Introduction
Atherosclerosis is the underlying cause of human
death, which is involved in the onset of many diseases, including
myocardial infarction, stroke, ischemic heart pain, and sudden
cardiac death (1,2). It has been widely reported that
atherosclerosis mainly occurs in the intima of medium arteries at
the blood flow disorganized regions and can be triggered by the
interaction of endothelial dysfunction and subendothelial
lipoprotein retention (3). A
non-resolving inflammatory response is stimulated by the
atherosclerotic triggering process and thus induces intimal
destruction, end-organ ischemia and arterial thrombosis. Therefore,
studies on the pathophysiology of atherosclerosis are urgently
needed. Metabolomics has been reported to be used to identify the
specific biomarkers and discover the possible metabolic pathways,
which contributes to better exploration of the pathology of
diseases.
Metabolites as the ultimate products of cellular
regulatory process, are generally regarded as the final response of
biological systems to genetic or environmental changes. It is well
known that metabolite levels can be used to evaluate the current
physiological state of human body. The identification of
disease-associated metabolites has been demonstrated to be
important for better understanding the pathophysiology of
metabolites and enhancing the clinical diagnosis of diseases
(4,5). Metabolomics/metabonomics is a rapidly
evolving field of biochemical research following genomics,
transcriptomics and proteomics, which is used to quantitatively
measure the metabolic response to pathophysiological stimuli. The
study of metabolomics not only contributes to our understanding of
the underlying molecular mechanisms of diseases, but also helps us
for exploring the biomarkers of disease diagnosis (6,7). Up to
now, thousands of metabolites have been identified by metabolomics
technologies, including gas chromatography-mass spectrometry
(GC-MS), liquid chromatography-mass spectrometry (LC-MS), and
nuclear magnetic resonance (NMR) (8). However, investigation of the
identification and prioritization of high-risk metabolites
associated with atherosclerosis are still a challenging task.
Generally, metabolites hardly function alone, and
would be affected by genome and phenome when they are regarded as
the link of genotypes and phenotypes. As known, one or two
metabolites usually do not have a serious effect on the onset of a
disease, but they diffuse among function-associated metabolites and
genes organized into a complex network. Therefore, metabolites
related to adjacent functions generally tend to be associated with
phenotypically same or similar diseases (9). Furthermore, there are different
functions for the metabolites in the network, and in the same
functional module, some metabolites with strong correlation would
play a special role in biological function. Currently, with the
development of various ‘omics’ analysis, such as genomics,
phenomics, and proteomics, it is beneficial to provide valuable
information for prioritizing the high-risk metabolites associated
with diseases. Therefore, a comprehensive and accurate information
of disease-related metabolites can be provided by studying a
multi-omics network involved in genes, metabolites and
phenotypes.
In this study, an algorithm named MetPriCNet was
used to predict the prioritization of atherosclerosis-related risk
metabolites, which was performed by integrating multi-omics data.
Yao et al have demonstrated that MetPriCNet not only has a
high prediction validity in overall performance but also possesses
an excellent identification ability of disease types (10). Consequently, a global metabolic
network is involved in multi-omics network data, including
phenotypic networks, genetic network, metabolic network and
interaction network. Considering the global functional correlations
of metabolites in this intricate network, the prioritization of key
metabolites related to a certain phenotype can be confirmed. The
freely accessible web of MetPriCNet is available at http://www.bio-bigdata.net/MetPriCNet/.
Materials and methods
Obtaining multi-omics integrated
information
A multi-omics composite network involved in genes,
metabolites, phenotypes, gene-metabolite interactions,
phenotype-gene interactions and phenotype-metabolite interactions
was constructed. The data sources of relative information are
described in the sections.
Gene network (AG)
The human protein interacted network containing
1,048.576 interaction relationships were obtained from STRING
(http://string-db.org/) database (11). We transformed the protein ID and gene
name for removing the repetitive interaction relationships.
Consequently, a PPI gene network with 16,785 nodes and 1,515.370
interaction relationships was obtained.
Metabolite network
(AM)
Firstly, 4,994 human metabolites were collected from
the metabolite pathways of KEGG and HMDB databases, the human
pathways of MSEA, SMPDB and Reactome databases (12–16).
Subsequently, STITCH database was selected to collect the human
metabolites and metabolite interaction relationships that were
contained in the 4,994 human metabolites (17). Eventually, a metabolite network
containing 3764 human metabolites and 74,667 metabolite interaction
relationships was constructed.
Phenotype network (AP)
We constructed the phenotype network containing
5,080 phenotypes that have similarity scores through applying the
phenotype-phenotype similarity relationships from van Driel et
al (18).
Gene-metabolite interaction networks
(AGM)
The chemicals, human genes and STITCH-associated
information were extracted for obtaining the interaction between
genes and metabolites. Based on the 4,994 human metabolites, we
removed the genes that were not contained in the gene network and
the metabolites that were not included in the metabolite network.
Ultimately, a total of 192,763 gene-metabolite interactions
involved in 12,342 genes and 3,278 metabolites were obtained.
Phenotype-gene interaction networks
(AGP)
The interactions between phenotypes and genes were
collected from the Morbid Map file from OMIM. Similarly, the
phenotypes that were not included in the phenotype network and the
genes that were not involved in the gene network were removed.
Consequently, we obtained 2,603 phenotype-gene interactions with
1,715 genes and 1,886 phenotypes. Notably, the weighted score of
each phenotype-gene association was defined as 1.
Phenotype-metabolite interaction
network (APM)
The associations between phenotypes and metabolites
can be extracted from HMDB database. Similarly, we removed the
needless phenotypes and metabolites that were not found in the
phenotype or metabolite network, respectively. Finally, 664
phenotype-metabolite associations containing 149 phenotypes and 388
metabolites were preserved.
Collecting disease-related gene
expression data
Gene expression profiles associated with
atherosclerosis were collected from ArrayExpress database
(http://www.ebi.ac.uk/arrayexpress/)
the access number is E-GEOD-57691. Then 10 samples called normal
aortic tissue and 9 specimens named aortic atheroma tissue in
expression sets were selected for further analysis. For improving
the quality of the data, several standard pretreatments were
performed, including probe correction, background correction,
normalization and summarization of expressed value (19–21). As
a result, an expression profile dataset containing 19,211 genes was
obtained for subsequent analysis. The atherosclerosis-associated
phenotype information with access number 143890 was obtained from
Online Mendelian Inheritance in Man (OMIM; http://omim.org/) database. Integrating the gene
expression profiles obtained from ArrayExpress and phenotype
information containing related disease genes obtained from the
Morbid Map file in OMIM database, the seed genes with differential
expression were selected. To correctly screen the differentially
expressed genes (DEGs), limma package was used to compute the
differential expression (22).
Moreover, t-test and F-test were conducted to analyze the gene
expression matrix. Furthermore, lmFit function was used to perform
the empirical Bayes (eBayes) statistics and a false discovery rate
(FDR) calibration of P-values for the data (23,24).
Eventually, we obtained the candidate genes with large difference
for the establishment of the gene network.
Constructing an intricate multi-omics
network
The six networks (AG,
AM, AP, AGM,
AGP and APM) obtained above
from multiple data sets were integrated into one weighted adjacency
matrix A for constructing an intricate multi-omics network.
Subsequently, based on the adjacency matrix, a column-normalized
adjacency matrix W can be acquired, which is a transition matrix of
the intricate network, Wij can be used to calculate the
transition probability of each node from i to j. Ultimately, six
networks were merged into a weighted composite network, and Yao
et al have described the specific algorithm (10).
Identification of candidate metabolite
prioritization based on the weighted multi-omics composite
network
To acquire the global omics information in the
intricate network and to identify the prioritization of candidate
metabolites, the random walk with restart (RWR) method was
introduced into the intricate multi-omics network (25). On account of the proximity of each
metabolite to the seed nodes, the prioritization of candidate
metabolites was identified by the use of the RWR method, which was
achieved through simulating the random walk processes of
metabolites starting from a seed node. For each step, metabolite
shifts from the current node(s) to its adjacent node(s) with
probability 1-α or goes back to the seed node(s) with probability
α. The probability is calculated as follows:
Pk+1=(1-α)WPk+αP0
Where P° is the initial probability vector,
Pk is the probability vector that the i-th element keeps
in the node i at step k, W is the transition matrix of the
composite network. Notably, the obtained DEGs were used to provide
the gene node in the intricate multi-omics network. It is worth
mentioning that a stable probability calculated with the RWR method
would be achieved by multiple iterations, and the probability
reaches the steady state and terminates the iteration processes, as
the change between Pk+1 and Pk is lower than
10−10. Eventually, the relative probability of each
metabolite in the composite network was obtained.
Results
The construction of an
atherosclerosis-related composite multi-omics network
Atherosclerosis-related genes with larger
differential expression were used to construct the gene network. In
this study, we screened the atherosclerosis-related DEGs with limma
package, and then applied t-test, F-test, eBayes statistics as well
as FDR calibration. Ultimately, DEGs with the criteria of the
adjusted P<0.05 were selected for establishing the gene network.
Furthermore, constructing an atherosclerosis-related composite
multi-omics network containing the interactions of gene-gene,
phenotype-phenotype, metabolite-metabolite, gene-phenotype,
gene-metabolite and phenotype-metabolite by importing the gene
network. Consequently, we obtained 9,993 nodes and 10,271.126 edges
involved in atherosclerosis-related DEGs, phenotypes and
metabolites.
Identifying the
atherosclerosis-related metabolite prioritization
The prioritization of the atherosclerosis-related
metabolites can be identified by evaluating the relation score of
each metabolite in the composite network. Through combining the
original weight score that was calculated with the RWR method in
equation (1), the new relation score
of each metabolite was computed, and further ranked the metabolite
by the corresponding score. In the present study, we screened out
50 metabolites as the prioritizing candidate metabolites which
ranked in the top 50 of relation score (Table I). Then, we searched for the
prioritizing candidate metabolites from the multi-omics composite
network, and thus extracting the atherosclerosis-related
prioritizing metabolite composite network. The associations of
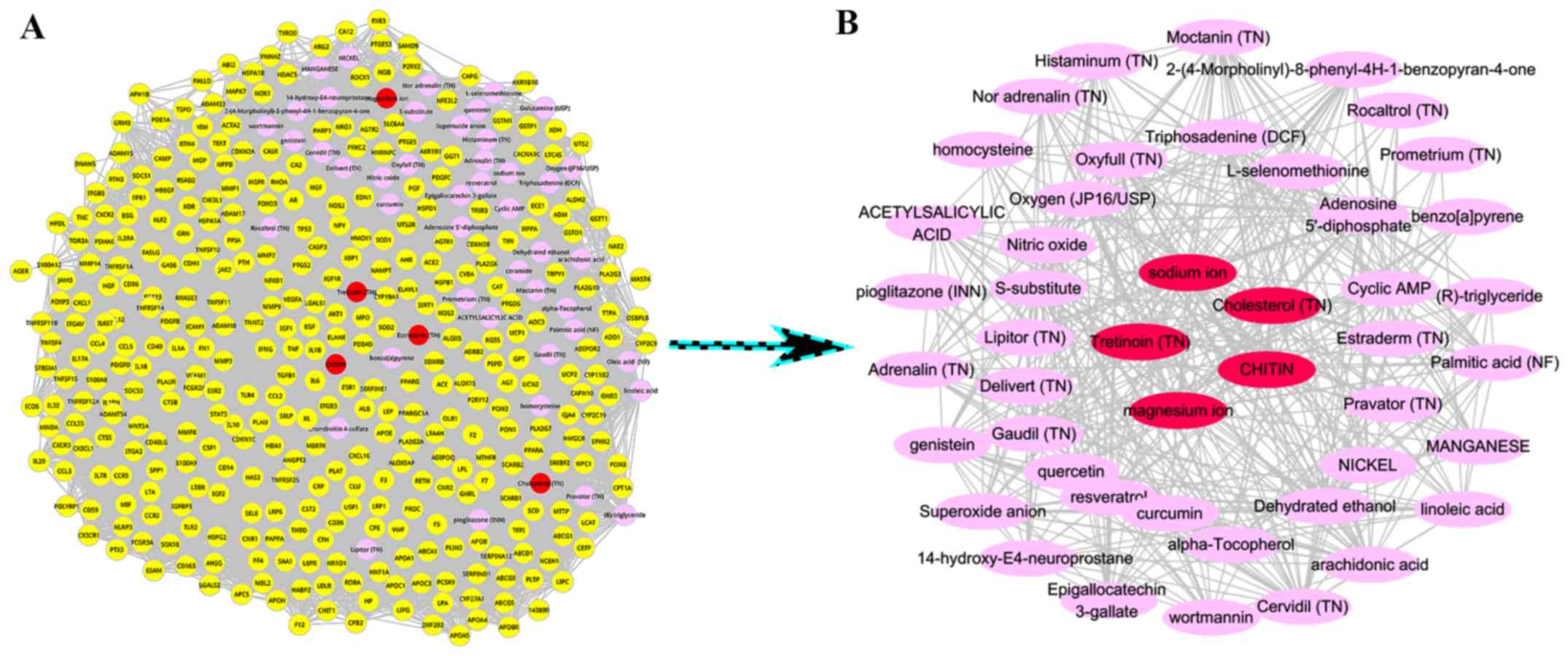
candidate metabolites are shown in Fig.
1, where the top 5 metabolites with higher score were marked in
red, including Tretinoin (TN) (score=0.000677293), Estraderm (TN)
(score=0.000622015), Cholesterol (TN) (score=0.000548486), CHITIN
(score=0.000537016), magnesium ion (score=0.000473373). Seed nodes
were marked in yellow and other metabolites were in pink.
 | Table I.The parameter of top 50 metabolites
identified using the RWR method in the weighted composite
network. |
Table I.
The parameter of top 50 metabolites
identified using the RWR method in the weighted composite
network.
| Rank | Metabolite CID | Metabolite
name | Score |
|---|
| 1 | 444795 | Tretinoin (TN) | 0.000677293 |
| 2 | 5757 | Estraderm (TN) | 0.000622015 |
| 3 | 5997 | Cholesterol
(TN) | 0.000548486 |
| 4 | 24139 | CHITIN | 0.000537016 |
| 5 | 888 | Magnesium ion | 0.000473373 |
| 6 | 6022 | Adenosine
5′-diphosphate | 0.000465706 |
| 7 | 5280360 | Cervidil (TN) | 0.000462843 |
| 8 | 5994 | Prometrium
(TN) | 0.000450763 |
| 9 | 5957 | Triphosadenine
(DCF) | 0.000426371 |
| 10 | 753 | Moctanin (TN) | 0.000415301 |
| 11 | 945 | Nitric oxide | 0.000375328 |
| 12 | 5280961 | Genistein | 0.00035226 |
| 13 | 784 | Oxyfull (TN) | 0.000345901 |
| 14 | 60823 | Lipitor (TN) | 0.00034207 |
| 15 | 5280453 | Rocaltrol (TN) | 0.000339252 |
| 16 | 923 | Sodium ion | 0.000331388 |
| 17 | 172198 | Delivert (TN) | 0.000330823 |
| 18 | 445154 | Resveratrol | 0.000317558 |
| 19 | 77999 | Gaudil (TN) | 0.000314271 |
| 20 | 444899 | Arachidonic
acid | 0.000314223 |
| 21 | 702 | Dehydrated
ethanol | 0.000295643 |
| 22 | 5280343 | Quercetin | 0.000294725 |
| 23 | 3973 |
2-(4-Morpholinyl)-8-phenyl-4H-1-benzopyran-4-one | 0.000276657 |
| 24 | 65064 | Epigallocatechin
3-gallate | 0.000273649 |
| 25 | 312145 | Wortmannin | 0.000250849 |
| 26 | 2336 | Benzo[a]pyrene | 0.000248994 |
| 27 | 2244 | Acetylsalicylic
acid | 0.000233603 |
| 28 | 14985 | α-Tocopherol | 0.000233033 |
| 29 | 105024 |
L-selenomethionine | 0.000229817 |
| 30 | 445639 | Oleic acid
(NF) | 0.00022218 |
| 31 | 935 | Nickel | 0.000203525 |
| 32 | 4829 | Pioglitazone
(INN) | 0.000203337 |
| 33 | 12035 | S-substitute | 0.000201323 |
| 34 | 5816 | Adrenalin (TN) | 0.000197363 |
| 35 | 24766 | Chondroitin
4-sulfate | 0.000196775 |
| 36 | 439260 | Nor adrenalin
(TN) | 0.000194196 |
| 37 | 6076 | Cyclic AMP | 0.000189559 |
| 38 | 5359597 | Superoxide
anion | 0.000189271 |
| 39 | 5280450 | Linoleic acid | 0.000188959 |
| 40 | 774 | Histaminum
(TN) | 0.000188733 |
| 41 | 5460048 |
(R)-triglyceride | 0.000187959 |
| 42 | 158 |
14-hydroxy-E4-neuroprostane | 0.000184986 |
| 43 | 778 | Homocysteine | 0.000183673 |
| 44 | 977 | Oxygen
(JP16/USP) | 0.000180497 |
| 45 | 969516 | Curcumin | 0.000177388 |
| 46 | 54687 | Pravator (TN) | 0.000174654 |
| 47 | 33032 | Gulutamine
(USP) | 0.000173862 |
| 48 | 23930 | Manganese | 0.000172384 |
| 49 | 17756770 | Ceramide | 0.000172203 |
| 50 | 985 | Palmitic acid
(NF) | 0.000168691 |
Identification of co-expression genes
in the composite network
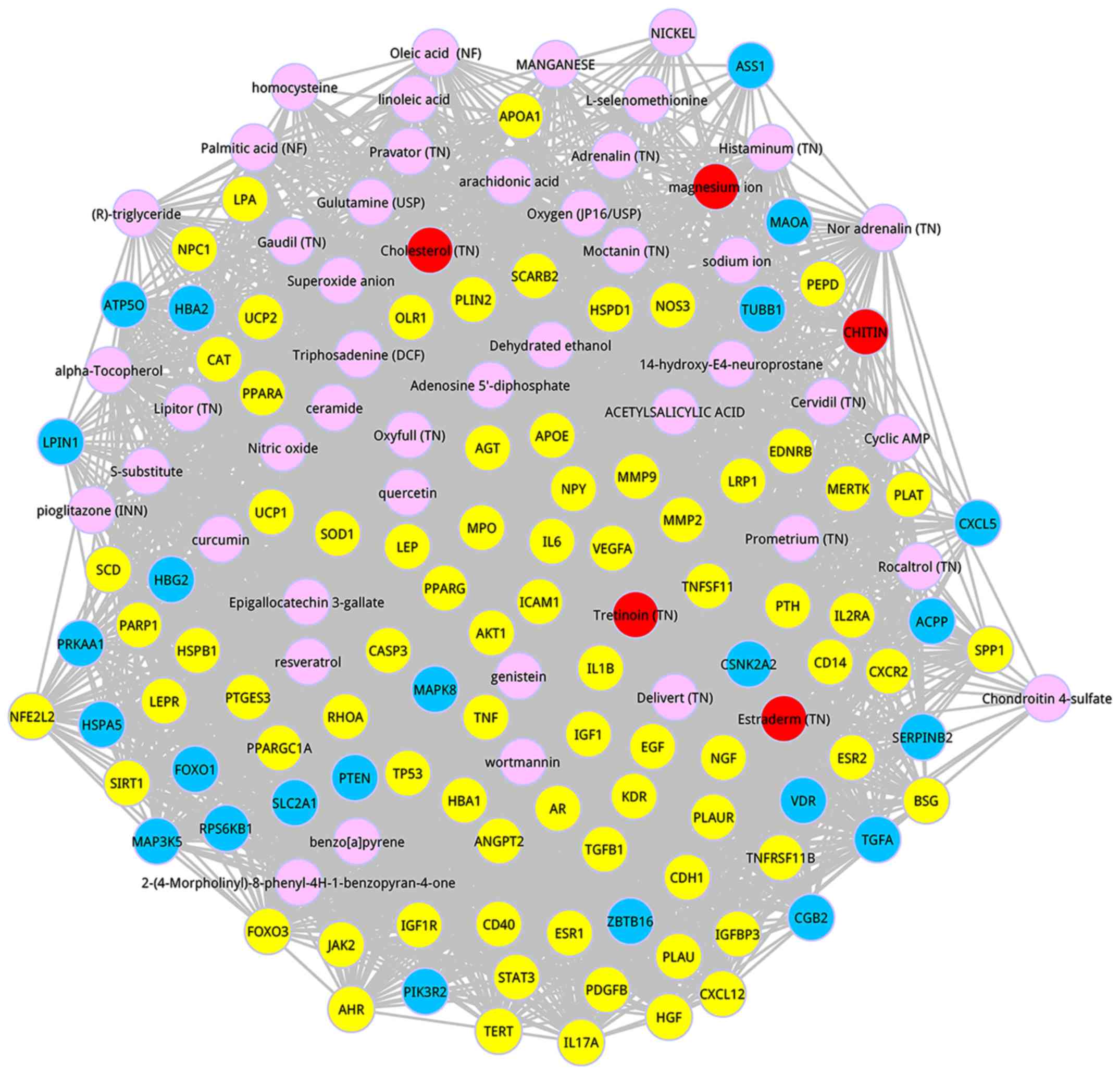
Based on the 50 prioritizing candidate metabolites,
genes that were interacted with metabolites in the composite
network were chosen. Furthermore, we analyzed the closeness between
the known genes and candidate metabolites and screened out the
co-expression genes, the closeness of which achieved a greater
score than the average and ranked in top 100. Consequently, a
co-expression network that was involved in the composite network
was obtained (Fig. 2). In this
network, yellow nodes represent the seed nodes, blue nodes
represent the co-expression genes, pink nodes are the prioritizing
candidate metabolites and red nodes are the top 5 metabolites.
Ultimately, 24 co-expression genes marked in blue were identified,
including 16 genes with degree greater than 30 and 8 genes with
degree lower than 30. The parameter information of the 24
co-expression genes is listed in Table
II. Significantly, MAPK8 (also known as JNK1), that has the
highest degree among those genes, was reported to be necessary for
EC apoptosis and lipid deposition in the early atherogenesis
(26). Furthermore, FOXO1 gene has
been demonstrated to be expressed strongly in atherosclerotic
plaques, and to participate in multiple atherogenic pathways of
endothelial cells (27). By
analyzing the degree of metabolites in the co-expression network,
it can be found that 9 metabolites with degree more than 70 were
confirmed, that is, Tretinoin (TN), Estraderm (TN), genistein,
Oxyfull (TN), Dehydrated ethanol, quercetin,
2-(4-Morpholinyl)-8-phenyl-4H-1-benzopyran-4-one, resveratrol and
Nitric oxide. It is worth mentioning that Tretinoin (TN) was the
metabolite that had the highest score, and Estraderm (TN) was the
second metabolite followed by Tretinoin (TN).
 | Table II.Co-expression genes associated with
the top 50 metabolites of atherosclerosis in the multi-omics
composite network. |
Table II.
Co-expression genes associated with
the top 50 metabolites of atherosclerosis in the multi-omics
composite network.
| Co-expression
gene | Degree | Closeness | adj.P.Val |
|---|
| MAPK8 | 100 | 3.644899487 | 8.74E-05 |
| PTEN | 81 | 3.526471283 | 2.45E-06 |
| FOXO1 | 59 | 2.841944445 | 2.20E-06 |
| SLC2A1 | 57 | 4.021206485 | 6.39E-05 |
| TGFA | 50 | 3.38473277 | 5.27E-05 |
| VDR | 47 | 3.99371055 | 3.87E-05 |
| HSPA5 | 46 | 2.673613247 | 8.11E-05 |
| CGB2 | 42 | 2.298998928 | 6.32E-05 |
| RPS6KB1 | 41 | 2.920705668 | 6.56E-05 |
| MAP3K5 | 41 | 3.568998598 | 1.67E-05 |
| HBG2 | 39 | 2.68166783 | 5.61E-05 |
| ATP5O | 38 | 3.171190898 | 6.13E-06 |
| ACPP | 36 | 3.916740409 | 6.60E-05 |
| ZBTB16 | 35 | 2.764784562 | 1.44E-06 |
| PIK3R2 | 34 | 3.981092177 | 1.04E-04 |
| SERPINB2 | 33 | 3.450172722 | 7.11E-05 |
| PRKAA1 | 29 | 3.124904434 | 9.48E-05 |
| LPIN1 | 24 | 3.82077733 | 1.41E-06 |
| TUBB1 | 22 | 3.474172 | 9.47E-05 |
| CXCL5 | 21 | 3.173767298 | 1.91E-05 |
| HBA2 | 18 | 2.356349598 | 7.23E-05 |
| ASS1 | 17 | 3.587780531 | 5.29E-06 |
| MAOA | 15 | 3.373168305 | 3.44E-05 |
| CSNK2A2 | 13 | 3.148020812 | 3.90E-05 |
Discussion
Atherosclerosis, a progressive disease, is the major
cause of cardiovascular disease and stroke, and the leading cause
of death worldwide. It generally occurs in the large arteries that
accumulate a large number of lipids and fibrous elements (28). In the early lesions of
atherosclerosis, numerous cholesterol-engorged macrophages were
accumulated in the subendothelial tissues, which is called ‘foam
cells’. The accumulation of lipids leads to more advanced lesions
based on the foam cells. Plaques containing multiple compositions,
such as calcification, fibrosis, cholesterol and hemorrhage, are
formed, and thus lead to the acute occlusion of vessels. The
pathogenesis of atherosclerosis is a complex and manifold process.
The identification of disease-related metabolite prioritizations
contributes to improving diagnosis of atherosclerosis-associated
diseases, and helps exploring the metabolic pathological
process.
Metabolites are the final products of cellular
regulation processes, and can be used to assess the state of
biological systems in genetic or environmental changes. As known, a
metabolite is always affected by the combination of genes and
phenotypes when it is considered as the link between genes and
phenotypes. Furthermore, metabolites affected by one disease
contain not only one or two, and some metabolites may play
important biological functions in the development of a disease.
Therefore, the identification of disease-related metabolite
prioritization is necessary for improving the therapeutic effect of
the disease. In this study, we integrated the multi-omics
information that contains genes, metabolites, and phenotype related
to pathogenesis, for predicting the atherosclerosis-related
metabolites. Moreover, the RWR method was used to compute the
probability of each metabolite in the composite network for
identifying the candidate metabolite prioritization. Consequently,
several atherosclerosis-related key metabolites were confirmed,
including the top 5 metabolites with high relation score [Tretinoin
(TN), Estraderm (TN), Cholesterol (TN), Chitin and magnesium ion]
and other 7 metabolites with high degree [Nitric oxide, genistein,
Oxyfull (TN), resveratrol, Dehydrated ethanol, quercetin and
2-(4-Morpholinyl)-8-phenyl-4H-1-benzopyran-4-one)].
Numerous studies indicate that antioxidants, such as
vitamins A and E, can decrease the severity of
atherosclerosis-related hypercholesterolemia and coronary heart
disease (29,30). Significantly, Tretinoin (retinoic
acid), a metabolite of vitamin A, plays a critical role in cell
survival, differentiation and death, and has particular functions
on neurogenesis and angiogenesis (31,32).
Tretinoin has been demonstrated to decrease the expression and
activity of tissue factor in cultured human endothelial cells
(33). Jiang et al have
reported that Tretinoin can improve the atherogenic effects of
C. pneumoniae by decreasing the damage of C.
pneumoniae infection, and reduce the development of
atherosclerotic lesions (34).
Estraderm as a class of steroid hormone, is the most biologically
active estrogen. Estrogen is reported to suppress the proliferation
of vascular smooth muscle cells, the formation of atherosclerotic
plaques, and oxidative stress. Investigation conducted by Li et
al shows that low-concentration estrogen can function as an
inhibitor for inhibiting the development of atherosclerosis through
the mediation of cystathionine lyase-generated hydrogen sulfide
(35). Furthermore, Dai et al
demonstrated that estradiol and testosterone affect synergistically
the early stage atherosclerosis, and estradiol/testosterone with an
appropriate ratio can markedly inhibit the progression of
atherosclerosis by decreasing foam cell formation, lipid lesions
and endothelial damage (36).
Additionally, the functional roles of other metabolites on the
progression of atherosclerosis were also investigated, such as
genistein suppress the atherosclerotic lesions by inhibiting the
expression of matrix metalloproteinase-3 protein for protecting the
vascular endothelial wall, Nitric oxide is regarded as a key
modulator of vascular disease and the activation of endothelial
nitric oxide synthase can improve atherosclerosis, and resveratrol
functions as atheroprotective activity since the antioxidant,
anti-inflammatory, antifibrotic, and cardioprotective properties
(37–40).
In addition, we identified 24 co-expression genes
that are closely related to the important
atherosclerosis-associated metabolites. Moreover, some
co-expression genes have been demonstrated to play a crucial role
in the development of atherosclerosis, especially for genes with a
high degree. For example, MAPK8 (degree: 100) as a member of the
JNK family that play an important role in cardiovascular lesion and
disease, has been reported to be required for apoptosis of
endothelial cells and the deposition of lipids during early
atherosclerosis (26,41). Thus, inhibitors of MAPK8 may decrease
atherosclerosis by reducing endothelial cell damage and preventing
foam cell formation. PTEN gene (degree: 81) is reported to inhibit
the expression of VCAM-1 molecule that is highly expressed in
atherosclerotic lesions, suggesting that modulation of PTEN
activity may contribute to reduce atherosclerosis (42). Furthermore, FOXO1 (degree: 59), a
frequent transcription factor expressed frequently in mammalian
cells, plays a crucial role in metabolic homeostasis regulation,
cell differentiation, oxidative stress and autophagy (43,44).
Qiang et al studied the role of FOXO1 on atherosclerosis and
endothelial cells, the results indicate that FOXO1 deacetylation
can enhance the change of vascular endothelial cells, and thus
contributing to the formation of atherosclerotic plaques (45). The functional effects of other
co-expression genes on atherosclerosis are also reported, including
HSPA5, CXCL5 and MAOA genes (46–48).
Therefore, these results suggest that the metabolites and genes
extracted by multi-omics composite network are important for the
therapy of atherosclerosis.
In summary, atherosclerosis-related metabolites can
be identified by the multi-omics composite network, and
prioritization of metabolites can be confirmed by the use of the
RWR method. Several key metabolites were found, including Tretinoin
(TN), Estraderm (TN), Cholesterol (TN), Chitin, magnesium ion,
Nitric oxide, genistein, Oxyfull (TN), resveratrol, dehydrated
ethanol, quercetin and
2-(4-Morpholinyl)-8-phenyl-4H-1-benzopyran-4-one, which could be
considered as biomarkers of atherosclerosis diagnosis. Furthermore,
24 co-expression genes, identified by analyzing the metabolites in
composite network, may be available to be targets of
atherosclerosis therapy. However, for the analysis of metabolites
and genes, some limitations are still present. Firstly, there are
many diseases associated with atherosclerosis, whereas we only
selected one dataset (GEOD-57691). Additionally, the candidate
metabolites extracted by the multi-omics composite network have not
been verified by clinical experiments. Nonetheless, the results
obtained from this study are still able to provide some preliminary
evidence to explore the potential therapeutic strategies for
atherosclerosis.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JQC, CXL and SMM performed the experiments and
analyzed the data. JQC was also a major contributor in writing the
manuscript. RYW and JJC made a substantial contribution to the
study concept and design. JQC, CXL and WYW performed the
statistical analysis. LJM was involved in the conception and design
of the study as well as the drafting of the manuscript. All authors
read and approved the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lusis AJ: Atherosclerosis. Nature.
407:233–241. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Akerele OA and Cheema SK: Fatty acyl
composition of lysophosphatidylcholine is important in
atherosclerosis. Med Hypotheses. 85:754–760. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tabas I, García-Cardeña G and Owens GK:
Recent insights into the cellular biology of atherosclerosis. J
Cell Biol. 209:13–22. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Holmes E, Wilson ID and Nicholson JK:
Metabolic phenotyping in health and disease. Cell. 134:714–717.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Iqbal O, Ottman A, Gaynes J, De Alba F,
Gaynes BI, Fareed J and Bouchard CS: The effects of aspirin and its
metabolites on peripheral blood mononuclear cells and human retinal
pigment epithelial cells - implications in the pathophysiology of
age-related macular degeneration. IOVS. 55:6342014.
|
|
6
|
Kell DB: Metabolomics and systems biology:
Making sense of the soup. Curr Opin Microbiol. 7:296–307. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nicholson JK and Lindon JC: Systems
biology: Metabonomics. Nature. 455:1054–1056. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Moco S, Bino RJ, De Vos RCH and Vervoort
J: Metabolomics technologies and metabolite identification. Trends
Analyt Chem. 26:855–866. 2007. View Article : Google Scholar
|
|
9
|
Lee DS, Park J, Kay KA, Christakis NA,
Oltvai ZN and Barabási AL: The implications of human metabolic
network topology for disease comorbidity. Proc Natl Acad Sci USA.
105:9880–9885. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yao Q, Xu Y, Yang H, Shang D, Zhang C,
Zhang Y, Sun Z, Shi X, Feng L, Han J, et al: Global prioritization
of disease candidate metabolites based on a multi-omics composite
network. Sci Rep. 5:172012015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
von Mering C, Huynen M, Jaeggi D, Schmidt
S, Bork P and Snel B: STRING: A database of predicted functional
associations between proteins. Nucleic Acids Res. 31:258–261. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wishart DS, Knox C, Guo AC, Eisner R,
Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, et al:
HMDB: A knowledgebase for the human metabolome. Nucleic Acids Res.
37:D603–D610. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kanehisa M, Goto S, Sato Y, Furumichi M
and Tanabe M: KEGG for integration and interpretation of
large-scale molecular data sets. Nucleic Acids Res. 40:D109–D114.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
D'Eustachio P: Reactome knowledgebase of
human biological pathways and processes. Methods Mol Biol.
694:49–61. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xia J and Wishart DS: MSEA: a web-based
tool to identify biologically meaningful patterns in quantitative
metabolomic data. Nucleic Acids Res. 38 (Suppl 2):W71–W77. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jewison T, Su Y, Disfany FM, Liang Y, Knox
C, Maciejewski A, Poelzer J, Huynh J, Zhou Y, Arndt D, et al: SMPDB
2.0: Big improvements to the Small Molecule Pathway Database.
Nucleic Acids Res. 42:D478–D484. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kuhn M, Szklarczyk D, Franceschini A,
Campillos M, von Mering C, Jensen LJ, Beyer A and Bork P: STITCH 2:
An interaction network database for small molecules and proteins.
Nucleic Acids Res. 38 (Suppl 1):D552–D556. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
van Driel MA, Bruggeman J, Vriend G,
Brunner HG and Leunissen JA: A text-mining analysis of the human
phenome. Eur J Hum Genet. 14:535–542. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bolstad BM, Irizarry RA, Åstrand M and
Speed TP: A comparison of normalization methods for high density
oligonucleotide array data based on variance and bias.
Bioinformatics. 19:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Irizarry RA, Bolstad BM, Collin F, Cope
LM, Hobbs B and Speed TP: Summaries of Affymetrix GeneChip probe
level data. Nucleic Acids Res. 31:e152003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Belitskaya-Lévy I, Zeleniuch-Jacquotte A,
Russo J, Russo IH, Bordás P, Ahman J, Afanasyeva Y, Johansson R,
Lenner P, Li X, et al: Characterization of a genomic signature of
pregnancy identified in the breast. Cancer Prev Res (Phila).
4:1457–1464. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Reiner A, Yekutieli D and Benjamini Y:
Identifying differentially expressed genes using false discovery
rate controlling procedures. Bioinformatics. 19:368–375. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Datta S, Satten GA, Benos DJ, Xia J,
Heslin MJ and Datta S: An empirical bayes adjustment to increase
the sensitivity of detecting differentially expressed genes in
microarray experiments. Bioinformatics. 20:235–242. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu X, Jiang R, Zhang MQ and Li S:
Network-based global inference of human disease genes. Mol Syst
Biol. 4:1892008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Amini N, Boyle JJ, Moers B, Warboys CM,
Malik TH, Zakkar M, Francis SE, Mason JC, Haskard DO and Evans PC:
Requirement of JNK1 for endothelial cell injury in atherogenesis.
Atherosclerosis. 235:613–618. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kedenko L, Lamina C, Kedenko I, Kollerits
B, Kiesslich T, Iglseder B, Kronenberg F and Paulweber B: Genetic
polymorphisms at SIRT1 and FOXO1 are associated with carotid
atherosclerosis in the SAPHIR cohort. BMC Med Genet. 15:1122014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hadi NR: Effect of vildagliptin on
atherosclerosis progression in high cholesterol - fed male rabbits.
J Clin Exp Cardiolog. 04:150252016.
|
|
29
|
Gey KF and Puska P: Plasma vitamins E and
A inversely correlated to mortality from ischemic heart disease in
cross-cultural epidemiology. Ann NY Acad Sci. 570:268–282. 1989.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hornig B: Vitamins, antioxidants and
endothelial function in coronary artery disease. Cardiovasc Drugs
Ther. 16:401–409. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Környei Z, Gócza E, Rühl R, Orsolits B,
Vörös E, Szabó B, Vágovits B and Madarász E: Astroglia-derived
retinoic acid is a key factor in glia-induced neurogenesis. FASEB
J. 21:2496–2509. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bowles J, Secker G, Nguyen C, Kazenwadel
J, Truong V, Frampton E, Curtis C, Skoczylas R, Davidson TL, Miura
N, et al: Control of retinoid levels by CYP26B1 is important for
lymphatic vascular development in the mouse embryo. Dev Biol.
386:25–33. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim YM, Kim JH, Park SW, Kim HJ and Chang
KC: Retinoic acid inhibits tissue factor and HMGB1 via modulation
of AMPK activity in TNF-α activated endothelial cells and
LPS-injected mice. Atherosclerosis. 241:615–623. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jiang SJ, Campbell LA, Berry MW, Rosenfeld
ME and Kuo CC: Retinoic acid prevents Chlamydia pneumoniae-induced
foam cell development in a mouse model of atherosclerosis. Microbes
Infect. 10:1393–1397. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li H, Mani S, Wu L, Fu M, Shuang T, Xu C
and Wang R: The interaction of estrogen and CSE/H2S pathway in the
development of atherosclerosis. Am J Physiol Heart Circ Physiol.
312:H406–H414. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dai W, Ming W, Li Y, Zheng HY, Wei CD, Rui
Z and Yan C: Synergistic effect of a physiological ratio of
estradiol and testosterone in the treatment of early-stage
atherosclerosis. Arch Med Res. 46:619–629. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lee CS, Kwon SJ, Na SY, Lim SP and Lee JH:
Genistein supplementation inhibits atherosclerosis with
stabilization of the lesions in hypercholesterolemic rabbits. J
Korean Med Sci. 19:656–661. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Voloshyna I, Hussaini SM and Reiss AB:
Resveratrol in cholesterol metabolism and atherosclerosis. J Med
Food. 15:763–773. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tin A, Grams ME, Maruthur NM, Astor BC,
Couper D, Mosley TH, Selvin E, Coresh J and Kao WH: Results from
the Atherosclerosis Risk in Communities study suggest that low
serum magnesium is associated with incident kidney disease. Kidney
Int. 87:820–827. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Förstermann U, Xia N and Li H: Roles of
vascular oxidative stress and nitric oxide in the pathogenesis of
atherosclerosis. Circ Res. 120:713–735. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Babaev VR, Yeung M, Erbay E, Ding L, Zhang
Y, May JM, Fazio S, Hotamisligil GS and Linton MF: Jnk1 deficiency
in hematopoietic cells suppresses macrophage apoptosis and
increases atherosclerosis in low-density lipoprotein receptor null
mice. Arterioscler Thromb Vasc Biol. 36:1122–1131. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yuan M, Wang X, Zhan Q, Duan X, Yang Q and
Xia J: Association of PTEN genetic polymorphisms with
atherosclerotic cerebral infarction in the Han Chinese population.
J Clin Neurosci. 19:1641–1645. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Accili D and Arden KC: FoxOs at the
crossroads of cellular metabolism, differentiation, and
transformation. Cell. 117:421–426. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sengupta A, Molkentin JD and Yutzey KE:
FoxO transcription factors promote autophagy in cardiomyocytes. J
Biol Chem. 284:28319–28331. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Qiang L, Tsuchiya K, Kim-Muller JY, Lin
HV, Welch C and Accili D: Increased atherosclerosis and endothelial
dysfunction in mice bearing constitutively deacetylated alleles of
Foxo1 gene. J Biol Chem. 287:13944–13951. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhao J, Forsberg CW, Goldberg J, Smith NL
and Vaccarino V: MAOA promoter methylation and susceptibility to
carotid atherosclerosis: Role of familial factors in a monozygotic
twin sample. BMC Med Genet. 13:1002012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Rousselle A, Qadri F, Leukel L, Yilmaz R,
Fontaine JF, Sihn G, Bader M, Ahluwalia A and Duchene J: CXCL5
limits macrophage foam cell formation in atherosclerosis. J Clin
Invest. 123:1343–1347. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Orešič M, Clish CB, Davidov EJ, Verheij E,
Vogels J, Havekes LM, Neumann E, Adourian A, Naylor S, van der
Greef J and Plasterer T: Phenotype characterisation using
integrated gene transcript, protein and metabolite profiling.
Applied Bioinformatics. 3:205–217. 2004. View Article : Google Scholar : PubMed/NCBI
|