Introduction
Hepatitis C virus (HCV) is considered to be one of
the major causes of liver fibrosis and cirrhosis. Estimating the
stage of disease is essential for determining disease prognosis and
the appropriate antiviral therapy (1). Although therapy is not essential to
treat insignificant fibrosis [Meta-analysis of Histological Data in
Viral Hepatitis (METAVIR stage F1)] in patients with chronic
hepatitis C (CHC), significant fibrosis (METAVIR stage F≥2) must be
treated to avoid progression to cirrhosis or hepatocellular
carcinoma (HCC) (2–4). Therefore, diagnosis of the fibrosis
stage is crucial in routine clinical practice. Liver biopsy is the
gold standard method for the staging of fibrosis; however, this
invasive technique has a mortality rate of 0.01–0.1% and harbors
the risk of severe complications (5). Therefore, accurate, non-invasive and
readily available methods for identifying fibrosis in patients with
HCV are urgently required.
In the past decade, microRNAs (miRNAs or miRs) have
been proposed as useful biomarkers for predicting the presence and
severity of various pathologies (6,7).
Numerous studies have identified specific miRNAs that serve
important roles in a variety of cellular processes, including
metabolism, immune function, cell proliferation and apoptosis
(8–12). The circulating miRNA profile is
altered during the initiation and progression of various liver
diseases (13), and there are
correlations between circulating miRNA levels and various
clinicopathological endpoints (14,15).
This, in addition to the remarkable stability of miRNAs in plasma,
serum and other body fluids, emphasizes their significance as a
novel class of blood-based biomarkers and offers novel strategies
for the development of non-invasive prognostic tests (16–18).
Various non-invasive diagnostic and prognostic
methods, ranging from serum biomarker assays to advanced imaging
techniques, are being developed (19,20).
Analysis of liver stiffness measurements (LSM), the aspartate
aminotransferase (AST)-to-platelet (PLT) ratio index (APRI) and
fibrosis 4 score (FIB-4) are now routinely performed to assess
fibrosis in patients with liver disease (12,21). The
majority of these non-invasive tests are primarily used to
distinguish the presence of cirrhosis from minimal or absent
fibrosis; however, they have a low diagnostic performance,
particularly for the diagnosis of significant fibrosis (19). These non-invasive tests are also
affected by the weight of the patient, the presence of ascites, and
the transaminase and bilirubin levels (22–24).
The aim of the present study was to evaluate the
performance of miR-1273g-3p levels as a diagnostic tool for
fibrosis (METAVIR stage F≥2 and F=4), using liver histology as the
gold standard for diagnosis. Additionally, the diagnostic
performance of miR-1273g-3p was compared with LSM, APRI and FIB-4,
and the value of using miR-1273g-3p to predict the fibrosis stage
in patients with CHC was also determined.
Materials and methods
Patients
A total of 112 patients (56 males and 56 females)
with CHC infection underwent liver biopsies and provided blood
samples at the Third Hospital of Hebei Medical University
(Shijiazhuang, China) from January 2014 to May 2016. CHC infection
was diagnosed on the basis of positive tests for serum antibodies
against HCV and the presence of HCV RNA in the plasma in the
previous 6 months. Eligible patients were >18 years of age.
Patients with any of the following were excluded from the present
study: Presence of decompensated cirrhosis, co-infection with human
immunodeficiency virus, hepatitis A, B or D virus infection, other
causes of chronic liver disease or co-morbidities precluding
interferon therapy (25,26). Written informed consent was obtained
from all patients, and the study was approved by the Ethics
Committee of the Third Hospital of Hebei Medical University,
according to the Declaration of Helsinki and Good Clinical Practice
guidelines.
Detection of antibodies, viral load
and genotypes of HCV
Serum HCV antibodies were detected using commercial
anti-HCV ELISA kits (cat. nos. 2013040508, 2013111208, 2014030608
and 2014081408; Zhuhai Livzon Diagnostics, Inc., Zhuhai, China),
which are approved by China's State Food and Drug Administration
(SFDA: S10950020). Plasma HCV RNA was determined by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
using a COBAS TAQman HCV test (Roche Molecular Diagnostics,
Pleasanton, CA, USA), which included three major processes:
specimen preparation to isolate HCV RNA; reverse transcription of
the target RNA to generate cDNA and simultaneous PCR amplification
of target cDNA, and detection of cleaved dual-labeled
oligonucleotide detection probes specific to the target. The lowest
limit of quantification was 15 IU/ml. HCV genotypes were identified
using the HCV genotyping oligochip (Tianjin Third Central Hospital,
Tianjin, China) as previously described (27).
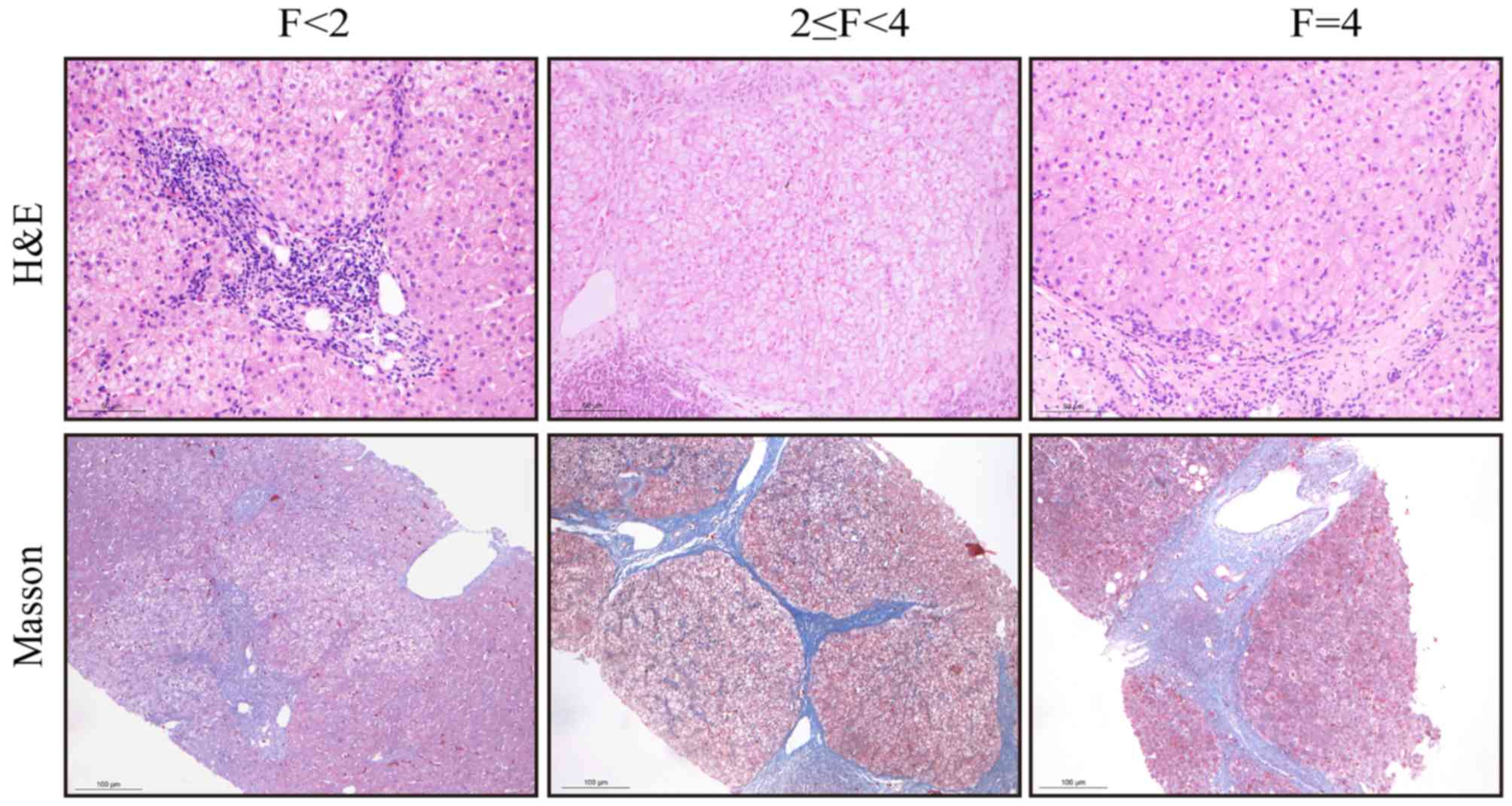
METAVIR fibrosis stages
Needle biopsies of the liver were performed by a
trained hepatologist, with a 16-G cutting needle (Bard Peripheral
Vascular, Inc., Tempe, AZ, USA). The samples were fixed in 10%
formalin for 48 h at room temperature and embedded in paraffin.
Liver tissue sections (4-µm thick) were stained with hematoxylin
and eosin (H&E) and Masson's trichrome at room temperature. For
H&E staining the sections were deparaffinized and rehydrated
with xylene and a decreasing graded ethanol series, stained in
hematoxylin solution for 5 min, differentiated in 0.5% acid alcohol
for 30 sec and stained blue in ammonia water. Counterstaining was
performed in eosin solution for 1 min. Masson's trichrome staining
was performed in ponceau-picric acid saturated solution for 10 min,
rinsed in 1% acetic acid water, differentiated in 1%
phosphomolybdic acid for 1 min, rinsed in distilled water, stained
in toluidine blue for 3 min, rinsed in 1% acetic acid-water,
differentiated in 95% alcohol, hydrated in absolute alcohol,
cleared with xylene and mounted with neutral balsam. The liver
sections were observed at ×100 to ×400 magnification using a Leica
DM 2000 microscope (Leica Microsystems, Inc., Buffalo Grove, IL,
USA).
All liver biopsies were evaluated by expert
pathologists, who were blinded to the clinical history of the
patients. Fibrosis was classified into five stages according to the
METAVIR scoring system (28) as
follows: 0, no fibrosis; 1, portal fibrosis without septa; 2,
portal fibrosis with rare septa; 3, many septa without cirrhosis;
and 4, cirrhosis. Liver fibrosis was evaluated according to the
fibrosis METAVIR staging, with significant fibrosis defined as
METAVIR stages ≥2 (29).
Biochemical assays
Serum alanine aminotransferase (ALT) and AST were
detected by optimized International Federation of Clinical
Chemistry and Laboratory Medicine reference method (30), and total bilirubin (TBIL) was
detected using the vanadate-oxidation method (31) with an Olympus AUS5400 automatic
chemical analyzer (Olympus Corporation, Tokyo, Japan) according to
the manufacturer's protocol. Blood platelet counts (PLT) were
analyzed by an automated hematology analyzer using the Hydro
Dynamic Focusing method (32)
(XS-1000i; Sysmex Corporation, Kobe, Japan).
APRI and FIB-4 biomarker panels
APRI was calculated using the following formula:
APRI=[AST (U/l)/upper limit of normal (U/l)] ×100/PLT
(109/l). The FIB-4 index was calculated using the
following formula: FIB-4=Age (years)xAST (U/l)/PLT
(109/l)x[ALT (U/l)]1/2 (33).
LSM tests
Patients with CHC infection at baseline underwent
LSM using FibroTouch (HISKY Medical Technologies Co., Ltd.,
Beijing, China) on the right lobe of the liver as previously
described (34); the procedure was
conducted by a technician who had performed >10 LSMs. A liver
biopsy analysis was subsequently performed as described above. The
results were expressed in kilopascals (kPa) and the median value of
10 acquisitions was used for analysis, including only cases with a
success rate >60% and an interquartile range/median ratio of
<0.3. The same professionally trained physician performed all of
these procedures.
Sample preparation
According to METAVIR fibrosis stages, 112 serum
samples were collected from patients with CHC (genotype 1b or 2a)
and fibrosis. Peripheral blood samples (5 ml) were collected at the
baseline and serum was separated by centrifugation at 1,200 × g for
10 min at 4°C. The supernatant was transferred into a new
microcentrifuge tube and further processed by an additional
centrifugation step at 12,000 × g for 15 min at 4°C and stored at
−80°C prior to further analysis, as previously described (35).
RNA isolation and spike-in
control
Total RNA was isolated from 250 µl serum using
TRIpure Reagent LS (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) according to the manufacturer's protocol. For
each RNA sample, the relative miR-1273g-3p expression levels were
normalized to that of C. elegans miR-39-3p (cel-miR-39-3p;
miRB0000010, Guangzhou RiboBio Co., Ltd., Guangzhou, China) because
of its wide and stable expression in circulation (36,37).
Prior to RNA isolation, 40 fmol cel-miR-39 was added to each sample
as a spike-in control. Total RNA was resuspended in nuclease-free,
PCR-grade water and the RNA concentration was determined using the
NanoDrop 2000C spectrophotometer (NanoDrop; Thermo Fisher
Scientific Inc., Wilmington, DE, USA).
miR-1273g-3p quantification by
RT-qPCR
Reverse transcription reactions were performed using
the miScript-Reverse Transcription kit (Takara Biotechnology Co.,
Ltd., Dalian, China), cDNA was synthesized using reverse
transcriptase with miR-1273g-3p specific stem-loop primers (forward
primer, ssD1381210710; reverse primer, ssD089261711; RT-primer,
ssD1381210709; Guangzhou RiboBio Co., Ltd.) and cel-miR-39-3p
specific stem-loop primer (forward primer, ssD1083145002; reverse
primer, ssD089261711; RT primer, ssD1083145001, Guangzhou RiboBio
Co., Ltd.). The RT reaction was performed under the conditions of
37°C for 15 min and 85°C for 5 sec. Differential expression
analysis was performed using RT-qPCR on an ABI 7500 Real-Time PCR
system (Applied Biosystems; Thermo Fisher Scientific, Inc.) using
SYBR Green Master Mix (Beijing CoWin Biotech Co., Ltd., Beijing,
China). The amplification conditions were as follows: Initial
denaturation at 95°C for 30 sec, then 40 cycles of 95°C for 5 sec
and 60°C for 30 sec. The relative abundance of miRNA was normalized
to that of cel-miR-39 and the relative amount of each miRNA was
measured using the 2−ΔΔCq method (38). All RT-qPCR reactions were conducted
in triplicate. All data were obtained using Sequence Detector
Software v2.0.4 (Applied Biosystems; Thermo Fisher Scientific
Inc.).
Statistical analysis
Quantitative variables were expressed as the median
(range) or mean ± standard deviation and qualitative variables were
expressed as percentages. Correlations between variables were
calculated using Spearman rank order correlations and the
diagnostic performance of non-invasive markers were evaluated by
receiver operating characteristic (ROC) curves. Data were analyzed
using SPSS software (version 16.0; SPSS, Inc., Chicago, IL, USA).
All P-values were two-tailed and P<0.05 was considered to
indicate a statistically significant difference.
Results
Patient characteristics
The clinical characteristics of the cohort of 112
patients with CHC are summarized in Table I. There were 39 patients with mild
liver fibrosis (F<2), including 17 males and 22 females, with a
mean age of 43±12.42 years. There were 47 subjects with moderate to
severe liver fibrosis (2≤F<4), including 22 males and 25
females, with a mean age of 52.29±9.54 years. There were 26
patients with cirrhosis (F=4), including 17 male and 9 female, with
a mean age of 55.19±10.18 years. There were no significant
differences in patient sex, body mass index (BMI), HCV RNA load,
HCV genotype, or ALT and AST levels at baseline between different
grades of liver fibrosis. At higher fibrosis stages, the age
(P<0.001) and TBIL (P=0.035) were significantly increased,
whereas the PLT was decreased (P<0.001; Table I). The different stages of fibrosis
were assessed by histology and were distributed as follows among
the samples: F<2, 34.82%; 2≤F<4, 41.96%; and F=4, 23.21%.
 | Table I.Baseline characteristics of patients
included in the present study (n=112). |
Table I.
Baseline characteristics of patients
included in the present study (n=112).
| Parameter | F<2 | 2≤F<4 | F=4 | P-value |
|---|
| Patients, n
(%) | 39 (34.82) | 47 (41.96) | 26 (23.21) |
|
| Sex, m/f | 17/22 | 22/25 | 17/9 | 0.341 |
| Age, years, mean ±
SD | 43±12.42 |
52.29±9.54a |
55.19±10.18a | <0.001 |
| BMI,
kg/m2, mean ± SD | 24.2±2.69 | 25.39±3.12 | 25.35±2.58 | 0.21 |
| ALT, U/l
(range) | 31.5
(9.0–199.0) | 33.0
(5.0–363.0) | 38.4
(13.0–345.0) | 0.199 |
| AST, U/l
(range) | 34.0
(16.0–150.0) | 39.0
(18.0–256.0) | 48.5
(16.0–299.0) | 0.245 |
| TBIL, µmol/l
(range) | 12.70
(4.90–58.00) | 13.90
(6.71–32.60) | 17.31
(3.55–85.50)a,b | 0.035 |
| Platelets,
×109/l (range) | 183.0
(78.3–334.0) | 149.6
(64.0–277.0)a | 125.0
(31.0–304.0)a,c | <0.001 |
| HCV RNA,
log10 IU/ml (range) | 6.02
(4.48–7.99) |
6.31(3.60–7.84) |
6.15(3.63–7.18) | 0.532 |
| HCV genotypes,
1b/2a/n.d | 22/11/6 | 29/11/7 | 8/8/10 | 0.174 |
Correlations between non-invasive
models and histological findings
According to METAVIR fibrosis staging, patients were
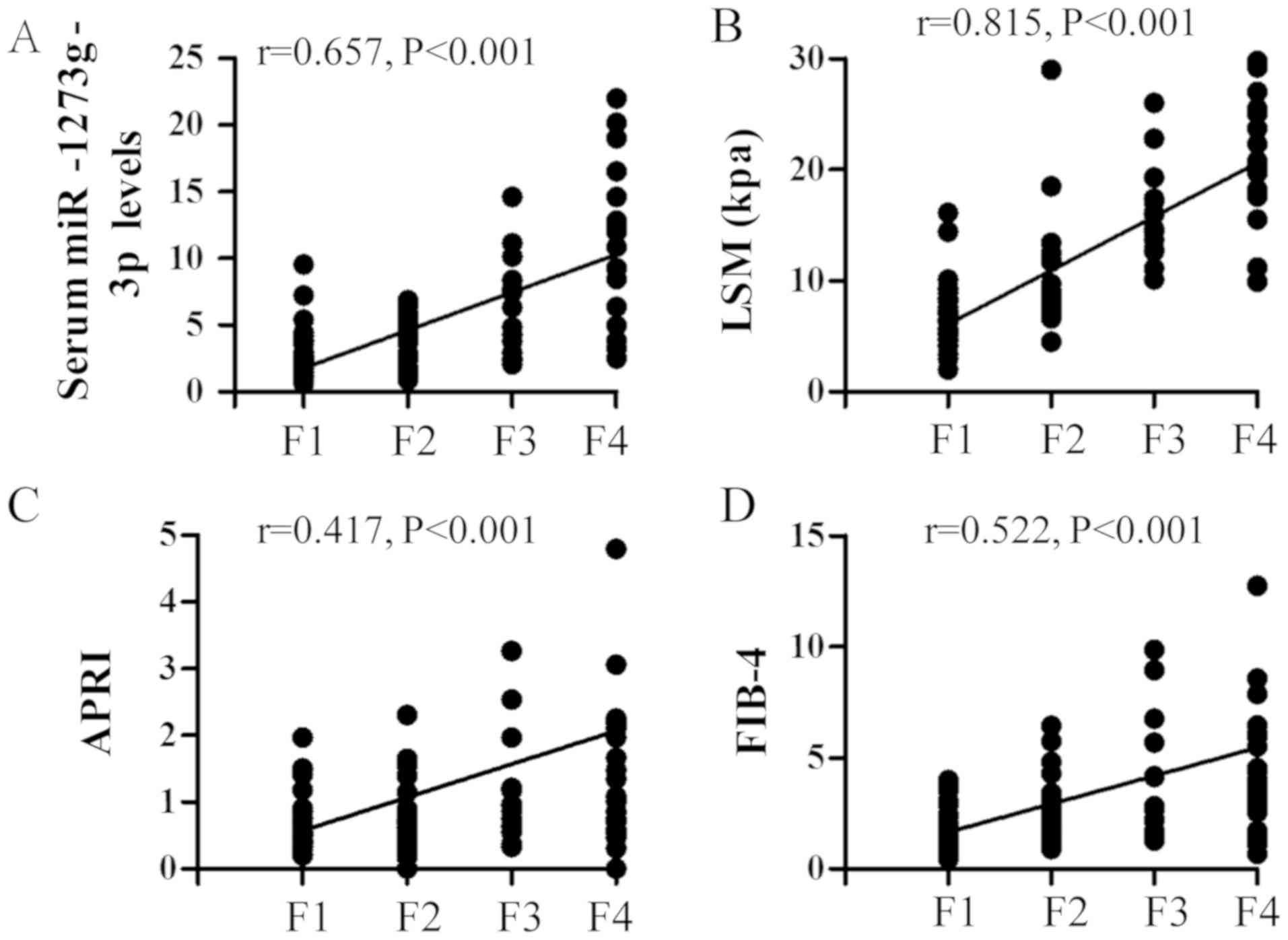
categorized as F1 to F4 as presented in Fig. 1. The Spearman's correlation
coefficient results indicated that fibrosis stage was significantly
correlated with miR-1273g-3p levels (r=0.657, P<0.001; Fig. 2A). This correlation was lower
compared with that between fibrosis stage and LSM (r=0.815,
P<0.001; Fig. 2B) and higher
compared with that between fibrosis stage and APRI (r=0.417,
P<0.001; Fig. 2C) or FIB-4
(r=0.522, P<0.001; Fig. 2D).
Performance of miR-1273g-3p, LSM, APRI
and FIB-4 in fibrosis stage assessment
Patients were divided into three groups according to
their METAVIR stage: F<2, 2≤F<4 and F=4. The area under the
ROC curve of miR-1273g-3p was 0.841 (95% CI, 0.761–0.921) for the
significant fibrosis and early fibrosis groups (2≤F<4 and
F<2, respectively), which was lower compared with that of LSM
(0.890; 95% CI, 0.825–0.955), and higher compared with that of
FIB-4 (0.791; 95% CI, 0.701–0.881) and APRI (0.719; 95% CI,
0.612–0.826; Fig. 3A; Table II). The sensitivity and specificity
values of the four analyses are presented in Table II. Using an optimal cut-off of 2.67,
miR-1273g-3p had a sensitivity of 85% and specificity of 69% in
predicting significant fibrosis. In addition, in ROC analysis of
the performance of these markers in the diagnosis of cirrhosis (F=4
vs. F<4), the AUC of miR-1273g-3p (0.933; 95% CI, 0.874–0.993)
was lower compared with that of LSM (0.937; 95% CI, 0.887–0.987),
and higher compared with values for FIB-4 (0.766; 95% CI,
0.639–0.881) and APRI (0.760; 95% CI, 0.649–0.871). miR-1273g-3p
had 80% sensitivity and 95% specificity for predicting cirrhosis
with a cut-off value of 8.36 (Fig.
3B; Table III).
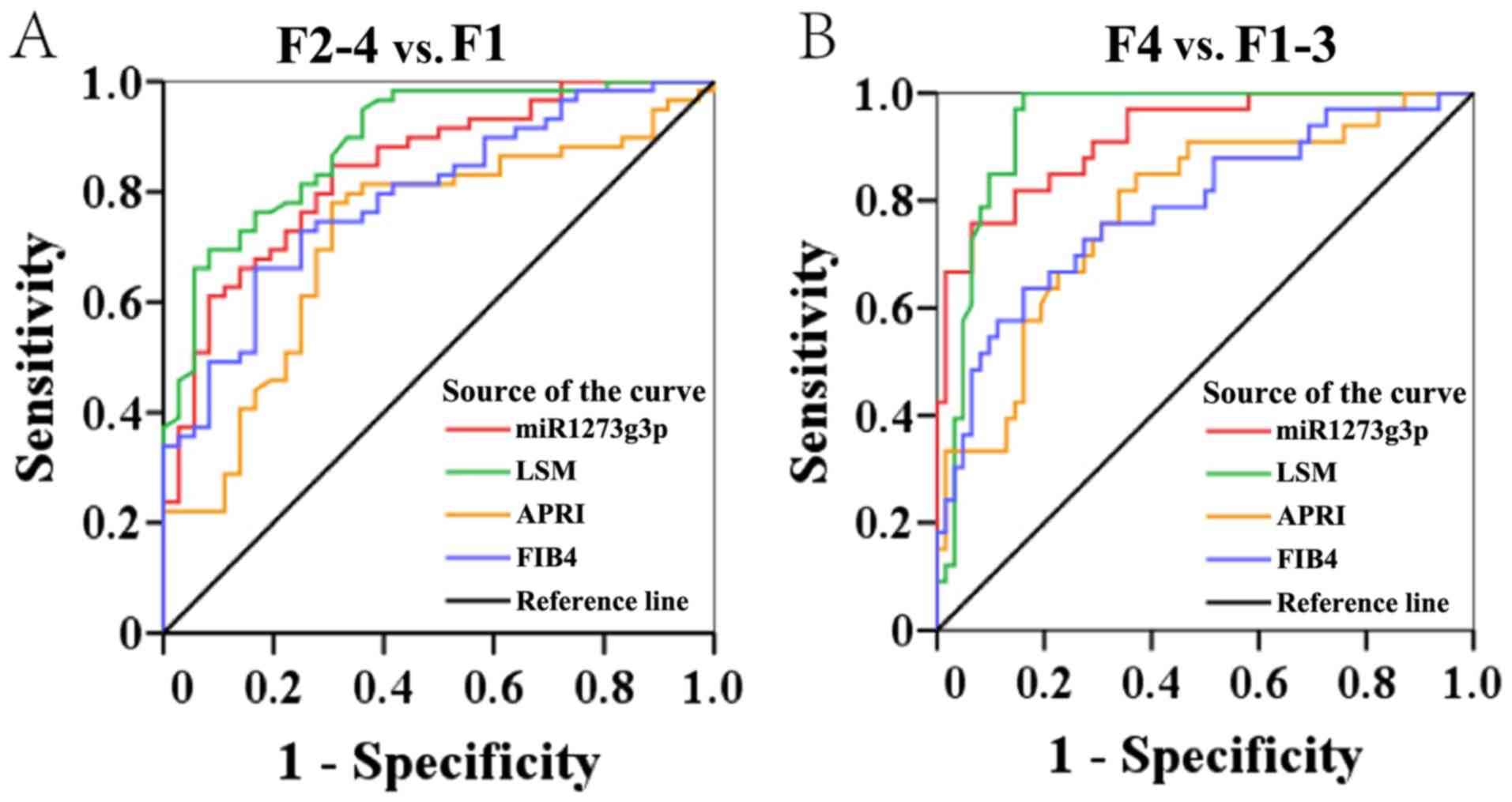 | Figure 3.Receiver operating characteristic
curve analysis of serum miR-1273g-3p, LSM, APRI and FIB-4 in
distinguishing (A) 4>F≥2 from F1, and (B) F=4 from F1-3. F
values correspond to Meta-analysis of Histological Data in Viral
Hepatitis scoring. miR, microRNA; LSM, liver stiffness
measurements; APRI, aspartate aminotransferase-to-platelet ratio
index; FIB-4, Fibrosis-4 score; 4>F≥2, significant liver
fibrosis; F1, non-significant liver fibrosis; F=4, cirrhosis. |
 | Table II.Validation and comparison of
non-invasive methods for the prediction of liver fibrosis in
chronic hepatitis C (Meta-analysis of Histological Data in Viral
Hepatitis stage F2-4 vs. F1). |
Table II.
Validation and comparison of
non-invasive methods for the prediction of liver fibrosis in
chronic hepatitis C (Meta-analysis of Histological Data in Viral
Hepatitis stage F2-4 vs. F1).
| Assay | AUC | 95% CI | Cut-off | Se | Sp |
|---|
| miR-1273g-3p | 0.841 | 0.761–0.921 | 2.67 | 0.85 | 0.69 |
| LSM | 0.890 | 0.825–0.955 | 9.50 | 0.70 | 0.92 |
| APRI | 0.719 | 0.612–0.826 | 0.55 | 0.78 | 0.69 |
| FIB-4 | 0.791 | 0.701–0.881 | 2.49 | 0.66 | 0.83 |
 | Table III.Validation and comparison of
non-invasive models for the prediction of liver fibrosis in chronic
hepatitis C (Meta-analysis of Histological Data in Viral Hepatitis
F4 vs. F1-3). |
Table III.
Validation and comparison of
non-invasive models for the prediction of liver fibrosis in chronic
hepatitis C (Meta-analysis of Histological Data in Viral Hepatitis
F4 vs. F1-3).
| Assay | AUC | 95% CI | Cut-off | Se | Sp |
|---|
| miR-1273g-3p | 0.933 | 0.874–0.993 | 8.36 | 0.80 | 0.95 |
| LSM | 0.937 | 0.887–0.987 | 15.09 | 0.90 | 0.89 |
| APRI | 0.760 | 0.649–0.871 | 0.67 | 0.85 | 0.63 |
| FIB-4 | 0.766 | 0.639–0.881 | 2.95 | 0.80 | 0.75 |
Predictive factors of
miR-1273g-3p
The influence of certain factors on the expression
of miR-1273g-3p was also assessed. It was observed that the serum
levels of miR-1273g-3p were significantly positively correlated
with age, BMI, and ALT, AST and TBIL levels (r=0.396, 0.219, 0.215,
0.228 and 0.225, respectively; all P<0.05; Fig. 4), whereas a significant negative
correlation with PLT count was observed (r=−0.318; P=0.001;
Fig. 4). No significant correlation
was observed between miR-1273g-3p and baseline HCV RNA loads and
genotype (Fig. 4).
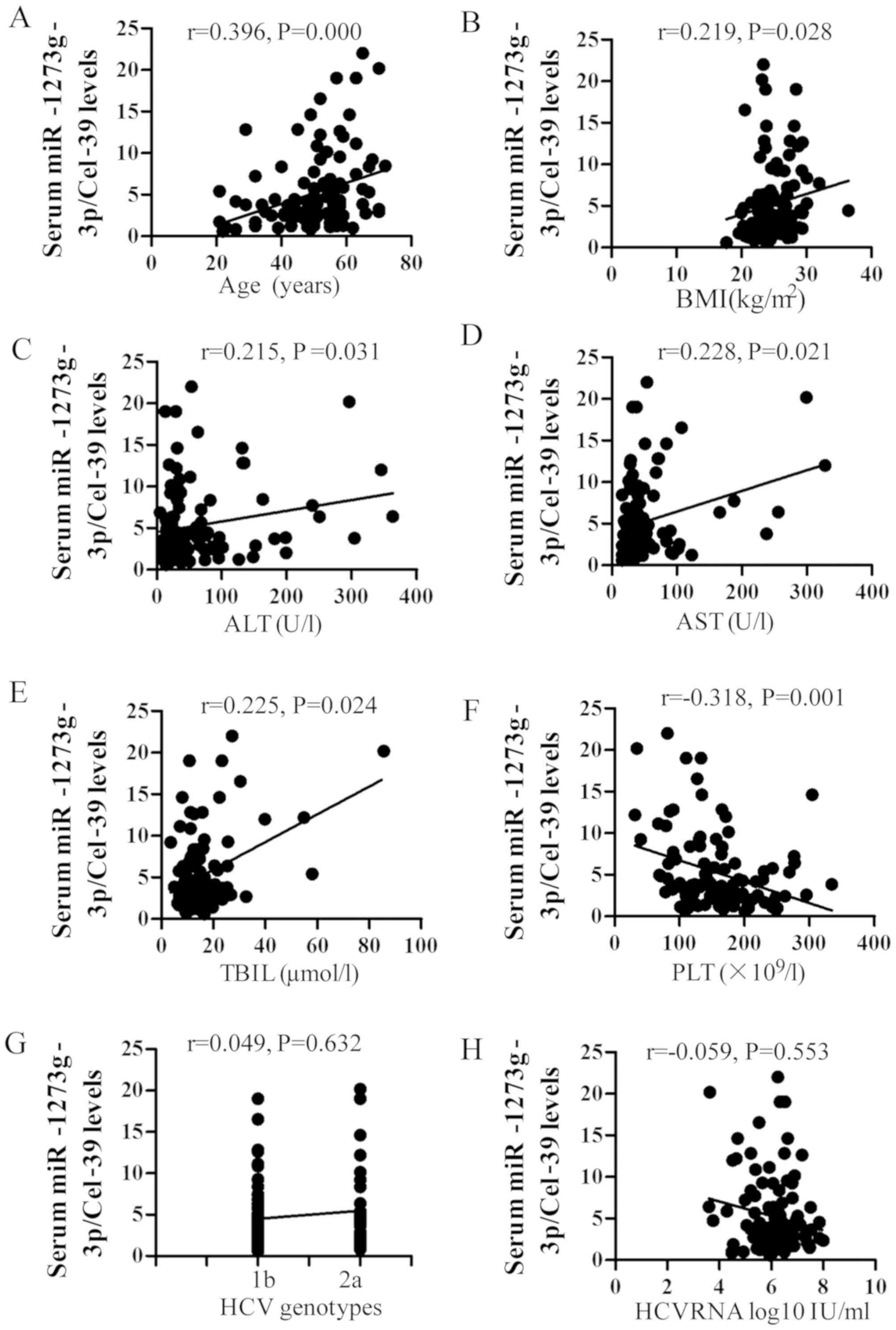 | Figure 4.Correlation between miR-1273g-3p,
baseline characteristics and biochemical assays. The Spearman
analysis demonstrated a positive correlation between miR-1273g-3p
and (A) age, (B) BMI, (C) ALT, (D) AST and (E) TBIL and a negative
correlation between miR-1273g-3p and (F) PLT. No correlation was
observed between miR-1273g-3p and (G) HCV RNA viral load or (H) HCV
genotype. miR, microRNA; BMI, body mass index; ALT, alanine
aminotransferase; AST, aspartate aminotransferase; TBIL, total
bilirubin; HCV, hepatitis C virus. |
Discussion
Liver biopsies are considered the gold standard for
the diagnosis of liver fibrosis; however, this is an invasive
procedure, which is subject to complications and high costs
(39). Therefore, it is important to
identify molecular markers that are able to predict disease
progression. Increasing evidence suggests that miRNA profiling is a
promising approach to facilitate the development of novel
diagnostic biomarkers and the identification of therapeutic targets
(40).
The results of a previous study by the present
authors demonstrated that miR-1273g-3p was significantly
upregulated in the fibrotic liver of patients with CHC and
associated with higher fibrosis stages (41). In situ hybridization revealed
that miR-1273g-3p was present in hepatocytes or hepatic stellate
cells around the portal area (41).
Additionally, it has been demonstrated that miR-1273g-3p modulates
the activation and apoptosis of hepatic stellate cells via
phosphatase and tensin homologs in HCV-associated liver fibrosis.
The effects of various miRNAs on liver fibrosis have been
illustrated in previous studies (11,41–43). The
miR-199 and −200 families are upregulated with the progression of
liver fibrosis (44). miR-29 family
members, including miR-29b and miR-29c, have been reported to be
associated with the occurrence of fibrosis via regulating the
synthesis of extracellular matrix components, particularly collagen
(45). Previous studies have
demonstrated that hepatic levels of miR-122 decrease significantly
as the severity of fibrosis increases (14,42).
miRNA levels in HCC samples with or without previous HCV infection
have also been reported; thus, determination of HCV-specific
effects in these studies is complicated by the overall
dysregulation of miRNAs observed in tumors (46). Taken together, changes in miRNA
expression appear to be sensitive indicators of hepatic injury and
are potentially associated with the development of liver fibrosis
(14).
In the present study, the pathological results of
patients with CHC were used as reference standards, and additional
analyses were performed to evaluate the association between
miR-1273g-3p and LSM, APRI and FIB-4. Acoustic radiation force
impulse, APRI and FIB-4 have been previously reported to be
elevated in patients with chronic hepatitis B infection and
correlated with the stage of fibrosis (47). In the present study, levels of
miR-1273g-3p were increased with the stage of liver fibrosis, and
the correlation coefficient between fibrosis stage and miR-1273g-3p
was higher compared with that for APRI and FIB-4, whereas it was
lower compared with that for LSM. The results of the present study
demonstrate that miR-1273g-3p, LSM, APRI and FIB-4 may be used to
determine the stage of fibrosis according to the METAVIR score. The
potential of miR-1273g-3p as an indicator of fibrosis progression
in CHC was evaluated. ROC analysis demonstrated that the
performance of miR-1273g-3p in differentiating between F=4 and
4>F≥2 stages was superior compared with that of FIB-4 and APRI;
however, it was inferior compared with that of LSM for the
diagnosis of significant fibrosis. The cut-off value of
miR-1273g-3p was increased with the stage of fibrosis, suggesting
that cut-off values may be used to predict the stage of fibrosis.
Our previous study demonstrated that, compared with APRI and FIB-4,
LSM is a more convenient and reliable diagnostic indicator of liver
fibrosis in patients with chronic liver disease (20). However, LSM may be affected by liver
inflammation, increased transaminase levels and TBIL.
Non-invasive assessment of liver fibrosis is an
important goal for the treatment of patients with CHC (48). In the present study, miR-1273g-3p
levels were low in the early stages and increased in the later
stages of fibrosis (F≥2), suggesting a changing pattern of
circulating miR-1273g-3p levels as the disease progresses. Further
analysis of the factors that affect the miR-1273g-3p expression
levels revealed that age, BMI, and ALT, AST and TBIL levels were
positively correlated with miR-1273g-3p expression, whereas a
negative correlation was observed between miR-1273g-3p expression
and PLT. These results indicate that the expression of miR-1273g-3p
may be affected by liver inflammation, increased transaminase
levels, TBIL and PLT.
Several limitations of the present study should be
noted. The study was conducted at a single center and the small
proportion of patients with significant fibrosis may be a source of
selection bias. To corroborate these results, further studies
involving large patient populations are required to analyze the
association between serum miR-1273g-3p levels and HCV-induced
hepatic steatosis, inflammation and cholestasis, as well as other
etiologies of liver disease.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Chinese
Foundation for Hepatitis Prevention and Control's Wang Bao-en
Hepatic Fibrosis Foundation (grant no. 20140018), the Graduate
Student Innovation Fund Project in Hebei Province (2015), the Key
Science and Technology Project of Hebei Province (grant no.
14277746D), the Hebei Province Key Laboratory of Research and
Development for Chinese Medicine (2014) and the Government-Funded
Clinical Medical Talents Projects in Hebei Province (2016).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
YN designed the research; XN, NF, JD and BW
performed the experiments; YW, YZ and SZ analyzed data; XN, RW and
YN wrote the paper.
Ethics approval and consent to
participate
Written informed consent was obtained from all
patients and the study was approved by the Ethics Committee of the
Third Hospital of Hebei Medical University, according to the
Declaration of Helsinki and Good Clinical Practice guidelines.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wilkins T, Akhtar M, Gititu E, Jalluri C
and Ramirez J: Diagnosis and management of hepatitis C. Am Fam
Phys. 91:835–842. 2015.
|
|
2
|
WHO guidelines approved by the guidelines
review committee: In: Guidelines for the screening, care and
treatment of persons with hepatitis C infection. Geneva: 2014
|
|
3
|
Kayadibi H, Yasar B, Ozkara S, Serdar MA,
Kurdas OO and Gonen C: The diagnostic accuracy of the Forns index,
platelet count and AST to platelet ratio index derived fibrosis
index for the prediction of Hepatitis C virus-related significant
liver fibrosis and cirrhosis. Scand J Clin Lab Invest. 74:240–247.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ghany MG, Strader DB, Thomas DL and Seeff
LB: American Association for the Study of Liver Diseases:
Diagnosis, management, and treatment of hepatitis C: An update.
Hepatology. 49:1335–1374. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bravo AA, Sheth SG and Chopra S: Liver
biopsy. N Engl J Med. 344:495–500. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Noetel A, Kwiecinski M, Elfimova N, Huang
J and Odenthal M: microRNA are central players in anti- and
profibrotic gene regulation during liver fibrosis. Front Physiol.
3:492012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kosaka N, Iguchi H and Ochiya T:
Circulating microRNA in body fluid: A new potential biomarker for
cancer diagnosis and prognosis. Cancer Sci. 101:2087–2092. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee HM, Nguyen DT and Lu LF: Progress and
challenge of microRNA research in immunity. Front Genet. 5:1782014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Othman N and Nagoor NH: The role of
microRNAs in the regulation of apoptosis in lung cancer and its
application in cancer treatment. Biomed Res Int. 2014:3180302014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shenoy A and Blelloch RH: Regulation of
microRNA function in somatic stem cell proliferation and
differentiation. Nat Rev Mol Cell Biol. 15:565–576. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Du J, Niu X, Wang Y, Kong L, Wang R, Zhang
Y, Zhao S and Nan Y: MiR-146a-5p suppresses activation and
proliferation of hepatic stellate cells in nonalcoholic fibrosing
steatohepatitis through directly targeting Wnt1 and Wnt5a. Sci Rep.
5:161632015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Abdollahi M, Pouri A, Ghojazadeh M,
Estakhri R and Somi M: Non-invasive serum fibrosis markers: A study
in chronic hepatitis. Bioimpacts. 5:17–23. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bala S, Petrasek J, Mundkur S, Catalano D,
Levin I, Ward J, Alao H, Kodys K and Szabo G: Circulating microRNAs
in exosomes indicate hepatocyte injury and inflammation in
alcoholic, drug-induced, and inflammatory liver diseases.
Hepatology. 56:1946–1957. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Halász T, Horváth G, Pár G, Werling K,
Kiss A, Schaff Z and Lendvai G: miR-122 negatively correlates with
liver fibrosis as detected by histology and FibroScan. World J
Gastroenterol. 21:7814–7823. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mahgoub A and Steer CJ: MicroRNAs in the
evaluation and potential treatment of liver diseases. J Clin Med.
5:E522016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cortez MA and Calin GA: MicroRNA
identification in plasma and serum: A new tool to diagnose and
monitor diseases. Expert Opin Biol Ther. 9:703–711. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cortez MA, Bueso-Ramos C, Ferdin J,
Lopez-Berestein G, Sood AK and Calin GA: MicroRNAs in body
fluids-the mix of hormones and biomarkers. Nat Rev Clin Oncol.
8:467–477. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Etheridge A, Lee I, Hood L, Galas D and
Wang K: Extracellular microRNA: A new source of biomarkers. Mutat
Res. 717:85–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Castera L, Winnock M, Pambrun E, Paradis
V, Perez P, Loko MA, Asselineau J, Dabis F, Degos F and Salmon D:
Comparison of transient elastography (FibroScan), FibroTest, APRI
and two algorithms combining these non-invasive tests for liver
fibrosis staging in HIV/HCV coinfected patients: ANRS CO13 HEPAVIH
and FIBROSTIC collaboration. HIV Med. 15:30–39. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang R, Ren W, Zhao S, Niu X, Tan P, Du H
and Nan Y: Clinical study on FibroTouch and multi-parameter model
for diagnosis of hepatic fibrosis in patients with chronic liver
disease. Zhonghua Gan Zang Bing Za Zhi. 23:265–269. 2015.(In
Chinese). PubMed/NCBI
|
|
21
|
Crespo G, Fernández-Varo G, Mariño Z,
Casals G, Miquel R, Martínez SM, Gilabert R, Forns X, Jiménez W and
Navasa M: ARFI, FibroScan, ELF, and their combinations in the
assessment of liver fibrosis: A prospective study. J Hepatol.
57:281–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Friedrich-Rust M, Ong MF, Martens S,
Sarrazin C, Bojunga J, Zeuzem S and Herrmann E: Performance of
transient elastography for the staging of liver fibrosis: A
meta-analysis. Gastroenterology. 134:960–974. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fraquelli M, Rigamonti C, Casazza G,
Donato MF, Ronchi G, Conte D, Rumi M, Lampertico P and Colombo M:
Etiology-related determinants of liver stiffness values in chronic
viral hepatitis B or C. J Hepatol. 54:621–628. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Baranova A, Lal P, Birerdinc A and
Younossi ZM: Non-invasive markers for hepatic fibrosis. BMC
Gastroenterol. 11:912011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
European Association for the Study of the
Liver: EASL clinical practice guidelines: Management of hepatitis C
virus infection. J Hepatol. 55:245–264. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chinese Society of Hepatology, Chinese
Society of Infectious Diseases, Parasitology of Chinese Medical
Association. The guideline for prevention and treatment of
hepatitis C. Zhonghua Gan Zang Bing Za Zhi. 12:194–198. 2004.(In
Chinese).
|
|
27
|
Sun ZH, Yang HL, Wei M, Wang SY, Wang CR,
Shi YL and Ma WL: Preparation and application of oligo microarrays
for hepatitis virus detection and genotyping. Zhonghua Gan Zang
Bing Za Zhi. 15:816–820. 2007.(In Chinese). PubMed/NCBI
|
|
28
|
Intraobserver and interobserver variations
in liver biopsy interpretation in patients with chronic hepatitis
C. The French METAVIR Cooperative Study Group = Hepatology.
20:15–20. 1994.
|
|
29
|
Amorim TG, Staub GJ, Lazzarotto C, Silva
AP, Manes J, Ferronato Mda G, Shiozawa MB, Narciso-Schiavon JL,
Dantas-Correa EB and Schiavon Lde L: Validation and comparison of
simple noninvasive models for the prediction of liver fibrosis in
chronic hepatitis C. Ann Hepatol. 11:855–861. 2012.PubMed/NCBI
|
|
30
|
Schumann G, Bonora R, Ceriotti F, Férard
G, Ferrero CA, Franck PF, Gella FJ, Hoelzel W, Jørgensen PJ, Kanno
T, et al: IFCC primary reference procedures for the measurement of
catalytic activity concentrations of enzymes at 37 degrees C.
International federation of clinical chemistry and laboratory
medicine. Part 4. Reference procedure for the measurement of
catalytic concentration of alanine aminotransferase. Clin Chem Lab
Med. 40:718–724. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ye YW, Wang YS and Shen ZY: National guide
to clinical laboratory procedures. (3rd). Southeast University
Press. (Nanjing). 2006.(In Chinese).
|
|
32
|
Hill VL, Simpson VZ, Higgins JM, Hu Z,
Stevens RA, Metcalf JA and Baseler M: Evaluation of the performance
of the Sysmex XT-2000i hematology analyzer with whole bloods stored
at room temperature. Lab Med. 40:709–718. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Teshale E, Lu M, Rupp LB, Holmberg SD,
Moorman AC, Spradling P, Vijayadeva V, Boscarino JA, Schmidt MA and
Gordon SC: CHeCS Investigators: APRI and FIB-4 are good predictors
of the stage of liver fibrosis in chronic hepatitis B: The Chronic
Hepatitis Cohort Study (CHeCS). J Viral Hepat. 21:917–920. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Castéra L, Vergniol J, Foucher J, Le Bail
B, Chanteloup E, Haaser M, Darriet M, Couzigou P and De Lédinghen
V: Prospective comparison of transient elastography, Fibrotest,
APRI, and liver biopsy for the assessment of fibrosis in chronic
hepatitis C. Gastroenterology. 128:343–350. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mummadi RR, Petersen JR, Xiao SY and
Snyder N: Role of simple biomarkers in predicting fibrosis
progression in HCV infection. World J Gastroenterol. 16:5710–5715.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kroh EM, Parkin RK, Mitchell PS and Tewari
M: Analysis of circulating microRNA biomarkers in plasma and serum
using quantitative reverse transcription-PCR (qRT-PCR). Methods.
50:298–301. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Arroyo JD, Chevillet JR, Kroh EM, Ruf IK,
Pritchard CC, Gibson DF, Mitchell PS, Bennett CF,
Pogosova-Agadjanyan EL, Stirewalt DL, et al: Argonaute2 complexes
carry a population of circulating microRNAs independent of vesicles
in human plasma. Proc Natl Acad Sci USA. 108:5003–5008. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bedossa P, Dargère D and Paradis V:
Sampling variability of liver fibrosis in chronic hepatitis C.
Hepatology. 38:1449–1457. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yokoi T and Nakajima M: Toxicological
implications of modulation of gene expression by microRNAs. Toxicol
Sci. 123:1–14. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Niu X, Fu N, Du J, Wang R, Wang Y, Zhao S,
Du H, Wang B, Zhang Y, Sun D and Nan Y: miR-1273g-3p modulates
activation and apoptosis of hepatic stellate cells by directly
targeting PTEN in HCV-related liver fibrosis. FEBS Lett.
590:2709–2724. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Trebicka J, Anadol E, Elfimova N, Strack
I, Roggendorf M, Viazov S, Wedemeyer I, Drebber U, Rockstroh J,
Sauerbruch T, et al: Hepatic and serum levels of miR-122 after
chronic HCV-induced fibrosis. J Hepatol. 58:234–239. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang J, Chu ES, Chen HY, Man K, Go MY,
Huang XR, Lan HY, Sung JJ and Yu J: microRNA-29b prevents liver
fibrosis by attenuating hepatic stellate cell activation and
inducing apoptosis through targeting PI3K/AKT pathway. Oncotarget.
6:7325–7338. 2015.PubMed/NCBI
|
|
44
|
Murakami Y, Toyoda H, Tanaka M, Kuroda M,
Harada Y, Matsuda F, Tajima A, Kosaka N, Ochiya T and Shimotohno K:
The progression of liver fibrosis is related with overexpression of
the miR-199 and 200 families. PloS One. 6:e160812011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Roderburg C, Urban GW, Bettermann K, Vucur
M, Zimmermann H, Schmidt S, Janssen J, Koppe C, Knolle P, Castoldi
M, et al: Micro-RNA profiling reveals a role for miR-29 in human
and murine liver fibrosis. Hepatology. 53:209–218. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zekri AN, Youssef AS, El-Desouky ED, Ahmed
OS, Lotfy MM, Nassar AA and Bahnassey AA: Serum microRNA panels as
potential biomarkers for early detection of hepatocellular
carcinoma on top of HCV infection. Tumour Biol. 37:12273–12286.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu J, Liu Y, Dong C, Yao S, Li S, Yuan J,
Chen C, Zhao M, Lin Y and Peng Z: ARFI, Forns index, FIB-4 and APRI
diagnosis liver fibrosis in patients with chronic liver diseases.
Zhongguo Gan Zang Bing Za Zhi. 6:18–21. 2014.(In Chinese).
|
|
48
|
Vergniol J, Boursier J, Coutzac C,
Bertrais S, Foucher J, Angel C, Chermak F, Hubert IF, Merrouche W,
Oberti F, et al: Evolution of noninvasive tests of liver fibrosis
is associated with prognosis in patients with chronic hepatitis C.
Hepatology. 60:65–76. 2014. View Article : Google Scholar : PubMed/NCBI
|