Introduction
Colorectal cancer (CRC) is a leading cause of cancer
and associated mortalities worldwide. With the improvement in
living standards and change in lifestyle and dietary structure, the
incidence of CRC has demonstrated annual increases. CRC arises from
the accumulation of genetic alterations in colonic epithelial cell
and in the majority of cases, the cancer develops via the normal
adenoma-carcinoma sequence (1).
MicroRNAs (miRNAs/miRs) are a class of small, endogenous,
non-coding, regulatory RNA molecules that inhibit gene translation
after forming a complex referred to as the RNA-interference-induced
silencing complex or after induction by mRNA cleavage (2). miRNAs may function as oncogenes by
targeting tumor suppressor genes or as tumor suppressors by either
inhibiting cellular oncogene expression or regulating cell
apoptosis, as well as tumor occurrence and development (3,4).
DNA methylation is a type of epigenetic modification
that refers to changes in gene expression without alteration of the
gene sequence. Under DNA methyltransferase (DNMT) catalysis, a
methyl group is covalently bound to the 5′carbon of cytosine in the
genomic CpG dinucleotide, which is unstable and may spontaneously
undergo deamination to form thymine, thereby affecting gene
expression (5). Aberrant
methylation, including DNA hypomethylation and/or promoter gene CpG
hypermethylation, is implicated in the development of a variety of
tumor types, including CRC (6). The
expression of miR-145 in CRC cells has been reported to be either
activated or inhibited by the methylation of histones at the core
promoter regions in the upper regulatory sequence of pre-miR-145
(7). miR-663 expression was
downregulated and the promoter was consistently and significantly
methylated in human myeloid leukemia cell lines; following
treatment with the demethylation reagent 5-aza-2′-deoxycytidine
(5-Aza-dC), miR-663 expression was significantly upregulated
(8). Another epigenetic phenomenon
is acetylation of histones, which is regulated by histone
acetyltransferase (HAT) and histone deacetylase (HDAC). HDAC is
able to remove the acetyl group from the histone lysine residue,
increase the DNA-binding ability of histones and make the promoter
less accessible to transcriptional regulatory elements, finally
causing transcriptional repression. The effect of HAT is, however,
just the opposite. Together, they synergistically maintain the
normal acetylation level of histone (9). HDAC inhibition with suberoylanilide
hydroxamic acid increased acetylated histone enrichment on the
Cdc20b/miR-449a promoter, consequently upregulating miR-449a levels
during diabetes (10). Together,
these studies suggested the susceptibility of miRNAs to epigenetic
regulation.
miR-32 is an intronic miRNA, located within intron
14 of C9orf5, the gene encoding transmembrane protein 245
(TMEM245). In previous studies by our group, miR-32 was identified
to be upregulated in CRC tissues (11,12).
Overexpression of miR-32 led to increased cell proliferation,
migration and invasion, and reduced apoptosis of CRC cells, by
inhibiting the target gene phosphatase and tensin homolog. However,
the mechanism underlying the increase in miR-32 levels remains
elusive. In the present study, the methylation status of miR-32
promoter in CRC and normal colonic epithelial cells was
investigated using bisulfate sequencing polymerase chain reaction
(PCR) (BSP), and the role of methylation and histone acetylation in
the regulation of miR-32 expression in CRC cells was examined.
Materials and methods
Cell culture
The CRC cell lines HT-29 and HCT-116 (Type Culture
Collection of the Chinese Academy of Sciences, Shanghai, China)
were cultured in RPMI-1640 medium (GE Healthcare Life Sciences,
Little Chalfont, UK) and the normal colonic epithelial cell line
NCM460 (BeNa Culture Collection, Jiangsu, China) was cultured in
Dulbecco's modified Eagle's medium/F12 (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), each supplemented with 10%
fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 100
IU/ml penicillin and 100 µg/ml streptomycin, at 37°C in a
humidified atmosphere with 5% CO2.
BSP of CpG islands in the miR-32
promoter
Genomic DNA was extracted from the HT-29, HCT-116
and NCM460 cells using the TIANamp Genomic DNA kit (cat. no. DP304;
Tiangen Biotech, Beijing, China). It was subjected to bisulfite
modification using the EpiTect Bisulfite kit (cat. no. 59826;
Qiagen GmBH, Hilden, Germany), according to the manufacturer's
protocol. The target region of the promoter of TMEM245, the host
gene of miR-32, containing 94 CpG sites, located between −195 and
+521 bp numbered from the start codon ATG, was amplified by PCR
using DreamTaq Hot Start DNA Polymerase (Thermo Fisher Scientific,
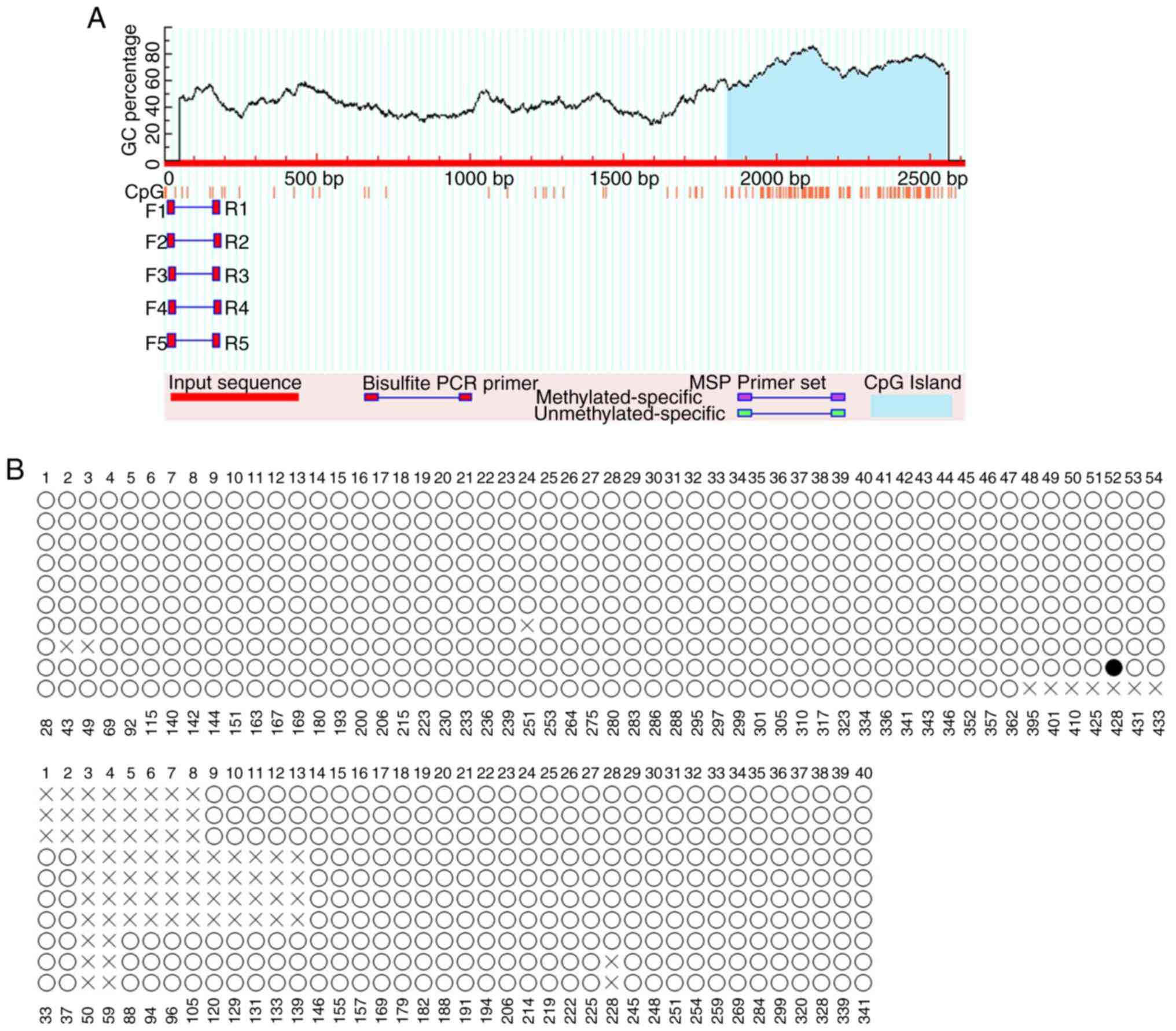
Inc.) and specific primers (Fig. 1;
Table I). The primers were designed
by MethPrimer (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi)
and synthesized by YINGBIO technology Co., Ltd. (Shanghai, China).
The PCR products were purified and sub-cloned to T vector (T-Vector
pMD 19; cat. no. 3271, Takara Bio Inc., Otsu, Japan). Colony PCR
was performed using M13 primers (forward,
5′-CGCCAGGGTTTTCCCAGTCACGAC-3′ and reverse,
5′-AGCGGATAACAATTTCACACAGGA-3′). A total of 10 independent colonies
for each tested region were selected and sequenced, and the extent
of methylation was assessed by identifying the number and position
of methylated cytosine residues.
 | Table I.Primer sequences for bisulfite
sequencing. |
Table I.
Primer sequences for bisulfite
sequencing.
| Name | Sequence |
|---|
| F1 |
5′-TTTTGTAAATTTGTTAGTAGGATTTTG-3′ |
| R1 |
5′-TCCAAAATAAAATAAACCAACACC-3′ |
| F2 |
5′-TGTTGGTTTATTTTATTTTGGAGG-3′ |
| R2 |
5′-TCACCCACAAACTACTAAAATAATCC-3′ |
5-Aza-dC and trichostatin A (TSA)
treatment
HT-29 cells were incubated for 72 h in media
containing 10 µM of the demethylating agent 5-Aza-dC
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). The culture medium
was replaced with new medium containing 5-Aza-dC every 24 h. For
inhibition of histone deacetylation, 1 µM TSA (Sigma-Aldrich; Merck
KGaA), which specifically inhibits HDAC, was added to the medium
for 24 h. For combined inhibition, the cells were incubated with 10
µM 5-Aza-dC for 48 h, followed by an additional 24 h with 1 µM TSA,
as described previously (13,14).
Untreated HT-29 cells were used as a control. Total RNA was
purified and subjected to reverse transcription-quantitative
(RT-q)PCR for evaluating miR-32 expression.
Cell transfection
DNMT1 overexpression plasmid pRP-hDNMT1 was
synthesized by VectorBuilder Inc. (Guangzhou, China). HT-29 cells
were seeded onto 6-well plates, at a density of 3×105 cells per
well. The cells were cultured at 37°C in a 5% CO2
incubator in antibiotic-free medium (RPMI-1640 medium with 10%
fetal bovine serum) 24 h prior to transfection. HT-29 cells were
transfected with 2 µg of pRP-hDNMT1 or pRP vector as a negative
control. Cells were transfected using Lipofectamine 2000™ reagent
(Thermo Fisher Scientific, Inc.) in Opti-MEM I (Gibco; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
The relative levels of miR-32 in transfected cells were examined by
RT-qPCR.
RT-qPCR
The expression of miR-32 in various treated cells
(as described above) was evaluated using RT-qPCR. Total RNA was
extracted with RNAiso Plus (Takara Bio Inc.), and reverse
transcribed using the Mir-X miRNA First Strand Synthesis kit (cat.
no. 638315; Clontech Laboratories, Inc., Mountainview, CA, USA). U6
small nuclear RNA was used as an internal control for miR-32. PCR
was performed using TB Green Premix Ex Taq II (Tli RNaseH Plus;
cat. no. RR820A; Takara Bio, Inc.) in a LightCycler 480 II system
(Roche Diagnostics, Basel, Switzerland). Sequence-specific forward
primers for mature miR-32 and U6 internal control were
5′-CGGTATTGCACATTACTAAGTTGCA-3′ and 5′-CTCGCTTCGGCAGCACA-3′,
respectively. The primers were synthesized by Sangon Biotech
(Shanghai, China); mRQ 3′ Primer was included in the Mir-X miRNA
First Strand Synthesis kit. The PCR conditions were 30 sec at 95°C,
followed by 40 cycles of 5 sec at 95°C and 20 sec at 60°C. The
relative levels of miR-32 were determined using the
2−∆∆Cq method (15).
Statistical analysis
5-Aza-dC and TSA treatments as well as cell
transfections were repeated in triplicate. Values are expressed as
the mean ± standard deviation. Student's t-test was used for
comparison between two groups, while multiple-group comparisons
were performed using analysis of variance. All analyses were
performed with SPSS 19.0 (IBM Corp., Armonk, NY, USA). P<0.05
was considered to indicate statistical significance.
Results
Methylation analysis of the miR-32
promoter in CRC cells and normal colonic epithelial cells
To identify promoters harboring CpG islands, a
Bioinformatics search of the promoter of TMEM245, the host gene of
miR-32, was performed. Analysis of the TMEM245 promoter sequence,
predicted by MethPrimer, identified one CpG island (Fig. 2A). The methylation status of the CpG
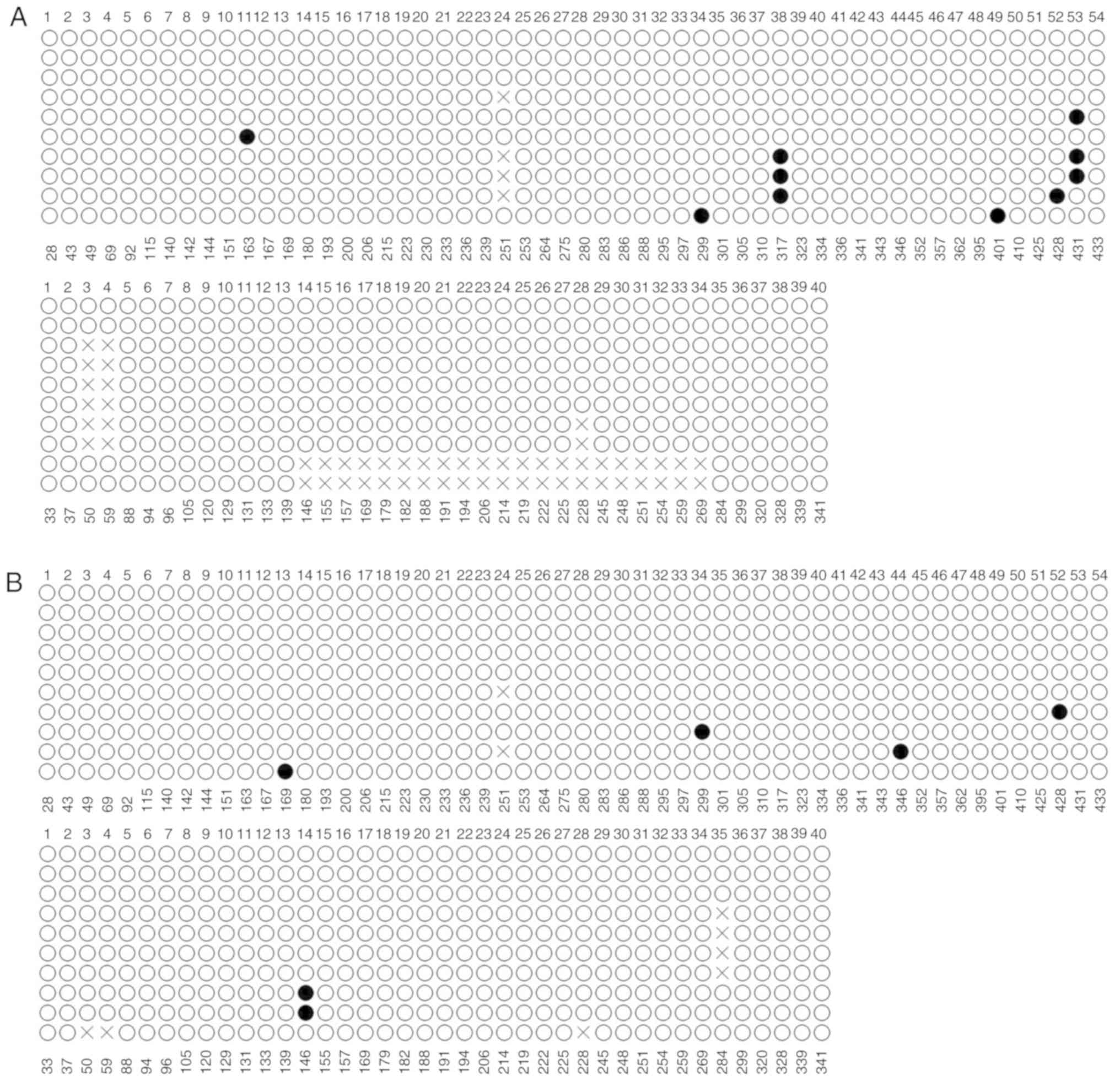
sites was examined by BSP. As presented in Fig. 2B, CpG sites in the promoter region
were rarely methylated (solid circles) in HCT-116 cells in all of
the randomly selected clones, with a methylation rate of 0.12%. The
CpG sites were also observed to be rarely methylated in clones
derived from HT-29 and NCM460 cells, with methylation rates of 1.14
and 0.64 %, respectively (Fig. 3A and
B).
miR-32 expression following 5-Aza-dC
and/or TSA treatment
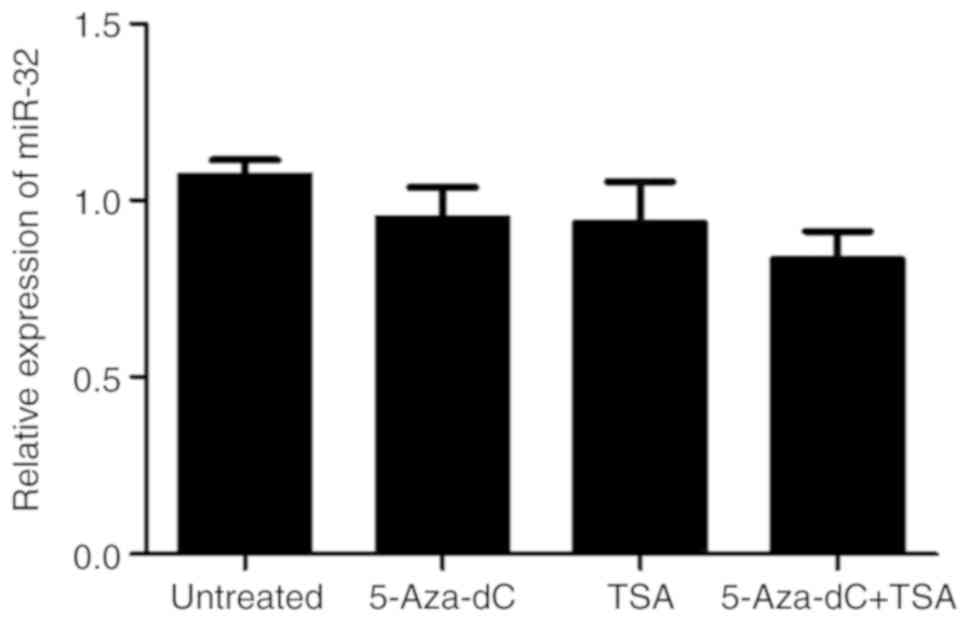
To determine the effect of methylation and histone
deacetylation on the expression levels of miR-32, HT-29 cells were
treated with 5-Aza-dC and/or TSA. Compared with that in untreated
controls, treatment with 5-Aza-dC or TSA alone or in combination
exerted no marked effect on the expression of miR-32 (P>0.05;
Fig. 4).
miR-32 expression following
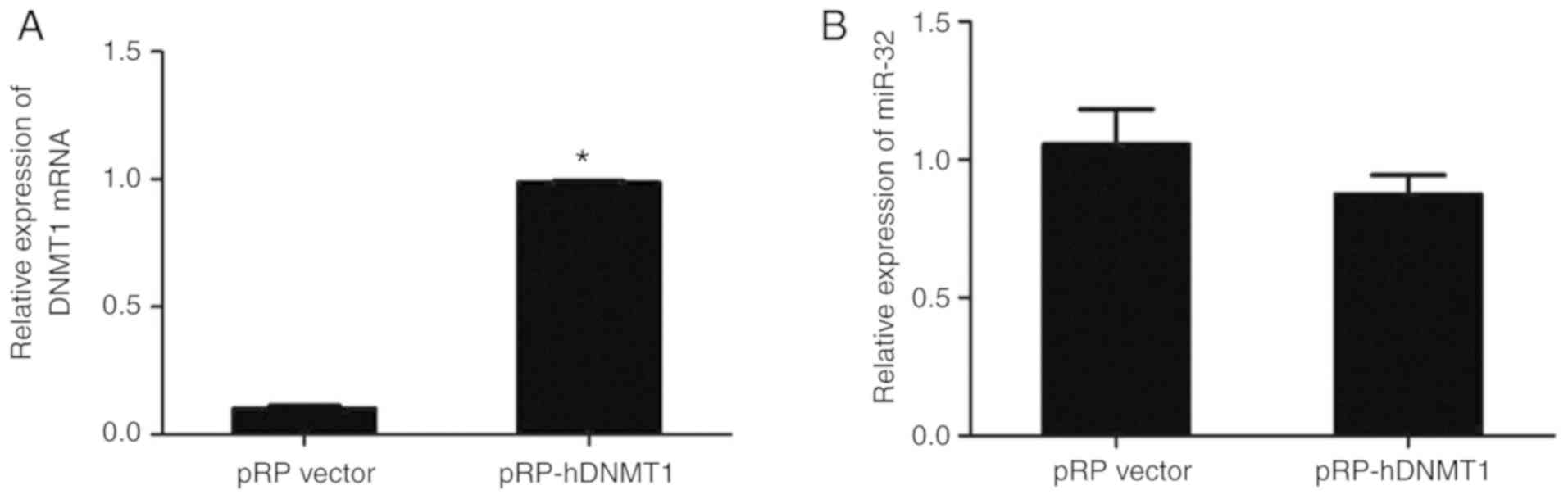
transfection with DNMT1 plasmid
HT-29 cells were transfected with DNMT1 plasmid to
enhance methylation. The RT-qPCR results indicated that DNMT1
expression was significantly higher in HT-29 cells transfected with
pRP-DNMT1 compared with that in cells transfected with pRP vector
(P<0.05; Fig. 5A). However,
transfection with pRP-DNMT1 did not change the expression level of
miR-32. Together, the results suggested that miR-32 expression is
not markedly affected by DNA methylation or histone deacetylation
in HT-29 cells (P>0.05; Fig.
5B).
Discussion
DNA methylation and histone acetylation represent
two types of epigenetic modification that regulate chromatin
structure and gene transcription. The CpG dinucleotides tend to
cluster in regions called CpG islands. DNA methylation mostly
occurs on CpG islands, with 60% of genes in mammalian genomes
containing promoter-proximal CpG islands (16). CpG island methylation in those
regions is an important mechanism of gene expression regulation
(16), and may be involved in the
regulation of miRNA expression.
Dysregulation of miRNA expression in different
cancer types may be attributed to the improper binding of
transcription factors to response elements of the promoter regions,
and to epigenetic changes, including aberrant DNA methylation and
histone modification (17). Numerous
miRNAs are epigenetically regulated by DNA methylation and/or
histone modification (18). Xia
et al (19) reported that
decreased miR-98 levels in glioma tissues and cell lines are
associated with DNA methylation. Treatment with 5-Aza-dC
significantly increased the expression of miR-98 in glioma cells.
Furthermore, downregulation of miR-200b was reported to be
associated with CpG island hypermethylation in bladder cancer cells
and pharmacological de-methylation using 5-Aza-dC was able to
restore miR-200b expression (20).
miR-21 upregulation, induced by histone acetylation, was even
significantly elevated after TSA treatment (21). Another study indicated that miR-214
was upregulated in HeLa cells upon treatment with 5-Aza-dC and/or
TSA; in fact, the most significant increase of miR-214 was seen in
HeLa cells treated with 5-Aza-dC and TSA simultaneously (22).
miR-32 is an intronic miRNA encoded by TMEM245
(23). Intragenic miRNAs are
reported to be embedded within the host gene and thought to be
regulated by the host gene promoter (24). They are transcribed in parallel with
their host transcripts (25). Human
papillomavirus type16 protein E6 may indirectly regulate miR-23b,
an intronic miRNA, through the methylation of its host gene C9orf3
(26). Chen et al (27) indicated that TSA caused an
upregulation of the expression of miR-15a/16-1, residing in the
host tumor suppressor Dleu2 gene, by increasing the histone
acetylation in the promoter region of Dleu2/miR-15a/16-1 in lung
cancer cells. Yang et al (28) indicated that SAHA, an HDAC inhibitor,
repressed the transcription of miR-20a, miR-93 and miR-106b by
repressing their host genes (miR-17–92 cluster and minichromosome
maintenance complex component 7) in hepatocellular carcinoma cells.
The intronic miRNA miR-449a and the host gene cell division cycle
(Cdc)20b were highly upregulated in response to HDAC inhibition.
Bioinformatics analyses identified a common promoter for Cdc20b and
miR-449a, featuring significant histone acetylation, which was
further verified by augmented occupancy of acetylated histones on
the Cdc20b/miR-449a promoter in SAHA-treated C2C12 cells (10). It was also reported that the
transcript levels of C9orf5 (TMEM245) and miR-32 are positively and
significantly correlated (23).
The present study examined whether miR-32 expression
is affected by epigenetic mechanisms, namely DNA methylation and
histone acetylation. The level of miR-32 promoter methylation was
identified to be low, with a methylation rate of 0.12, 1.14 and
0.64% in HCT-116, HT-29 and NCM460 cells, respectively. Regardless
of whether normal colonic mucosal cells or CRC cells were observed,
the promoter of miR-32 was hypomethylated, and the present study
aimed to further clarify whether methylation has a significant
effect on the expression of miR-32. For this, the levels of miR-32
expression after treatment with 5-Aza-dC and/or TSA were determined
by RT-qPCR, and the results demonstrated no significant effect.
Furthermore, DNMT1 plasmid transfection had no effect on the
expression levels of miR-32. It therefore appears unlikely that
these epigenetic modifiers are involved in the regulation of miR-32
expression, but these results require to be verified in other CRC
cell lines and in CRC tissues in future studies. Since this type of
epigenetic modification appears to have no effect on the expression
of miR-32, it is further required to clarify whether other
regulatory pathways, including transcription factors or interaction
with non-coding RNAs, are able to regulate miR-32 expression.
Acknowledgements
Not applicable.
Funding
This study was supported by the Guangdong Natural
Science Foundation of China (grant no. 2017A030313546).
Availability of data and materials
Available from the corresponding authors on
reasonable request.
Authors' contributions
WW and ZY designed the experiments. WW, YS and TW
performed the experiments. WW and QJ performed statistical analysis
of the data obtained and drafted the manuscript. ZY revised the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Vogelstein B, Fearon ER, Hamilton SR, Kern
SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM and Bos
JL: Genetic alterations during colorectal-tumor development. N Engl
J Med. 319:525–532. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang W, Liu J and Wang G: The role of
microRNAs in human breast cancer progression. Tumour Biol.
35:6235–6344. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zheng Q, Chen C, Guan H, Kang W and Yu C:
Prognostic role of microRNAs in human gastrointestinal cancer: A
systematic review and meta-analysis. Oncotarget. 8:46611–46623.
2017.PubMed/NCBI
|
|
5
|
Moore LD, Le T and Fan G: DNA methylation
and its basic function. Neuropsychopharmacology. 38:23–38. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tse JWT, Jenkins LJ, Chionh F and
Mariadason JM: Aberrant DNA Methylation in colorectal cancer: What
should we target? Trends Cancer. 3:698–712. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang W, Ji G, Xiao X, Chen X, Qin WW, Yang
F, Li YF, Fan LN, Xi WJ, Huo Y, et al: Epigenetically regulated
miR-145 suppresses colon cancer invasion and metastasis by
targeting LASP1. Oncotarget. 7:68674–68687. 2016.PubMed/NCBI
|
|
8
|
Yan-Fang T, Jian N, Jun L, Na W, Pei-Fang
X, Wen-Li Z, Dong W, Li P, Jian W, Xing F and Jian P: The promoter
of miR-663 is hypermethylated in Chinese pediatric acute myeloid
leukemia (AML). BMC Med Genet. 14:742013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Spange S, Wagner T, Heinzel T and Krämer
OH: Acetylation of non-histone proteins modulates cellular
signalling at multiple levels. Int J Biochem Cell Biol. 41:185–198.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Poddar S, Kesharwani D and Datta M:
Histone deacetylase inhibition regulates miR-449a levels in
skeletal muscle cells. Epigenetics. 11:579–587. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu W, Yang J, Feng X, Wang H, Ye S, Yang
P, Tan W, Wei G and Zhou Y: MicroRNA-32 (miR-32) regulates
phosphatase and tensin homologue (PTEN) expression and promotes
growth, migration, and invasion in colorectal carcinoma cells. Mol
Cancer. 12:302013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu W, Yang P, Feng X, Wang H, Qiu Y, Tian
T, He Y, Yu C, Yang J, Ye S and Zhou Y: The relationship between
and clinical significance of MicroRNA-32 and phosphatase and tensin
homologue expression in colorectal cancer. Genes Chromosomes
Cancer. 52:1130–1140. 2013. View Article : Google Scholar
|
|
13
|
Mizuno Y, Maemura K, Tanaka Y, Hirata A,
Futaki S, Hamamoto H, Taniguchi K, Hayashi M, Uchiyama K, Shibata
MA, et al: Expression of delta-like 3 is downregulated by aberrant
DNA methylation and histone modification in hepatocellular
carcinoma. Oncol Rep. 39:2209–2216. 2018.PubMed/NCBI
|
|
14
|
Guo LH, Li H, Wang F, Yu J and He JS: The
tumor suppressor roles of miR-433 and miR-127 in gastric cancer.
Int J Mol Sci. 14:14171–14184. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Weber M and Schubeler D: Genomic patterns
of DNA methylation: Targets and function of an epigenetic mark.
Curr Opin Cell Biol. 19:273–280. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lopez-Serra P and Esteller M: DNA
methylation-associated silencing of tumor-suppressor microRNAs in
cancer. Oncogene. 31:1609–1622. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang Z, Yao H, Lin S, Zhu X, Shen Z, Lu G,
Poon WS, Xie D, Lin MC and Kung HF: Transcriptional and epigenetic
regulation of human microRNAs. Cancer Lett. 331:1–10. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xia Z, Qiu D, Deng J, Jiao X, Yang R, Sun
Z, Wan X and Li J: Methylation-induced downregulation and
tumor-suppressive role of microRNA-98 in glioma through targeting
Sal-like protein 4. Int J Mol Med. 41:2651–2659. 2018.PubMed/NCBI
|
|
20
|
Shindo T, Niinuma T, Nishiyama N, Shinkai
N, Kitajima H, Kai M, Maruyama R, Tokino T, Masumori N and Suzuki
H: Epigenetic silencing of miR-200b is associated with cisplatin
resistance in bladder cancer. Oncotarget. 9:24457–24469. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Song WF, Wang L, Huang WY, Cai X, Cui JJ
and Wang LW: MiR-21 upregulation induced by promoter zone histone
acetylation is associated with chemoresistance to gemcitabine and
enhanced malignancy of pancreatic cancer cells. Asian Pac J Cancer
Prev. 14:7529–7536. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang F, Liu M, Li X and Tang H: MiR-214
reduces cell survival and enhances cisplatin-induced cytotoxicity
via down-regulation of Bcl2l2 in cervical cancer cells. FEBS Lett.
587:488–495. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ambs S, Prueitt RL, Yi M, Hudson RS, Howe
TM, Petrocca F, Wallace TA, Liu CG, Volinia S, Calin GA, et al:
Genomic profiling of microRNA and messenger RNA reveals deregulated
microRNA expression in prostate cancer. Cancer Res. 68:6162–6170.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Erdogan B, Bosompem A, Peng D, Han L,
Smith E, Kennedy ME, Alford CE, Wu H, Zhao Z, Mosse CA, et al:
Methylation of promoters of microRNAs and their host genes in
myelodysplastic syndromes. Leuk Lymphoma. 54:2720–2727. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rodriguez A, Griffiths-Jones S, Ashurst JL
and Bradley A: Identification of mammalian microRNA host genes and
transcription units. Genome Res. 14:1902–1910. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yeung CL, Tsang TY, Yau PL and Kwok TT:
Human papillomavirus type 16 E6 suppresses microRNA-23b expression
in human cervical cancer cells through DNA methylation of the host
gene C9orf3. Oncotarget. 8:12158–12173. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen CQ, Chen CS, Chen JJ, Zhou LP, Xu HL,
Jin WW, Wu JB and Gao SM: Histone deacetylases inhibitor
trichostatin A increases the expression of Dleu2/miR-15a/16-1 via
HDAC3 in non-small cell lung cancer. Mol Cell Biochem. 383:137–148.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang H, Lan P, Hou Z, Guan Y, Zhang J, Xu
W, Tian Z and Zhang C: Histone deacetylase inhibitor SAHA
epigenetically regulates miR-17–92 cluster and MCM7 to upregulate
MICA expression in hepatoma. Br J Cancer. 112:112–121. 2015.
View Article : Google Scholar : PubMed/NCBI
|