Introduction
Heart failure is a costly, and fatal health problem
in the world (1). In 2015, this
disease affected approximately 40 million people in the world
(2). Approximately 50% of the heart
failure patients die in 5 years after diagnosis, but more than half
of the advanced heart failure patients die within the first year
following diagnosis (3). The cost to
health-care of heart failure is up to 30 billion dollars per year
in US alone. According to the statistical data, an urgent need is
highlighted for identifying more accurate and available heart
failure signatures with diagnostic and prognostic values.
Identification of gene signatures for complicated
diseases such as heart failure and cancer is greatly important for
diagnosis, prognosis, as well as developing personalized medicine
and treatment strategy. In recent years, an increasing number of
studies have proposed many biomarkers associated with heart
failure. For example, cardiac troponin T, a biomarker of
cardiomyocyte injury, has been demonstrated to be a predictor of
side effects for heart failure patients (4). Moreover, Kusumoto et al
(4) demonstrated that MYH7, ANXA2,
IGFBP7, as well as DESM are promising candidate signatures of heart
failure both in human and mouse. Frequently, these biomarkers
cannot reflect the substrate relationship between genotypes and
phenotypes, therefore they offer little information in the etiology
of disorders, resulting in the urgent need of elucidating the
potential pathogenesis of the disease as well as developing
treatments. Additionally, reliable and simple measurements to
detect the disorder earlier as well as to efficiently predict the
prognosis still remain insufficient.
Thus, detecting novel biomarkers is of critical
importance to understand and prevent heart failure. Long non-coding
RNAs (lncRNAs) are a class of ncRNAs with >200 nucleotides
(5). LncRNAs have been implicated to
play an important role, such as cell cycle, differentiation, and
apoptosis (6,7). Moreover, lncRNAs have been reported to
competitively regulate biological pathways and to be closely
related to heart-related diseases, for example, heart failure, and
hypertrophy (8–10). Therefore, it is necessary to
integratively analyze the joint effect of lncRNAs and pathway
topologies. Moreover, extracting pathways competitively regulated
by lncRNAs not only can provide novel insights into the potential
mechanisms, but also help us to explore the functions of lncRNAs in
a given disease (11). Currently,
the functions of lncRNAs under a given disease condition have not
been well revealed and a method for uncovering the lncRNA role is
needed. Fortunately, key local subregions, not complete pathways,
have been proposed to be more explainable to the molecular
mechanisms of diseases (12,13). It is noteworthy that detecting the
sub-pathways instead of entire pathways might be more relevant for
interpreting the related biological meaning and extracting the
functions of lncRNAs. No report on lncRNA-modulated sub-pathways in
heart failure patients is available.
In this study, to explore the etiology of heart
failure, the expression of lncRNA-mRNA and pathway topologies were
profiled to extract the lncRNAs competitively regulated
sub-pathways in heart failure using the sub-pathway based
strategy.
Materials and methods
Gene expression data acquisition and
pretreating
The gene expression profile on heart failure with
the accession number E-GEOD-26887 (14) was recruited from Arrayexpress
database, which was deposited in the platform of
A-Affy-141-Affymetrix GeneChip Human Gene 1.0 AT Array
[HuGene-1-0-st-v1]. In the E-GEOD-26887 (14), there were 19 patients with heart
failure, and 5 controls from left ventricle cardiac biopsies. In
the present work, a total of 24 subjects were used for subsequent
analysis.
Then, pre-treatment for the original microarray
profile was conducted. The specific steps contained background
correction using robust multiarray average (RMA) approach (15), normalization by means of quartile
(16), probe correction via MAS, and
summarization of probe values through Medianpolish method. After
probes were transformed to the human gene symbols, the final gene
expression matrix (covering 18,715 genes) was established.
Proposed scheme
Sub-pathway based method was used to detect
sub-pathways competitively regulated by lncRNA in heart failure.
This method included four steps: i) collection of candidate
lncRNA-mRNA interactions; ii) using KEGG pathways as backbone,
construction of condition-specific lncRNA competitively regulated
pathways (LCRP) on the basis of lncRNA data, mRNA expression data,
and lncRNA-mRNA interactions; iii) aligning competing lncRNAs and
the interesting genes into the condition-specific LCRP, then
locating sub-pathways in pathways and iv) evaluation of the
significance of candidate sub-pathways.
Collection of lncRNA-mRNA
interactions
Firstly, lncRNA-miRNA interactions were recruited
from StarBase database (version 2.0) (17), and mRNA-miRNA interactions were
collected from the databases of mir2Disease, mirTarBase, TarBase,
and miRecords (V4.0). On the basis of the common miRNAs, we
collected candidate lncRNA-mRNA competitively regulated
relationships. With the goal of guaranteeing data reliability, we
filtered and reserved several candidate competing mRNAs for each
lncRNA when they met the following criteria. The first criterion
was that we utilized hypergeometric test to estimate the importance
of the common miRNAs, and that the threshold of false discovery
rate (FDR) was defined as 0.05. The second criterion was that
Jaccard Coefficient of the common miRNAs were ordered at top 20%.
Using these criteria, 7,693 candidate lncRNA-mRNA interactions
covering 835 lncRNAs, and 1,749 mRNAs were obtained.
After that, we took the intersection between 18,715
genes of the microarray profile and 7,693 lncRNAs-mRNA
relationships, we got a total of 1,605 mRNAs as well as 94
lncRNAs.
Co-expression analysis for the
lncRNA-mRNA interactions using Pearson correlation coefficient
(PCC)
As known, PCC is an index of correlation between two
variables, which ranges from −1 to 1 (18). Hence, in our study, PCC was used to
assess the co-expression possibility for any pair of lncRNA-mRNA
interactions on the basis of the matched lncRNA and mRNA data.
After analysis using Fisher's r-to-Z transformation (19), those lncRNA-mRNA interactions that
possess r value reaching a significant positive threshold were
reserved (P<0.05).
Identification of informative
pathways
KEGG is a database, which can provide reference
knowledge to comprehensively understand cellular processes using
pathway aligning (20). Thus, in our
study, we downloaded all the KEGG background pathways based on KEGG
database. After that, the mRNAs of the reconstructed lncRNA-mRNA
interactions were mapped to the KEGG reference pathways, and then
informative pathways were obtained. We used FDR to adjust the raw
P-values by means of Benjamini-Hochberg procedure (21). We selected informative pathways using
the criteria of FDR <0.01.
Establishment of condition-specific
LCRP and hubs selection
Then, R package was used to transform the identified
informative pathways above into undirected graphs which kept the
structure of the original pathway (12). The lncRNAs in the lncRNA-mRNA
interactions reweighted by PCC were aligned into the pathway
graphs, which were as nodes via connecting to the mRNAs regulated
by lncRNAs. Ultimately, the condition-specific LCRP were
constructed, which contained lncRNA nodes as well as lncRNA-mRNA
competitively regulated edges.
To our knowledge, hubs were frequently considered
important within the biological network. Therefore, in order to
extract significant lncRNAs, we concentrated on the hub lncRNAs in
the LCRP. In this study, the top 10% of lncRNAs having the highest
degree of connectivity were extracted and determined as the hub
lncRNAs.
Positioning sub-pathways competing
regulated by lncRNAs
LncRNAs were considered as signature nodes, since
these lncRNAs referred to the genes of interest and competing
regulation. Integration of these lncRNAs with the topology features
of LCRP is beneficial to effectively locate lncRNA-mediated
subregions. The lncRNAs were embedded in the LCRP, and then
‘lenient distance’ similarity was used to identify the
lncRNA-regulated sub-pathways based on the topology characteristics
of the LCRP. In the current study, n (the number of
molecules) = 1 and s (the number of nodes in the molecules
in a pathway) = 8 were employed to extract the candidate
sub-pathways.
Assessment of significance of
candidate sub-pathway
Wallenius approximation approach was applied to
assess the significance of candidate sub-pathways based on
BiasedUrn of R package (22), to
further evaluate whether the candidate sub-pathways were
competitively regulated by lncRNAs. During the evaluation process,
the parameters below were used: the count of mRNAs of interest; the
count of background mRNAs; the count of background mRNAs in a given
sub-pathways; the number of interesting mRNAs enriched in this
given sub-pathway; and the weight score of the sub-pathway. The
weight was the strength of competing regulation by lncRNAs locating
in this sub-pathway, which was computed using the equation
described in a previous study (11).
Next, FDR was used to correct the original P-values, and the
significant sub-pathways were selected when on the threshold of FDR
was set as 0.01.
Results
Co-expression analysis for lncRNA-mRNA
interactions and informative pathways identification
Before identifying informative pathways, PCC was
used to evaluate the co-expression possibility for lncRNA-mRNA
interactions according to matched lncRNA as well as mRNA data. When
the criteria were set at a P<0.05, overall 44 lncRNAs, 165 mRNAs
and 224 co-expressed interactions were extracted (data not shown).
In order to detect seed pathways, these 165 mRNAs were then
embedded into the background pathways. Finally, we obtained a total
of 56 informative pathways when the FDR was set at 0.01 (Table I). Of note, the three most
significant pathways were PI3K-Akt signaling pathway
(FDR=2.81E-10), pathways in cancer (FDR=3.64E-10), and glioma
(FDR=1.02E-09).
 | Table I.The seed pathways. |
Table I.
The seed pathways.
| Pathways | False discovery rate
(FDR) |
|---|
| hsa05211: Renal cell
carcinoma | 8.81E-05 |
| hsa04066: HIF-1
signaling pathway | 7.99E-05 |
| hsa05169:
Epstein-Barr virus infection | 7.13E-08 |
| hsa05210: Colorectal
cancer | 7.07E-06 |
| hsa05215: Prostate
cancer | 5.94E-11 |
| hsa05219: Bladder
cancer | 5.44E-09 |
| hsa05220: Chronic
myeloid leukemia | 4.75E-08 |
| hsa04110: Cell
cycle | 4.56E-07 |
| hsa04510: Focal
adhesion | 4.45E-05 |
| hsa04012: ErbB
signaling pathway | 4.20E-08 |
| hsa05200: Pathways in
cancer | 3.64E-10 |
| hsa05222: Small cell
lung cancer | 3.64E-08 |
| hsa05218:
Melanoma | 3.31E-07 |
| hsa05223: Non-small
cell lung cancer | 2.92E-07 |
| hsa04151: PI3K-Akt
signaling pathway | 2.81E-10 |
| hsa05203: Viral
carcinogenesis | 2.32E-06 |
| hsa04115: p53
signaling pathway | 2.09E-07 |
| hsa05120: Epithelial
cell signaling in Helicobacter pylori infection | 1.90E-06 |
| hsa05161: Hepatitis
B | 1.55E-08 |
| hsa05212: Pancreatic
cancer | 1.52E-07 |
| hsa05213: Endometrial
cancer | 1.50E-05 |
| hsa04722:
Neurotrophin signaling pathway | 1.12E-05 |
| hsa05214: Glioma | 1.02E-09 |
| hsa04150: mTOR
signaling pathway | 9.92E-03 |
| hsa04620: Toll-like
receptor signaling pathway | 8.55E-03 |
| hsa04810: Regulation
of actin cytoskeleton | 7.55E-03 |
| hsa05133:
Pertussis | 5.90E-03 |
| hsa05152:
Tuberculosis | 5.67E-03 |
| hsa04520: Adherens
junction | 5.17E-03 |
| hsa04662: B cell
receptor signaling pathway | 4.83E-03 |
| hsa04666: Fc gamma
R-mediated phagocytosis | 3.73E-03 |
| hsa04540: Gap
junction | 3.29E-03 |
| hsa04914:
Progesterone-mediated oocyte maturation | 3.09E-03 |
| hsa05162:
Measles | 2.66E-03 |
| hsa05160: Hepatitis
C | 2.52E-03 |
| hsa03008: Ribosome
biogenesis in eukaryotes | 2.37E-03 |
| hsa04740: Olfactory
transduction | 2.36E-03 |
| hsa04728:
Dopaminergic synapse | 2.28E-03 |
| hsa04730: Long-term
depression | 1.92E-03 |
| hsa04350: TGF-β
signaling pathway | 1.79E-03 |
| hsa05166: HTLV–I
infection | 1.66E-03 |
| hsa04910: Insulin
signaling pathway | 9.71E-04 |
| hsa04912: GnRH
signaling pathway | 8.83E-04 |
| hsa04062: Chemokine
signaling pathway | 8.78E-04 |
| hsa05168: Herpes
simplex infection | 7.27E-04 |
| hsa04720: Long-term
potentiation | 6.21E-04 |
| hsa04330: Notch
signaling pathway | 5.84E-04 |
| hsa04660: T cell
receptor signaling pathway | 4.37E-04 |
| hsa05142: Chagas
disease (American trypanosomiasis) | 4.37E-04 |
| hsa05216: Thyroid
cancer | 3.72E-04 |
| hsa05145:
Toxoplasmosis | 2.67E-04 |
| hsa04310: Wnt
signaling pathway | 2.38E-04 |
| hsa04621: NOD-like
receptor signaling pathway | 2.27E-04 |
| hsa05221: Acute
myeloid leukemia | 2.27E-04 |
| hsa04725:
Cholinergic synapse | 1.74E-04 |
| hsa04010: MAPK
signaling pathway | 1.52E-04 |
Selecting lncRNA-regulated
sub-pathways
Then, the 56 informative pathways were embedded in
undirected graphs, and the 44 lncRNAs were mapped to the pathway
graphs as nodes. After that, a condition-specific LCRP was built
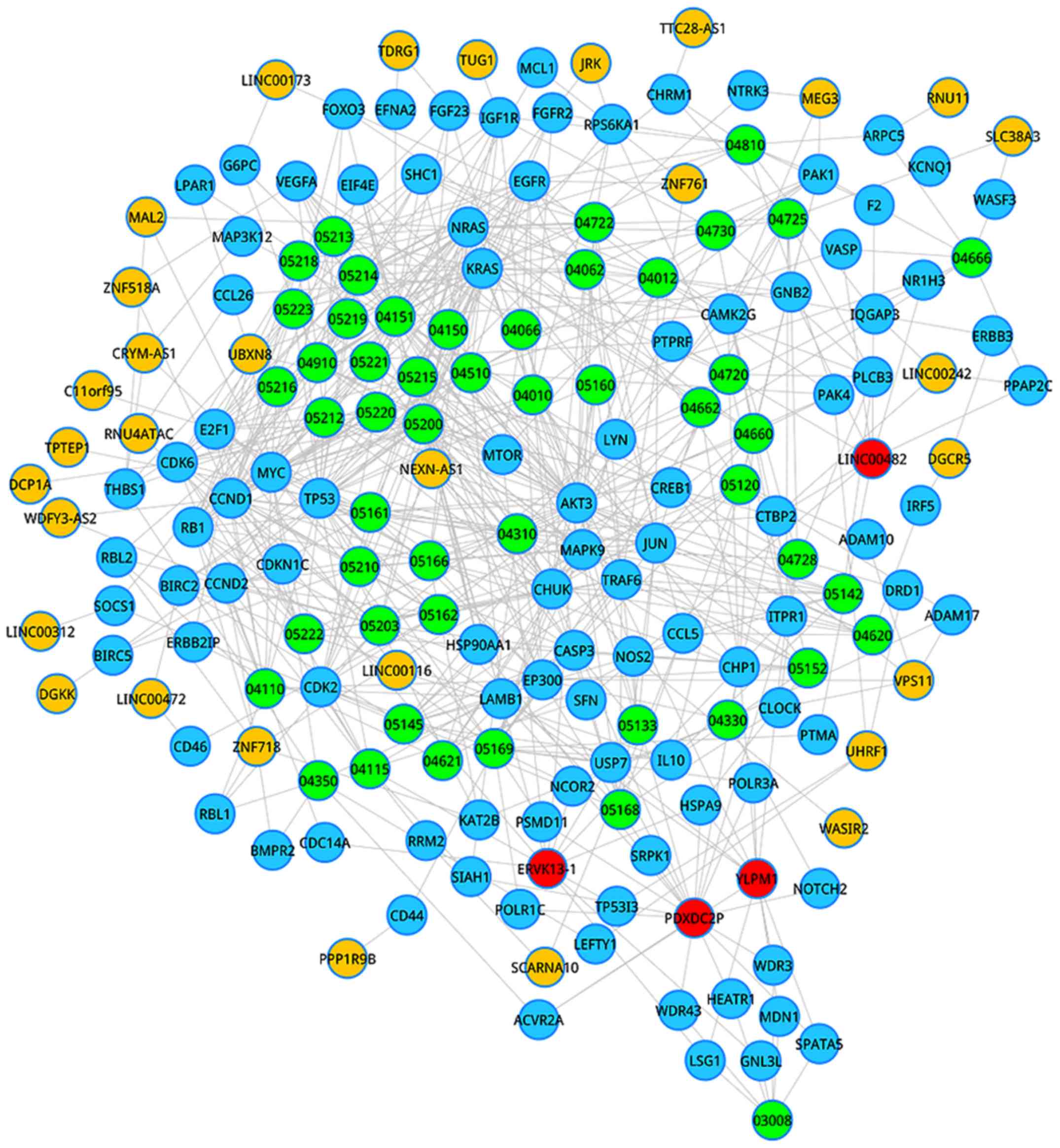
(Fig. 1). To extract crucial lncRNAs
of heart failure, a degree analysis was implemented for all nodes
of the LCRP. According to degree distribution, a total of 4 hub
lncRNAs were selected, including ERVK13-1, YLPM1, PDXDC2P, and
LINC00482.
Based on the information of the LCRP, we discovered
a total of 36 lncRNAs competitively regulating sub-pathways
involved in 40 complete pathways when the FDR was <0.01
(Table II). In advanced
investigation, we mainly concentrated on the top three sub-pathways
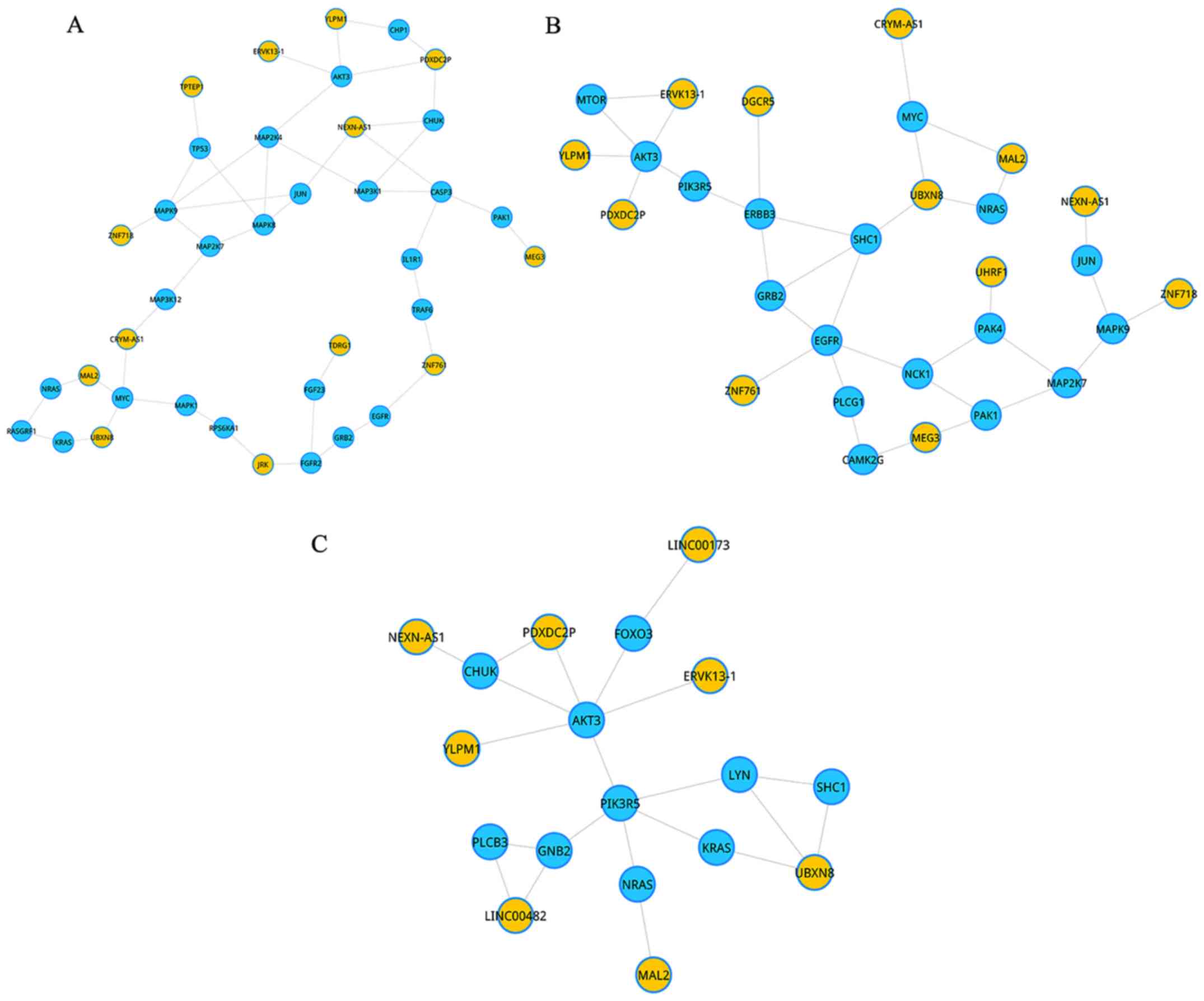
(Fig. 2). The first most significant
sub-pathway was path: 04010_1, which was a subpart of MAPK
signaling pathway (Fig. 2A).
Subsequently, we further analyzed this sub-pathway, and we found
that a total of 13 lncRNAs competitively regulated this subpart
synergistically. Within these lncRNAs, two lncRNAs (PDXDC2P and
NEXN-AS1), directly regulated three genes. In addition, MYC was
coordinately regulated by three lncRNAs (CRYM-AS1, MAL2, and
UBXN8). Moreover, AKT3 was also simultaneously regulated by three
lncRNAs (ERVK13-1, YLPM1, and PDXDC2P).
 | Table II.Sub-pathways list based on
FDR<0.01 |
Table II.
Sub-pathways list based on
FDR<0.01
| Pathway IDs | Pathway name | FDR |
|---|
| 04010_1 | MAPK signaling
pathway | 2.80E-16 |
| 04012_1 | ErbB signaling
pathway | 5.35E-16 |
| 04062_2 | Chemokine signaling
pathway | 1.96E-15 |
| 04110_1 | Cell cycle | 5.16E-15 |
| 04115_1 | p53 signaling
pathway | 4.50E-15 |
| 04151_1 | PI3K-Akt signaling
pathway | 4.53E-15 |
| 04310_1 | Wnt signaling
pathway | 4.62E-15 |
| 04510_1 | Focal adhesion | 4.87E-15 |
| 04722_1 | Neurotrophin
signaling pathway | 1.88E-14 |
| 05161_1 | Hepatitis B | 3.28E-14 |
| 05166_1 | HTLV–I
infection | 5.96E-14 |
| 05168_1 | Herpes simplex
infection | 7.40E-14 |
| 05169_1 | Epstein-Barr virus
infection | 1.56E-13 |
| 05200_1 | Pathways in
cancer | 5.33E-13 |
| 05212_1 | Pancreatic
cancer | 5.92E-13 |
| 05214_1 | Glioma | 2.06E-12 |
| 05215_1 | Prostate
cancer | 5.99E-12 |
| 05218_1 | Melanoma | 6.87E-11 |
| 05222_1 | Small cell lung
cancer | 6.85E-11 |
| 05223_1 | Non-small cell lung
cancer | 2.16E-10 |
| 04728_1 | Dopaminergic
synapse | 5.61E-10 |
| 05203_2 | Viral
carcinogenesis | 7.29E-10 |
| 04066_1 | HIF-1 signaling
pathway | 1.61E-09 |
| 05152_2 | Tuberculosis | 2.54E-09 |
| 04720_1 | Long-term
potentiation | 7.51E-09 |
| 04810_3 | Regulation of actin
cytoskeleton | 8.92E-09 |
| 04662_1 | B cell receptor
signaling pathway | 2.21E-08 |
| 05145_1 | Toxoplasmosis | 3.08E-08 |
| 03008_1 | Ribosome biogenesis
in eukaryotes | 7.25E-08 |
| 05210_1 | Colorectal
cancer | 1.79E-07 |
| 05220_1 | Chronic myeloid
leukemia | 9.38E-07 |
| 05221_1 | Acute myeloid
leukemia | 2.29E-06 |
| 04620_1 | Toll-like receptor
signaling pathway | 1.61E-06 |
| 04330_1 | Notch signaling
pathway | 3.67E-06 |
| 04660_2 | T cell receptor
signaling pathway | 8.54E-06 |
| 05213_1 | Endometrial
cancer | 9.12E-06 |
| 04910_1 | Insulin signaling
pathway | 3.27E-05 |
| 05160_1 | Hepatitis C | 1.59E-05 |
| 05162_2 | Measles | 9.14E-05 |
| 04350_1 | TGF-β signaling
pathway | 1.09E-04 |
The second significant subregion was path: 04012-1,
an important subpart in ErbB signaling pathway (Fig. 2B). Similar to the first significant
sub-pathway, gene AKT3 was also simultaneously regulated by three
lncRNAs (ERVK13-1, YLPM1, and PDXDC2P).
The third sub-pathway, path: 04062_2, was a part of
chemokine signaling pathway (Fig.
2C). Similar to the first and the second significant
sub-pathways, gene AKT3 was also simultaneously regulated by three
lncRNAs (ERVK13-1, YLPM1, and PDXDC2P). Notably, lncRNA UBXN8
competitively regulated three genes (KRAS, LYN, and SHC1). Thus,
these results demonstrated that this approach could identify
biological meaningful sub-pathways, and highlighted some critical
lncRNAs in heart failure condition.
Discussion
Currently, alteration of lncRNA competitively
regulated functions may result in the occurrence and progression of
diseases, but, a better understanding of this regulatory mechanism
may give more and better opportunities for developing new
therapies. Hence we constructed an LCRP for the informative
pathways, and integrated the lncRNAs with the topology features of
LCRP. Normally, comparing with the entire pathways, sub-pathways
may be more explainable to the molecular mechanisms underlying
disorders. Consequently, detecting the sub-pathways which were
regarded to be more possibly disrupted pathways from the LCRP would
be the best means to discriminate the heart failure group from the
control group.
Thus, an LCRP was constructed with 36 lncRNAs, 102
mRNA nodes and 40 pathways by combining gene expression profile,
lncRNA data, pathway information and PCC related analyses. Finally,
based on the information of the LCRP, a total of 36 lncRNAs
competitively regulating sub-pathways involved in 40 complete
pathways were identified. Among these 40 pathways, we mainly
concentrated on the top three sub-pathways that competitively
regulated by lncRNAs, including path: 04010_1, which was a subpart
of MAPK signaling pathway, path: 04012-1, an important subpart in
ErbB signaling pathway, and path: 04062_2, a part of chemokine
signaling pathway.
Taking MAPK signaling pathways as examples to
discuss, the MAPK signaling pathway has been implicated to control
myocyte growth that responds to pathological stimuli in the heart
(23). Heart failure is considered
to be a remodeling process of myocyte cells, and cellular as well
as extracellular factors participate in this remodeling process to
further cause the changes in myocardial structures and functions.
Matrix metalloproteinases (MMPs) are the factors participating in
this remodeling process. Significantly, MMPs secretion has been
reported to be dependent on MAPK or MAPK signaling pathway
(24,25). Thus, we speculate that a close
relationship exists between the heart failure and MAPK signaling
pathway.
Of note, in the first, second, and third most
significant sub-pathways, gene AKT3 was simultaneously regulated by
three lncRNAs (ERVK13-1, YLPM1, and PDXDC2P). Moreover, ERVK13-1,
YLPM1, and PDXDC2P were all hubs in the LCRP. YLPM1 was also termed
as ZAP3. A former study has demonstrated that the highest
expression of ZAP3 seems to be in brain, heart and kidneys
(26). Of note, ZAP3 has been
implicated to decrease telomerase activity (27). Further, telomerase ablation can cause
the occurrence of heart failure (28). Thus, we infer that lncRNA YLPM1 might
play crucial roles in the development of heart failure, partially
via regulating the telomerase activity. Although few reports have
suggested the link between lncRNAs (ERVK13-1, or PDXDC2P) and heart
failure, we found these lncRNAs regulated the common gene AKT3 in
our study. AKT3 is one of the three isoforms of AKT which mediates
cardiac growth, cell death, and myocardial angiogenesis in cardiac
myocytes (29). Moreover, coronary
vasculature, and cell death are essential for the maintenance of
the contractile function, especially in heart failure (30). Accordingly, we infer that lncRNAs
(ERVK13-1 and PDXDC2P) have indirect relationships with heart
failure.
The current study had several limitations. The study
population was small. The lack of verification of microarray
results by means of quantitative real-time PCR technique or western
blot based on animal or patient tissues was another important
drawback. Thus, further studies in larger study samples are
necessary to confirm these findings in terms of the efficiency as
diagnostic or prognostic biomarkers of heart failure.
In conclusion, we applied sub-pathway-based analysis
by introducing the concept of pathway topology, lncRNA data, and
subregion of pathways. The importance of this method is that it can
help to understand the etiology of diseases, and provide important
insight into the functions of lncRNAs in heart failure. Based on
the results, we showed that MAPK signaling pathway, ErbB signaling
pathway, as well as chemokine signaling pathway were the
significant sub-pathways for heart failure. Nevertheless, the
verification using other datasets is implemented to demonstrate
that these sub-pathways are useful in classifying heart failure and
normal group.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
DMH conceived the study, analyzed the data and
drafted the manuscript. The author has read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The author declares that he has no competing
interests.
References
|
1
|
McMurray JJ and Pfeffer MA: Heart failure.
Lancet. 365:1877–1889. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Charlson FJ, Erskine HE, Ferrari AJ, Leung
J, Whiteford HA, Abajobir AA, Knibbs LD, Lalloo R, Scott JG and Guo
Y; GBD 2015 Disease and Injury Incidence and Prevalence
Collaborators, : Global, regional, and national incidence,
prevalence, and years lived with disability for 310 diseases and
injuries, 1990–2015: A systematic analysis for the Global Burden of
Disease Study 2015. Lancet. 388:1545–1602. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Go AS, Mozaffarian D, Roger VL, Benjamin
EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, et al
American Heart Association Statistics Committee and Stroke
Statistics Subcommittee, : Executive summary: heart disease and
stroke statistics - 2013 update: a report from the American Heart
Association. Circulation. 127:143–152. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kusumoto A, Miyata M, Kubozono T, Ikeda Y,
Shinsato T, Kuwahata S, Fujita S, Takasaki K, Yuasa T, Hamasaki S,
et al: Highly sensitive cardiac troponin T in heart failure:
Comparison with echocardiographic parameters and natriuretic
peptides. J Cardiol. 59:202–208. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Di Gesualdo F, Capaccioli S and Lulli M: A
pathophysiological view of the long non-coding RNA world.
Oncotarget. 5:10976–10996. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhao W, Luo J and Jiao S: Comprehensive
characterization of cancer subtype associated long non-coding RNAs
and their clinical implications. Sci Rep. 4:65912014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gomes da Silva AM and Silbiger VN: miRNAs
as biomarkers of atrial fibrillation. Biomarkers. 19:631–636. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liang WC, Fu WM, Wong CW, Wang Y, Wang WM,
Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF, et al: The lncRNA H19
promotes epithelial to mesenchymal transition by functioning as
miRNA sponges in colorectal cancer. Oncotarget. 6:22513–22525.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Papait R, Kunderfranco P, Stirparo GG,
Latronico MVG and Condorelli G: Long noncoding RNA: A new player of
heart failure? J Cardiovasc Transl Res. 6:876–883. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Han P, Li W, Lin CH, Yang J, Shang C,
Nuernberg ST, Jin KK, Xu W, Lin CY, Lin CJ, et al: A long noncoding
RNA protects the heart from pathological hypertrophy. Nature.
514:102–106. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shi X, Xu Y, Zhang C, Feng L, Sun Z, Han
J, Su F, Zhang Y, Li C and Li X: Subpathway-LNCE: Identify
dysfunctional subpathways competitively regulated by lncRNAs
through integrating lncRNA-mRNA expression profile and pathway
topologies. Oncotarget. 7:69857–69870. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li C, Han J, Yao Q, Zou C, Xu Y, Zhang C,
Shang D, Zhou L, Zou C, Sun Z, et al: Subpathway-GM: Identification
of metabolic subpathways via joint power of interesting genes and
metabolites and their topologies within pathways. Nucleic Acids
Res. 41:e1012013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li X, Shen L, Shang X and Liu W:
Subpathway analysis based on signaling-pathway impact analysis of
signaling pathway. PLoS One. 10:e01328132015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Greco S, Fasanaro P, Castelvecchio S,
D'Alessandra Y, Arcelli D, Di Donato M, Malavazos A, Capogrossi MC,
Menicanti L and Martelli F: MicroRNA dysregulation in diabetic
ischemic heart failure patients. Diabetes. 61:1633–1641. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Irizarry RA, Bolstad BM, Collin F, Cope
LM, Hobbs B and Speed TP: Summaries of Affymetrix GeneChip probe
level data. Nucleic Acids Res. 31:e152003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bolstad BM, Irizarry RA, Astrand M and
Speed TP: A comparison of normalization methods for high density
oligonucleotide array data based on variance and bias.
Bioinformatics. 19:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang HB, Jiang ZB and Li M: Research on
the typical miRNA and target genes in squamous cell carcinoma and
adenocarcinoma of esophagus cancer with DNA microarray. Pathol
Oncol Res. 20:245–252. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nahler G: Pearson correlation coefficient.
Dictionary of Pharmaceutical Medicine. Springer-Verlag Vienna;
Austria: pp. 1322009, View Article : Google Scholar
|
|
19
|
Best DJ and Roberts DE: Algorithm AS 89:
The upper tail probabilities of Spearman's Rho. Appl Stat.
24:377–379. 1975. View
Article : Google Scholar
|
|
20
|
Kanehisa M, Araki M, Goto S, Hattori M,
Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T,
et al: KEGG for linking genomes to life and the environment.
Nucleic Acids Res. 36:D480–D484. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Stat Soc B. 57:289–300. 1995.
|
|
22
|
Epstein MP, Duncan R, Jiang Y, Conneely
KN, Allen AS and Satten GA: A permutation procedure to correct for
confounders in case-control studies, including tests of rare
variation. Am J Hum Genet. 91:215–223. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Molkentin JD and Dorn GW II: Cytoplasmic
signaling pathways that regulate cardiac hypertrophy. Annu Rev
Physiol. 63:391–426. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kim ES, Kim MS and Moon A:
TGF-beta-induced upregulation of MMP-2 and MMP-9 depends on p38
MAPK, but not ERK signaling in MCF10A human breast epithelial
cells. Int J Oncol. 25:1375–1382. 2004.PubMed/NCBI
|
|
25
|
Qiu Q, Yang M, Tsang BK and Gruslin A:
EGF-induced trophoblast secretion of MMP-9 and TIMP-1 involves
activation of both PI3K and MAPK signalling pathways. Reproduction.
128:355–363. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ulke-Lemée A, Trinkle-Mulcahy L, Chaulk S,
Bernstein NK, Morrice N, Glover M, Lamond AI and Moorhead GBG: The
nuclear PP1 interacting protein ZAP3 (ZAP) is a putative nucleoside
kinase that complexes with SAM68, CIA, NF110/45, and HNRNP-G.
Biochim Biophys Acta. 1774:1339–1350. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Armstrong L, Lako M, van Herpe I, Evans J,
Saretzki G and Hole N: A role for nucleoprotein Zap3 in the
reduction of telomerase activity during embryonic stem cell
differentiation. Mech Dev. 121:1509–1522. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Leri A, Franco S, Zacheo A, Barlucchi L,
Chimenti S, Limana F, Nadal-Ginard B, Kajstura J, Anversa P and
Blasco MA: Ablation of telomerase and telomere loss leads to
cardiac dilatation and heart failure associated with p53
upregulation. EMBO J. 22:131–139. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shiojima I and Walsh K: Regulation of
cardiac growth and coronary angiogenesis by the Akt/PKB signaling
pathway. Genes Dev. 20:3347–3365. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chaanine AH and Hajjar RJ: AKT signalling
in the failing heart. Eur J Heart Fail. 13:825–829. 2011.
View Article : Google Scholar : PubMed/NCBI
|