statistically processed using GraphPad software (GraphPad Software,
Inc., La Jolla, CA, USA). t-test was used for comparison between
two groups. P<0.05, p<0.01 and p<0.001 were considered to
be statistically significant, as indicated in the figures.
Results
Detection of microRNA expression in
bone tissues and serum samples
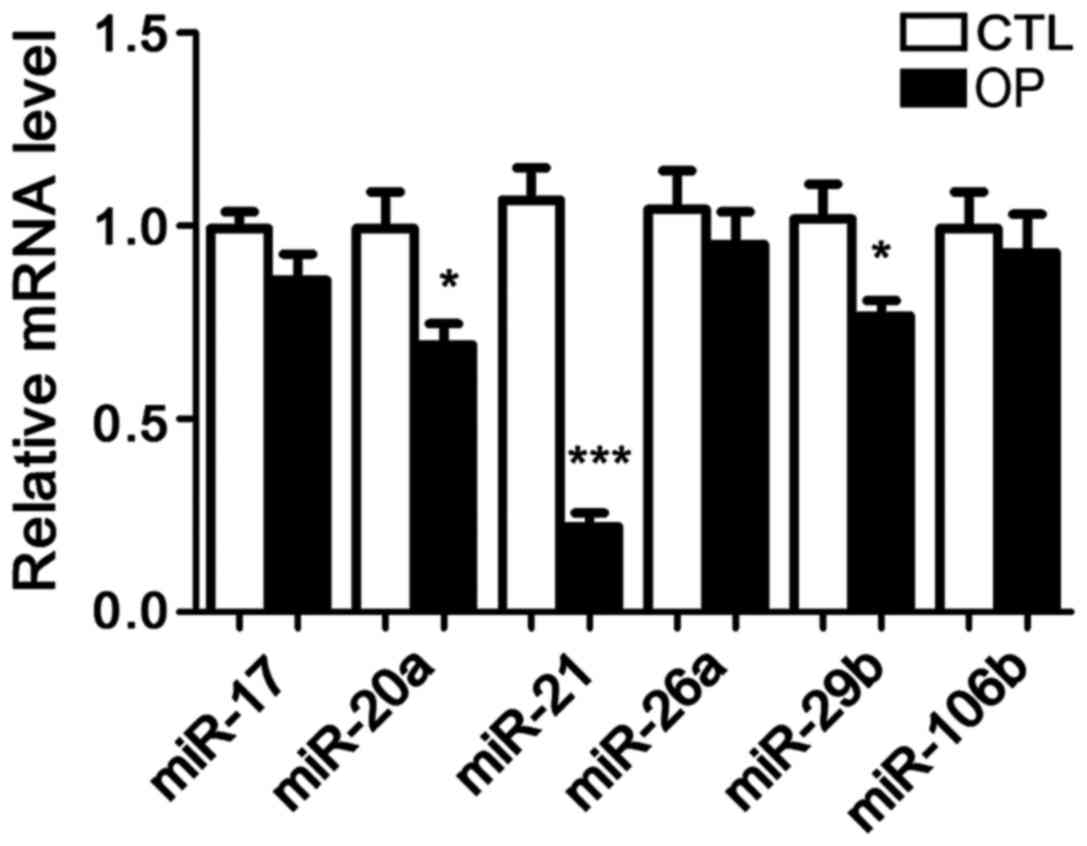
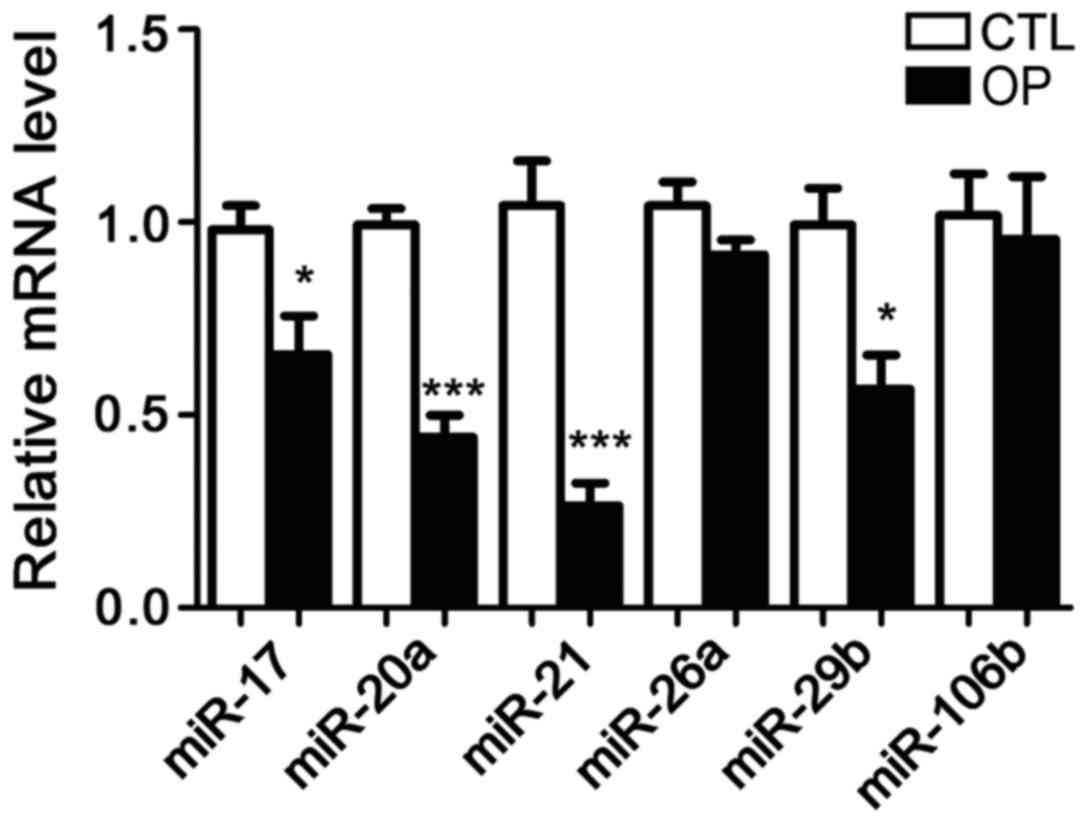
Following extraction of total RNA from bone tissue
of patients and healthy controls, the expression of six microRNAs
including microRNA-17, microRNA-20a, microRNA-21, microRNA-26a,
microRNA-29b, and microRNA-106b was detected. It has been reported
that these six microRNAs are related to OP in mice, but no studies
have shown whether they are differentially expressed in patients
with OP. The results showed that compared with control group, the
expression of microRNA-21 in patients with OP was significantly
reduced. In addition, the serum levels of microRNA-21 were also
significantly lower in patients than in controls. Therefore,
microRNA-21 was selected as the object of our study (Figs. 1 and 2).
Detection of microRNA-21 expression in
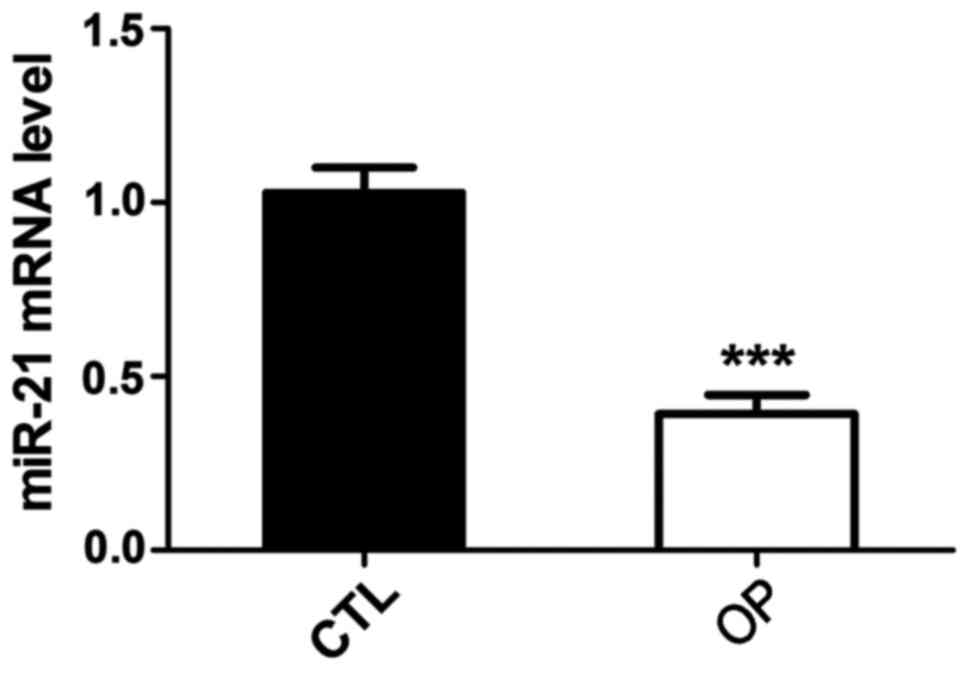
BMSCs of osteoporotic rats
BMSCs of normal and osteoporotic rats were isolated
and total RNA was extracted to detect the expression of
microRNA-21. Results showed that compared with the control group,
the expression level of microRNA-21 in BMSCs of osteoporotic rats
was significantly reduced (Fig.
3).
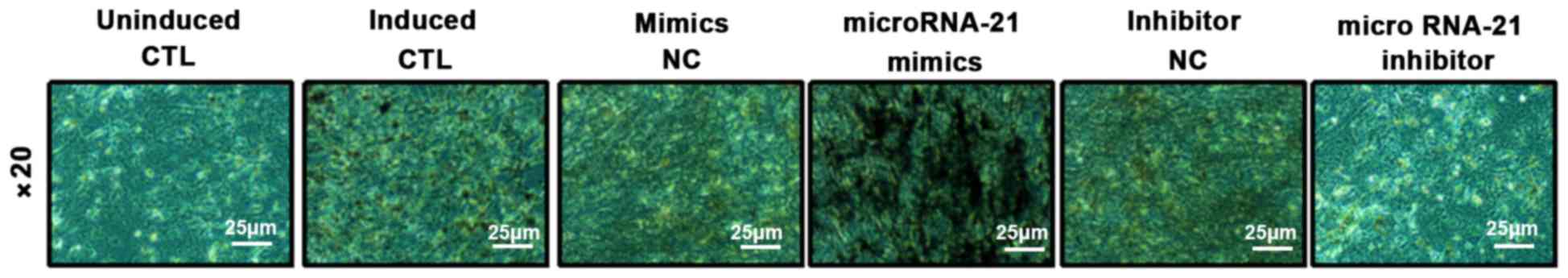
Identification and comparison of
osteogenic differentiation
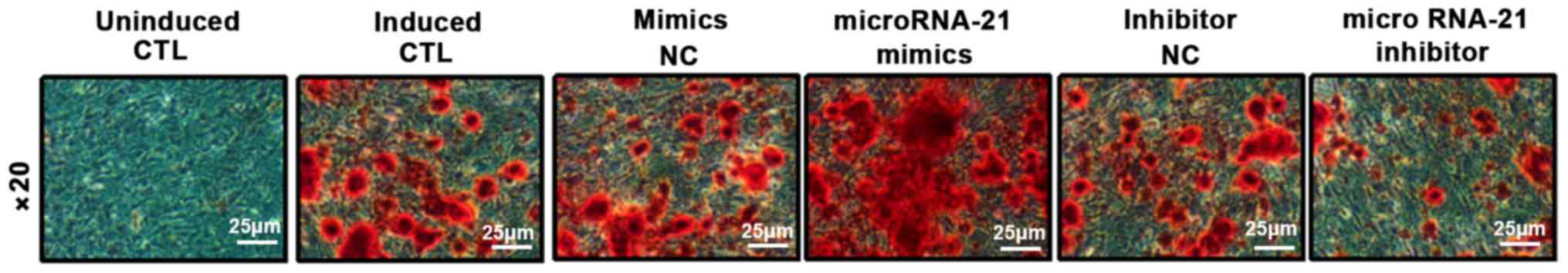
BMSCs were transfected with microRNA-21 mimics,
inhibitors and their corresponding NCs. ALP staining and alizarin
red staining were performed 10 days after osteogenic induction to
detect the area and number of nodules in the mineralized area, and
calcium deposition was assessed. ALP results showed that compared
with the control group, mineralized nodule staining in the
microRNA-21 mimics group was significantly deepened, and the area
of mineralized nodules was increased. However, the mineralized
nodules in microRNA-21 inhibitor group became lighter and the area
of mineralized nodules decreased. Alizarin red S (ARS) results
showed that compared with control group, orange-red color was
significantly deepened and calcium deposition was significantly
increased in microRNA-21 mimics group. However, ARS staining in
microRNA-21 inhibitor group was lighter and calcium deposition was
significantly reduced. These results indicate that microRNA-21 has
the ability to promote the differentiation of BMSCs into
osteoblasts (Figs. 4 and 5).
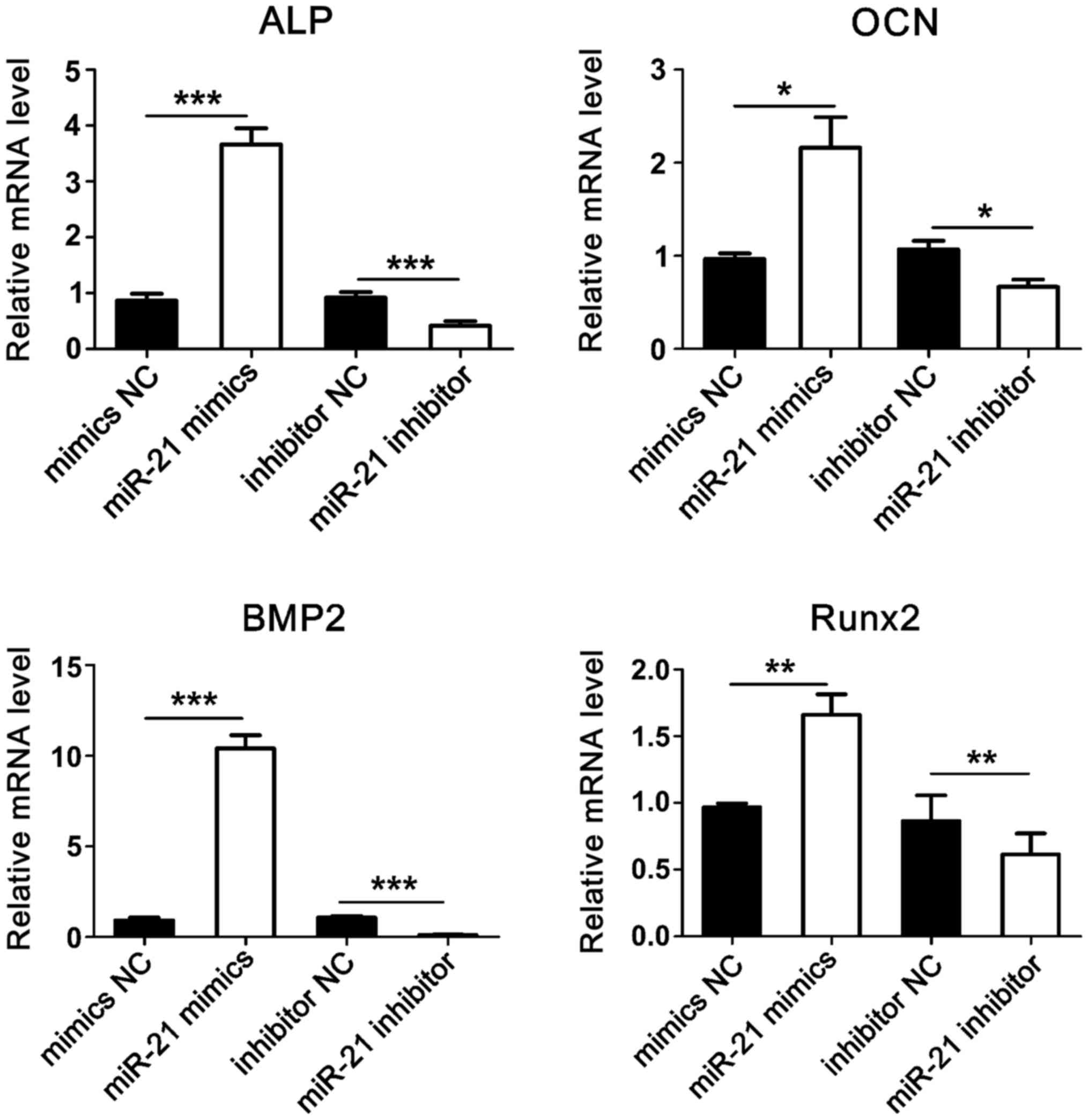
Expression of osteogenesis-related
genes
After transfection and osteogenic induction, total
RNA was extracted and RT-qPCR was used to detect the expression of
ALP, OCN, BMP2, and Runx2 genes. Results showed that compared with
the control group, the expression levels of osteogenesis-related
genes were significantly increased in the microRNA-21 mimics group.
Conversely, the expression levels of osteogenesis-related genes
were significantly reduced in microRNA-21 inhibitor group (Fig. 6).
Discussion
OP is a systemic metabolic disease characterized by
reduced bone mass, reduced bone density, and reduced bone
microstructure (14,15). Patients with OP have reduced
trabecular number, increased brittleness of bones, and markedly
decreased bone strength, resulting in increased fracture risk,
which brings pain and financial burden to patients and their
families (16). At present,
incidence of OP is increasing year by year, and it has become one
of the most common diseases in the world. Therefore, studies on OP
have attracted increasing attention (17).
BMSCs are a type of adult stem cells that exist in
the bone marrow and have self-renewal ability and multi-directional
differentiation potential. Under certain induction conditions, they
can differentiate into many kinds of cells, such as osteoblasts,
adipocytes, chondrocytes, cardiomyocytes and nerve cells (18). The osteogenic potential of BMSCs
plays an important role in the treatment of OP, bone fractures,
bone necrosis, osteoarthritis and bone defects (19,20).
Therefore, to study the differentiation of BMSCs is of great
significance for the treatment of OP.
An increasing number of studies have shown that
microRNA can be used as a key regulator of the osteogenesis of
BMSCs. For example, it has been reported that microRNA-320a can
inhibit the differentiation of human BMSCs into osteoblasts
(21). In another study,
microRNA-196a was reported to promote osteogenic differentiation of
human adipocytes and inhibited their proliferation (22). Recently, it has been reported that
microRNA-21 can promote osteogenic differentiation of human
umbilical cord stem cells (23).
Based on these studies, we believe that microRNAs also play a role
in the osteogenic differentiation of BMSCs. However, no studies
have reported the changes in thr expression of microRNA-21 in bone
tissue and serum of OP patients and the effects on osteogenic
differentiation of rat BMSCs.
Our results suggest that the expression of
microRNA-21 in bone tissue and serum is lower in osteoporotic
patients than in healthy controls. In addition, we extracted and
isolated, purified BMSCs from SD rats, and transfected microRNA-21
mimics, inhibitors and their corresponding NCs into the cells, and
induced osteogenic differentiation of BMSCs by osteogenic induction
for 10 days. After transfection of microRNA-21 mimics, ARS and ALP
staining showed a significant increase in the number of mineralized
nodules, stained calcified area, depth of color, and the expression
levels of ALP, Runx2, OCN, and BMP2 genes related to osteogenic
differentiation, indicating that microRNA-21 has the potential to
promote the differentiation of BMSCs into osteoblasts. After
transfection with microRNA-21 inhibitor, osteogenic differentiation
ability of BMSCs was decreased, and the ARS and ALP staining showed
that the number of mineralized nodules, the area of calcified area
stained, and the degree of color depth were significantly reduced.
Besides that, the expression levels of ALP, Runx2, OCN, and BMP2
genes related to osteogenic differentiation also decreased,
indicating that microRNA-21 inhibitor can inhibit the
differentiation of BMSCs into osteoblasts. These results indicate
that microRNA-21 has the potential to promote the differentiation
of BMSCs into osteoblasts.
In summary, microRNA-21 is expressed at low level in
patients with OP, and microRNA-21 can promote the differentiation
of BMSCs into osteogenic cells. MicroRNA-21 may serve as a
potential therapeutic target for OP.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZZ, XL, DZ, YL, ST and ZD drafted the manuscript. ZZ
was responsible for the collection of the clinical samples and
microRNA detection. XL, DZ and YL were mainly responsible for the
cell culture and transfection. XL, DZ, ST and ZD assisted with
osteogenic differentiation of rat BMSCs. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
the Second Affiliated Hospital of Qiqihar Medical University
(Qiqihar, China). Signed informed consents were obtained from the
patients or the guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Anagnostis P, Vakalopoulou S, Christoulas
D, Paschou SA, Papatheodorou A, Garipidou V, Kokkoris P and Terpos
E: The role of sclerostin/dickkopf-1 and receptor activator of
nuclear factor κB ligand/osteoprotegerin signalling pathways in the
development of osteoporosis in patients with haemophilia A and B: A
cross-sectional study. Haemophilia. 24:316–322. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Armstrong DJ: Comment on: Patients'
preferences for anti-osteoporosis drug treatment: A cross-European
discrete choice experiment. Rheumatology (Oxford). 57:583–584.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Paccou J and Cortet B: Bisphosphonate drug
holidays in postmenopausal osteoporosis: Effect on clinical
fracture risk. Response to comments by Bredemeier. Osteoporos Int.
29:5212018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hiligsmann M, Dellaert BG, Watson V and
Boonen A: Comment on: Patients' preferences for anti-osteoporosis
drug treatment: A cross-European discrete choice experiment: Reply.
Rheumatology (Oxford). 57:584–585. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nogués X, Nolla JM, Casado E, Jódar E,
Muñoz-Torres M, Quesada-Gómez JM, Canals L, Balcells M and Lizán L:
Spanish consensus on treat to target for osteoporosis. Osteoporos
Int. 29:489–499. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cano A, Chedraui P, Goulis DG, Lopes P,
Mishra G, Mueck A, Senturk LM, Simoncini T, Stevenson JC, Stute P,
et al: Calcium in the prevention of postmenopausal osteoporosis:
EMAS clinical guide. Maturitas. 107:7–12. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li NB, Sun SJ, Bai HY, Xiao GY, Xu WH,
Zhao JH, Chen X, Lu YP and Zhang YL: Preparation of
well-distributed titania nanopillar arrays on Ti6Al4V surface by
induction heating for enhancing osteogenic differentiation of stem
cells. Nanotechnology. 29:0451012018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tian Y, Cui LH, Xiang SY, Xu WX, Chen DC,
Fu R, Zhou CL, Liu XQ, Wang YF and Wang XT: Osteoblast-oriented
differentiation of BMSCs by co-culturing with composite scaffolds
constructed using silicon-substituted calcium phosphate, autogenous
fine particulate bone powder and alginate in vitro.
Oncotarget. 8:88308–88319. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
McAlinden A and Im GI: MicroRNAs in
orthopaedic research: Disease associations, potential therapeutic
applications, and perspectives. J Orthop Res. 36:33–51.
2018.PubMed/NCBI
|
|
10
|
Raghuwanshi S, Gutti U, Kandi R and Gutti
RK: MicroRNA-9 promotes cell proliferation by regulating RUNX1
expression in human megakaryocyte development. Cell Prolif.
51:512018. View Article : Google Scholar
|
|
11
|
Wu DM, Zhang YT, Lu J and Zheng YL:
Effects of microRNA-129 and its target gene c-Fos on proliferation
and apoptosis of hippocampal neurons in rats with epilepsy via the
MAPK signaling pathway. J Cell Physiol. 233:6632–6643. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tokar T, Pastrello C, Rossos AEM, Abovsky
M, Hauschild AC, Tsay M, Lu R and Jurisica I: mirDIP
4.1-integrative database of human microRNA target predictions.
Nucleic Acids Res. 46(D1): D360–D370. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Qi M, Zhang L, Ma Y, Shuai Y, Li L, Luo K,
Liu W and Jin Y: Autophagy maintains the function of bone marrow
mesenchymal stem cells to prevent estrogen deficiency-induced
osteoporosis. Theranostics. 7:4498–4516. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hu Y, Tan LJ, Chen XD, Liu Z, Min SS, Zeng
Q, Shen H and Deng HW: Identification of novel potentially
pleiotropic variants associated with osteoporosis and obesity using
the cFDR method. J Clin Endocrinol Metab. 103:125–138. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Collison J: Osteoporosis: Teriparatide
preferable for fracture prevention. Nat Rev Rheumatol. 14:42018.
View Article : Google Scholar
|
|
17
|
Fukushima H, Shimizu K, Watahiki A,
Hoshikawa S, Kosho T, Oba D, Sakano S, Arakaki M, Yamada A,
Nagashima K, et al: NOTCH2 Hajdu-Cheney mutations escape
SCFFBW7-dependent proteolysis to promote osteoporosis. Mol Cell.
68(645–658): e52017.
|
|
18
|
Xu TB, Li L, Luo XD and Lin H: BMSCs
protect against liver injury via suppressing hepatocyte apoptosis
and activating TGF-β1/Bax singling pathway. Biomed Pharmacother.
96:1395–1402. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen L, Lu FB, Chen DZ, Wu JL, Hu ED, Xu
LM, Zheng MH, Li H, Huang Y, Jin XY, et al: BMSCs-derived
miR-223-containing exosomes contribute to liver protection in
experimental autoimmune hepatitis. Mol Immunol. 93:38–46. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Y, Luo S, Zhang D, Qu X and Tan Y:
Sika pilose antler type I collagen promotes BMSC differentiation
via the ERK1/2 and p38-MAPK signal pathways. Pharm Biol.
55:2196–2204. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang J, Meng Y, Liu Y, Chen Y, Yang H,
Chen D, Shi J and Guo Y: MicroRNA-320a regulates the osteogenic
differentiation of human bone marrow-derived mesenchymal stem cells
by targeting HOXA10. Cell Physiol Biochem. 38:40–48. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim YJ, Bae SW, Yu SS, Bae YC and Jung JS:
miR-196a regulates proliferation and osteogenic differentiation in
mesenchymal stem cells derived from human adipose tissue. J Bone
Miner Res. 24:816–825. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Meng YB, Li X, Li ZY, Zhao J, Yuan XB, Ren
Y, Cui ZD, Liu YD and Yang XJ: microRNA-21 promotes osteogenic
differentiation of mesenchymal stem cells by the PI3K/β-catenin
pathway. J Orthop Res. 33:957–964. 2015. View Article : Google Scholar : PubMed/NCBI
|