Introduction
p-Hydroxycinnamic acid is a type of aromatic
acid found in medicinal plants, exhibiting anticancer, antioxidant,
antivirus, immunomodulatory, antiallergic, and other forms of
pharmacological activity (1). Owing
to these numerous biological activities, p-hydroxycinnamic
acid is a useful medicine and is widely used in the chemical
industry and in organic synthesis to develop a variety of aromatic
amides with important activities (2). Substantial evidence exists to show that
p-hydroxycinnamic derivatives are promising and leading
compounds for the development of chemopreventive/anti-inflammatory
agents (3,4). p-hydroxycinnamic amides more
effectively inhibit the growth of cancer cells than
p-hydroxycinnamic acid (5).
Furthermore, the p-hydroxycinnamic amides possess higher
antioxidant activity against lipoperoxidation, thus increasing
their capability to reduce carcinogenesis (6). One category of p-hydroxycinnamic
derivatives takes advantage of sulfonamides as an amine for drug
synthesis (7). Sulfonamides are a
family of synthetic drugs used for the treatment of infections, and
the amide moiety may be involved by changing the metabolic activity
of the invading pathogenic microorganisms.
Previous studies have focused only on the
bioactivities of p-hydroxycinnamic and its derivatives, with
less attention focused on its ability to target its effects in
human tissues. It is well established that the interaction between
biomacromolecules and drugs can determine their distribution,
effective concentration and metabolism in plasma, in addition to
drug stability and safety during chemotherapeutic processes
(8). In pharmacology, interactions
between drugs and proteins can affect the bioactivities and
toxicity of a drug (9). Therefore,
the interaction of p-hydroxycinnamic derivatives with
protein require consideration, as such an interaction is associated
with drug transport, biological activity and clearance (10).
Serum albumin (SA) is the most abundant protein
within the blood, and it considered to have a leading role in drug
combination, processing and delivery (11). A previous study have investigated the
interactions of various active molecules with SA or other serum
components, which serve as carriers for these molecules in the
blood (12). Following entry to the
body, the drugs depend on SA to reach the lesion site, and then
fulfil their therapeutic effect. The binding and dissociating
constant of drugs and SA determines their in vivo
activities. Human SA (HSA) is usually selected as a target protein
molecule due to its pharmacological significance, stability,
binding and transport properties.
In the present study, a novel
p-hydroxycinnamic acid derivative,
(E)-3-(4-hydroxyphenyl)-N-(4-(N-(5-meth
oxypyrimidin-2-yl)-sulfamoyl)phenyl)acrylamide (HMSP) was
synthesized and characterized, using sulfonamide drugs as an amine
compound. Toxic experiments showed that HMSP inhibited cancer cell
proliferation at micromolar concentrations. The thermodynamics of
the interaction between HMSP and HSA were examined using
fluorescence, UV-Vis absorbance spectroscopy, and molecular
modeling methods. The binding site of HMSP was designated to site
I, namely, the warfarin site on the HSA molecule, and the van der
Waals interaction was found to be vital for the binding of HMSP to
HSA. However, several hydrogen bonds existed. Synchronous
fluorescence and circular dichroism (CD) spectra indicated that the
binding of HMSP onto HSA altered the conformation of HSA, and this
was further supported by molecular dynamic (MD) simulation results.
The present study provided accurate and complete data to clarify
the binding mechanisms of HMSP with HSA, which may prove valuable
for understanding the effects of p-hydroxycinnamic acid
derivative on serum function during the transportation process, and
provide insight into its toxicity in vivo.
Materials and methods
Materials
HSA was obtained from Shanghai Yuanju Biological
Technology Co., Ltd. (Shanghai, China) and stored under
refrigeration at 4.0°C. The p-hydroxycinnamic acid (chemical
reagent grade) and sulfamethoxydiazine (chemical reagent grade)
were purchased from Shenzhen Yuan Cheng Science and Technology Co.,
Ltd. (Shenzhen, China). A p-hydroxycinnamic acid derivative
was synthesized through a condensation reaction of
p-hydroxycinnamic acid with sulfamethoxydiazine. All other
reagents were commercial products of analytical grade, and
ddH2O was used for all solution preparations.
Apparatus and measurements
The X-ray diffraction data for the compound was
recorded on a Bruker SMART CCD diffractometer and solved using the
SHELXTL-97 program (13). The
measurement of fluorescence spectra was completed on a RF-5301PC
model spectrofluorometer (Shimadzu Corporation, Kyoto, Japan)
equipped with a water bath (T±0.1°C). The UV-Vis spectrum was
recorded on a U-2000 spectrophotometer (Pharmacia; GE Healthcare
Life Sciences, Uppsala, Sweden). The NMR spectra for compounds were
collected on a Bruker 300 MHz. The MS spectra were collected on a
Varian Saturn 2200 GCMS spectrometer. The IR spectra were analyzed
using a Fourier transform infrared spectrophotometer (Spectrum 100;
PerkinElmer, Inc., Waltham, MA, USA).
General procedure for the synthesis of
HMSP
The amide compounds of HMSP were prepared from the
condensation and deacetylation reaction of p-acetyl
cinnamoyl chloride and sulfonamide. The p-acetyl cinnamoyl
chloride was obtained as reported previously (8). At room temperature, condensation was
completed in tetrahydrofuran, and concentrated hydrochloric acid
(0.5 ml) was used to remove the acetyl group at 60°C, resulting in
the corresponding amide compound.
HMSP is a white powder, yield 70%, mp: 229–232°C,
MS: 467.2 m/z, calcd 468.11, 1H-NMR (300 MHz,
DMSO-d6, δppm): δ: 2.280 (s, 3H, -CO-CH3); 3.00
(s, 3H, -OCH3); 6.607–6.660 (d, 1H, -C=C-H,
J=15.9 Hz); 7.201–7.229 (d, 2H, Ar-H, J=8.4 Hz);
7.622–7.675 (d, 1H, -C=C-H, J=15.9 Hz); 7.675–7.703 (d, 2H,
Ar-H, J=8.4 Hz); 7.853–7.984 (m, 4H, Ar-H); 8.300 (s, 2H,
Py-H); 10.604 (s, 1H, -CO-NH); 11.431 (s, 1H, -SO2-NH).
Anal. calcd. for
C22H20N4O6S: C, 56.40;
H, 4.30; N, 11.96. Found: C, 56.50; H, 4.33; N, 11.89. IR data (KBr
pellets, cm−1): 3,361 (m), 3,104 (w), 3,029 (w), 1,681
(m), 1,590 (s), 1,511 (s), 1,287 (s) and 1,092 (m).
Cytotoxicity assay
Human cervical carcinoma cell lines (HeLa), gastric
cancer cell lines (SGC-7901), and pulmonary adenocarcinoma cell
lines (A549) were maintained in DMEM supplemented (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) with 10% heat-inactive
fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) at 37°C.
The cells were seeded into 96-well culture plates at 104
cells per well and adhered overnight. Different concentrations (0,
10, 20, 40, 60, 80, 100, 150 and 200 µM) of HMSP were added to the
cells. The cells were further incubated for 48 h, following which
the media was discarded, and
3-(4,5-dimethylthiazol)-2,5-diphenyltetrazolium bromide (MTT)
solution (0.5 mg/ml) was added to each well. After 4 h, the
supernatant was discarded and 150 µl DMSO was added into each well.
Following incubation at 40°C and shaking for 10 min, the absorbance
of the plates was recorded at a wavelength of 570 nm.
Spectroscopic experiments
The fluorescence emission spectra of HSA in each
case was recorded with a 1-cm quartz cell using 15/4 nm slit
widths. The emission spectra of HSA with or without HMSP were
recorded at 310–450 nm. A trace syringe was used to titrate HSA
solutions (1 µM) with HMSP. The final concentration of HMSP was in
the range of 0–2 µM.
Synchronous fluorescence spectra of the HSA-HMSP
systems were monitored under the same conditions as the emission
spectra. The spectra of the HSA-HMSP systems were scanned in the
wavelength ranges of 260–340 nm and 260–370 nm, and the differences
between the excitation and emission wavelengths were 15 and 60 nm,
respectively.
The CD spectra of the HSA-HMSP systems were measured
at room temperature under constant nitrogen flush (260-200 nm). The
concentration of HSA was 1 µM and the molar ratio of HSA to HMSP
was set to 1:0.6, 1:1, 1:2 and 1:5. The α-helix, β-sheet and
unordered percentages were calculated using SELCON3 software
(14).
Site marker competitive
experiments
The titration fluorescence spectra of the HSA-HMSP
system in the presence of the two typical site markers (warfarin
and ibuprofen) were monitored using the same methods as described
above. The concentrations of warfarin or ibuprofen were 1 µM.
Increasing quantities of HMSP were then added into the HSA-warfarin
or HSA-ibuprofen mixtures.
Molecular modeling
Molecular docking experiments were performed within
the Surflex-Dock program implicated in the ssybyl 8.1 software
package (15). The high-resolution
crystal structures of HSA were retrieved from the RSCB Protein Data
Bank (code: 1h9z) (16). The
molecular structure of HMSP was constructed and minimized using the
Tripos force field with Gasteiger-Marsili charges. The results of
the best-fit binding conformations were analyzed and mapped using
Visual Molecular Dynamics 1.9.1 software (17).
The MD simulation was performed in the present study
using Amber 14 software and Amber ff99SB force field (18). The MD simulations were started from
the best-fit docking conformation. A periodic cubic TIP3P water box
was used for the HSA-HMSP complex. The electroneutrality of the
solvated system was then maintained by adding sodium ions. Prior to
the MD simulations, the HSA-HMSP system was submitted to a two-step
energy minimization procedure. First, a 2,000-step minimization of
the solvent and ions only was performed, followed by a 2,000-step
minimization of the entire system. The HSA-HMSP complex was
gradually heated in an isothermal-isochoric (NVT) ensemble to 300 K
with a period of 200 ps. The systems were then submitted to a 200
ps MD simulation in an isothermal-isobaric (NPT) ensemble at 1 atm
and 300 K. The protein backbone was harmonically constrained with a
weak restraint (10 kcal/mol Å2) in the minimization,
heating, and equilibrating phases. Finally, 50 ns production MD
simulation was performed. A cutoff of 10 Å was set for short-range
interactions, whereas long-range electrostatic interactions beyond
the cutoff were handled with the particle mesh Ewald method.
Covalent bonds involving hydrogen atoms were constrained by the
SHAKE algorithm. The time step was set to 2 fs, and the trajectory
was recorded every 2 ps.
Results and Discussion
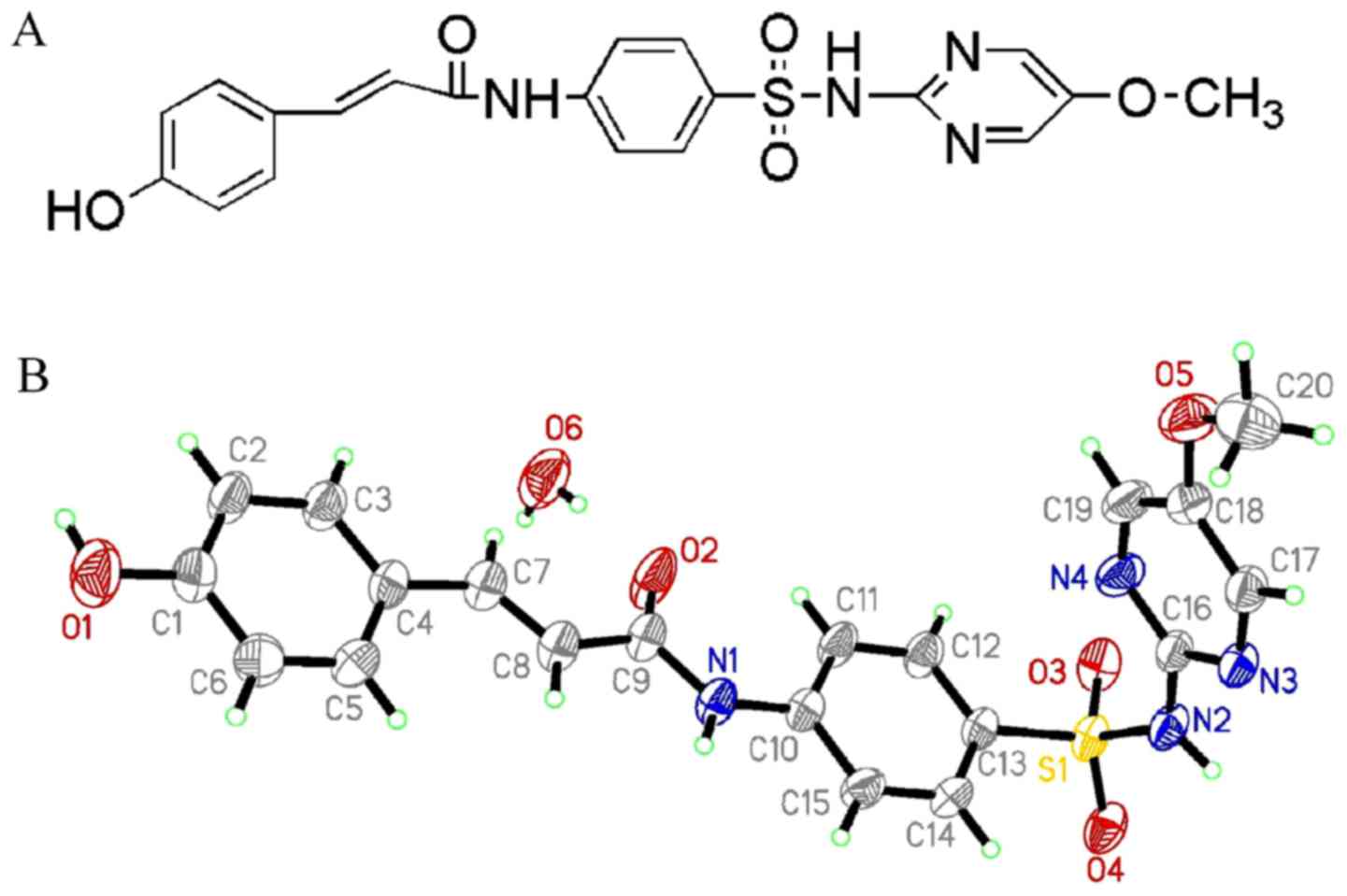
Characterization of HMSP
HMSP was characterized by X-ray diffraction
measurements. A perspective view of these two compounds in the
atomic labeling system is shown in Fig.
1. The crystal in the space group P2(1)/c had the
following unit cell parameters: a=8.188 (5) Å, b=13.656 (8) Å, c=18.821 (11) Å, β=96.781 (7)°, V=2090 (2) Å3, R=0.0468,
wR2=0.1425 and Z=4. The X-ray data have
been deposited at the Cambridge Crystallographic Data Centre (CCDC
no. 838365).
Cytotoxicity assay
The cytotoxicity of HMSP was investigated using a
typical MTT method. Three cancer cell lines, human cervical
carcinoma cell lines (HeLa), gastric cancer cell lines (SGC-7901),
and pulmonary adenocarcinoma cell lines (A549) were selected for
investigation. The cytotoxic results of HMSP towards three
different cancer cell lines are shown in Table I. It was observed that HMSP
efficiently inhibited the proliferation of each of the cancer cell
lines at micromolar concentration. In addition, as HMSP was
synthesized by modifying p-hydroxycinnamic acid, its
cellular toxicity was compared with its precursor. It was observed
that HMSP exhibited improved anticancer capability, compared with
p-hydroxycinnamic acid. These results demonstrated that HMSP
may be used as a potential anticancer agent.
 | Table I.IC50 values (µM) of HMSP
towards cancer cell lines. |
Table I.
IC50 values (µM) of HMSP
towards cancer cell lines.
| Cell | IC50
value |
|---|
| HeLa | 22.2 |
| SGC7901 | 17.5 |
| A549 | 37.8 |
Fluorescence quenching and quenching
mechanism
The binding of a drug on serum proteins brings about
increased drug solubility in blood, reduced toxicity, and
protection against oxidation of the bound drug (19). The present study investigated the
binding parameters of HMSP to HSA. Proteins possess intrinsic
fluorescence due to the existence of tryptophan, tyrosine and
phenylalanine. The involvement of tyrosine and tryptophan residues
in drug-HSA interactions can be evaluated by different excitation
wavelengths. At an excitation wavelength of 280 nm, the tryptophan
and tyrosine residues are excited, whereas a wavelength of 295 nm
excites tryptophan residues (20).
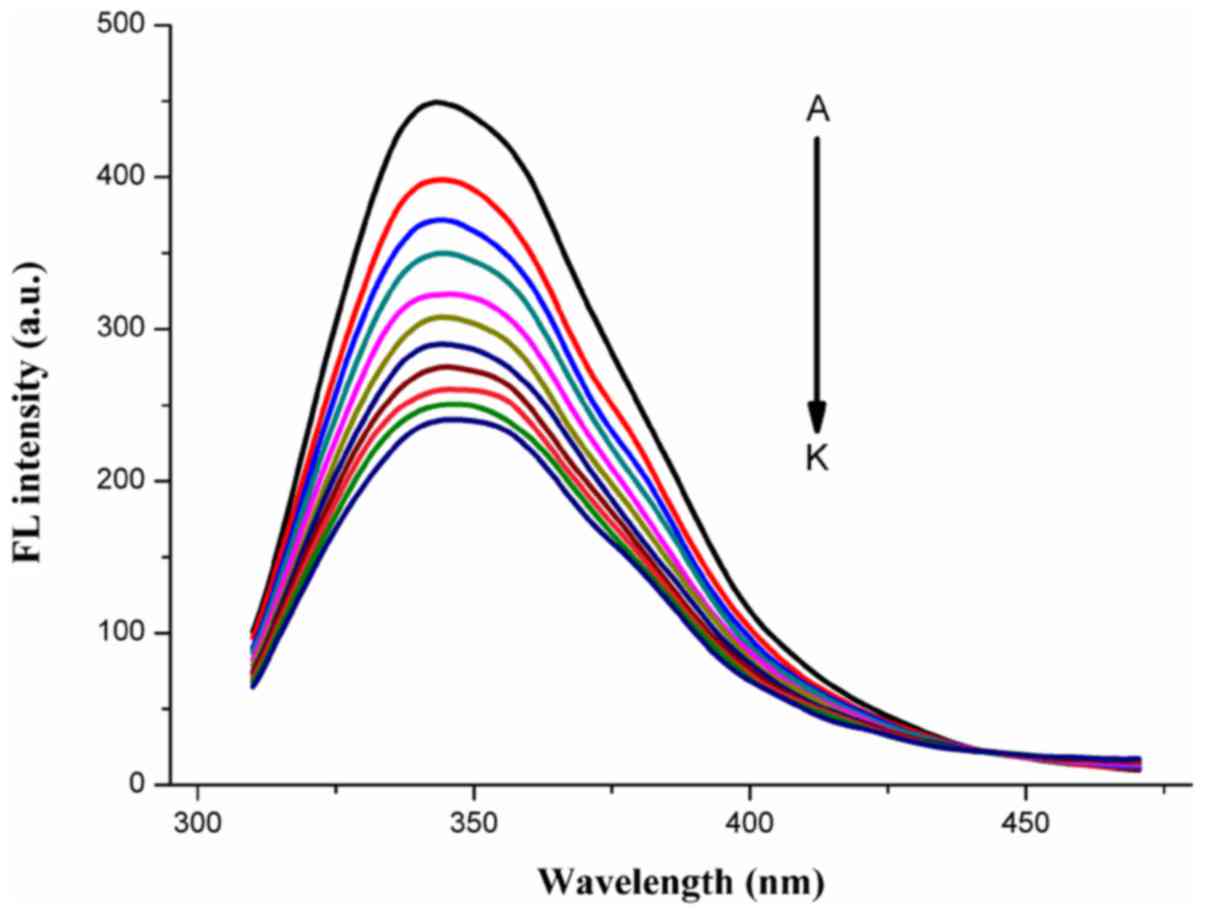
As shown in Fig. 2, HMSP quenched
HSA fluorescence excited at 295 nm. The intrinsic fluorescence of
HSA was gradually quenched compared with that of pure HSA solution,
which indicated that the compounds were able to interact with HSA
to form a compound-HSA complex.
It is known that there are two types of quenching
mechanisms of protein fluorescence, namely static quenching and
dynamic quenching (21). In static
quenching, the quencher binds to Phe, Tyr, or Trp residues of HSA,
and a nonfluorescent fluorophore-quencher complex is formed,
resulting in decreased fluorescence intensity of HSA. Dynamic
quenching represents the diffusion of the quencher towards the
fluorophore in the lifetime of the excited state and, in the
process of contact, the fluorophore returns to the ground state
without photon emission (22). The
quenching mechanism between compounds and HSA can be investigated
by the Stern-Volmer equation (23):
F0/F=1+kqτ0[Q]=1+KSV[Q](1)
where F0 and F are the
fluorescence intensities of HSA with or without HMSP;
KSV is the Stern-Volmer quenching constant;
kq represents the quenching rate constant of HSA;
τ0 is the average life-time of HSA without the
compound (τ0=10−8 s) (19); and [Q] is the concentration of
compound. In a dynamic quenching process, the quenching rate
constant of quenchers with the biopolymer is
<2.0×1010 l·mol−1·s−1. If the
Kq is substantially higher than this value, it
can be inferred that the quenching process is not caused by dynamic
quenching, but may be due to the static quenching as a result of
the complex formation.
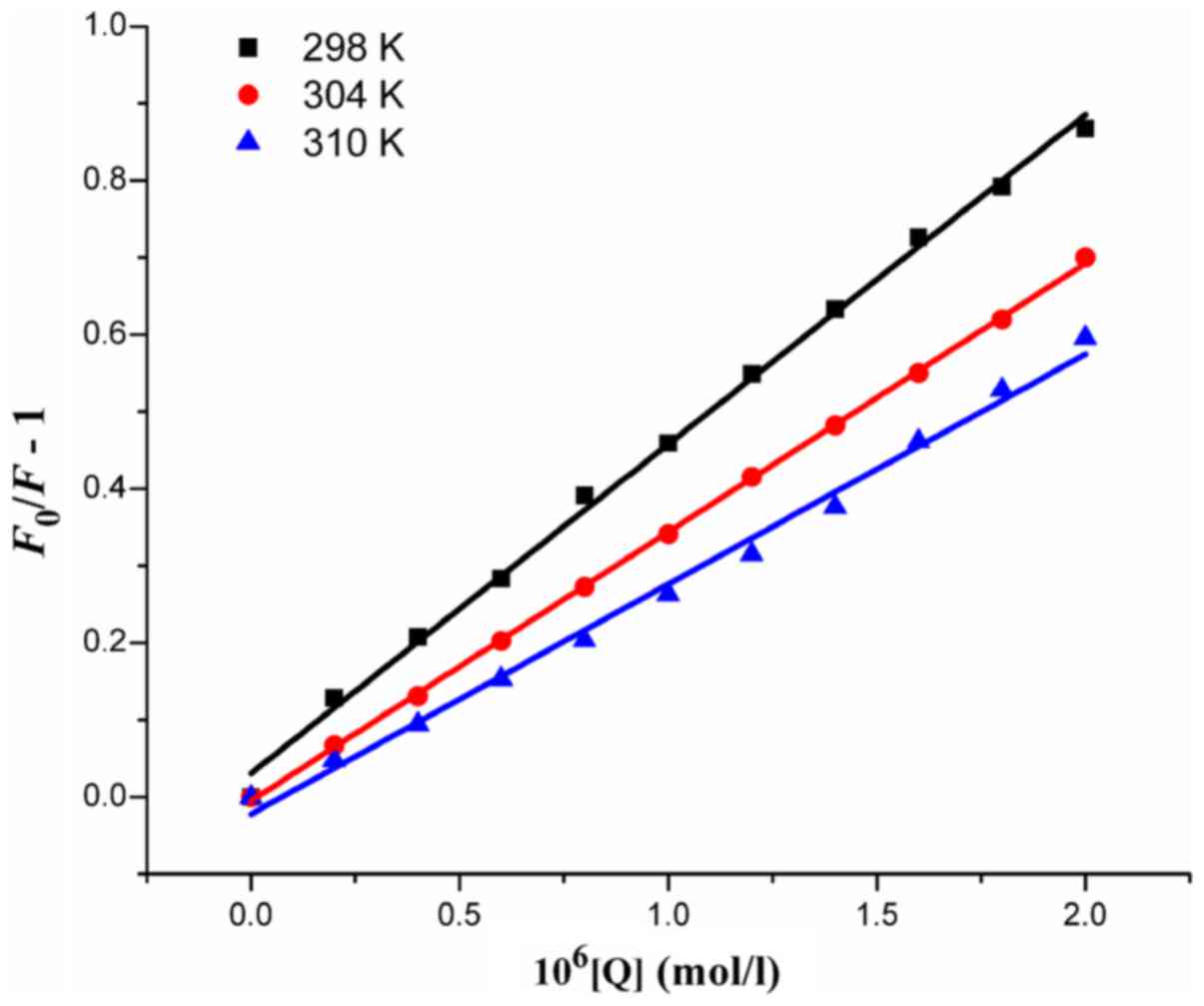
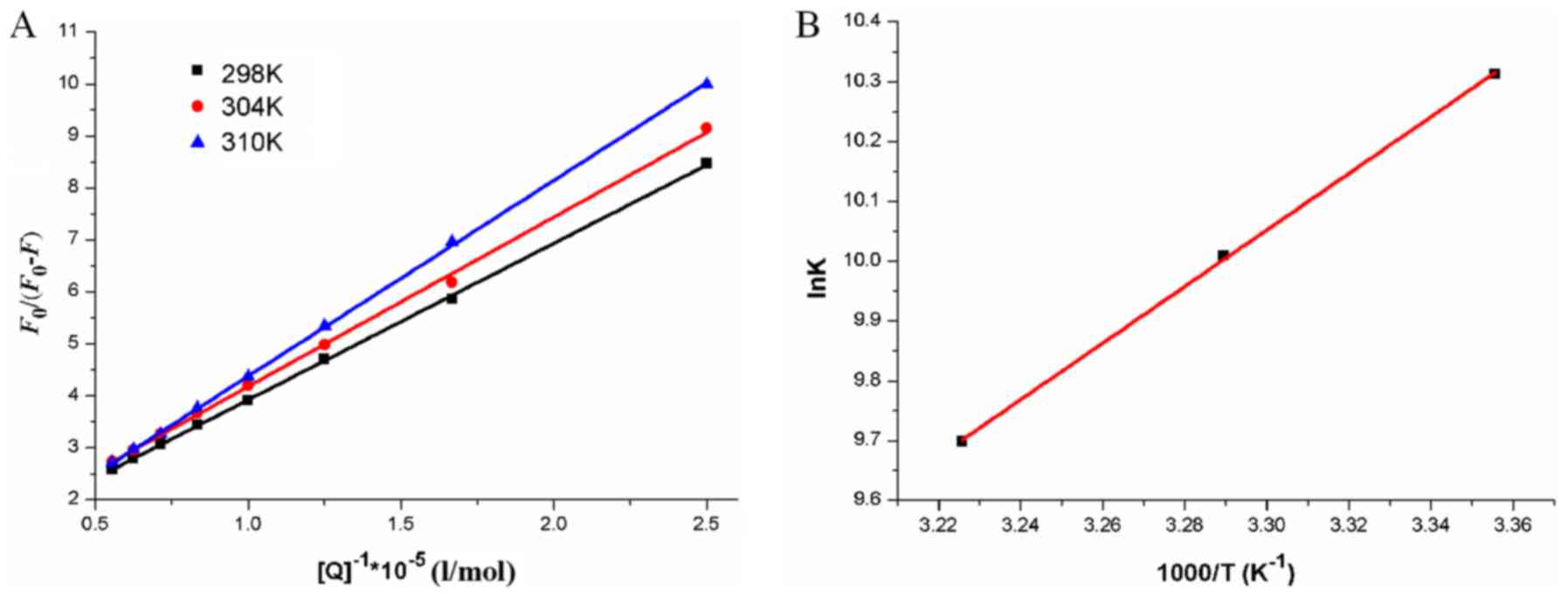
The Stern-Volmer plots of
F0/F for HSA, vs. [Q] at the
concentrations of 0–2.0×10−6 mol·l−1 based on
equation (1) at 298, 304 and 310 K
are shown in Fig. 3. The
comparatively good linear associations
(R2=0.9971, 0.9997 and 0.9930) indicate that a
single quenching mechanism, whether dynamic or static quenching,
occurred at 37°C. The values of KSV and
kq (=KSV/τ0)
obtained from the plots are listed in Table II. The values of
kq were all >2.0×1010
l·mol−1·s−1, which suggested that the
quenching mechanism arose mainly from the formation of the HSA-HMSP
complex and was controlled by a static mechanism. The results also
suggested that the binding between HMSP and HSA was strong, and
that HMSP was stored and carried by HSA in the plasma.
 | Table II.Quenching constants for the
interaction of p-hydroxycinnamic acid with human serum
albumin at different temperatures and pH 7.4. |
Table II.
Quenching constants for the
interaction of p-hydroxycinnamic acid with human serum
albumin at different temperatures and pH 7.4.
| Temperature
(K) |
KSV/(×105
l·mol−1) |
kq/(×1013
l·mol−1 S−1) |
R2 | SD |
|---|
| 298 | 4.27 | 4.27 | 0.9972 | 0.07 |
| 304 | 3.48 | 3.48 | 0.9997 | 0.02 |
| 310 | 2.98 | 2.98 | 0.9930 | 0.08 |
The dynamic and static quenching processes can be
differentiated by their disparate dependence on temperature. Higher
temperatures lead to a high rate of diffusion, resulting in
increased dynamic quenching. By contrast, higher temperatures
usually cause the decomposition of the weakly-bound complex,
resulting in less static quenching (24). To clarify the quenching mechanism,
fluorescence data at three different temperatures were analyzed
using the Stern-Volmer equation. As shown in Fig. 3, the KSV was
negatively correlated with temperature, suggesting that the
possible quenching mechanism of the HMSP-HSA binding process was
caused by complex formation, rather than by dynamic collision.
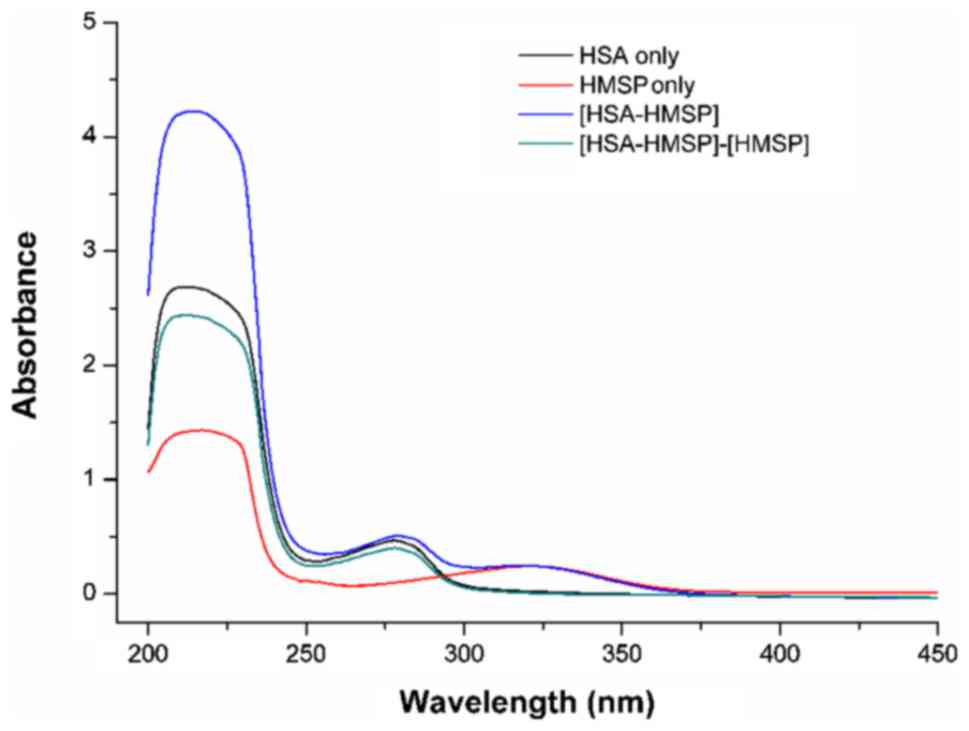
To further determine the static quenching mechanism,
the UV-Vis absorption spectra of HSA, HMSP and HSA-HMSP systems
were measured. It is known that the formation of a protein-compound
complex causes a change of the absorbance spectra. By contrast, a
collisional quenching process only alters the excited states of the
fluorophores, and does not alter the absorption spectra (25). As shown in Fig. 4, the difference in absorption spectra
between HSA-HMSP and HMSP were not superimposed with the absorption
spectra of HSA. Therefore, the fluorescence quenching was caused by
the formation of complex HSA-compounds, and this indicated
primarily a static quenching mechanism.
Determination of binding modes
For the static quenching interaction, the quenching
data were analyzed according to a modified Stern-Volmer equation
(26):
F0/ΔF=1/faKa[Q]+1/fa(2)
where Ka is the binding constant
of the protein and quencher; and fa is the
fraction of the initial fluorescence that can be quenched. The
thermodynamic parameters can be calculated by the van't Hoff
equation:
lnKa=-(ΔH/RT)+(ΔS/R)(3)
where Ka is the binding constant,
R is the gas constant and ∆S is the entropy change. The
enthalpy change (ΔH) can be estimated from the plot of
lnKa, vs. 1/T. The free energy change
(ΔG) can be calculated from the following equation:
ΔG=ΔH-TΔS=-RTlnKa(4)
Modified Stern-Volmer plots for the quenching of
HSA by HMSP at different temperatures are shown in Fig. 5A and plots of the van't Hoff equation
are shown in Fig. 5B. The binding
parameters and the thermodynamic parameters of binding of HMSP to
HSA are shown in Table III. The
negative values for ΔG indicate that the binding process was
spontaneous.
 | Table III.Thermodynamic parameters of the
p-hydroxycinnamic acid-human serum albumin system. |
Table III.
Thermodynamic parameters of the
p-hydroxycinnamic acid-human serum albumin system.
| Temperature
(K) |
Ka (×104
l·mol−1) |
Ra |
ΔHθ
(kJ·mol−1) |
ΔGθ
(kJ·mol−1) |
ΔSθ
(J·mol−1·K−1) |
Rb |
|---|
| 298 | 4.801 | 0.983 | −31.01 | −35.29 | 14.38 | 0.9993 |
| 304 | 3.696 | 0.999 |
| −35.38 |
|
|
| 310 | 1.983 | 0.999 |
| −35.46 |
|
|
There are four types of non-covalent forces that
contribute to the binding of ligands to biomacromolecules,
including hydrogen bonds, van der Waals forces, electrostatic
forces, and hydrophobic interactions. The details of these forces
can be determined through thermodynamic experiments. Leckband and
Subramanian (27) suggested the
values and magnitude of thermodynamic parameters associated with
various interactions. The negative ΔH and ΔS in the
present study indicated that the main forces for the HMSP-HSA
complex were van der Waals forces and hydrogen bonds.
Identification of binding site and binding
distance. The crystal structure of HSA consists of three domains
sequentially named I–III, each including two sub-domains (A and B).
Several studies have shown that the regions of ligands bound to HSA
are mainly located in hydrophobic cavities in sub-domains IIA and
IIIA. These have been labeled site I and site II (28). To identify the compound binding sites
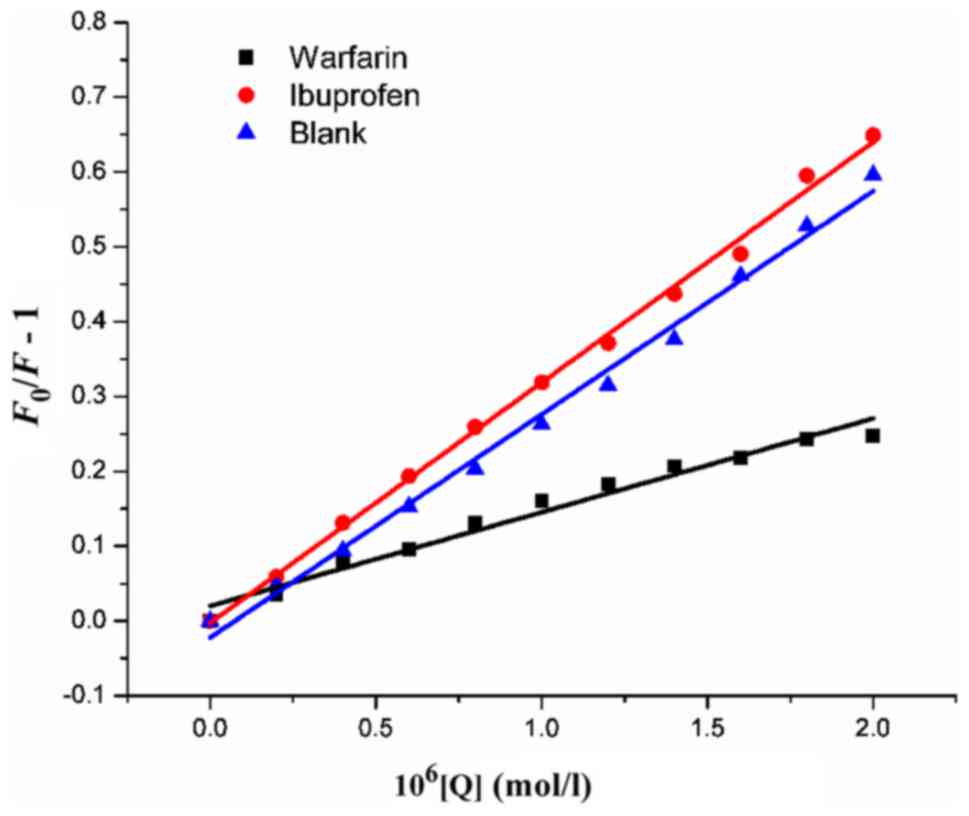
on HSA, site marker competitive experiments were performed by
utilizing warfarin and ibuprofen as site markers. The fluorescence
spectra were recorded upon excitation at 295 nm and the
Stern-Volmer quenching constant (Ksv) was
calculated according to equation (2). As shown in Fig. 6, the quenching constants of the
system with warfarin showed a marked decrease for HMSP, whereas the
constants obtained from the system with ibuprofen remained
unchanged. This suggested that the binding of compounds to HSA were
affected by adding warfarin, which indicated that the binding of
HMSP to HSA was mainly located within site I (sub-domain IIA).
The spectral results indicated that HMSP can form
complexes with HSA. For more information on the HMSP-HSA system,
the distance r between derivatives and HSA, according to
Föster's non-radioactive energy transfer theory (29), were examined. Based on this theory,
the efficiency (E) of energy transfer between the donor and
acceptor can be calculated as follows:
E=1-(F/F0)=R06/(R06+r6)(5)
where R0 is the critical distance
when the transfer efficiency is 50%. R06 is
calculated using the following equation:
R06=8.8x10-25K2N-4ΦJ(6)
where K2 is the spatial
orientation factor between the emission dipole of the donor and the
absorption dipole of the acceptor; N is the refractive index
of the medium; Ф is the fluorescence quantum yield of the
donor; and J is the overlap integral of the fluorescence
emission spectrum of the donor and the absorption spectrum of the
acceptor, determined according to the following equation:
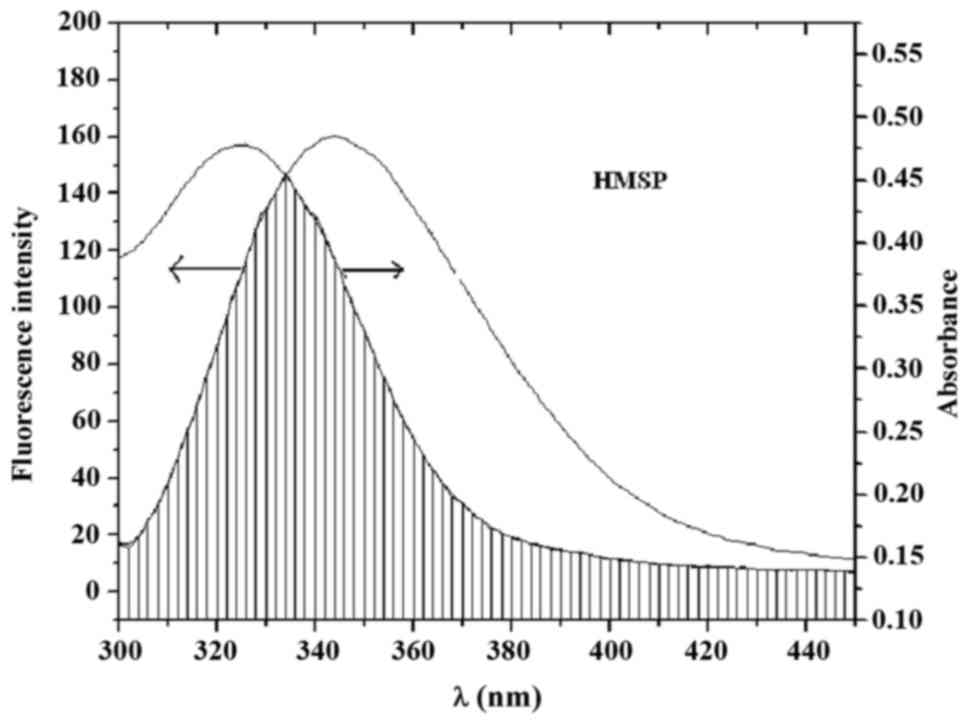
J=[∑F(λ)ε(λ)λ4Δλ]/∑F(λ)Δλ(7)
where F(λ) and ε(λ) are the
fluorescence intensity of HSA and molar absorption coefficient of
HMSP at wavelength λ, respectively. J can be evaluated by
integrating spectra, as shown in Fig.
7. It has been reported for HSA that K2=2/3,
Ф=0.15 and N=1.36 (22). According to equations (5–7), it was
calculated that the value of r between HMSP and HSA was 0.58
nm, and this indicated that the probability of energy transfer from
HSA to HMSP was high, and the quenching process was a
non-radioactive transfer process.
Conformation investigation
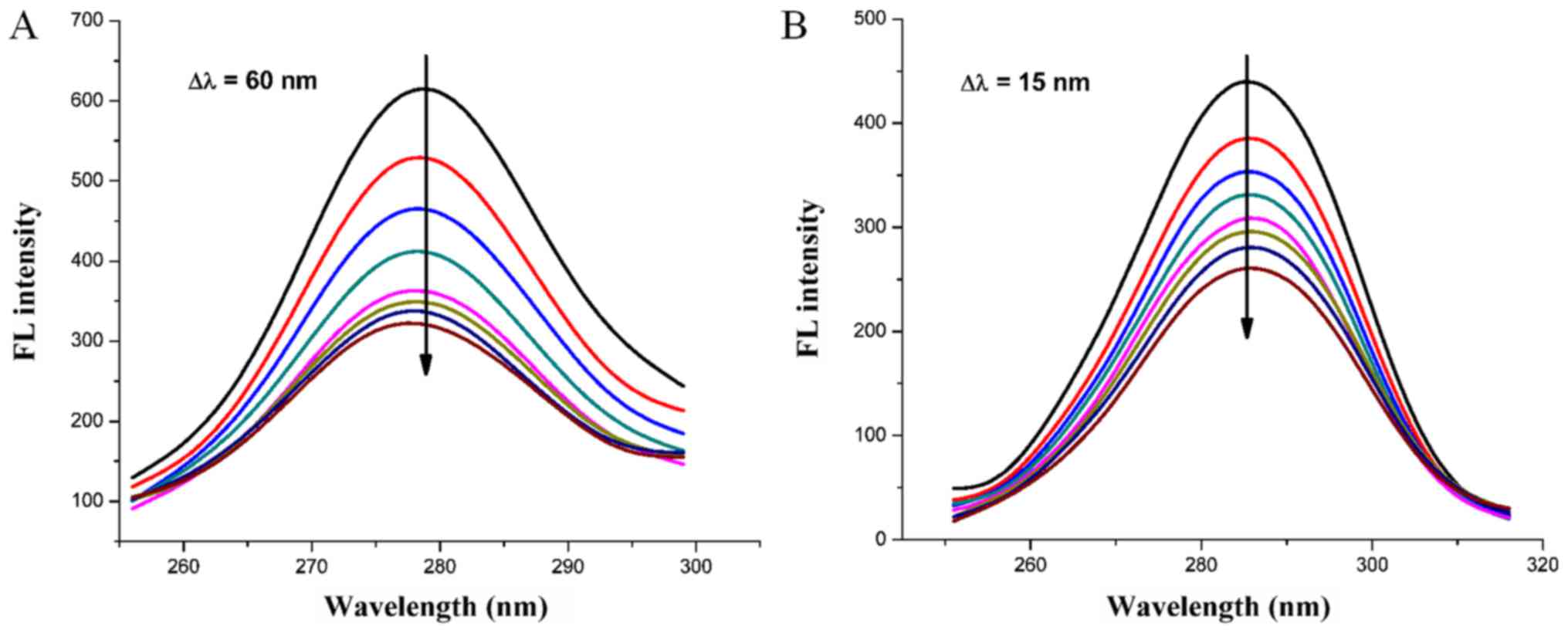
Synchronous fluorescence spectra can provide
information on the microenvironments close to the vicinity of the
fluorophore molecules at a molecular level (30). The possible shift in position of
maximum emission wavelength of biomolecules indicates changes of
polarity around the chromospheres molecule. When the difference
(Δλ) between the wavelength of excitation and the emission is set
to 15 nm, the synchronous fluorescence spectrum gives the typical
information of tyrosine. When Δλ=60 nm, the synchronous
fluorescence presents the typical information of tryptophan
(12). As shown in Fig. 8A and B, the synchronous fluorescence
spectra for Δλ=15 nm exhibited no marked shift at maximum emission
peak, however, for Δλ=60 nm the maximum emission wavelength
exhibited a blue-shift, suggesting the binding of HMSP increased
the hydrophobicity of the microenvironment around the Trp residue,
whereas the microenvironment around the Tyr residue was not
affected.
To further investigate the effects of HMSP on the
conformation of HSA, CD experiments were performed. The CD spectrum
is widely used to investigate the conformational changes of
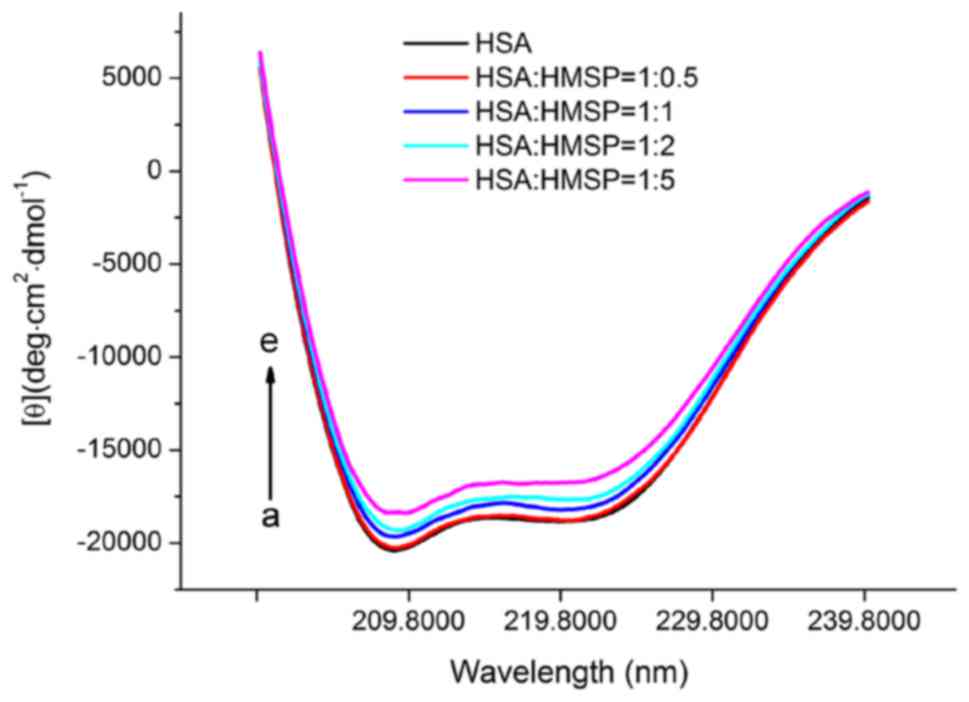
proteins or peptides in solution (31). As shown in Fig. 9, HSA exhibited two typical negative
peaks of 208 and 222 nm, representing the α-helix structure of HSA
(32). HMSP was then added in
increasing concentrations to measure the CD spectra of HSA. The
contents of α-helix, β-strand and other structures were calculated
using SELCON3 software (Table IV).
The free HSA in the absence of HMSP consisted of ~58% α-helix, ~20%
β-sheet and β-turn, and ~22% random coils, and this was in
agreement with a previous report (33). When the concentration of HMSP
increased, the proportion of the secondary structural underwent
marginal variations. At the molar ratio of 5:1, the α-helix content
decreased to 51.8% whereas he ratio of β-sheets and random coils
increased to 23.1 and 25.1%, respectively. These results indicated
that the secondary structures of HSA were gradually disordered due
to the formation of the HSA-HMSP complex.
 | Table IV.Secondary structure of HSA with
different concentrations of HMSP. |
Table IV.
Secondary structure of HSA with
different concentrations of HMSP.
| Structural
components | α-helix | β-sheet | β-turn | Random coil |
|---|
| HSA | 0.583 | 0.064 | 0.138 | 0.222 |
| HMSP:HSA=0.5:1 | 0.572 | 0.065 | 0.139 | 0.224 |
| HMSP:HSA=1:1 | 0.561 | 0.069 | 0.146 | 0.240 |
| HMSP:HSA=2:1 | 0.535 | 0.069 | 0.153 | 0.246 |
| HMSP:HSA=5:1 | 0.518 | 0.072 | 0.159 | 0.251 |
Computational experiment
A docking experiment was used to investigate the
binding modes between HMSP and HSA, particularly the binding sites
and the binding conformations of the compounds. In the present
study, the Surflex-Dock program (sybyl 8.1, tripos) was selected to
identify the binding conformations of HMSP at the active HSA sites.
In total, 20 conformations were obtained, and the conformation with
the lowest free energy was selected for further analysis. The
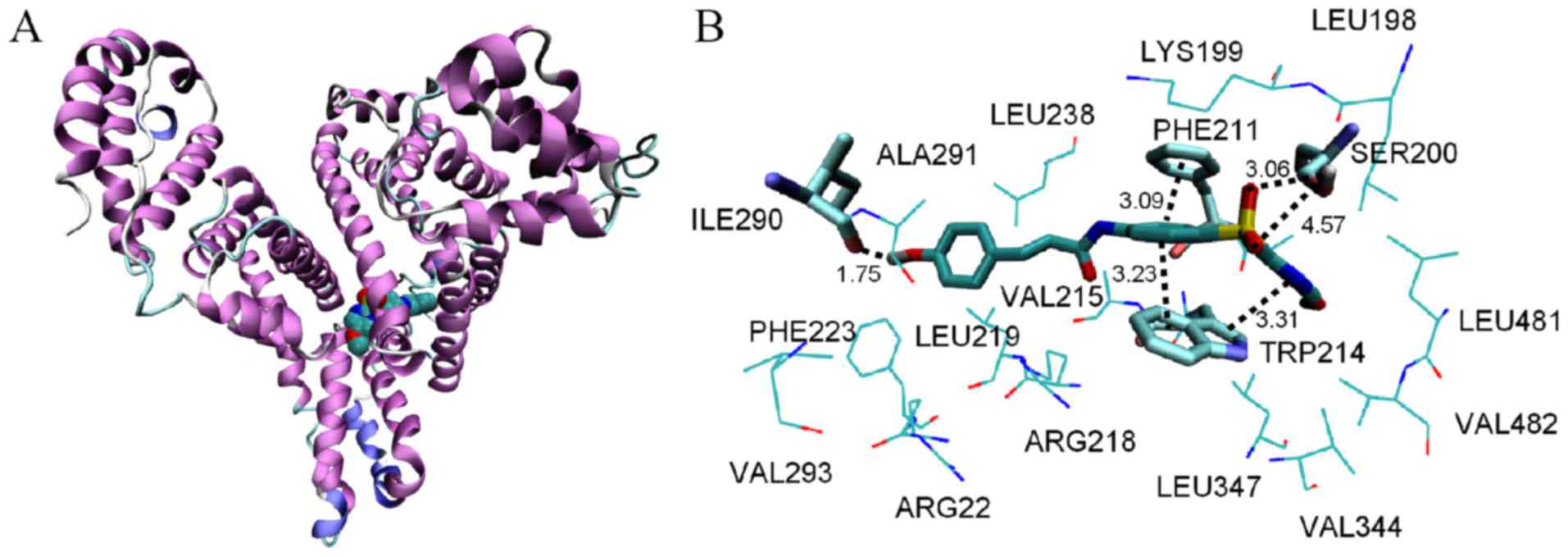
highest score-ranked result is shown in Fig. 10. The docking results indicated that
HMSP bound in the hydrophobic pocket of sub-domain IIA (site I).
The surrounding residues within 0.5 nm of HMSP were LEU198, LYS199,
SER200, PHE211, TRP214, VAL215, ARG218, LEU219, ARG222, PHE223,
LEU238, ILE290, and ALA291. As HMSP interacted mainly with
hydrophobic side chains, it was inferred that the interaction
between HMSP and HSA was mainly hydrophobic. This was consistent
with the thermodynamic results. The phenyl and pyrimidine ring
formed π-π stacking with sidechains of PHE211 and TRP214.
Furthermore, several hydrogen bonds existed between HMSP and
sidechains of SER200 and a backbone of ILE290. The molecule docking
results supported the indication that the binding of HMSP with HSA
was stabilized by van der Waals and hydrogen bonds
interactions.
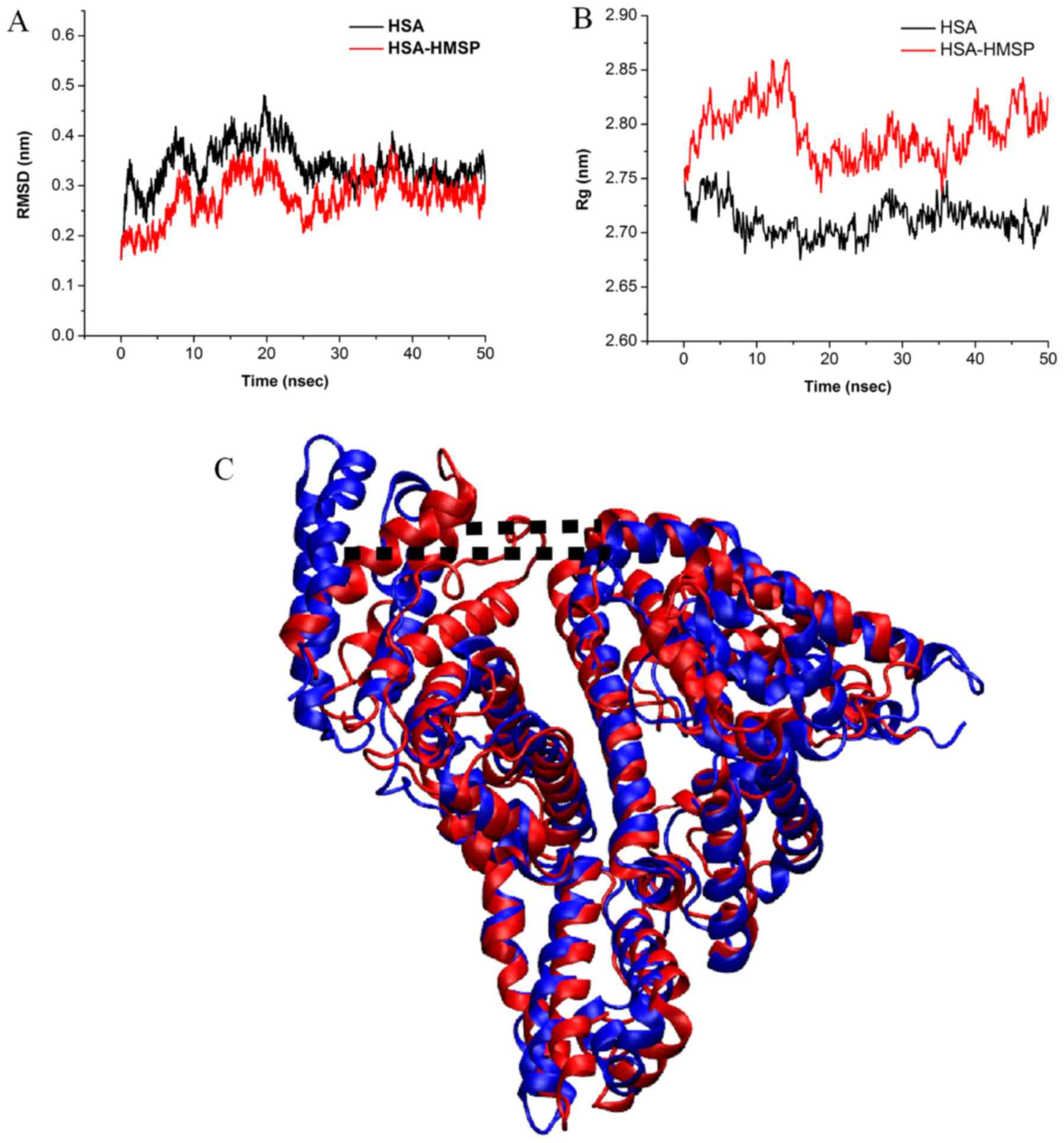
MD simulation was performed on the HSA-HMSP complex
to investigate the binding modes of HMSP and its effect on
conformations of HSA. The convergence of the system equilibrium was
examined by the time-dependent evolution of the root-mean-square
deviation (RMSD) of backbone atoms to its initial structure. It was
observed that the HSA was stable as the fluctuation of RMSD was
small (Fig. 11A). The radius of
gyration (Rg) of free HSA and the HSA-HMSP complex were also
calculated, as shown in Fig. 11B.
The Rg values achieved equilibrium at ~2.72 and 2.85 nm,
respectively, suggesting that the Rg increased upon binding of HMSP
to HSA. By comparing the final protein structure of the 50-ns MD
simulation in the absence and presence of HMSP, it was found that
the length of the cleft of HSA increased from 3.79 to 7.85 nm
(Fig. 11C). This type of
conformation change may have be caused by the insertion of HMSA in
the center of the protein.
In conclusion, the interaction of a novel
p-hydroxycinnamic amide HMSP with HSA was investigated under
simulated physiological conditions by fluorescence and UV-Vis
spectra, in addition to other molecular simulations. The binding
constants, binding forces, binding distances and energy transfer
parameters between HMSP and HSA were obtained. The results
indicated that HMSP was able to bind to HSA molecules, the
fluorescence quenching mechanism was static, and the binding
reaction was spontaneous. The interactions were mainly
enthalpy-driven, and hydrogen bonding, hydrophobic and van der
Waals interactions were involved in the binding. The binding
distance (r) between HSA and HMSP was calculated as 0.58 nm,
indicating a high probability of energy transfer from HSA to HMSP.
The binding site of HMSP on HSA was close to site I (sub-domain
IIA). The synchronous fluorescence spectra indicated that HMSP
affected the microenvironment of Tyr residues upon binding to HSA
molecules. The CD spectrum and MD simulation results showed that
the conformation of HSA became partially disordered upon binding of
HMSP. The present study provided accurate and comprehensive data
for identifying the binding mechanism of HMSP with HSA, and assists
in understanding its effects on protein function during its
transportation and distribution in plasma.
Acknowledgements
Not applicable.
Funding
The present study was financially supported by the
Natural Science Foundation of Guangxi (grant no.
2014GXNSFBA118053), the College Student Innovation and
Entrepreneurship Training Program of Hubei Province (grant no.
201710920034) and the Natural Science Foundation of Hubei Province
of China (grant no. 2017CFB582).
Availability of data and materials
The datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
YHJ and FYM conceived the research. GBZ, LZR and
YFJ conducted the experiments. QZ analyzed the data. YHJ and FYM
wrote the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Calabriso N, Scoditti E, Massaro M,
Pellegrino M, Storelli C, Ingrosso I, Giovinazzo G and Carluccio
MA: Multiple anti-inflammatory and anti-atherosclerotic properties
of red wine polyphenolic extracts: Differential role of
hydroxycinnamic acids, flavonols and stilbenes on endothelial
inflammatory gene expression. Eur J Nutr. 55:477–489. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Maury DK and Devasagayam TP: Antioxidant
and prooxidant nature of hydroxycinnamic acid derivatives ferulic
and caffeic acids. Food Chem Toxicol. 48:3369–3373. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Marques MPM, Borges F, Sousa J, Calheiros
R, Garrido J, Gaspar A, Diniz C and Fresco P: Cytotoxic and COX-2
inhibition properties of hydroxycinnamic derivatives. Lett Drug Des
Dis. 3:316–320. 2006. View Article : Google Scholar
|
|
4
|
Mazzone C, Russo N and Toscano M:
Antioxidant properties comparative study of natural hydroxycinnamic
acids and structurally modified derivatives: Computational
insights. Comput Theor Chem. 1077:39–47. 2016. View Article : Google Scholar
|
|
5
|
Tavares-da-Silva EJ, Varela CL, Pires AS,
Encarnação JC, Abrantes AM, Botelho MF, Carvalho RA, Proença C,
Freitas M, Fernandes E and Roleira FM: Combined dual effect of
modulation of human neutrophils' oxidative burst and inhibition of
colon cancer cells proliferation by hydroxycinnamic acid
derivatives. Bioorg Med Chem. 24:3556–3564. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Esteves M, Siquet C, Gaspar A, Rio V,
Sousa JB, Reis S, Marques MP and Borges F: Antioxidant versus
cytotoxic properties of hydroxycinnamic acid derivatives-a new
paradigm in phenolic research. Arch Pharm (Weinheim). 341:164–173.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bailey D, Kirby BP, Atkinson J, Fixon-Owoo
S, Henman MC, Shaw GG and Doyle KM: Hydroxycinnamic acid amide
derivatives of polyamines reverse spermine-induced CNS excitation.
Pharmacol Biochem Behav. 133:57–64. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yeggoni DP, Gokara M, Manidhar DM,
Rachamallu A, Nakka S, Reddy CS and Subramanyam R: Binding and
molecular dynamics studies of 7-hydroxycoumarin derivatives with
human serum albumin and its pharmacological importance. Mol Pharm.
11:1117–1131. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Urtti A: Challenges and obstacles of
ocular pharmacokinetics and drug delivery. Adv Drug Deliv Rev.
58:1131–1135. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Meng FY, Zhu JM, Zha AR, Yu SR and Lin CW:
Synthesis of p-hydroxycinnamic acid derivatives and investigation
of fluorescence binding with bovine serum albumin. J Lumin.
132:1290–1298. 2012. View Article : Google Scholar
|
|
11
|
Qi ZD, Zhang Y, Liao FL, Ou-Yang YW, Liu Y
and Yang X: Probing the binding of morin to human serum albumin by
optical spectroscopy. J Pharm Biomed Anal. 46:699–706. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
He W, Li Y, Xue C, Hu Z, Chen X and Sheng
F: Effect of Chinese medicine alpinetin on the structure of human
serum albumin. Bioorg Med Chem. 13:1837–1845. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sheldrick GM: SHELXS97 and SHELXL97.
program for crystal structure solution and refinement. University
of Göttingen; Göttingen. Germany: 1997
|
|
14
|
He H, Li WD, Yang LY, Fu L, Zhu XJ, Wong
WK, Jiang FL and Liu Y: A novel bifunctional mitochondria-targeted
anticancer agent with high selectivity for cancer cells. Sci Rep.
5:135432015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
He H, Xu J, Cheng DY, Fu L, Ge YS, Jiang
FL and Liu Y: Identification of binding modes for amino naphthalene
2-cyanoacrylate (ANCA) probes to amyloid fibrils from molecular
dynamics simulations. J Phys Chem B. 121:1211–1221. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Petitpas I, Bhattacharya AA, Twine S, East
M and Curry S: Crystal structure analysis of warfarin binding to
human serum albumin: Anatomy of drug site I. J Mol Biol.
276:22804–22809. 2001.
|
|
17
|
Humphrey W, Dalke A and Schulten K: VMD:
Visual molecular dynamics. J Mol Graph. 14:33–38. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
He H, Xu J, Xie W, Guo QL, Jiang FL and
Liu Y: Reduced state transition barrier of CDK6 from open to closed
state induced by Thr177 phosphorylation and its implication in
binding modes of inhibitors. Biochim Biophys Acta. 1862:501–512.
2018. View Article : Google Scholar
|
|
19
|
Tong J, Tian F, Liu Y and Jiang F:
Comprehensive study of the adsorption of an acylhydrazone
derivative by serum albumin: unclassical static quenching. RSC Adv.
4:59686–59696. 2014. View Article : Google Scholar
|
|
20
|
Cao H, Jia X, Shi J, Xiao J and Chen X:
Non-covalent interaction between dietary stilbenoids and human
serum albumin: Structure–affinity relationship, and its influence
on the stability, free radical scavenging activity and cell uptake
of stilbenoids. Food Chem. 202:383–388. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hu YJ, Liu Y, Pi ZB and Qu SS: Specific
activation of mGlu2 induced IGF-1R transactivation in vitro through
FAK phosphorylation. Bioorg Med Chem. 13:6609–6614. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bhogale A, Patel N, Sarpotdar P, Mariam J,
Dongre PM, Miotello A and Kothari DC: Systematic investigation on
the interaction of bovine serum albumin with ZnO nanoparticles
using fluorescence spectroscopy. Colloids Surf B Biointerfaces.
102:257–264. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nishijima M, Pace TC, Nakamura A, Mori T,
Wada T, Bohne C and Inoue Y: Supramolecular photochirogenesis with
biomolecules. Mechanistic studies on the enantiodifferentiation for
the photocyclodimerization of 2-anthracenecarboxylate mediated by
bovine serum albumin. J Org Chem. 72:2707–2715. 2017. View Article : Google Scholar
|
|
24
|
Anand U, Jash C, Boddepalli RK,
Shrivastava A and Mukherjee S: Exploring the mechanism of
fluorescence quenching in proteins induced by tetracycline. J Phys
Chem B. 115:6312–6320. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mi R, Hu YJ, Fan XY, Ouyang Y and Bai AM:
Exploring the site-selective binding of jatrorrhizine to human
serum albumin: Spectroscopic and molecular modeling approaches.
Spectrochim Acta A Mol Biomol Spectrosc. 117:163–169. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gao H, Lei L, Liu J, Kong Q, Chen X and Hu
Z: The study on the interaction between human serum albumin and a
new reagent with antitumour activity by spectrophotometric methods.
J Photochem Photobiol A Chem. 167:213–221. 2004. View Article : Google Scholar
|
|
27
|
Ross PD and Subramanian S: Thermodynamics
of protein association reactions: Forces contributing to stability.
Biochemistry. 20:3096–3102. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yue Y, Liu R, Liu J, Dong Q and Fan J:
Experimental and theoretical investigation on the interaction
between cyclovirobuxine D and human serum albumin. Spectrochim Acta
A Mol Biomol Spectrosc. 128:552–558. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang Q, Xiao Y, Huang Y and Li H: An
important prerequisite for efficient Förster resonance energy
transfer (FRET) from human serum albumin to alkyl gallate. RSC Adv.
6:36146–36151. 2016. View Article : Google Scholar
|
|
30
|
Fan XY, Zhang Y, Wang J, Yang LY, Jiang FL
and Liu Y: Exploring the interaction between rotenone and human
serum albumin. J Chem Thermodyn. 69:186–192. 2014. View Article : Google Scholar
|
|
31
|
Pescitelli G, Di Bari L and Berova N:
Application of electronic circular dichroism in the study of
supramolecular systems. Chem Soc Rev. 43:5211–5233. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang HX and Liu E: Spectroscopic and
molecular modeling investigation on the binding of a synthesized
steroidal amide to protein. J Lumin. 153:182–187. 2014. View Article : Google Scholar
|
|
33
|
Hou H, Qu X, Li Y, Kong Y, Jia B, Yao X
and Jiang B: Binding of citreoviridin to human serum albumin:
multispectroscopic and molecular docking. Biomed Res Int.
2015:1623912015. View Article : Google Scholar : PubMed/NCBI
|