Introduction
It is a common phenomenon that the symptoms and
signs are slightly different among patients with the same disease
(1). For instance, coronary heart
disease (CHD) manifests as chest pain (2). However, certain patients suffer from a
stabbing pain, while others have complaints of a burning pain
(3). Within the framework of
Traditional Chinese Medicine (TCM) theory, practitioners always
prescribe a medical formula based on symptoms and signs (4). In fact, TCM practitioners may divide
patients with the same disease into several subtypes according to
these slight differences and prescribe the corresponding formulae.
In TCM theory, these subtypes are known as Zheng (5). It is renowned that the advancement of
Zheng depends on the genes inherited from the parents, the
lifestyle and the environmental conditions, which may differ
between individual patients, and therefore, different types of
Zheng exist (6). The first rule of
Zheng is that one type of disease may be divided into several
different Zheng (7).
A disequilibrium of the body's original condition is
accountable for the development of Zheng, and certain deviations
from the equilibrium may lead to certain types of disease and
manifestations thereof (8). For
instance, Qi stagnation and blood stasis syndrome (QSBSS), a type
of Zheng, describes the condition of unsmooth flow of Qi and blood,
which may occur in diseases including CHD, chronic gastritis (CG)
and rheumatoid arthritis (RA) (9).
This exemplifies the second rule of Zheng, namely that one type of
Zheng may be the basis of several different types of disease
(7). However, the western scientific
basis for this theory, including the substantial essence of this
Zheng, remains to be fully elucidated. Previous studies in the
field mainly focus on the subtypes of one disease based on Zheng,
while those on the biological basis of the same Zheng in different
diseases are scarce (10,11).
The genetic profile of an individual provides
information on what could theoretically happen in the body, while
protein expression profiles are representative of the processes
that are active in the body at the time-point of determination
(12). Of note, RNAs are the
connection between DNAs and proteins, which reflect the interaction
of the genotype and the environment (8). Among the different types of RNAs,
messenger RNAs (mRNAs) are transcripts of DNA and the information
they carry is transferred by their translation into proteins in
accordance with the central dogma (13). Long non-coding RNAs (lncRNAs) are a
class of non-protein-coding RNAs of >200 nucleotides in length,
and they are known to have roles in controlling chromatin
structure, transcriptional regulation and post-transcriptional
processing (14,15). Furthermore, circular RNAs (circRNAs),
which have drawn an increasing amount of attention recently, are a
type of non-canonical form of alternative splicing and are more
stable than linear RNAs (16,17).
Network pharmacology studies based on the theory of systems biology
may be used for the preliminary validation of the interaction
between TCM herbs and RNA targets (18). Due to its core methods based on the
holistic view and the dynamic balance of the human body, network
pharmacology is useful for the study of TCM herbs with complex
mechanisms of action (19). In the
present study, a network pharmacology analysis was employed to
preliminarily validate the differentially expressed (DE) RNAs in
subjects with QSBSS. The purpose of the present study was to
explore the biological basis of one common Zheng, QSBSS, from the
perspective of ncRNAs and mRNAs in a diversity of associated
diseases.
Materials and methods
Patients and samples
The protocol of the present study was approved by
the Ethics Committee of Guang'anmen Hospital (Beijing, China). The
study included 5 patients with CHD and QSBSS (group a), 5 patients
with CG and QSBSS (group b), 5 patients with RA and QSBSS (group c)
and 5 completely healthy subjects (control) who presented at
Guang'anmen Hospital affiliated to the China Academy of Chinese
Medical Sciences (Beijing, China) between October 2016 and December
2016. Informed consent was provided by all of the participants
prior to the study.
The diagnosis of QSBSS was based on the judgement of
an expert clinician according to the standard criteria approved by
the China Food and Drug Administration (20) and the scaled diagnosis criteria,
which is used to diagnose Qi Stagnation and Blood Stasis Syndrome
(21). The criteria for QSBSS
included primary symptoms and signs [i) distending pain; ii)
tingling sensation anywhere on the body; iii) pain in a fixed
place; iv) pain aggravated by touch; v) lumps in the body; vi)
self-reported irritability; vii) clinical depression diagnosed by a
psychiatrist], secondary symptoms and signs [i) loss of appetite;
ii) paleness of the face; iii) hemorrhagic spots on any part of the
body], TCM tongue presentations [i) purple, dusky tongue; ii)
stasis macules or stasis spots on the tongue] and pulse
presentation [i) choppy pulse; ii) wiry pulse] (22). A patient was diagnosed with QSBSS if
they had >2 of the primary symptoms and signs, >1 of the
secondary symptoms and signs, and 1 of the tongue or pulse
presentations. The diagnosis of CHD, RA and CG were based on
guidelines for the diagnosis and management of patients with stable
ischaemic heart disease (23), the
diagnosis and treatment for rheumatoid arthritis (24) and the consensus of chronic gastritis
(25), respectively. From each of
the patients, blood samples (3–4 ml) were collected in
EDTA-containing tubes.
RNA isolation and library
preparation
RNA degradation and contamination was monitored on
1% agarose gels. RNA purity was confirmed using the Agencourt
AMPure XP (cat. no. A63881; Beckman Coulter, Brea, CA, USA). RNA
integrity was assessed using the RNA Nano 6000 Assay Kit for the
Bioanalyzer 2100 system (cat. no. 5067-1511; Agilent Technologies,
Inc., Santa Clara, CA, USA). The methods of lncRNA quantification
were identical to the conventional methods for mRNAs.
Quantification of circRNAs was performed with exonuclease to
exclude non-circRNAs. After RNA extraction, the total RNA was
digested with TruSeq Stranded Total RNA with Ribo-Zero Gold (cat.
no. 15021048; Illumina, Inc., San Diego, CA, USA). The sample was
then subjected to reverse transcription using SuperScript II
Reverse Transcriptase (cat. no. 18064014; Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA).
A total of 3 µg RNA per sample was used as input
material for the RNA sample preparations. First, ribosomal (r)RNA
was removed using Agencourt RNAClean XP (cat. no. A63987; Beckman
Coulter), and rRNA-free material was cleaned up by ethanol
precipitation. Subsequently, sequencing libraries were generated
using the rRNA-depleted RNA. Library preparation was performed
according to manufacturer's protocol (Illumina). In brief,
fragmentation was performed using divalent cations at 94°C in 5 µl
rRNA Binding Buffer and 5 µl rRNA Removal Mix-Gold. First-strand
complementary (c)DNA was synthesized using 8 µl First Strand
Synthesis Act D Mix and SuperScript II Reverse Transcriptase.
Second-strand cDNA synthesis was performed using 5 µl End Repair
Control (2 µl End Repair Control + 98 µl Resuspension Buffer). The
reaction buffer contained deoxynucleoside triphosphate where
thymine was replaced by uracil. After adenylation of the 3′ends of
the DNA fragments, an adaptor with a hairpin loop structure was
ligated to prepare for hybridization. To select cDNA fragments of
preferentially 150–200 bp in length, the library fragments were
purified using the AMPure XP system (cat. no. A63881; Beckman
Coulter). The DNA fragment was enriched by using 5 µl polymerase
chain reaction (PCR) Primer Cocktail and 25 µl PCR Master Mix prior
to performing PCR. The mixture was incubated in the following
conditions: 98°C for 30 sec, 15 cycles 98°C for 10 sec, 60°C for 30
sec, 72°C for 30 sec, then 72°C for 5 min and held at 10°C.
Finally, products were purified (AMPure XP system) and library
quality was assessed on the Agilent Bioanalyzer 2100 system.
Sequencing of RNAs and DE RNAs
analysis
The clustering of the index-coded samples was
performed on a cBot Cluster Generation System using TruSeq PE
Cluster Kit v3cBot-HS (Illumina) according to the manufacturer's
protocols. After cluster generation, the libraries were sequenced
on an Illumina HiSeq 2,500 platform, and 125 bp paired-end and 50
bp single-end reads were generated. The number of reads for each
sample was 8×107. The number of clean bases of lncRNAs,
circRNAs and mRNAs for each sample in filtered data was 12–13 Gb.
The error rate of each base in lncRNAs, circRNAs and mRNAs for each
sample was <0.001. The spliced transcription products of each
sample were combined and screened as lncRNAs with Cuffmerge program
in the Cufflinks 2.2.1 software package. All transcripts that
overlapped with known mRNAs, other non-coding RNA and non-lncRNA
were discarded. Next, the transcripts that longer than 200 bp and
>2 exons were obtained, and CPC (version 0.9-r2) (26), PLEK (version 1.2) (27), CNCI (version 1.0) (28) and Pfam (version 30) (29) software packages were used to predict
transcripts with coding potential. The characteristics (including
length, type and number of exons) of lncRNA were analysed after
screening by the Cuffcompare program in the Cufflinks 2.2.1
software package (30). CircRNAs
were identified using the CircRNA Identifier tool (31). The expression of lncRNAs were
calculated using fragments per kb per million reads (FPKM), while
the expression of circRNAs were calculated by spliced reads per
million reads (RPM). Then, DESeq2 with negative binomial
distribution was used to analyse DE of RNAs (32). All sequencing processes and analyses
were performed by Shanghai OE Biotech Co., Ltd. (Shanghai,
China).
Gene ontology (GO) annotations and
Kyoto encyclopedia of genes and genomes (KEGG) pathway
analysis
GO annotations and KEGG pathway analysis were
performed to investigate the possible roles of the DE ncRNAs. GO
annotations were performed to identify regulatory networks of DE
genes (http://geneontology.org). KEGG analysis
was also performed to explore the enriched pathways of the DE RNAs
based on the KEGG database (http://www.genome.jp/kegg/). To reveal the role and
interactions among the DE ncRNAs and mRNAs in QSBSS patients, an
ncRNAs regulatory network was constructed using Cytoscape software
version 3.2.1 (http://www.cytoscape.org/).
Network pharmacology validation
The process of network pharmacology validation was
performed in three steps. First, according to the sequencing
results of DE RNAs, the RNA targets associated with QSBSS were
prepared by ID normalization at http://www.uniprot.org/ (33). Next, the active components of Xuefu
Zhuyu decoction, a classic and CFDA-approved TCM formula for
treating QSBSS, were retrieved from the TCM Systems Pharmacology
Database (TCMSP) and the TCM Integrated Database (34,35). The
compounds were then screened according to parameters including
drug-likeness (DL) and oral bioavailability (OB), and the specific
ingredients were selected if their DL ≥0.18 and OB ≥30%, suggested
criteria given by the TCMSP database (34). Finally, the targets of the compounds
were predicted using the TCMSP analysis platform and the merged
compound-RNA target network was constructed using Cytoscape
(version 3.2.1), in order to determine whether any of the DE RNAs
associated with QSBSS were targets of the components of Xuefu Zhuyu
decoction.
Statistical analysis
The data regarding the clinical characteristics of
the participants are expressed as the mean ± standard error of the
mean and the corresponding results were statistically analyzed
using one-way analysis of variance. The results of DE RNAs were
analyzed by the algorithm of DESeq2 in R software (32). The regulatory network for ncRNAs was
analyzed by the algorithm of ClueGo in Cytoscape software.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Clinical characteristics of
participants
A total of 20 participants were recruited for the
present study and were divided into 4 groups. The clinical
characteristics of these 4 groups were listed in Table I. The 4 groups were matched in terms
of sex, as well as body height and weight, and there were no
significant differences in terms of the frequency of hypertension,
diabetes mellitus and cancer between the groups (P>0.05).
 | Table I.Clinical characteristics of
sequencing subjects. |
Table I.
Clinical characteristics of
sequencing subjects.
| Characteristic | Control (n=5) | Group a (n=5) | Group b (n=5) | Group c (n=5) | P-value |
|---|
| Males | 3 | 3 | 3 | 1 | 0.051 |
| Age (years) | 50.40±7.26 | 63.60±5.12 | 64.40±6.38 | 58.60±5.94 | 0.010 |
| Height (cm) | 163.00±9.89 | 162.40±8.01 | 169.20±5.80 | 165.80±5.35 | 0.482 |
| Weight (kg) | 66.60±11.28 | 70.30±9.91 | 75.80±12.37 | 60.60±11.26 | 0.225 |
| Hypertension | 0 | 2 | 1 | 1 | 0.337 |
| Diabetes
mellitus | 0 | 0 | 2 | 1 | 0.058 |
| Cancer | 0 | 0 | 1 | 0 | 0.059 |
DE ncRNAs and mRNAs, and their overlap
among patient groups with different types of disease
The results of the DE ncRNAs and mRNAs in the
comparisons were as follows: A total of 678 DE lncRNAs (448
upregulated and 230 downregulated), 166 DE circRNAs (59 upregulated
and 107 downregulated), and 2,676 DE mRNAs (1,287 upregulated and
1,389 downregulated) were identified in group a (CHD with QSBSS vs.
controls), respectively. In group b (CG with QSBSS vs. control),
1,158 DE lncRNAs (901 upregulated and 257 downregulated), 116 DE
circRNAs (38 upregulated and 78 downregulated) and 2,880 DE mRNAs
(1,336 upregulated and 1,544 downregulated) were detected. A total
of 697 DE lncRNAs (470 upregulated and 227 downregulated), 120 DE
circRNAs (49 upregulated and 71 downregulated) and 2,776 DE mRNAs
(1,420 upregulated and 1,356 downregulated) were identified in
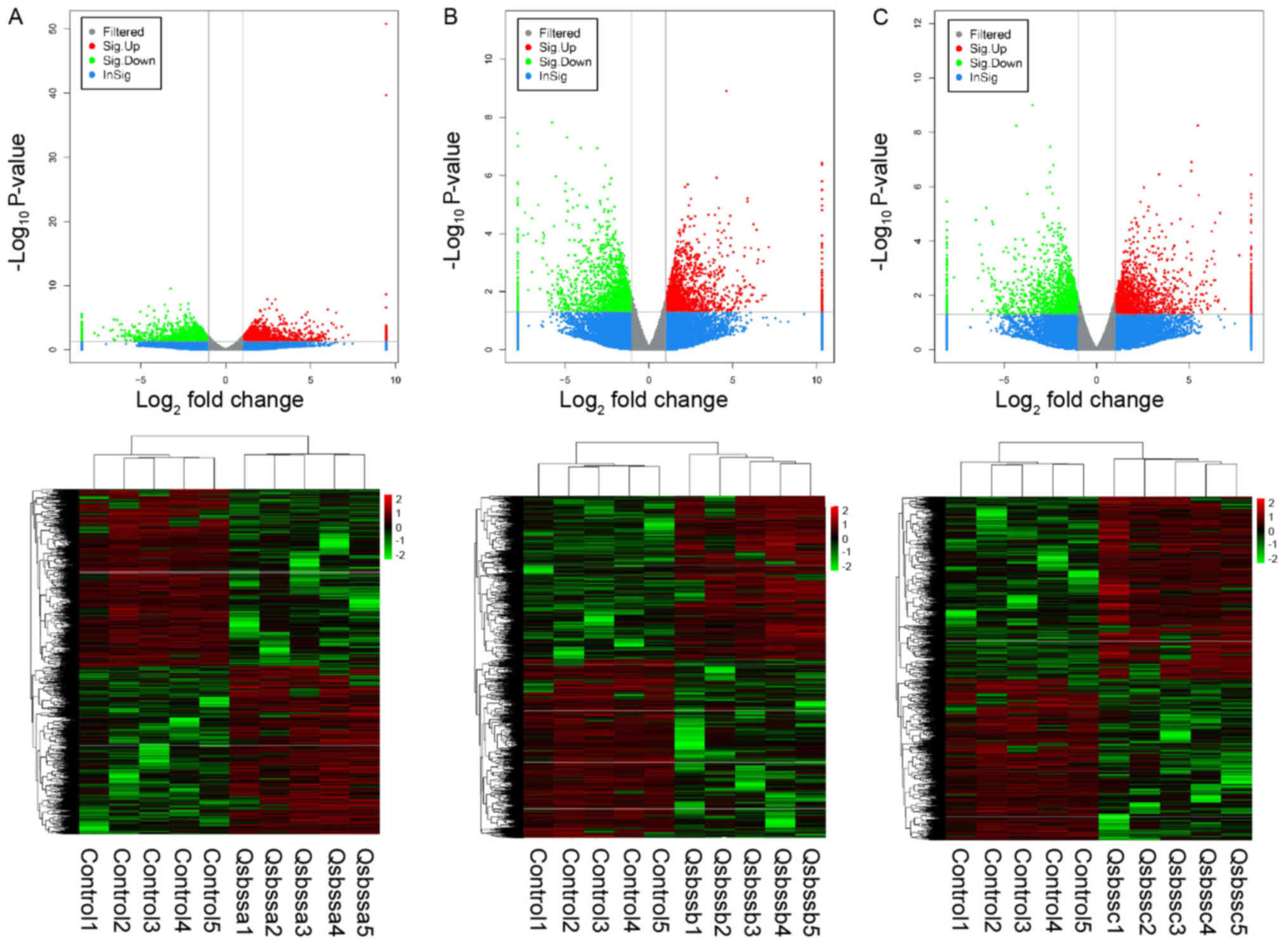
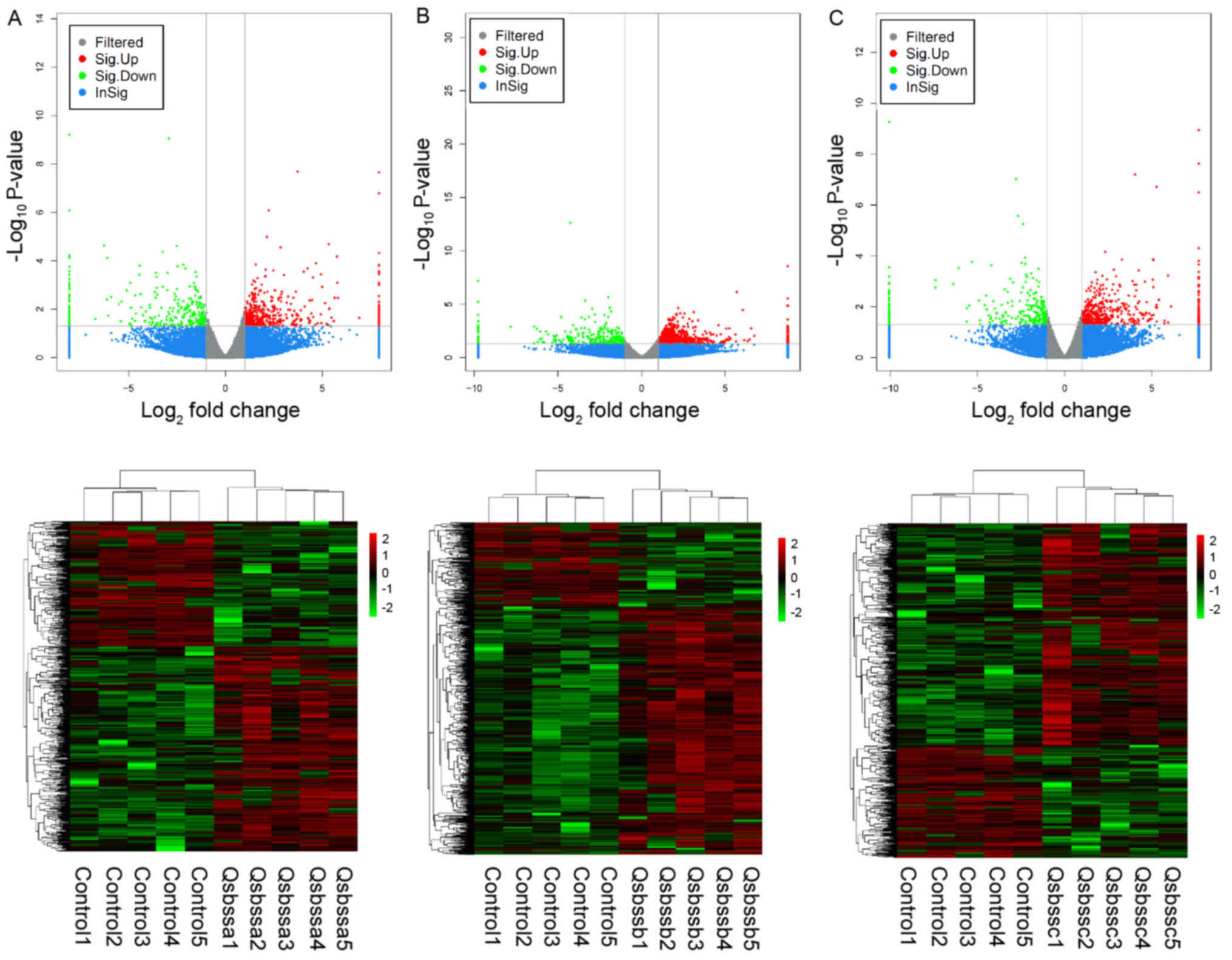
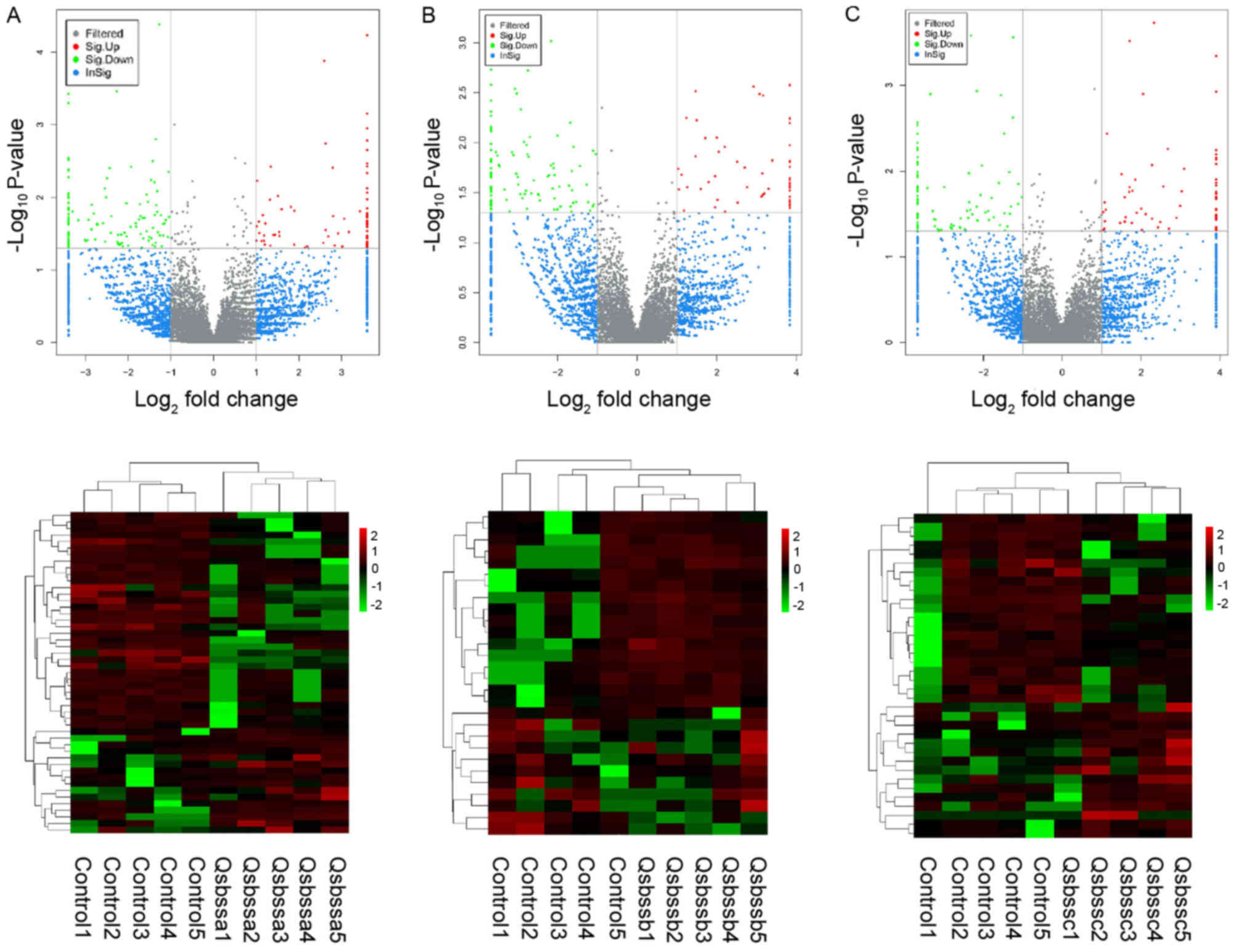
group c (RA with QSBSS vs. control). DE ncRNAs (Figs. 1 and 2) and mRNAs (Fig. 3) in the samples were displayed using
Volcano plot and clustering maps for groups a, b and c
individually. The top 10 upregulated and top 10 downregulated
lncRNAs, circRNAs and mRNAs in groups a, b and c are listed in
Tables II, III and IV, respectively. Overlapped DE lncRNAs,
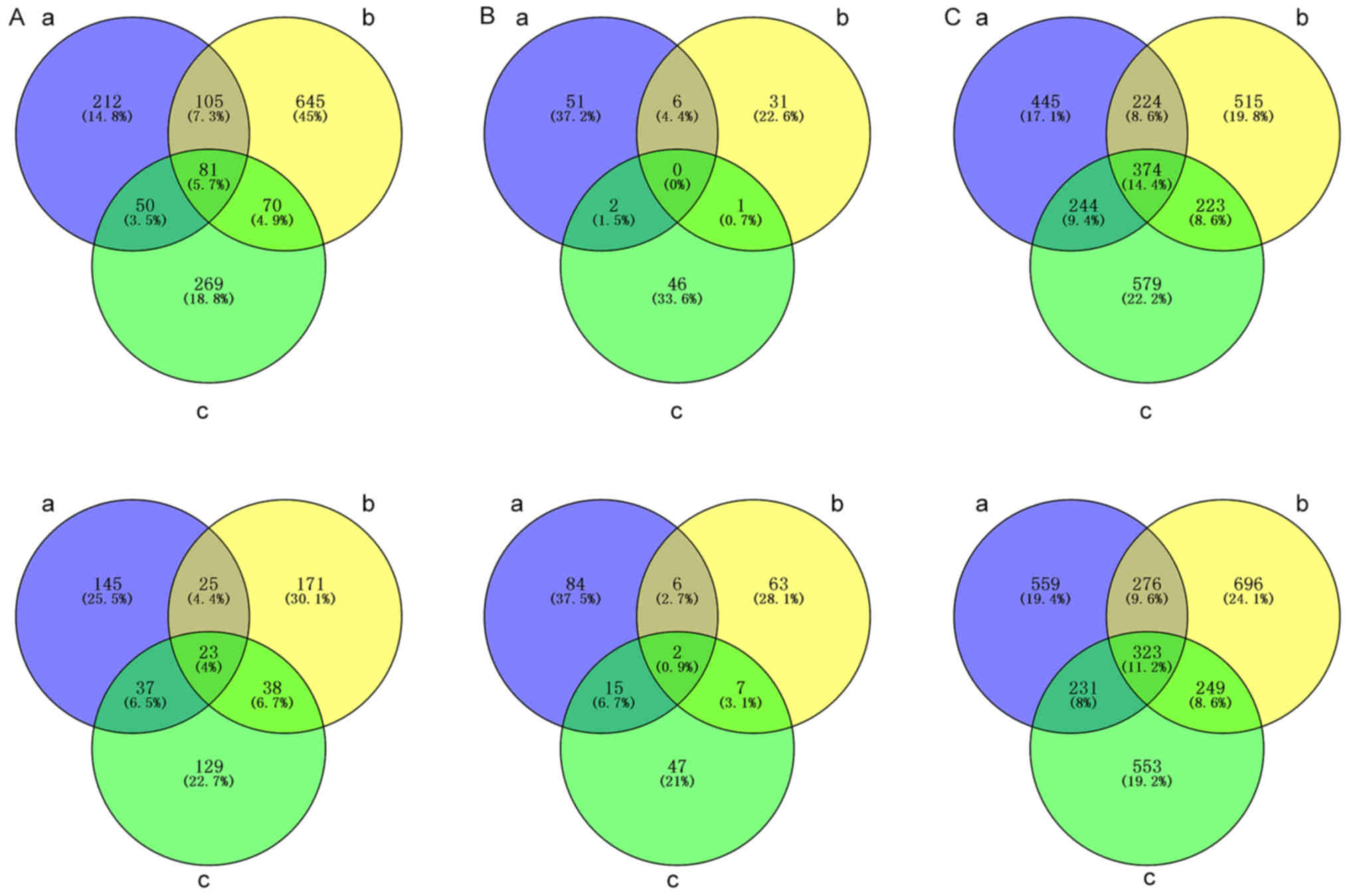
circRNAs and mRNAs among groups a, b and c were illustrated in a
Venn diagram (Fig. 4). A total of
374 up-regulated DE mRNAs, 323 down-regulated DE mRNAs, 81
up-regulated DE lncRNAs, 23 down-regulated DE lncRNAs and 2
down-regulated DE circRNAs overlapped. No up-regulated DE circRNAs
overlapped.
 | Table II.Detailed information of the top 10
upregulated and 10 downregulated lncRNAs. |
Table II.
Detailed information of the top 10
upregulated and 10 downregulated lncRNAs.
| A, Group a |
|---|
|
|---|
| Gene ID | Gene name | BM_control | BM_QSBSS | log2 (fold
change) | P-value | Regulation |
|---|
|
lnc-SUPT16H-3:1 | Novel_lncRNA | 28.45432035 | 374.5259054 | 3.718345886 |
2.06×10−8 | Up |
|
lnc-RALGAPA1-4:1 | Novel_lncRNA | 76.75550691 | 359.25044 | 2.226647757 |
8.13×10−7 | Up |
| LINC01619:1 | Novel_lncRNA | 44.81014998 | 196.9603127 | 2.136007497 |
1.03×10−5 | Up |
| lnc-PTP4A2-3:4 | Novel_lncRNA | 7.397430792 | 297.6452117 | 5.330425581 |
2.06×10−5 | Up |
| lnc-PKDCC-4:1 | Novel_lncRNA | 13.12539052 | 94.73577234 | 2.851548944 |
2.76×10−5 | Up |
| lnc-SOX4-2:1 | Novel_lncRNA | 0.187964496 | 45.41608604 | 7.916599391 |
4.77×10−5 | Up |
| LINC-PINT:20 | Novel_lncRNA | 0.68761825 | 37.12070593 | 5.7544725 |
6.62×10−5 | Up |
| lnc-FAN1-6:1 | Novel_lncRNA | 3.007431931 | 76.98804768 | 4.678030497 |
1.25×10−4 | Up |
| lnc-NEDD9-2:3 | Novel_lncRNA | 204.8829122 | 607.9154915 | 1.569071119 |
1.40×10−4 | Up |
| lnc-UACA-6:3 | Novel_lncRNA | 2.85167033 | 53.72383561 | 4.235683197 |
2.03×10−4 | Up |
| lnc-CFLAR-1:1 | Novel_lncRNA | 317.5740647 | 42.23690812 | −2.910516963 |
8.76×10−10 | Down |
|
lnc-C18orf54-3:1 | Novel_lncRNA | 17.59737855 | 0.229176766 | −6.262755929 |
2.35×10−5 | Down |
| lnc-PTPN6-1:2 | Novel_lncRNA | 73.0536666 | 12.89197843 | −2.502483005 |
2.46×10−5 | Down |
| lnc-TOB1-4:1 | Novel_lncRNA | 49.503338 | 5.280343401 | −3.228822147 |
4.21×10−5 | Down |
| lnc-NINJ2-4:3 | Novel_lncRNA | 15.78127325 | 0.229176766 | −6.105609007 |
7.60×10−5 | Down |
| lnc-VSTM5-1:12 | Novel_lncRNA | 88.5702391 | 19.58027393 | −2.177421065 |
1.42×10−4 | Down |
|
lnc-AC092329.1–1:5 | Novel_lncRNA | 20.45877718 | 0.948506969 | −4.430917734 |
1.61×10−4 | Down |
| lnc-MDM1-1:10 | Novel_lncRNA | 54.70206769 | 9.823915046 | −2.477225377 |
1.65×10−4 | Down |
|
lnc-CRYBA4-1:53 | Novel_lncRNA | 121.6228189 | 28.76734593 | −2.079909903 |
2.02×10−4 | Down |
| lnc-PTP4A2-3:3 | Novel_lncRNA | 244.8594302 | 0.916707064 | −8.061277253 |
2.65×10−4 | Down |
|
| B, Group
b |
|
| Gene ID | Gene
name |
BM_control |
BM_QSBSS | log2 (fold
change) | P-value |
Regulation |
|
| LINC-PINT:20 | Novel_lncRNA | 0.669136799 | 34.68062215 | 5.695684784 |
7.29×10−7 | Up |
| lnc-FGL2-3:1 | Novel_lncRNA | 2.497604805 | 1065.79556 | 8.737169794 |
1.34×10−5 | Up |
|
lnc-KIAA0748-2:1 | Novel_lncRNA | 38.88426047 | 178.264157 | 2.196758448 |
2.32×10−5 | Up |
| lnc-RCAN1-4:1 | Novel_lncRNA | 0.363197716 | 24.31360581 | 6.064864925 |
3.43×10−5 | Up |
|
lnc-RTEL1.1–3:5 | Novel_lncRNA | 4.653772495 | 40.51149143 | 3.121858609 |
5.39×10−5 | Up |
| lnc-INSL4-5:1 | Novel_lncRNA | 19.05513281 | 95.23213668 | 2.321268839 |
6.94×10−5 | Up |
| lnc-PKDCC-4:1 | Novel_lncRNA | 12.72486822 | 76.09097524 | 2.580074638 |
7.43×10−5 | Up |
| lnc-RIMS3-5:1 | Novel_lncRNA | 29.60597764 | 126.9453778 | 2.100247466 |
8.52×10−5 | Up |
| lnc-LYRM7-4:1 | Novel_lncRNA | 36.08770968 | 151.2244038 | 2.067111483 |
8.82×10−5 | Up |
| lnc-CTNNA3-3:1 | Novel_lncRNA | 32.24888452 | 139.4355372 | 2.112277136 |
1.02×10−4 | Up |
| lnc-CFLAR-1:1 | Novel_lncRNA | 307.456305 | 0.353369173 | −9.764989488 |
0.00×10−11 | Down |
| lnc-APEX1-3:1 | Novel_lncRNA | 211.245845 | 11.0242814 | −4.26016644 |
2.00×10−13 | Down |
|
lnc-PRKAR1A-2:1 | Novel_lncRNA | 1122.360252 | 283.5025529 | −1.985102191 |
2.10×10−6 | Down |
| lnc-RAG1-5:3 | Novel_lncRNA | 51.27926169 | 4.964879496 | −3.368544883 |
4.78×10−6 | Down |
| LINC00936:1 | Novel_lncRNA | 963.0937225 | 258.3817153 | −1.89817222 |
2.11×10−5 | Down |
| lnc-ZNF101-3:1 | Novel_lncRNA | 108.7867263 | 14.13476693 | −2.944182536 |
2.60×10−5 | Down |
| lnc-MB21D1-3:5 | Novel_lncRNA | 6778.947852 | 1925.503917 | −1.815825315 |
4.63×10−5 | Down |
|
lnc-CCDC111-2:1 | Novel_lncRNA | 87.39175981 | 12.76026571 | −2.775838883 |
9.09×10−5 | Down |
| lnc-UGDH-3:2 | Novel_lncRNA | 132.0477892 | 28.57958569 | −2.20800324 |
1.00×10−4 | Down |
| lnc-TOB1-4:1 | Novel_lncRNA | 47.99402748 | 5.477222185 | −3.131338574 |
1.43×10−4 | Down |
|
| C, Group
c |
|
| Gene ID | Gene
name |
BM_control |
BM_QSBSS | log2 (fold
change) | P-value |
Regulation |
|
|
lnc-SUPT16H-3:1 | Novel_lncRNA | 28.98663335 | 473.8065463 | 4.030838445 |
6.38×10−8 | Up |
| lnc-PTP4A2-3:4 | Novel_lncRNA | 7.51850112 | 290.4813765 | 5.271856785 |
1.90×10−7 | Up |
| PAX8-AS1:6 | Novel_lncRNA | 28.16175293 | 142.8994525 | 2.343191347 |
6.99×10−5 | Up |
|
lnc-ACTR1B-3:18 | Novel_lncRNA | 0.522432354 | 17.71173166 | 5.083317214 |
1.36×10−4 | Up |
| lnc-OR4F21-1:1 | Novel_lncRNA | 3.529033861 | 34.29076606 | 3.280474957 |
1.37×10−4 | Up |
| lnc-ANAPC7-3:1 | Novel_lncRNA | 0.511499328 | 17.62430933 | 5.106690566 |
1.47×10−4 | Up |
| SNHG4:19 | Novel_lncRNA | 5.177511537 | 36.41675096 | 2.814271445 |
3.52×10−4 | Up |
| lnc-HACL1-10:1 | Novel_lncRNA | 0.955682697 | 18.22549975 | 4.253282866 |
3.67×10−4 | Up |
| lnc-LYRM7-4:1 | Novel_lncRNA | 37.9577107 | 131.007337 | 1.787182722 |
4.35×10−4 | Up |
| lnc-UGT3A2-3:1 | Novel_lncRNA | 1.126190273 | 68.64681004 | 5.92967018 |
5.95×10−4 | Up |
| TCONS_00066371 | XLOC_034909 | 206.0849162 | 0.190001322 | −10.08301374 |
1.00×10−13 | Down |
| TCONS_00133003 | XLOC_069024 | 168.4915946 | 24.48540625 | −2.782682585 |
9.50×10−8 | Down |
| lnc-APEX1-3:1 | Novel_lncRNA | 221.6790465 | 35.26124522 | −2.652317082 |
2.66×10−6 | Down |
| lnc-GPR123-2:1 | Novel_lncRNA | 145.0012968 | 27.90207696 | −2.377621381 |
5.64×10−6 | Down |
| lnc-PTPN6-1:2 | Novel_lncRNA | 74.19783961 | 15.26041243 | −2.281583228 |
1.19×10−4 | Down |
| lnc-MBP-2:6 | Novel_lncRNA | 16.34600311 | 0.412915251 | −5.306948399 |
1.74×10−4 | Down |
| TCONS_00055344 | XLOC_028836 | 57.55245189 | 10.47564043 | −2.457838946 |
1.91×10−4 | Down |
| TCONS_00015300 | XLOC_007246 | 68.91323715 | 14.48951949 | −2.249771377 |
2.09×10−4 | Down |
| lnc-SPG7-1:6 | Novel_lncRNA | 20.96867059 | 1.140007934 | −4.201119628 |
2.34×10−4 | Down |
| TCONS_00113555 | XLOC_059030 | 13.52330416 | 0.199388024 | −6.083725124 |
2.93×10−4 | Down |
 | Table III.Detailed information of the top 10
upregulated and 10 downregulated circRNAs. |
Table III.
Detailed information of the top 10
upregulated and 10 downregulated circRNAs.
| A, Group a |
|---|
|
|---|
| Gene ID | Gene name | Type | BM_control | BM_QSBSS | log2 (fold
change) | P-value | Regulation |
|---|
|
circRNA_17075|NC_000011.10:43830976_43840064_+ | HSD17B12 |
Sense-overlapping | 1.32462559 | 7.971212934 | 2.589214633 |
1.31×10−4 | Up |
|
circRNA_11571|NC_000007.14:6046071_6050045_- | EIF2AK1 |
Sense-overlapping | 0.37603418 | 4.548890914 | 3.596579135 |
1.12×10−3 | Up |
|
circRNA_26415|NC_000020.11:37061103_37062270_- | RBL1 |
Sense-overlapping | 0.87921177 | 5.411278881 | 2.621686986 |
1.81×10−3 | Up |
|
circRNA_18831|NC_000012.12:94169153_94186473_+ | PLXNC1 |
Sense-overlapping | 127.149245 | 180.8054391 | 0.507915189 |
2.85×10−3 | Up |
|
circRNA_08586|NC_000004.12:177353308_177353728_+ | NEIL3 | Exonic | 80.5534029 | 134.3022057 | 0.737465559 |
3.36×10−3 | Up |
|
circRNA_07354|NC_000004.12:15625252_15644708_- | FBXL5 |
Sense-overlapping | 5.48874207 | 13.87619515 | 1.338064586 |
3.74×10−3 | Up |
|
circRNA_00060|NC_000001.11:2302978_2304585_+ | SKI |
Sense-overlapping | 0.67309 | 4.659019059 | 2.791154914 |
3.88×10−3 | Up |
|
circRNA_26127|NC_000020.11:3963875_3974400_- | RNF24 |
Sense-overlapping | 7.43905456 | 15.06876907 | 1.018370387 |
5.88×10−3 | Up |
|
circRNA_23305|NC_000016.10:84979189_84981994_- | ZDHHC7 |
Sense-overlapping | 2.64881177 | 7.528229138 | 1.506965212 |
9.52×10−3 | Up |
|
circRNA_07469|NC_000004.12:38790070_38791231_- | Novel_circRNA | Exonic | 17.4723667 | 28.21041441 | 0.691152823 |
9.78×10−3 | Up |
|
circRNA_11447|NC_000006.12:158583954_158589782_+ | TMEM181 |
Sense-overlapping | 36.7072718 | 15.15471652 | −1.276299027 |
4.15×10−5 | Down |
|
circRNA_08702|NC_000005.10:34179013_34182867_- | Novel_circRNA | Intergenic | 8.27722775 | 1.722922054 | −2.264290219 |
3.43×10−4 | Down |
|
circRNA_05856|NC_000003.12:51541498_51552063_+ | RAD54L2 |
Sense-overlapping | 57.6149601 | 30.5210945 | −0.916636766 |
9.97×10−4 | Down |
|
circRNA_26641|NC_000020.11:56381433_56384324_- | AURKA |
Sense-overlapping | 13.2686213 | 5.198360651 | −1.351889836 |
1.58×10−3 | Down |
|
circRNA_09849|NC_000005.10:155854549_155870424_+ | Novel_circRNA | Intergenic | 10.8990358 | 4.058654063 | −1.425127223 |
3.14×10−3 | Down |
|
circRNA_14375|NC_000009.12:33960826_33963791_- | UBAP2 |
Sense-overlapping | 10.1539178 | 2.658377854 | −1.933418403 |
3.77×10−3 | Down |
|
circRNA_05293|NC_000003.12:3167634_3175269_- | CRBN |
Sense-overlapping | 6.28998378 | 1.166948766 | −2.430315076 |
3.93×10−3 | Down |
|
circRNA_18046|NC_000012.12:10386903_10393867_- | KLRC4-KLRK1 |
Sense-overlapping | 24.8329163 | 11.97296213 | −1.052473572 |
4.43×10−3 | Down |
|
circRNA_17571|NC_000011.10:86250296_86256512_+ | EED |
Sense-overlapping | 4.04730028 | 0.383577527 | −3.399369787 |
5.26×10−3 | Down |
|
circRNA_17157|NC_000011.10:57491224_57491862_- | SLC43A1 |
Sense-overlapping | 4.85469127 | 0.862811798 | −2.492261744 |
5.42×10−3 | Down |
|
| B, Group
b |
|
| Gene ID | Gene
name | Type |
BM_control |
BM_QSBSS | log2 (fold
change) | P-value |
Regulation |
|
|
circRNA_21868|NC_000015.10:63213989_63214386_+ | RAB8B | Intronic | 0.34767227 | 4.947228119 | 3.830820527 |
2.62×10−3 | Up |
|
circRNA_05307|NC_000003.12:5170503_5174414_+ | ARL8B |
Sense-overlapping | 0.69534454 | 5.271795149 | 2.922494414 |
2.74×10−3 | Up |
|
circRNA_11390|NC_000006.12:154212770_154223243_- | IPCEF1 |
Sense-overlapping | 4.98927089 | 13.78793369 | 1.466505359 |
3.04×10−3 | Up |
|
circRNA_00019|NC_000001.11:1669664_1734835_- | SLC35E2B |
Sense-overlapping | 1.0430168 | 8.760815597 | 3.070302787 |
3.25×10−3 | Up |
|
circRNA_07459|NC_000004.12:38035588_38054338_+ | TBC1D1 |
Sense-overlapping | 0.45711334 | 4.093290549 | 3.162637257 |
3.36×10−3 | Up |
|
circRNA_17230|NC_000011.10:65430769_65437657_+ | Novel_circRNA | Intergenic | 5.53385736 | 13.02586425 | 1.235021733 |
5.65×10−3 | Up |
|
circRNA_07354|NC_000004.12:15625252_15644708_- | FBXL5 |
Sense-overlapping | 5.05562864 | 14.20073669 | 1.490003375 |
5.95×10−3 | Up |
|
circRNA_06938|NC_000003.12:182961226_182965753_- | DCUN1D1 |
Sense-overlapping | 1.48698271 | 5.932368937 | 1.996220447 |
8.90×10−3 | Up |
|
circRNA_26020|NC_000019.10:52588532_52592228_+ | Novel_circRNA | Intergenic | 2.46658164 | 8.047647043 | 1.706053993 |
9.01×10−3 | Up |
|
circRNA_17075|NC_000011.10:43830976_43840064_+ | HSD17B12 |
Sense-overlapping | 1.23520758 | 5.672992987 | 2.199356575 |
1.10×10−2 | Up |
|
circRNA_23131|NC_000016.10:68190768_68191775_+ | NFATC3 | Exonic | 8.7889668 | 1.963322605 | −2.162396327 |
9.56×10−4 | Down |
|
circRNA_10301|NC_000006.12:24416397_24418578_+ | MRS2 |
Sense-overlapping | 6.05013791 | 0.901748722 | −2.746170651 |
1.88×10−3 | Down |
|
circRNA_11368|NC_000006.12:152329730_152331890_- | SYNE1 | Exonic | 4.73930706 | 0.565708258 | −3.066545998 |
2.89×10−3 | Down |
|
circRNA_26904|NC_000021.9:39218152_39218660_- | BRWD1 |
Sense-overlapping | 4.60809631 | 0.565708258 | −3.026040733 |
3.20×10−3 | Down |
|
circRNA_15770|NC_000010.11:31908172_31910563_- | ARHGAP12 |
Sense-overlapping | 38.5982122 | 20.88132193 | −0.88632098 |
4.47×10−3 | Down |
|
circRNA_20028|NC_000013.11:95161189_95170628_- | ABCC4 |
Sense-overlapping | 4.31735406 | 0.568986286 | −2.923681624 |
4.66×10−3 | Down |
|
circRNA_26634|NC_000020.11:51516784_51524110_- | NFATC2 |
Sense-overlapping | 8.9643749 | 2.800608967 | −1.678462422 |
6.29×10−3 | Down |
|
circRNA_03181|NC_000002.12:32377588_32395593_+ | BIRC6 |
Sense-overlapping | 4.23291619 | 0.332762435 | −3.669087438 |
6.94×10−3 | Down |
|
circRNA_12096|NC_000007.14:72830692_72831798_+ | Novel_circRNA | Intergenic | 7.00764767 | 1.755228151 | −1.997271671 |
8.52×10−3 | Down |
|
circRNA_06618|NC_000003.12:143985543_143989837_+ | C3orf58 |
Sense-overlapping | 4.51771847 | 0.699135631 | −2.691950103 |
8.72×10−3 | Down |
|
| C, Group
c |
|
| Gene ID | Gene
name | Type |
BM_control |
BM_QSBSS | log2 (fold
change) | P-value |
Regulation |
|
|
circRNA_15618|NC_000010.11:24629996_24635103_- | ARHGAP21 |
Sense-overlapping | 1.88682423 | 9.46047819 | 2.325953078 |
1.86×10−4 | Up |
|
circRNA_12051|NC_000007.14:66286511_66286709_+ | TPST1 | Exonic | 3.05288673 | 10.0082037 | 1.712937089 |
3.04×10−4 | Up |
|
circRNA_09327|NC_000005.10:95755396_95763620_+ | RHOBTB3 |
Sense-overlapping | 25.9992577 | 45.80635744 | 0.817077408 |
1.10×10−3 | Up |
|
circRNA_12851|NC_000007.14:152263016_152315338_- | KMT2C |
Sense-overlapping | 1.78037988 | 7.382084284 | 2.051843109 |
1.26×10−3 | Up |
|
circRNA_22782|NC_000016.10:23988508_24035547_+ | PRKCB |
Sense-overlapping | 6.91237695 | 15.18024088 | 1.134940885 |
3.68×10−3 | Up |
|
circRNA_25741|NC_000019.10:21987216_21988909_- | ZNF208 |
Sense-overlapping | 0.67237467 | 4.30048113 | 2.677160804 |
5.51×10−3 | Up |
|
circRNA_03640|NC_000002.12:61498673_61499894_- | XPO1 |
Sense-overlapping | 1.16372918 | 5.641809329 | 2.277402555 |
8.49×10−3 | Up |
|
circRNA_07695|NC_000004.12:55893552_55899884_+ | EXOC1 |
Sense-overlapping | 0.35072822 | 2.994007933 | 3.093652605 |
9.37×10−3 | Up |
|
circRNA_03222|NC_000002.12:32633876_32640410_+ | TTC27 |
Sense-overlapping | 2.93624913 | 8.323607273 | 1.503234514 |
1.08×10−2 | Up |
|
circRNA_21325|NC_000015.10:34860042_34867609_- | AQR |
Sense-overlapping | 1.16372918 | 4.197502539 | 1.850775843 |
1.25×10−2 | Up |
|
circRNA_18044|NC_000012.12:10386903_10390027_- | KLRK1 |
Sense-overlapping | 9.31112428 | 1.861257049 | −2.322678066 |
2.62×10−4 | Down |
|
circRNA_18046|NC_000012.12:10386903_10393867_- | KLRC4-KLRK1 |
Sense-overlapping | 23.1502828 | 9.766171882 | −1.245164743 |
2.75×10−4 | Down |
|
circRNA_17950|NC_000012.12:350621_356537_- | KDM5A |
Sense-overlapping | 5.35061004 | 0.47990746 | −3.478875243 |
5.00×10−4 | Down |
|
circRNA_01807|NC_000001.11:155767425_155773210_- | GON4L |
Sense-overlapping | 7.78597137 | 1.731447168 | −2.168898673 |
1.16×10−3 | Down |
|
circRNA_05316|NC_000003.12:9388087_9390497_- | Novel_circRNA | Intergenic | 4.86113657 | 0.47990746 | −3.340465522 |
1.27×10−3 | Down |
|
circRNA_09849|NC_000005.10:155854549_155870424_+ | Novel_circRNA | Intergenic | 10.1352809 | 3.446893745 | −1.556017338 |
1.29×10−3 | Down |
|
circRNA_11131|NC_000006.12:130175841_130184623_- | SAMD3 |
Sense-overlapping | 14.8131577 | 6.226198349 | −1.25045577 |
2.38×10−3 | Down |
|
circRNA_15991|NC_000010.11:67988663_68014186_- | HERC4 |
Sense-overlapping | 8.52545734 | 3.069284144 | −1.473875016 |
3.67×10−3 | Down |
|
circRNA_24400|NC_000017.11:64052780_64053371_- | ERN1 |
Sense-overlapping | 4.25072746 | 0.333416255 | −3.672313416 |
4.69×10−3 | Down |
|
circRNA_12107|NC_000007.14:73225711_73226051_+ | Novel_circRNA | Intergenic | 4.18052411 | 0.328072145 | −3.671598812 |
6.84×10−3 | Down |
 | Table IV.Detailed information of the top 10
upregulated and 10 downregulated mRNAs. |
Table IV.
Detailed information of the top 10
upregulated and 10 downregulated mRNAs.
| A, Group a |
|---|
|
|---|
| Gene ID | Gene name | Gene location | BM_control | BM_QSBSS | log2 (fold
change) | P-value | Regulation |
|---|
|
| XM_011510258.1 | LOC100996763 | Chr
1:148595857-148712520 | 128.7757693 | 709.5584027 | 2.462060276 |
1.33×10−8 | Up |
| XM_011527030.1 | PLAUR | Chr
19:43646095-43670547 | 20.98685367 | 159.9897378 | 2.930421569 |
1.43×10−8 | Up |
| NM_014779.3 | TSC22D2 | Chr
3:150408335-150466431 | 32.78009306 | 192.5260299 | 2.554161659 |
3.34×10−7 | Up |
| XM_011533056.1 | SLC8A1 | Chr
2:40097270-40611053 | 0.531373673 | 34.14398459 | 6.005760869 |
5.52×10−7 | Up |
| XM_011539511.1 | RAB11FIP2 | Chr
10:118004916-118046603 | 10.20389314 | 82.18530749 | 3.009760806 |
6.06×10−7 | Up |
| NM_001190794.1 | NCF2 | Chr
1:183555563-183590876 | 486.8308955 | 1954.560939 | 2.005351932 |
7.19×10−7 | Up |
| XM_011541247.1 | AMPD2 | Chr
1:109616104-109632051 | 155.6506686 | 651.8712724 | 2.066275324 |
1.11×10−6 | Up |
| NM_145197.2 | LIPT1 | Chr
2:99154955-99163157 | 1.075086932 | 30.27521872 | 4.815612156 |
1.43×10−6 | Up |
| XM_011547051.1 | LILRB2 | Chr
19:54273821-54281184 | 0.503235941 | 46.09541145 | 6.517244373 |
1.68×10−6 | Up |
| XM_011515764.1 | DBF4 | Chr
7:87876216-87909541 | 4.68292893 | 47.08883617 | 3.329902018 |
4.28×10−6 | Up |
| XM_005271698.1 | MSANTD4 | Chr
11:105995623-106022403 | 194.4517168 | 20.76045887 | −3.227501735 |
2.96×10−10 | Down |
| XM_006714859.2 | GRK6 | Chr
5:177403204-177442901 | 1476.118081 | 321.2056531 | −2.200238944 |
6.66×10−8 | Down |
| XM_011523632.1 | SMYD4 | Chr
17:1779485-1830634 | 73.7875596 | 6.195994556 | −3.57396982 |
1.05×10−7 | Down |
| NM_001242851.1 | RNF146 | Chr
6:127266610-127288567 | 371.8379549 | 80.23879516 | −2.212302191 |
4.86×10−7 | Down |
| NM_001032373.1 | ZNF226 | Chr
19:44165073-44178381 | 170.6048738 | 26.99739109 | −2.659766959 |
5.40×10−7 | Down |
| NM_001243775.1 | LIMA1 | Chr
12:50175788-50283546 | 41.82845265 | 2.271853512 | −4.202542911 |
8.07×10−7 | Down |
| XM_006717434.2 | KIN | Chr
10:7750962-7787981 | 82.05054397 | 7.772315863 | −3.400096461 |
9.81×10−7 | Down |
| NM_001286646.1 | SLC45A4 | Chr
8:141207166-141308305 | 338.7563764 | 79.07800005 | −2.098899813 |
2.32×10−6 | Down |
| NM_002250.2 | KCNN4 | Chr
19:43766533-43781257 | 148.593054 | 28.77836175 | −2.368310307 |
3.38×10−6 | Down |
| NM_001698.2 | AUH | Chr
9:91213815-91361913 | 28.1687429 | 1.14588383 | −4.619562486 |
4.59×10−6 | Down |
|
| B, group
b |
|
| Gene ID | Gene
name | Gene
location |
BM_control |
BM_QSBSS | log2 (fold
change) | P-value |
Regulation |
|
| NM_001291963.1 | DAP | Chr
5:10679230-10761272 | 3.083042965 | 75.88918508 | 4.621467403 |
1.23×10−9 | Up |
| XM_005274953.3 | HLA-DQA1 | Chr
6:32628179-32647062 | 0.167025521 | 213.6609304 | 10.32103885 |
4.22×10−7 | Up |
| NM_001098272.2 | HMGCS1 | Chr
5:43289395-43313512 | 2.515427217 | 41.20482768 | 4.033938026 |
1.18×10−6 | Up |
| NM_173214.2 | NFAT5 | Chr
16:69565094-69704666 | 137.1028468 | 614.3398191 | 2.163778369 |
2.45×10−6 | Up |
| XM_005271523.3 | ZBTB44 | Chr
11:130226677-130314686 | 45.08295693 | 256.2092972 | 2.50666878 |
6.35×10−6 | Up |
| NM_001282441.1 | GNA12 | Chr
7:2728112-2844324 | 0.341979162 | 20.09576134 | 5.876839008 |
7.94×10−6 | Up |
| NM_014779.3 | TSC22D2 | Chr
3:150408335-150466431 | 31.77170716 | 167.9742018 | 2.402425158 |
1.05×10−5 | Up |
| XM_011514978.1 | ABCC10 | Chr
6:43427366-43450430 | 1.615876704 | 29.14153126 | 4.172687661 |
1.32×10−5 | Up |
| XM_006717383.2 | CREM | Chr
10:35126791-35212958 | 2.740860538 | 32.21154032 | 3.554876823 |
2.26×10−5 | Up |
| XM_011521157.1 | DENND4A | Chr
15:65658046-65792293 | 1.968440767 | 30.31479595 | 3.944896906 |
2.29×10−5 | Up |
| NM_003037.3 | SLAMF1 | Chr
1:160608100-160647295 | 42.62133994 | 0.782999144 | −5.766421409 |
1.49×10−8 | Down |
| NM_153373.3 | PHYKPL | Chr
5:178208497-178232791 | 58.97775271 | 2.014078279 | −4.87197919 |
4.93×10−8 | Down |
| XM_005262404.3 | HTATSF1 | Chr
X:136497079-136512346 | 51.81171726 | 3.116477327 | −4.055290279 |
1.14×10−7 | Down |
| NM_001099696.2 | REPIN1 | Chr
7:150368189-150374044 | 258.5384205 | 43.226086 | −2.580404575 |
4.48×10−7 | Down |
| XM_011512333.1 | STAG1 | Chr
3:136336233-136752403 | 27.59010032 | 0.596392567 | −5.531744615 |
1.06×10−6 | Down |
| XM_006713188.2 | SHISA5 | Chr
3:48467798-48504826 | 587.4466021 | 123.8085754 | −2.246346475 |
1.24×10−6 | Down |
| XM_011519861.1 | C11orf58 | Chr
11:16613132-16756881 | 668.0596614 | 132.9186866 | −2.329433007 |
1.96×10−6 | Down |
| XM_011523594.1 | TBCD | Chr
17:82752064-82945922 | 39.27908367 | 1.676943758 | −4.549855064 |
2.95×10−6 | Down |
| XM_005252018.1 | ODF2 | Chr
9:128455186-128501292 | 31.19036594 | 0.990939876 | −4.976159143 |
3.52×10−6 | Down |
| XM_006714696.2 | CAST | Chr
5:96525267-96779595 | 33.84494268 | 1.942626191 | −4.122860051 |
4.68×10−6 | Down |
|
| C, group
c |
|
| Gene ID | Gene
name | Gene
location |
BM_control |
BM_QSBSS | log2 (fold
change) | P-value |
Regulation |
|
| XM_011534376.1 | SCFD2 | Chr
4:52872982-53366075 | 1.559496265 | 69.36997091 | 5.475159277 |
5.61×10−9 | Up |
| XM_005259105.1 | GRAMD1A | Chr
19:34994784-35026471 | 2.850311301 | 99.56136105 | 5.126394554 |
1.23×10−7 | Up |
| XM_011515624.1 | PHF14 | Chr
7:10973872-11169630 | 0.979140157 | 33.97667549 | 5.1168855 |
2.58×10−7 | Up |
| XM_011547517.1 | FCAR | Chr
19:54874248-54890472 | 6.35636695 | 66.26373583 | 3.381945224 |
3.46×10−7 | Up |
| XM_011529325.1 | SIGLEC1 | Chr
20:3686970-3707128 | 5.397654189 | 124.1005265 | 4.523032875 |
9.33×10−7 | Up |
| NM_001276286.1 | BBX | Chr
3:107522936-107811324 | 53.75421935 | 316.8543227 | 2.559369792 |
1.05×10−6 | Up |
| XM_006722540.2 | TCF4 | Chr
18:55222331-55664787 | 0.55892364 | 25.69005846 | 5.522415166 |
2.04×10−6 | Up |
| NM_014779.3 | TSC22D2 | Chr 3:
150408335-150466431 | 33.32937132 | 172.858216 | 2.374723167 |
3.63×10−6 | Up |
| XM_011511806.1 | IL1R2 | Chr
2:101991844-102028544 | 11.97084228 | 84.73814635 | 2.823486905 |
5.41×10−6 | Up |
| XM_011537996.1 | GRASP | Chr
12:52006940-52015889 | 13.22125101 | 90.58703153 | 2.776445835 |
7.61×10−6 | Up |
| NM_184234.2 | RBM39 | Chr
20:35701347-35742312 | 311.6823768 | 25.6690681 | −3.601973759 |
1.00×10−12 | Down |
| XM_006711370.2 | UCHL5 | Chr
1:193012250-193060080 | 75.60834192 | 0.585379421 | −7.013029579 |
1.78×10−11 | Down |
| NM_001177676.1 | GPR68 | Chr
14:91232532-91253925 | 144.6164402 | 13.2274947 | −3.450619823 |
9.93×10−10 | Down |
| NM_001242599.1 | RBM39 | Chr
20:35701347-35742312 | 63.54456584 | 3.153938054 | −4.332542525 |
5.76×10−9 | Down |
| XM_005259103.2 | GRAMD1A | Chr
19:34994784-35026471 | 582.9135127 | 103.4609801 | −2.494195081 |
3.41×10−8 | Down |
| NM_021983.4 | HLA-DRB4 | Chr
1:32542598-32557561 | 1126.115406 | 223.72947 | −2.331527476 |
1.62×10−7 | Down |
| NM_001162953.2 | SYTL2 | Chr
11:85694224-85811159 | 231.7902901 | 40.56415454 | −2.514542807 |
2.95×10−7 | Down |
| NM_001287340.1 | NADK2 | Chr
5:36192592-36242279 | 179.4003614 | 31.63501319 | −2.503588694 |
1.02×10−6 | Down |
| XM_011547738.1 | LOC105369230 | Chr
1:32723350-32757450 | 4437.862978 | 689.6928005 | −2.685839312 |
1.07×10−6 | Down |
| XM_005266520.2 | ZMYM2 | Chr
13:19958670-20091829 | 207.5631578 | 40.26949202 | −2.365791212 |
1.76×10−6 | Down |
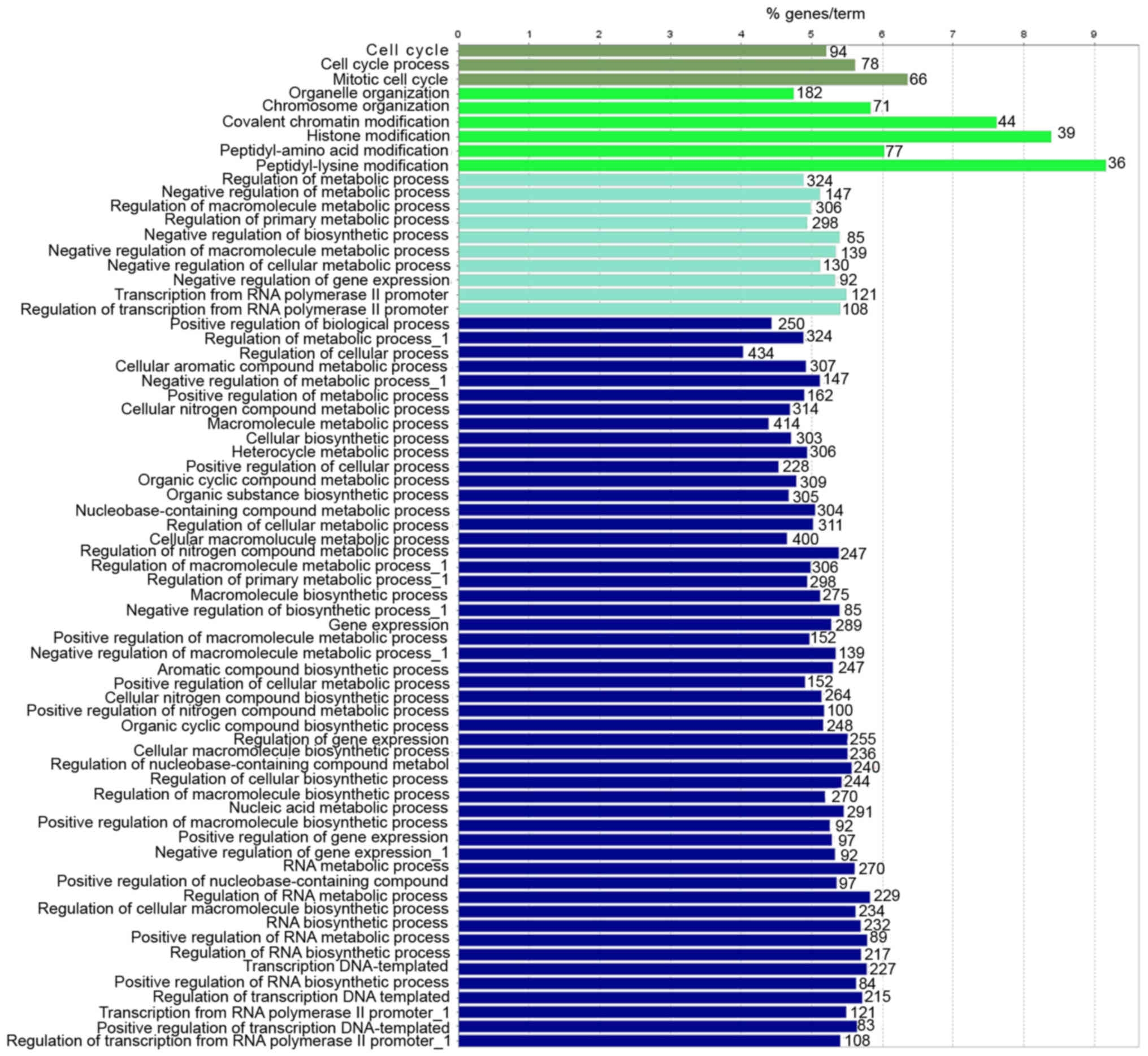
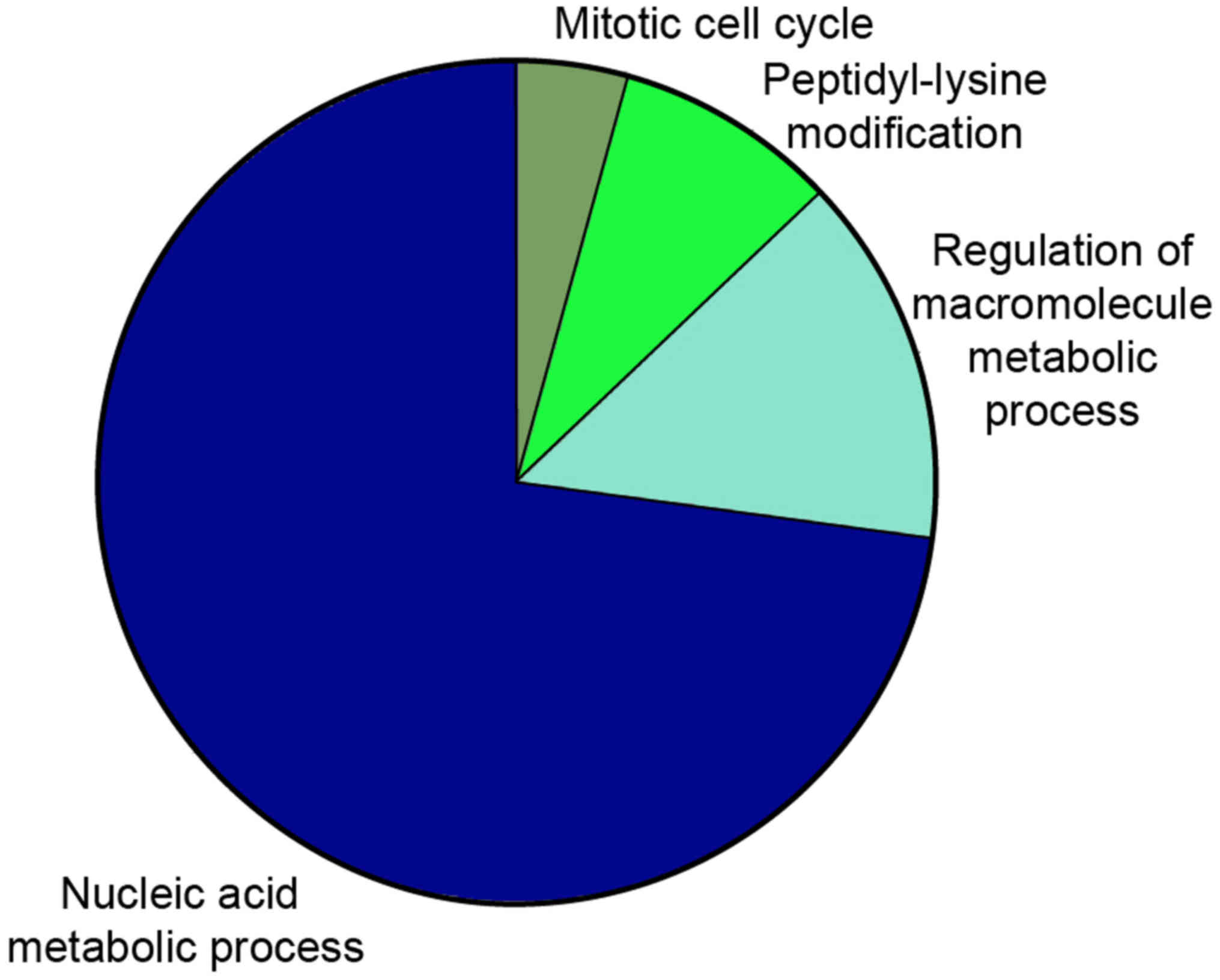
Functional prediction of ncRNAs
To further determine the possible functions of the
overlapped DE RNAs and investigate the association between the
functional groups and their underlying mechanisms in the biological
networks, the Cytoscape plugin ClueGo was utilized. Significant GO
terms in the categories MF and BP were specifically associated with
nucleic acid metabolic process, regulation of macromolecule
metabolic process, peptidyl-lysine modification and mitotic cell
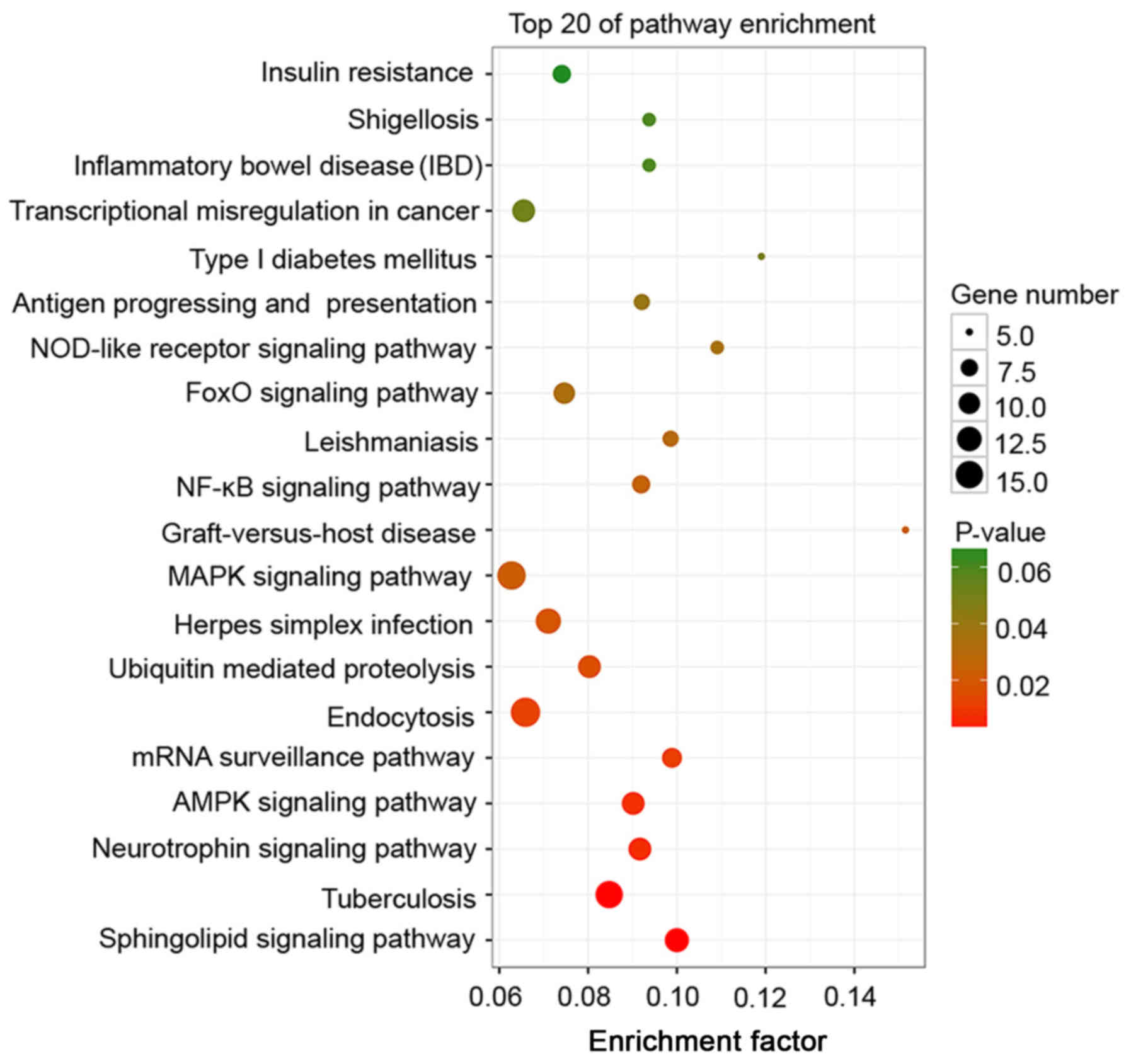
cycle (Figs. 5 and 6). Of note, these DE RNAs coordinate with
each other to perform their functions and the relevant major
biochemical pathways and signal transduction pathways were
predicted by enrichment analysis based on the KEGG database. A
scatterplot of the DE RNAs was employed to demonstrate the KEGG
enrichment analysis. In this plot, the degree of KEGG enrichment is
assessed by the enrichment factor, P-value and number of genes. A
greater enrichment factor is associated with a P-value closer to
zero and a greater number of genes, and therefore a more
significantly enriched pathway. When the data were analyzed
regarding the co-expression of genes of DE lncRNAs, DE mRNAs and
target genes of DE circRNAs, the most significantly enriched KEGG
pathways were sphingolipid, neurotrophin, AMP-activated protein
kinase (AMPK), mRNA surveillance pathway and endocytosis (Fig. 7). These major BPs and signal
transduction pathways provide insight regarding further directions
of research on the biological basis of QSBSS.
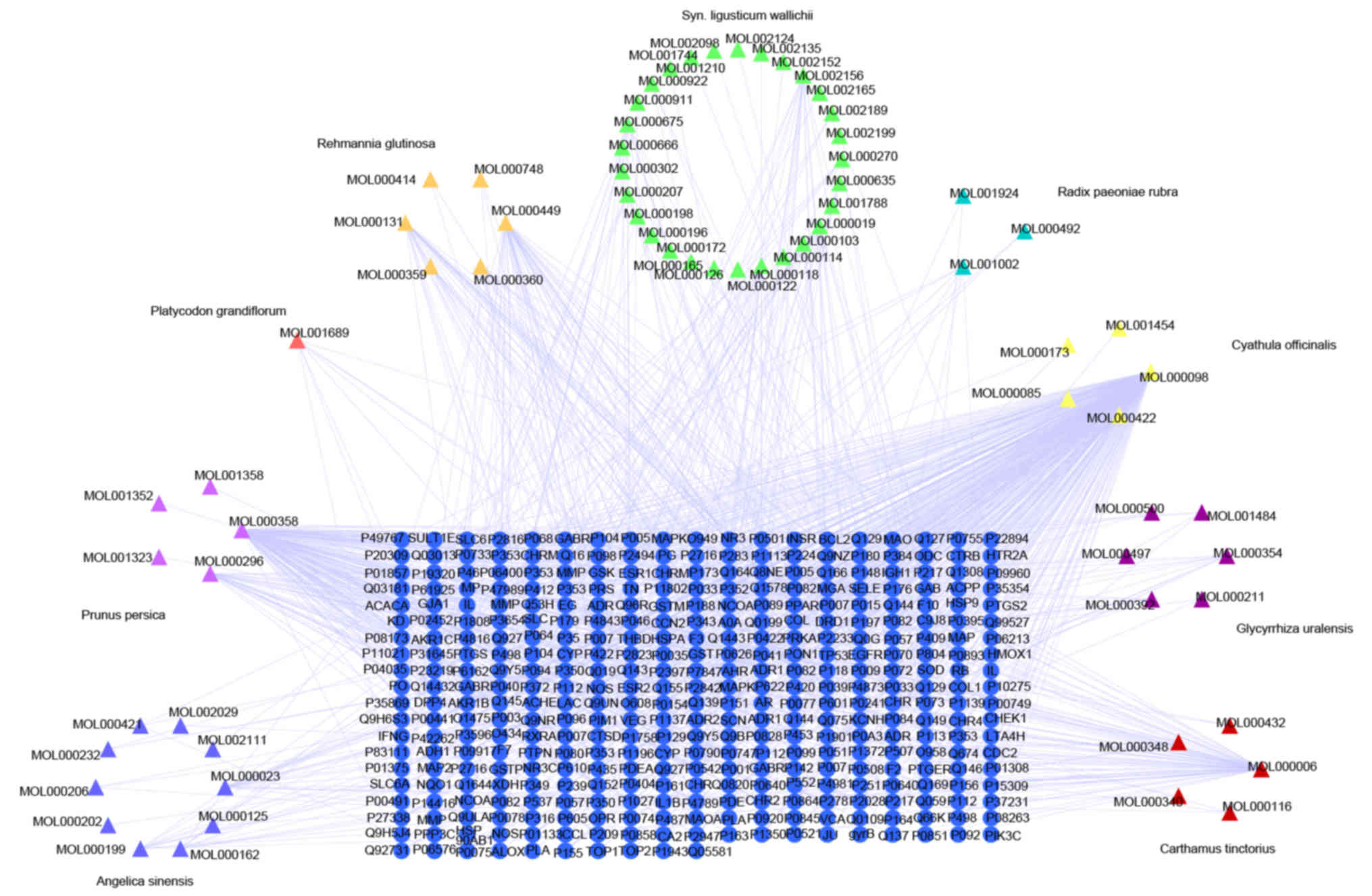
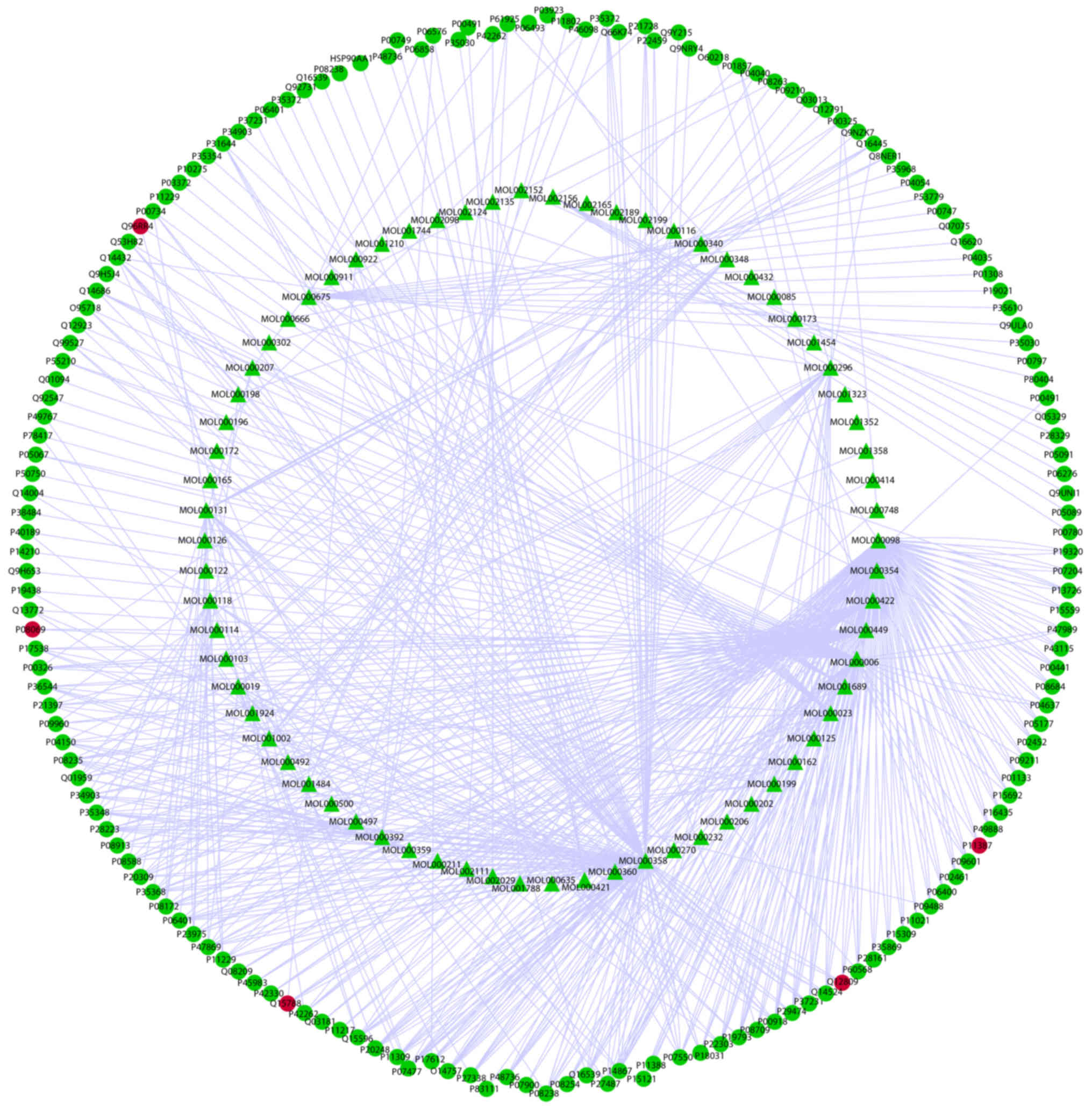
Merged network of DE RNAs and target
fishing
As presented in the network for compounds and
targets (Fig. 8) and the merged
network (Fig. 9), the targets of
active components of Xuefu Zhuyu decoction partly covered the
targets in the DE RNAs of QSBSS discovered by sequencing. The
shared targets of QSBSS and Xuefu Zhuyu Decoction were interleukin
1β, DNA topoisomerase I, potassium voltage-gated channel subfamily
H member 2, nuclear receptor coactivator 1, mitogen-activated
protein kinase 8, insulin-like growth factor 1 receptor and
calcium/calmodulin-dependent protein kinase kinase 2.
Discussion
In the present study, expression profiles of
lncRNAs, circRNAs and mRNA were obtained by next-generation
sequencing and compared in patients with QSBSS and three different
types of disease (CHD, CG or RA vs. normal controls). A total of
104 lncRNAs, 2 circRNAs and 697 mRNAs were finally identified to be
significantly DE among groups a, b and c in a Venn diagram. These
DE RNAs were subsequently displayed in a Volcano Plot and
integrated into hierarchical categories according to heat maps and
hierarchical clustering. GO and KEGG analyses were used to predict
their relevant functions by ClueGo. A network pharmacology study
was also performed to preliminarily confirm the DE RNAs with the
hypothesis that the DE RNAs of QSBSS may be regulated by the TCM
herbs with efficacy in treating QSBSS. These DE RNAs and their
corresponding pathways may be considered highly relevant to QSBSS,
and may contribute to the explanation of Zheng and prediction of
therapeutic targets of the TCM formula.
QSBSS is a common Zheng that is thought to be
associated with the manifestation of numerous types of disease,
including CHD, CG and RA, and it is always used to define the
condition of unsmooth flow of Qi and blood (36). In addition to distending pain in a
fixed place, one symptom of QSBSS is emotional problems, including
irritability and depression (21).
The development of QSBSS is complex (37) and the mechanism remains largely
elusive. The competing endogenous (ce)RNA hypothesis posits that
miRNA may bind to an argonaute protein and form the miRNA-guided
RNA-induced silencing complex (RISC) to target the relevant mRNA,
leading to accelerated degradation, blocked translation and
decreased expression, while ceRNA with shared target sites competes
for miRNA targeting and binding in the RISC, thus sequestering the
miRNA activity, resulting in repression of the expression the
target gene of the respective miRNA (14). Furthermore, lncRNA, circRNA and
pseudogene transcripts have gained considerable attention as they
may work together to regulate ceRNA mechanisms. Despite the growing
interest in exploring the biological basis of Zheng from DNA and
RNA (38–41), the molecular mechanisms underlying
the interaction of ncRNAs and mRNAs in QSBSS have remained largely
elusive. Therefore, for the first time, the present study provided
and compared expression profiles of lncRNAs, circRNAs and mRNAs in
QSBSS obtained by next-generation sequencing. These pioneering
discoveries may deepen the current understanding of the mechanisms
of Zheng as well as provide a foundation for further scientific
research into the molecular basis of TCM theory.
Previous studies have indicated the links between
QSBSS and the processes of inflammation, immune response and
sympathetic regulation (42–44). Of note, glycoprotein containing
disulfide bonds, G protein-coupled receptor signaling pathways and
catecholamine neurotransmitter activity were identified to
constitute the biological basis of QSBSS through methods of data
mining, which integrated the results of nearly all of the previous
research on QSBSS (45). Another
previous study suggested that QSBSS patients suffered from an
imbalance of lower pneumogastric nerve activity and higher
sympathetic nerve activity, as indicated by a heart rate
variability analysis (43). Although
evidence for the biological basis of QSBSS has accumulated, no
comprehensive analysis of the ncRNA and mRNA profile in QSBSS has
been performed to date, to the best of our knowledge. Thus,
next-generation sequencing was applied in the present study to
analyze DE ncRNAs and mRNAs in patients with QSBSS.
Bioinformatics analysis of the RNA sequencing
results of the present study indicated that DE RNAs in QSBSS were
most significantly enriched in pathways including the sphingolipid
signaling pathway, the neurotrophin signaling pathway, AMPK and
endocytosis. These results imply that QSBSS is not only associated
with an imbalance of neural function, but also with a deregulation
of energy metabolism. Specifically, sphingomyelin and its metabolic
products are known to have second messenger functions in a variety
of cellular signaling pathways (46,47).
Among its metabolic products, ceramide and S1P regulate cellular
responses to stress, with generally opposing effects (48,49).
Neurotrophins are a family of trophic factors involved in the
differentiation and survival of neural cells, which are central for
the development of diseases including hereditary sensory and
autonomic neuropathy, as well as congenital pain insensitivity with
anhidrosis (50). Endocytosis is a
mechanism for cells to remove ligands, nutrients, plasma membrane
proteins and lipids from the cell surface, which is essential for
the advance of hereditary sensory and autonomic neuropathy
(51). AMPK is a serine threonine
kinase that is highly conserved through evolution and the AMPK
system acts as a sensor of the cellular energy status (52,53).
These functions of enriched pathways may be indicative of why QSBSS
frequently manifests as pain accompanied with obscure sensations,
including a distending and tingling sensation, as well as emotional
problems, including irritability and depression (17). Of note, QSBSS is a stagnation of Qi
and the flow of blood, which is known to carry the substance,
including oxygen and energy in the body (54). Hence, it is likely that the
biological basis of QSBSS may be directly associated with a
dysfunction of energy metabolism and the autonomic nervous system,
which requires more in-depth study in the future.
To the best of our knowledge, the present study was
the first to explore the biological basis of Zheng in subjects with
different diseases rather than different Zheng in one disease and
it was attempted to interpret QSBSS, one of the common Zheng, from
the perspective of ncRNAs and mRNAs, which were determined by
next-generation sequencing technology. It is worth mentioning that
the grouping design in the present study may be implemented to
assess any type of Zheng in further studies. However, one of the
limitations of the present study is the lack of validation of DE
RNAs by quantitative (q)PCR as the cohort size was very small and
qPCR analysis was not performed, which is likely to be resolved in
a future study. Besides, the inter-group and inter-patient
variability should also be analysed in the future study.
Furthermore, one intended utility of these DE RNAs and enriched
pathways is to predict the potential targets of TCM formulae that
treating QSBSS, which mainly include interleukin 1β, DNA
topoisomerase I and potassium voltage-gated channel subfamily H
member 2. These putative targets may be candidates for experimental
validation of the mechanisms of action of the TCM formulae, which
may be essential for the optimization of ancient formulae and the
discovery of novel drugs.
Acknowledgements
Not applicable.
Funding
This work was supported by the China Food and Drug
Administration Project (grant no. 201207009).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GC designed the present study, and analyzed and
interpreted the data. JLG, HQH, CL, YML, JL and JW enrolled the
participants and examination of the blood samples. GC was a major
contributor in writing the manuscript. All authors read and
approved the final manuscript.
Ethical approval and consent to
participate
The protocol of the present study was approved by
the Ethics Committee of Guang'anmen Hospital (Beijing, China).
Informed consent was provided by all of the participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yuan R and Lin Y: Traditional Chinese
medicine: An approach to scientific proof and clinical validation.
Pharmacol Ther. 86:191–198. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Balta S, Cakar M, Demırkol S, Kucuk U, Ay
SA and Unlu M: May chest pain describe coronary heart disease?
Croat Med J. 54:4112013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang C, Niimi M, Watanabe T, Wang Y, Liang
J and Fan J: Treatment of atherosclerosis by traditional Chinese
medicine: Questions and quandaries. Atherosclerosis. 277:136–144.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zha LH, He LS, Lian FM, Zhen Z, Ji HY, Xu
LP and Tong XL: Clinical strategy for optimal traditional chinese
medicine (TCM) herbal dose selection in disease therapeutics:
Expert consensus on classic TCM herbal formula dose conversion. Am
J Chin Med. 43:1515–1524. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang YH and Xu AL: Zheng: A systems
biology approach to diagnosis and treatments. Sci Art Sci
Traditional Med (Suppl). S13–S15. 2014.
|
|
6
|
Blázovics A: The interpretation and
integration of traditional Chinese phytotherapy into Western-type
medicine with the possession of knowledge of the human genome. Orv
Hetil. 159:696–702. 2018.(In Hungarian). View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Josephine PB: A global scientific
challenge: Learning the right lessons from ancient healing
practices. Sci Art Scie Traditional Med (Suppl). S7–S9. 2014.
|
|
8
|
Chen G, Xue WD and Zhu J: Full genetic
analysis for genome-wide association study of Fangji: A powerful
approach for effectively dissecting the molecular architecture of
personalized traditional Chinese medicine. Acta Pharmacol Sin.
39:906–911. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gao JL, Chen G, He QY, Li J and Wang J:
Analysis of chinese patent medicine prescriptions for qi stagnation
and blood stasis syndrome. Zhongguo Zhong Yao Za Zhi. 42:187–191.
2017.(In Chinese). PubMed/NCBI
|
|
10
|
Li Y, Li R, Ouyang Z and Li S: Herb
Network Analysis for a Famous TCM Doctor's prescriptions on
treatment of rheumatoid arthritis. Evid Based Complement Alternat
Med. 2015:4513192015.PubMed/NCBI
|
|
11
|
Liu C, Chen G, Liu D, Nie S, Wang X, Huang
X, Li H, Li Y, Luo S, Zhao G, et al: Observational study of the
association between TCM zheng and types of coronary artery
stenosis: Protocol of a multicenter case series study. Evid Based
Complement Alternat Med. 2018:25649142018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guttman M and Rinn JL: Modular regulatory
principles of large non-coding RNAs. Nature. 482:339–346. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gilbert WV, Bell TA and Schaening C:
Messenger RNA modifications: Form, distribution, and function.
Science. 352:1408–1412. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Thomson DW and Dinger ME: Endogenous
microRNA sponges: Evidence and controversy. Nat Rev Genet.
17:272–283. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu J, Liu T, Wang X and He A: Circles
reshaping the RNA world: From waste to treasure. Mol Cancer.
16:582017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hopkins AL: Network pharmacology: The next
paradigm in drug discovery. Nat Chem Biol. 4:682–690. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li S and Zheng B: Traditional Chinese
medicine network pharmacology: Theory, methodology and application.
Chin J Nat Med. 11:110–120. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zheng XY: Guiding Principles for Clinical
Research of New Drugs in Traditional Chinese Medicine (Trial) 2002.
2. 1. China Med Sci Technol Press; BeiJing, BJ: pp. 69–126.
2002
|
|
21
|
Wang J, Gao JL, Chen G and Haoqiang H:
Diagnosis criteria on Qi Stagnation and Blood Stasis Syndrome. Chin
J Experimental Traditional Med Formulae. 24:16–20. 2018.(In
Chinese).
|
|
22
|
Shen CT, Zhang L, Wang Z, Chen QG, Chen
BW, Yuan Y, Zhong Y, Zhu J, Chen Y and Wang YY: Standard for
symptoms and signs of Traditional Chinese Medicine in new drug
clinical trials. J Chin Med. 54:1265–1267. 2013.
|
|
23
|
American College of Cardiology
Foundation/American Heart Association: 2012
ACCF/AHA/ACP/AATS/PCNA/SCAI/STS guideline for the diagnosis and
management of patients with stable ischemic heart disease. J Am
College Cardiololy. 60:2564–603. 2012. View Article : Google Scholar
|
|
24
|
Rheumatology Branch of Chinese Medical
Association: The diagnosis and treatment for rheumatoid arthritis.
Chin J Rheumatol. 14:265–269. 2010.(In Chinese).
|
|
25
|
Fang JY, Liu WZ, Li ZS, Du YQ, Ji XL, Ge
ZZ, Li YQ, Ni JM, Lv NH, Wu KC, et al: Digestive Disease Branch of
Chinese Medical Association: Consensus on chronic gastritis in
China (Shanghai 2012). Zhong Guo Yi Xue Qian Yan Za Zhi. 5:44–55.
2013.
|
|
26
|
Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ,
Wei L and Gao G: CPC: Assess the protein-coding potential of
transcripts using sequence features and support vector machine.
Nucleic Acids Res. 35:W345–W349. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li A, Zhang J and Zhou Z: PLEK: A tool for
predicting long non-coding RNAs and messenger RNAs based on an
improved k-mer scheme. BMC Bioinformatics. 15:3112014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun L, Luo H, Bu D, Zhao G, Yu K, Zhang C
and Zhao Y: Utilizing sequence intrinsic composition to classify
protein-coding and long non-coding transcripts. Nucleic Acids Res.
41:e1662013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Finn RD, Bateman A, Clements J, Coggill P,
Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J,
et al: Pfam: The protein families database. Nucleic Acids Res.
42:(Database Issue). D222–D230. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ghosh S and Chan CK: Analysis of RNA-Seq
data using TopHat and Cufflinks. Methods Mol Biol. 1374:339–361.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gao Y, Wang J and Zhao F: CIRI: An
efficient and unbiased algorithm for de novo circular RNA
identification. Genome Biol. 16:42015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Apweiler R, Bairoch A, Wu CH, Barker WC,
Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et
al: UniProt: The universal protein knowledgebase. Nucleic Acids
Res. 32:(Database Issue). D115–D119. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ru JL, Li P, Wang J, Zhou W, Li B, Huang
C, Li P, Guo ZH, Tao WY, Yang YF, et al: TCMSP: A database of
systems pharmacology for drug discovery from herbal medicines. J
Cheminform. 6:132014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xue R, Fang Z, Zhang M, Yi Z, Wen C and
Shi T: TCMID: Traditional Chinese medicine inegrative database for
herb molecular mechanism analysis. Nucleic Acids Res. 41:(Database
Issue). D1089–D1095. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu L, Liu JX, Guo H and Ren JX: Recent
advances on pericytes in microvascular dysfunction and traditional
Chinese medicine prevention. Zhongguo Zhong Yao Za Zhi.
42:3072–3077. 2017.(In Chinese). PubMed/NCBI
|
|
37
|
Wang T, Jia C, Chen Y, Li X and Cheng J:
Analysis on establishment and affecting factors of qi stagnation
and blood stasis rat model. Zhongguo Zhong Yao Za Zhi.
37:1629–1633. 2012.(In Chinese). PubMed/NCBI
|
|
38
|
Chen CS, Lin LW, Hsieh CC, Chen GW, Peng
WH and Hsieh MT: Differential gene expression in hemodialysis
patients with ‘cold’ zheng. Am J Chin Med. 34:377–385. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lu YY, Zhao Y, Song YN, Dong S, Wei B,
Chen QL, Hu YY and Su SB: Serum cytokine profiling analysis for
zheng differentiation in chronic hepatitis B. Chin Med. 10:242015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kiyama R: DNA microarray-based screening
and characterization of traditional chinese medicine. Microarrays
(Basel). 6:42017. View Article : Google Scholar
|
|
41
|
Hu XQ and Su SB: An overview of
epigenetics in Chinese medicine researches. Chin J Integr Med.
23:714–720. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li W, Liu P, Duan J, Ma CH, Shi XQ and Guo
JM: Effects of Xiangfu Siwu decoction on nerve endocrine immune
system in female rats with Qi Stagnation and Blood Stasis Syndrome.
Chin J Exp Trad Med Formulae. 20:99–104. 2014.
|
|
43
|
Yin CE, Zhang NN, Lv FF, Guan L and Wei
XJ: Study on the correlation of Qi Stagnation and Blood Stasis
Syndrome and heart rate variability in patients with Coronary Heart
Disease. J Sichuan Traditional Chin Med. 21:15–16. 2003.(In
Chinese).
|
|
44
|
Ren JX, Liu JX and Lin CR: Comparative
analysis on the biological basis of blood stasis syndrome induced
by Qi Stagnation and Qi deficiency in patients with unstable angina
pectoris. Zhongguo Zhong Xi Yi Jie He Za Zhi. 30:352–356. 2010.(In
Chinese). PubMed/NCBI
|
|
45
|
Liu JW: A preliminary bionetwork research
of Qi deficiency and Qi stagnation in the context of coronary heart
disease angina pectoris. Beijing Univ Chin Med,. 2:29–34. 2015.
|
|
46
|
Kolesnick RN: Sphingomyelin and
derivatives as cellular signals. Prog Lipid Res. 30:1–38. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hannun YA: The sphingomyelin cycle and the
second messenger function of ceramide. J Biol Chem. 269:3125–3128.
1994.PubMed/NCBI
|
|
48
|
Liu J, Beckman BS and Foroozesh M: A
review of ceramide analogs as potential anticancer agents. Future
Med Chem. 5:1405–1421. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Brunkhorst R, Vutukuri R and Pfeilschifter
W: Fingolimod for the treatment of neurological diseases-state of
play and future perspectives. Front Cell Neurosci. 8:2832014.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Arevalo JC and Wu SH: Neurotrophin
signaling: Many exciting surprises. Cell Mol Life Sci.
63:1523–1537. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Grant BD and Donaldson JG: Pathways and
mechanisms of endocytic recycling. Nat Rev Mol Cell Biol.
10:597–608. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Steinberg GR and Kemp BE: AMPK in health
and disease. Physiol Rev. 89:1025–1078. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Sanchez AM, Candau RB, Csibi A, Pagano AF,
Raibon A and Bernardi H: The role of AMP-activated protein kinase
in the coordination of skeletal muscle turnover and energy
homeostasis. Am J Physiol Cell Physiol. 303:C475–C485. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wang YY and Zhang HM: The scientific
explanation of Qi. J Chin Med. 58:811–813. 2017.(In Chinese).
|