Introduction
Bronchial asthma is a chronic airway inflammatory
disease characterized by airway hyperreactivity and reversible flow
limitation (1). Epidemiological
investigations indicate that ~0.3 billion patients currently suffer
from asthma and that 25,000 asthma-associated deaths occur per
annum worldwide. Furthermore, the morbidity and mortality of asthma
exhibit increasing trends, rendering asthma a major global public
health problem (2). Chronic airway
inflammation-induced airway remodeling is an important step in the
pathogenesis of asthma (3). Airway
remodeling mainly involves chronic inflammation-induced massive
proliferation of airway smooth muscle cells (ASMCs) and
extracellular matrix (ECM) deposition. The proliferation and
hypertrophy of ASMCs have a particularly important role in airway
remodeling, and are considered as important features thereof
(4). In addition, the extent of ASMC
proliferation is positively correlated with the severity of asthma
(5).
Various factors, including T-helper cell type 1/2
cytokine imbalances, cytokines, matrix metallopeptidase and tissue
inhibitor of metallopeptidases, genetic factors, neuromodulation
and gene mutation, have important roles in the occurrence and
development of asthma (6,7). MicroRNAs (miRNAs/miRs) are a class of
highly evolutionarily conserved, small-molecular, non-coding RNAs
with a length of 21–23 nt. They interact with the mRNAs of their
target genes to reduce their stability and/or inhibit translation,
thus negatively regulating target gene expression. miRNAs are
involved in a series of important pathophysiological processes,
including cell proliferation, differentiation and apoptosis, as
well as immune response and tumor formation (8,9). It is
estimated that miRNAs, which account for 1–3% of human genes,
regulate the expression of >30% of human genes (10). Recently, the role of miRNAs in the
genesis and development of asthma has attracted increasing
attention (11,12). Furthermore, abnormal miRNA expression
may be involved in asthmatic airway remodeling through regulating
the proliferation and apoptosis of ASMCs (13–15).
Let-7a, a member of the let-7 family, is one of the first
identified miRNAs and one of the most abundant miRNAs in lung
tissue (16). Let-7a regulates
interleukin (IL)-13 secretion, while the latter has a vital role in
the asthmatic inflammatory response and airway remodeling (17). Previous studies have indicated that
let-7a expression is downregulated in bronchial epithelial cells
and transbronchial lung biopsy tissues obtained during bronchoscopy
from asthmatic patients (18,19).
Furthermore, silencing of let-7a was demonstrated to significantly
alleviate airway inflammation and airway hyperreactivity in
asthmatic mice (20,21), suggesting that let-7a is associated
with asthma. However, let-7a expression in bronchial SMCs from
asthmatic patients as well as its regulatory actions have remained
to be fully elucidated. In the present study, changes in let-7a
expression levels in ASMCs from asthmatic vs. healthy patients were
detected using reverse transcription-quantitative polymerase chain
reaction (RT-qPCR), and the effect of let-7a on the proliferation
and apoptosis of ASMCs was assessed; furthermore, potential target
genes of let-7a were identified.
Materials and methods
Collection of patient samples and
primary culture of human ASMCs
ASMCs from asthmatic and non-asthmatic subjects were
derived from segmental bronchial biopsy specimens obtained through
fiberoptic bronchoscopy at Shengli Oil Field Central Hospital
(Dongying, China), including 15 asthmatic and 10 non-asthmatic
subjects. None of the subjects had a history of smoking or of
respiratory tract infection within the last 3 months. Asthmatic
subjects conformed to the diagnostic criteria in the Guidelines for
Prevention and Control of Bronchial Asthma in China (22). All subjects had provided written
informed consent to participate in the study, and the present study
was approved by the Ethics Committee of Shengli Oil Field Central
Hospital (Dongying, China). Epithelium, connective tissue, blood
vessels and cartilage were removed from the obtained bronchial
biopsy specimens (5×5 mm), followed by washing with the ice-cold
PBS containing penicillin-streptomycin 3 times. The specimens were
then placed in 5 ml tissue culture flasks. Fetal bovine serum (FBS;
1 ml; 10%; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
was added, and the smooth muscle tissue block was cut into small
pieces and repeatedly minced using iris scissors. The mixture was
evenly applied onto the bottom surface of the culture flask with a
sterile drinking straw. The culture flask was then inversely placed
in a 5% CO2 incubator at 37°C. After 2 h, when adherence
of the cells was achieved, the flask was turned over and Dulbecco's
modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.)
containing 10% FBS was added from one side. The medium was replaced
once every 3–4 days, and after 14 days of culture, hill and valley
cell fusion was observed. Following detachment with 0.125% trypsin,
the cells were passaged at a ratio of 1:2. Cells at passages 4–8
were used in the experiments. The ASMCs were identified by the
typical ‘hill and valley’ growth pattern and immunocytochemical
staining for α-smooth muscle actin (23).
RT-qPCR
Total RNA of was extracted from the cells using
TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.), and the purity
and content of the extracted RNA were determined using NanoDrop
2000 spectrophotometer (Thermo Fisher Scientific, Inc.) and RNA
samples with a A260/A280 ratio between 1.8 and 2.0 were used to
synthesize cDNA. Total RNA was reverse transcribed into cDNA using
the High-Capacity cDNA Reverse Transcription kit (Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. This
reaction was performed at 25°C for 5 min, 50°C for 20 min then 75°C
for 5 min. qPCR was performed with SYBR® Premix Ex Taq™
(Takara Bio Inc., Otsu, Japan; cat. no. DRR041A) according to the
manufacturer's instructions, using an ABI 7500 real-time
fluorescence qPCR machine (Applied Biosystems; Thermo Fisher
Scientific, Inc.). U6 and β-actin were used as the internal
reference genes for let-7a and signal transducer and activator of
transcription (STAT3) detection, respectively. The reaction
conditions for PCR were as follows: Initial denaturation at 95°C
for 3 min and 40 cycles of 95°C for 15 sec and 60°C for 30 sec. The
primers used for the detection of let-7a and STAT3 were as follows:
Let-7a forward, 5′-GCGCCTGAGGTAGTAGGTTG-3′ and reverse,
5′-CAGTGCAGGGTCCGAGGT-3′; STAT3 forward, 5′-TGCTGGAGGAGAGAATCGT-3′
and reverse, 5′-TAGTAGTGAACTGGACGCCG-3′; U6 forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′;
β-actin forward, 5′-GGTCATCACCATTGGCAA-3′ and reverse,
5′-GAGTTGAAGGTAGTTTCGTGGA-3′. The primers were designed and
synthesized by Shanghai Sangon Bioengineering Co., Ltd (Shanghai,
China). The relative mRNA expression levels of let-7a and STAT3
were calculated using the 2−∆∆Cq method (24).
Cell transfection
ASMCs from asthmatic subjects in the logarithmic
growth phase were inoculated into 6-well plates and divided into a
let-7a mimics group and a negative control (NC)-mimics group.
Let-7a mimics (5′-UGAGGUAGUAGGUUGUAUAGUU-3′; GenePharma, Shanghai,
China) and NC-mimics (5′-UUCUCCGAACGUGUCACGUTT-3′; GenePharma) were
transfected into the ASMCs of the corresponding groups using
Lipofectamine™ 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) in
strict accordance with the manufacturer's protocols. The
transfection medium was replaced with normal medium at 6 h after
transfection. The transfection efficiency was detected using
RT-qPCR.
Cell Counting Kit (CCK)-8 assay
After 24 h of transfection, the cells were collected
and seeded into the 96-well plates at a density of 3,000
cells/well. CCK-8 stain (10 µl; Dojindo Molecular Technologies,
Inc., Kumamoto, Japan) reaction liquid was added to designated
wells every 24 h for 2 h of incubation at 37°C, and the optical
density (OD) value of each well was detected at the wavelength of
450 nm by ELX800 Universal Microplate Reader (BioTek Instruments,
Inc., Winooski, VT, USA).
Flow cytometry
After 48 h of transfection, the cells were collected
and washed with PBS twice, digested with EDTA-free trypsin and
collected after centrifugation (111.8 × g, 5 min, 4°C) to prepare a
single-cell suspension (1×106 cells/ml) for each group
individually. Of this cell suspension, 5 ml was filled into a flow
tube, followed by addition of 5 µl Annexin V-fluorescein
isothiocyanate (BD Biosciences) and 10 µl propidium iodide (BD
Biosciences) in succession. The mixtures were incubated for 15 min
in the dark, followed by addition of 300 µl binding buffer. The
mixtures were then immediately subjected to flow cytometric
evaluation (BD FACSCanto II; BD Biosciences, Franklin Lakes, NJ,
USA) to determine the apoptotic rate.
Caspase-3/7 activity assay
After 48 h of transfection, the cells were harvested
and seeded onto the 96-well plates. Substrates and buffer solution
were defrosted at room temperature in strict accordance with the
manufacturer's protocol for the Apo-ONE Homogeneous Caspase-3/7
Assay (Promega Corporation, Madison, WI, USA; cat. no. G7790). A
total of 100 µl substrate was mixed with 9,900 µl buffer solution
to prepare the Apo-ONE Caspase-3/7 reagent. Subsequently, 100 µl of
this reaction reagent was added to each well, followed by agitation
using a shaker for 30 sec and incubation for 2 h at room
temperature in the dark. The fluorescence intensity of each well
was then detected using a Tecan Infinite M200 pro plate reader
(Tecan Trading AG, Maennedorf, Switzerland), from which the
caspase-3/7 activity was determined.
Dual-luciferase reporter gene
assay
Target genes of let-7a were predicted using the
TargetScan miRNA target gene prediction bioinformatics website
(http://www.targetscan.org). The
bioinformatics prediction suggested that STAT3 may be a potential
target gene for let-7a. The wild-type (WT) and mutant (MUT)
reporter gene plasmids were synthesized by GeneCopoeia Co., Ltd.
(Guangzhou, China). PCR amplification products of the
3′-untranslated region (3′-UTR) of STAT3 were connected to the
XbalI enzyme digestion site of the pGL3 plasmid, so as to
construct the wild-type pGL3-WT-STAT3 reporter gene plasmid
(5′-UGACCUCGGAGUGCGCUACCUCC-3′). Site-specific mutagenesis was
performed on the binding site in the 3′UTR of STAT3 that is
complementary to a sequence in let-7a, and the new plasmid obtained
after mutagenesis was the pGL3-MUT-STAT3 reporter gene plasmid
(5′-UGACCUCGGAGUGCGCAAGCACC-3′). 293 cells (ScienCell Research
Laboratories, Inc., San Diego, CA, USA) were divided into 4 groups,
namely the STAT3 WT plasmid control group (transfection with
pGL3-WT-STAT3 reporter gene plasmid and NC-mimics), STAT3 WT
plasmid experimental group (transfection with pGL3-WT-STAT3
reporter gene plasmid and let-7a mimics), STAT3 MUT plasmid control
group (transfection with pGL3-MUT-STAT3 reporter gene plasmid and
NC-mimics) and STAT3 MUT plasmid experimental group (transfection
with pGL3-MUT-STAT3 reporter gene plasmid and let-7a mimics). Cells
in each group were split after 24 h of transfection in accordance
with the manufacturer's protocol for the dual-luciferase reporter
gene system kit (Promega Corp.), and activities of firefly
luciferase and Renilla luciferase in each group were detected using
a GloMax 20/20 luminometer (Promega Corp.). Renilla luciferase gene
was used as the internal reference to verify the transfection
efficiency and calculate the relative luciferase activity as
follows: Relative luciferase activity = firefly luciferase
activity/Renilla luciferase activity.
Western blot analysis
Cells in each group were collected and washed with
PBS for 3 times, followed by addition of radioimmunoprecipitation
assay lysis buffer (Fermentas; Thermo Fisher Scientific, Inc.) and
1% protease inhibitor (cat. no. 78439; Pierce; Thermo Fisher
Scientific, Inc.), and incubation for 20 min on ice. The mixtures
were centrifuged (111.8 × g) at 4°C for 15 min and the supernatant
was collected to determine the protein concentration using the
bicinchoninic acid method. A total of 20 µg protein per lane was
separated by 10% SDS-PAGE, followed by transfer onto the
polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA,
USA). Subsequent to blocking with 5% skimmed milk at room
temperature for 6 h, the membranes were incubated with primary
antibodies against STAT3 (cat. no. sc-293151; 1:1,000 dilution) and
GAPDH (cat. no. sc-32233; 1:1,000 dilution; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) at 4°C overnight. The
membrane was washed prior to the addition of horseradish
peroxidase-conjugated secondary antibody (cat. no. sc-2354; 1:2,000
dilution; Santa Cruz Biotechnology, Inc.) and incubation at 37°C
for 1 h. The membrane was washed and protein bands were visualized
using the enhanced chemiluminescence reagent (Western Blotting
Detection kit; Applygen Technologies, Inc., Beijing, China)
according the manufacturer's protocol. Subsequently, QuantityOne
software (version 4.6; Bio-Rad Laboratories, Inc., Hercules, CA,
USA) was used for densitometric analysis, and GAPDH was used as the
internal reference to determine the relative expression of
STAT3.
Statistical analysis
Data were analyzed by SPSS 19.0 software (IBM Corp.,
Armonk, NY, USA). Values are expressed as the mean ± standard
deviation and comparisons between groups were performed using the
Student's t-test. All experiments were repeated at least 3 times.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Let-7a is downregulated in asthmatic
ASMCs
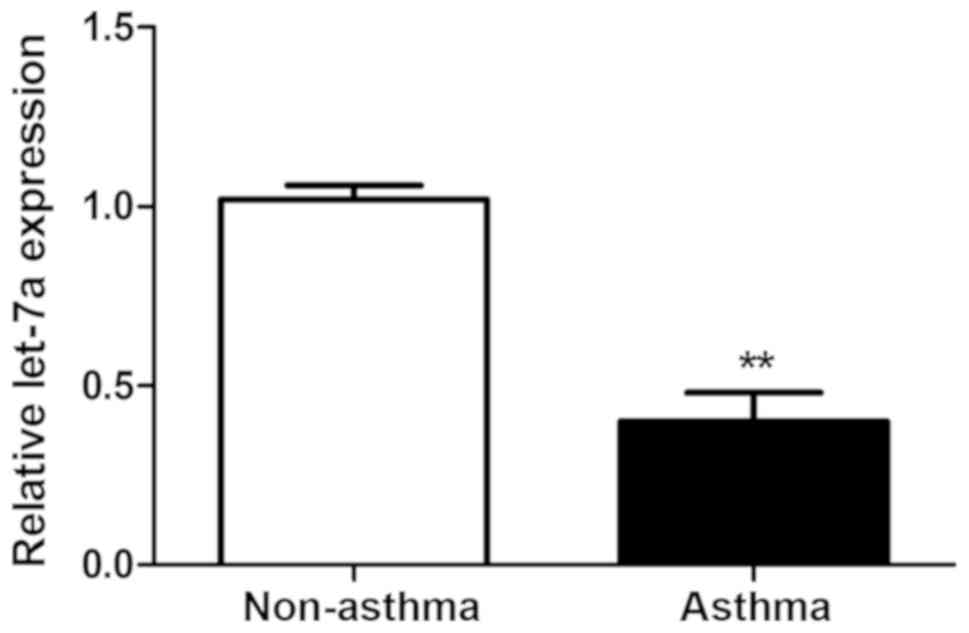
The relative expression levels of let-7a in ASMCs
from asthmatic and non-asthmatic subjects were detected using
RT-qPCR. As presented in Fig. 1, the
relative expression levels of let-7a in ASMCs from asthmatic
subjects were significantly decreased compared with those in the
ASMCs from non-asthmatic subjects (P<0.01).
Let-7a mimics inhibit the
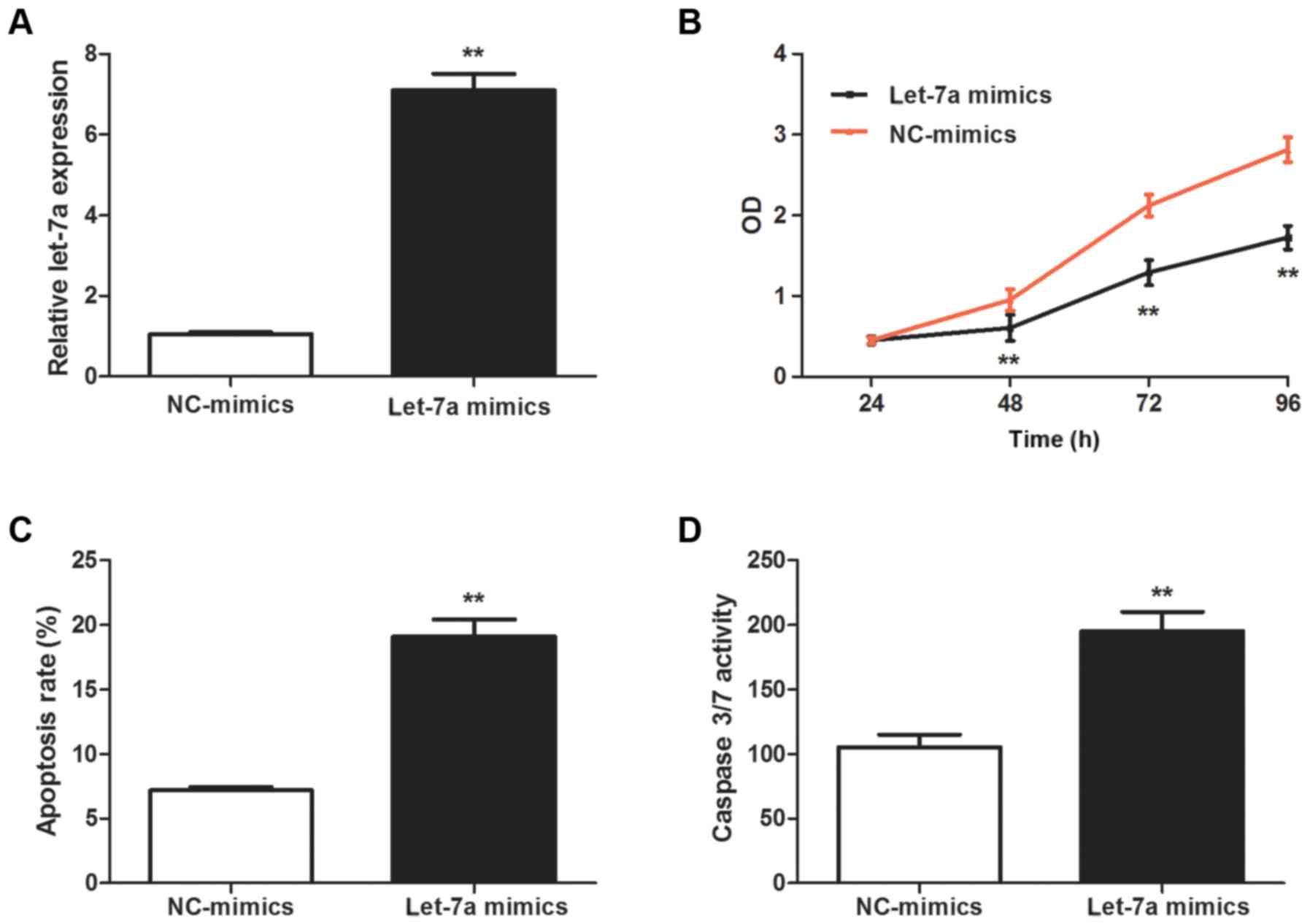
proliferation and promote the apoptosis of asthmatic ASMCs
ASMC proliferation has a vital role in airway
remodeling (4). In order to
determine the potential effect of let-7a on the proliferation of
asthmatic ASMCs, asthmatic ASMCs were transfection with let-7a
mimics, and the successful upregulation of let-7a was confirmed by
RT-qPCR (P<0.01; Fig. 2A). The
results of the CCK-8 assay indicated that let-7a mimics
significantly suppressed the proliferation of asthmatic ASMCs
(P<0.01; Fig. 2B). Flow
cytometric analysis indicated that let-7a mimics significantly
enhanced the apoptosis of asthmatic ASMCs (P<0.01; Fig. 2C). Furthermore, let-7a mimics
significantly increased the activity of caspase-3/7 in asthmatic
ASMCs (P<0.01; Fig. 2D).
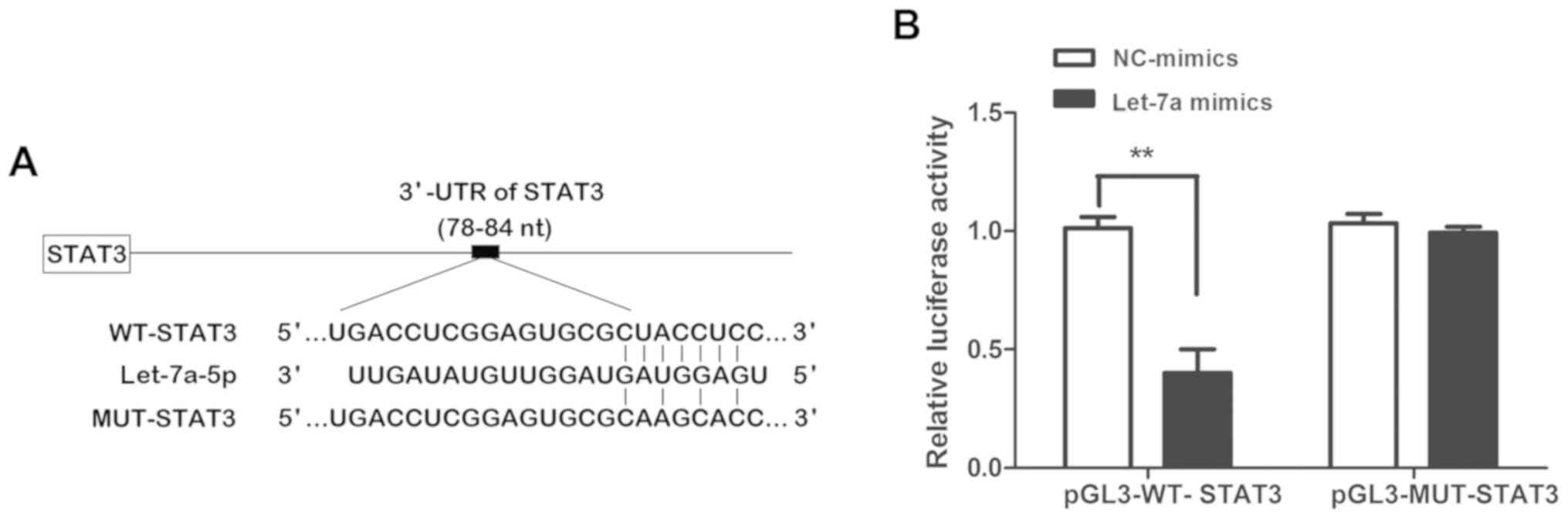
STAT3 is the direct regulatory target
gene of let-7a
To determine the precise molecular biological
mechanisms by which let-7a regulates the proliferation and
apoptosis of asthmatic ASMCs, potential target genes of let-7a were
analyzed and predicted using TargetScan. The analysis suggested
that STAT3 is a potential target gene of let-7a due to the presence
of theoretical complementary binding sites in the 3′-UTR of STAT3
mRNA with the seed sequence of let-7a (Fig. 3A). To further verify whether STAT3 is
a direct target gene of let-7a, a dual-luciferase reporter gene
assay was performed to confirm the effect of let-7a on the
luciferase activity of the 3′-UTR of STAT3 mRNA. As presented in
Fig. 3B, let-7a mimics evidently
suppressed the luciferase activity of the pGL3-WT-STAT3 reporter
gene vector, while they had no obvious inhibitory effect on the
luciferase activity of the pGL3-MUT-STAT3 reporter gene vector
(P<0.01). This revealed that let-7a specifically binds with the
3′-UTR of the STAT3 mRNA, and that STAT3 is a direct target gene of
let-7a.
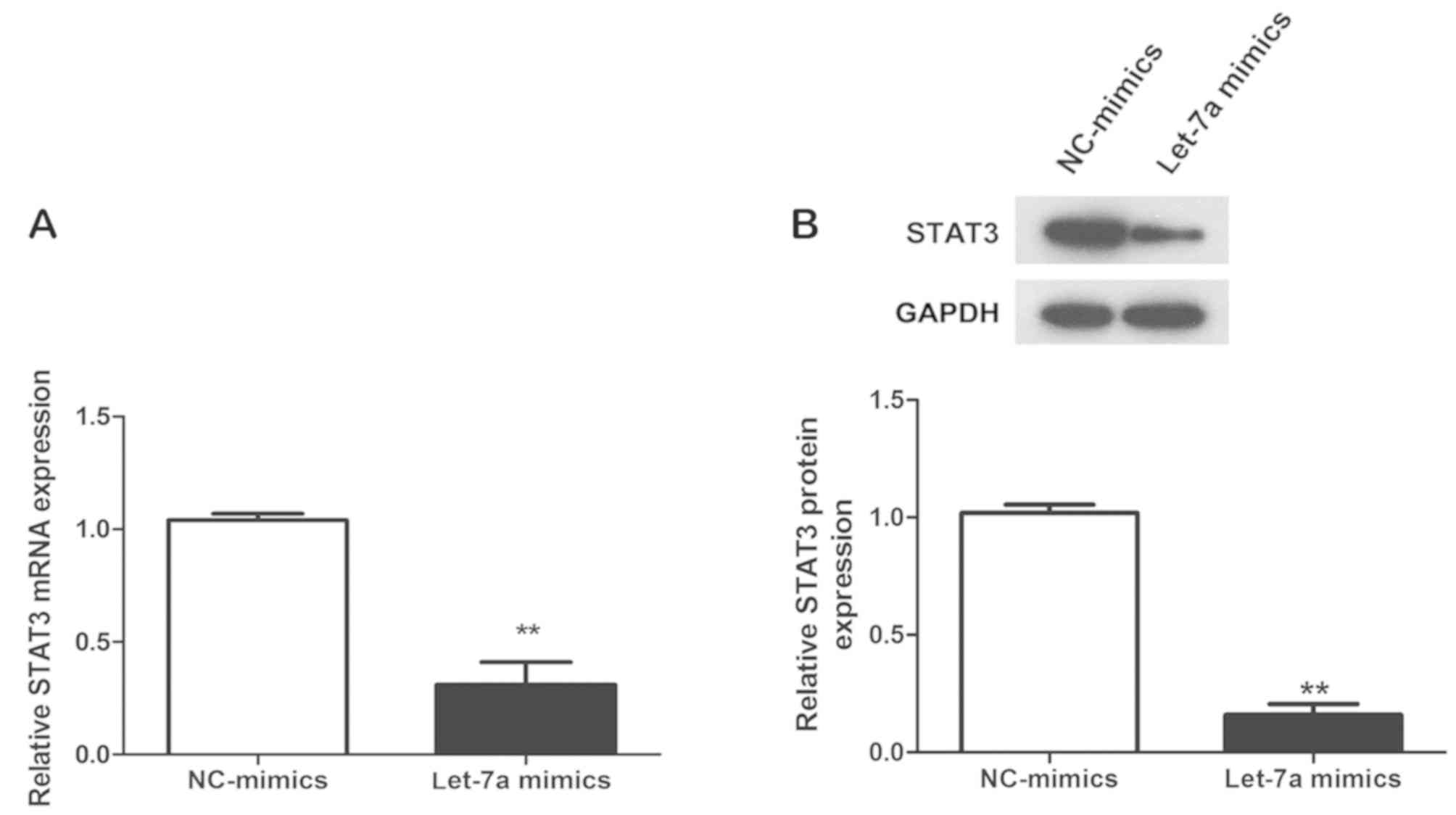
Let-7a mimics inhibit the mRNA and
protein expression of STAT3 in asthmatic ASMCs
Finally, the mRNA and protein expression of STAT3 in
asthmatic ASMCs transfected with let-7a mimics or NC-mimics was
detected by RT-qPCR and western blot analysis, respectively. The
expression levels of STAT3 mRNA and protein in the let-7a mimics
group were significantly lower than those in the NC-mics group
(P<0.01; Fig. 4A and B),
suggesting that upregulation of let-7a significantly inhibits the
expression of STAT3 mRNA and protein in asthmatic ASMCs, further
confirming that STAT3 is a target regulated by let-7a.
Discussion
Asthma is a common respiratory tract disease, which
is characterized by airway remodeling and airway hyperreactivity.
Airway remodeling refers to a series of structural changes in the
airway wall under the persistent stimulation of mitogens, which are
featured by epithelial damage, gland hyperplasia and hypertrophy,
ASMC proliferation, ECM deposition and basilar membrane thickening.
Of these, ASMC proliferation is the pathological foundation leading
to structural changes in airway tissues (25), which not only aggravate airway
stenosis but also increase airway hyperreactivity, thus having a
crucial role in the genesis and development of airway remodeling
(26). As a result, the prevention
of ASMC proliferation to control the genesis of asthmatic airway
remodeling has become a hotspot of current asthma research. In the
last decade, an increasing number of studies have focused on ASMCs
in asthma and have proposed to target them as a therapeutic
strategy (27,28).
Certain miRNAs have been verified to participate in
the regulation of various biological processes, including cell
proliferation, differentiation, apoptosis and tissue development
through regulating target gene expression, and to have a key role
in the genesis and development of multiple diseases (29,30).
Existing data regarding miRNAs and bronchial asthma suggest that
miRNAs are involved in almost all pathophysiological processes of
bronchial asthma, including allergic inflammation in the airways
dominated by eosinophil infiltration, ASMC hyperreactivity and
airway remodeling (11). In
addition, patients with different severities of asthma may exhibit
differences in their miRNA expression profile (31). Furthermore, multiple physiological
functions of ASMCs, including contraction, proliferation and
apoptosis, are also regulated by miRNAs. Hu et al (32) reported that overexpression of miR-10a
markedly reduced the proliferation of human ASMCs, while inhibition
of miR-10a promoted their proliferation. The underlying mechanisms
included the specific inhibition of the phosphatidylinositide-3
kinase catalytic subunit α by miR-10a, which blocked the AKT
signaling pathway, as well as cyclins and cyclin-dependent kinases.
Furthermore, another study identified that ras homolog family
member A (RhoA) expression in ASMCs is negatively regulated by
miR-133a, while IL-13 caused an upregulation of RhoA through
reducing the miR-133a content in ASMCs. RhoA was identified as the
key molecule to induce ASMC dysfunction, which may lead to
excessive contraction, abnormal proliferation and apoptotic
disorders of ASMCs (33). Therefore,
correcting the aberrant expression of critical miRNAs in ASMC may
potentially delay airway remodeling and the associated decline in
lung function, which may be a promising approach for treating
asthma.
Let-7a is a member of the let-7a family, the
aberrant expression of which has been identified in multiple
diseases, including malignancies, Alzheimer's disease, asthma,
allergic rhinitis and allergic dermatitis (34). Solberg et al (18) verified through miRNA microarrays
combined with RT-qPCR that let-7a is markedly downregulated in
bronchial epithelial cells of asthmatic patients. Rijavec et
al (19) reported that let-7a
was distinctly downregulated in transbronchial lung biopsy tissues
obtained during bronchoscopy from patients with severe asthma
compared with patients with mild asthma and non-asthmatic controls.
Levänen et al (35)
discovered that the levels of let-7a were obviously decreased in
the broncho-alveolar lavage fluid of asthmatic patients. However,
to the best of our knowledge, the expression of let-7a in asthmatic
ASMCs has not been previously reported. The present study indicated
that let-7a is notably downregulated in ASMCs from asthmatic
subjects. Therefore, it may be speculated that aberrant let-7a
expression is associated with the dysfunction of ASMCs in asthmatic
patients. Johnson et al (36)
suggested that Let-7a inhibited pathways controlling cell
proliferation, and may also be the major regulator of cell
proliferation. Furthermore, Cheng et al (37) discovered that upregulation of let-7a
expression in bone mesenchymal stem cells suppressed the
proliferation of pulmonary artery SMCs through the STAT3/bone
morphogenetic protein receptor type 2 signaling pathway, thus
delaying the progression of pulmonary hypertension. Furthermore, it
has been verified that upregulation of let-7a expression markedly
inhibits the proliferation of vascular SMCs (38); in addition, let-7a inhibits the
proliferation of multiple types of tumor cell and promotes tumor
cell apoptosis (39). In the present
study, to determine whether aberrant let-7a expression in asthmatic
ASMCs affects their proliferation and apoptosis, ASMCs from
asthmatic patients were transfected with let-7a mimics to
successfully upregulate let-7a expression. A CCK-8 assay indicated
that let-7a mimics markedly suppressed the proliferation of
asthmatic ASMCs. Furthermore, flow cytometric analysis and a
caspase-3/7 activity assay suggested that let-7a mimics markedly
enhanced the apoptosis and caspase-3/7 activity of asthmatic ASMCs,
thereby verifying that let-7a affects the remodeling process of
asthmatic airway tissues through suppressing the proliferation and
promoting the apoptosis of asthmatic ASMCs.
The miRNA target prediction database TargetScan was
then used to further explore the downstream targeted regulatory
mechanisms by which let-7a regulates the function of asthmatic
ASMCs. The bioinformatics prediction suggested that let-7a directly
and specifically regulates STAT3, as theoretical complementary
binding sites in the 3′-UTR of STAT3 mRNA with the seed sequence of
let-7a were identified. Furthermore, let-7a was confirmed in
previous studies to inhibit hepatoma cell proliferation through
specifically regulating STAT3 gene expression (40), which also enhanced the sensitivity of
hepatoma cells to cetuximab through specifically regulating the
expression of the STAT3 gene (41).
In the present study, the dual-luciferase assay indicated that
let-7a specifically binds with the 3′-UTR of STAT3. In addition,
RT-qPCR and western blot analysis indicated that upregulation of
let-7a evidently reduces the mRNA and protein expression levels of
STAT3 in asthmatic ASMCs, further suggesting that STAT3 is a target
gene regulated by let-7a. STAT3 is a member of the STAT family, and
is an important signal transcription factor in the Janus
kinase/STAT signal transduction pathway. It possesses multiple
important biological activities, and is involved in processes
including cell proliferation, differentiation, survival,
angiogenesis and cancer metastasis, through regulating the
expression of various genes and cytokines (42). STAT3 has been verified as a key
regulatory factor in airway remodeling. Allergic asthma and airway
hyperresponsiveness were significantly relieved in asthmatic mice
with STAT3 knockout (43).
Furthermore, STAT3 inhibitor was reported to significantly inhibit
airway remodeling and lung inflammation in asthmatic mice (44). STAT3 was demonstrated to promote ASMC
proliferation through inducing the expression of chemokines and
growth factors (45). Knockout of
STAT3 significantly inhibited the proliferation of SMCs in human
airway tissues (46).
Platelet-derived growth factor was reported to induce the
proliferation of human ASMCs through activation of STAT3 (47). Shi et al (48) also confirmed that upregulation of
STAT3 expression significantly promoted the proliferation of human
ASMCs.
In conclusion, the present study suggested that
let-7a is downregulated in ASMCs of asthmatic patients. In
addition, let-7a was indicated to inhibit the proliferation and
promote the apoptosis of human ASMCs, which may be associated with
the downregulation of STAT3 expression. Let-7a has a vital role
during asthmatic airway remodeling.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants of the
Natural and Science Foundation of Shandong Province (grant no.
ZR2014HL003). The funders had no role in the study design, data
collection and analysis, decision to publish or preparation of the
manuscript.
Availability of data and materials
The datasets generated and analyzed during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
HL designed the experiments. YC, LQ, ZZ, GH and JZ
performed the experiments. YC and HL analyzed the data. HL wrote
the manuscript.
Ethics approval and consent to
participate
All subjects had provided written informed consent
to participate in the study, and the present study was approved by
the Ethics Committee of Shengli Oil Field Central Hospital
(Dongying, China).
Patient consent for publication
Written informed consent was obtained from all
patients prior to publication.
Competing interests
The authors declare that they have no competing
interests
References
|
1
|
Bergmann KC: Bronchial asthma-many types,
different therapies. Dtsch Med Wochenschr. 141:687–692. 2016.(In
German). PubMed/NCBI
|
|
2
|
Wang HY, Wong GW, Chen YZ, Ferguson AC,
Greene JM, Ma Y, Zhong NS, Lai CK and Sears MR: Prevalence of
asthma among Chinese adolescents living in Canada and in China.
CMAJ. 179:1133–1142. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Miller M, Rosenthal P, Beppu A, Mueller
JL, Hoffman HM, Tam AB, Doherty TA, McGeough MD, Pena CA, Suzukawa
M, et al: ORMDL3 transgenic mice have increased airway remodeling
and airway responsiveness characteristic of asthma. J Immunol.
192:3475–3487. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Perry MM, Durham AL, Austin PJ, Adcock IM
and Chung KF: BET bromodomains regulate transforming growth
factor-β-induced proliferation and cytokine release in asthmatic
airway smooth muscle. J Biol Chem. 290:9111–9121. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bergeron C, Tulic MK and Hamid Q: Airway
remodelling in asthma: From benchside to clinical practice. Can
Respir J. 17:e85–93. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Balmasova IP, Sepiashvili RI, Sepiashvili
IaR and Malova ES: Bronchial asthma pathogenesis and genetic
prognosis development. Zh Mikrobiol Epidemiol Immunobiol. 3:60–67.
2014.(In Russian).
|
|
7
|
Hizawa N: Bronchial asthma: Progress in
diagnosis and treatments. Topics: II. Pathogenesis and
pathophysiology; 2. Genes associated with asthma and asthma-related
phenotypes. Nihon Naika Gakkai Zasshi. 102:1365–1369. 2013.(In
Japanese). View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: Are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Perry MM, Adcock IM and Chung KF: Role of
microRNAs in allergic asthma: Present and future. Curr Opin Allergy
Clin Immunol. 15:156–162. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kai W, Qian XU and Qun WU: MicroRNAs and
Asthma Regulation. Iran J Allergy Asthma Immunol. 14:120–125.
2015.PubMed/NCBI
|
|
13
|
Chen M, Shi J, Zhang W, Huang L, Lin X, Lv
Z, Zhang W, Liang R and Jiang S: MiR-23b controls TGF-β1 induced
airway smooth muscle cell proliferation via direct targeting of
Smad3. Pulm Pharmacol Ther. 42:33–42. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu Y, Yang K, Shi H, Xu J, Zhang D, Wu Y,
Zhou S and Sun X: MiR-21 modulates human airway smooth muscle cell
proliferation and migration in asthma through regulation of PTEN
expression. Exp Lung Res. 41:535–545. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang H, Sun Z, Yu L and Sun J: MiR-139-5p
inhibits proliferation and promoted apoptosis of human airway
smooth muscle cells by downregulating the Brg1 gene. Respir Physiol
Neurobiol. 246:9–16. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Takamizawa J, Konishi H, Yanagisawa K,
Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y,
et al: Reduced expression of the let-7 microRNAs in human lung
cancers in association with shortened postoperative survival.
Cancer Res. 64:3753–3756. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang FQ, Han XP, Zhang F, Ma X, Xiang D,
Yang XM, Ou-Yang HF and Li Z: Therapeutic efficacy of a co-blockade
of IL-13 and IL-25 on airway inflammation and remodeling in a mouse
model of asthma. Int Immunopharmacol. 46:133–140. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Solberg OD, Ostrin EJ, Love MI, Peng JC,
Bhakta NR, Hou L, Nguyen C, Solon M, Nguyen C, Barczak AJ, et al:
Airway epithelial miRNA expression is altered in asthma. Am J
Respir Crit Care Med. 186:965–974. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rijavec M, Korošec P, Žavbi M, Kern I and
Malovrh MM: Let-7a is differentially expressed in bronchial
biopsies of patients with severe asthma. Sci Rep. 4:61032014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Polikepahad S, Knight JM, Naghavi AO, Oplt
T, Creighton CJ, Shaw C, Benham AL, Kim J, Soibam B, Harris RA, et
al: Proinflammatory role for let-7 microRNAS in experimental
asthma. J Biol Chem. 285:30139–30149. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kumar M, Ahmad T, Sharma A, Mabalirajan U,
Kulshreshtha A, Agrawal A and Ghosh B: Let-7 microRNA-mediated
regulation of IL-13 and allergic airway inflammation. J Allergy
Clin Immunol. 128:1077–1085.e1-10. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Asthma Workgroup; Chinese Thoracic
Society; Chinese Societ of General Practitioners, : Chinese
guideline for the prevention and management of bronchial asthma
(Primary Health Care Version). J Thorac Dis. 5:667–677.
2013.PubMed/NCBI
|
|
23
|
Chen M, Huang L, Zhang W, Shi J, Lin X, Lv
Z, Zhang W, Liang R and Jiang S: MiR-23b controls TGF-β1 induced
airway smooth muscle cell proliferation via TGFβR2/p-Smad3 signals.
Mol Immunol. 70:84–93. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li LH, Lu B, Wu HK, Zhang H and Yao FF:
Apigenin inhibits TGF-β1-induced proliferation and migration of
airway smooth muscle cells. Int J Clin Exp Pathol. 8:12557–12563.
2015.PubMed/NCBI
|
|
26
|
Fahy JV, Corry DB and Boushey HA: Airway
inflammation and remodeling in asthma. Curr Opin Pulm Med. 6:15–20.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Baroffio M, Crimi E and Brusasco V: Airway
smooth muscle as a model for new investigative drugs in asthma.
Ther Adv Respir Dis. 2:129–139. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dowell ML, Lavoie TL, Solway J and
Krishnan R: Airway smooth muscle: A potential target for asthma
therapy. Curr Opin Pulm Med. 20:66–72. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Brase JC, Wuttig D, Kuner R and Sültmann
H: Serum microRNAs as non-invasive biomarkers for cancer. Mol
Cancer. 9:3062010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lovat F, Valeri N and Croce CM: MicroRNAs
in the pathogenesis of cancer. Semin Oncol. 38:724–733. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tsitsiou E, Williams AE, Moschos SA, Patel
K, Rossios C, Jiang X, Adams OD, Macedo P, Booton R, Gibeon D, et
al: Transcriptome analysis shows activation of circulating CD8+ T
cells in patients with severe asthma. J Allergy Clin Immunol.
129:95–103. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hu R, Pan W, Fedulov AV, Jester W, Jones
MR, Weiss ST, Panettieri RA Jr, Tantisira K and Lu Q: MicroRNA-10a
controls airway smooth muscle cell proliferation via direct
targeting of the PI3 kinase pathway. FASEB J. 28:2347–2357. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chiba Y, Tanabe M, Goto K, Sakai H and
Misawa M: Down-regulation of miR-133a contributes to up-regulation
of Rhoa in bronchial smooth muscle cells. Am J Respir Crit Care
Med. 180:713–719. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Russo F, Di Bella S, Nigita G, Macca V,
Laganà A, Giugno R, Pulvirenti A and Ferro A: miRandola:
Extracellular circulating microRNAs database. PLoS One.
7:e477862012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Levänen B, Bhakta NR, Torregrosa Paredes
P, Barbeau R, Hiltbrunner S, Pollack JL, Sköld CM, Svartengren M,
Grunewald J, Gabrielsson S, et al: Altered microRNA profiles in
bronchoalveolar lavage fluid exosomes in asthmatic patients. J
Allergy Clin Immunol. 131:894–903. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Johnson CD, Esquela-Kerscher A, Stefani G,
Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J,
Shingara J, et al: The let-7 microRNA represses cell proliferation
pathways in human cells. Cancer Res. 67:7713–7722. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cheng G, Wang X, Li Y and He L:
Let-7a-transfected mesenchymal stem cells ameliorate
monocrotaline-induced pulmonary hypertension by suppressing
pulmonary artery smooth muscle cell growth through STAT3-BMPR2
signaling. Stem Cell Res Ther. 8:342017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cao H, Hu X, Zhang Q, Wang J, Li J, Liu B,
Shao Y, Li X, Zhang J and Xin S: Upregulation of let-7a inhibits
vascular smooth muscle cell proliferation in vitro and in vein
graft intimal hyperplasia in rats. J Surg Res. 192:223–233. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lee YS and Dutta A: The tumor suppressor
microRNA let-7 represses the HMGA2 oncogene. Genes Dev.
21:1025–1030. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang Y, Lu Y, Toh ST, Sung WK, Tan P, Chow
P, Chung AY, Jooi LL and Lee CG: Lethal-7 is down-regulated by the
hepatitis B virus × protein and targets signal transducer and
activator of transcription 3. J Hepatol. 53:57–66. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xue F, Liu Y, Zhang H, Wen Y, Yan L, Tang
Q, Xiao E and Zhang D: Let-7a enhances the sensitivity of
hepatocellular carcinoma cells to cetuximab by regulating STAT3
expression. Onco Targets Ther. 9:7253–7261. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Aggarwal BB, Kunnumakkara AB, Harikumar
KB, Gupta SR, Tharakan ST, Koca C, Dey S and Sung B: Signal
transducer and activator of transcription-3, inflammation, and
cancer: How intimate is the relationship? Ann N Y Acad Sci.
1171:59–76. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Simeone-Penney MC, Severgnini M, Tu P,
Homer RJ, Mariani TJ, Cohn L and Simon AR: Airway epithelial STAT3
is required for allergic inflammation in a murine model of asthma.
J Immunol. 178:6191–6199. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gavino AC, Nahmod K, Bharadwaj U,
Makedonas G and Tweardy DJ: STAT3 inhibition prevents lung
inflammation, remodeling, and accumulation of Th2 and Th17 cells in
a murine asthma model. Allergy. 71:1684–1692. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Simeone-Penney MC, Severgnini M, Rozo L,
Takahashi S, Cochran BH and Simon AR: PDGF-induced human airway
smooth muscle cell proliferation requires STAT3 and the small
GTPase Rac1. Am J Physiol Lung Cell Mol Physiol. 294:L698–L704.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Redhu NS, Shan L, Movassagh H and Gounni
AS: Thymic stromal lymphopoietin induces migration in human airway
smooth muscle cells. Sci Rep. 3:23012013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Simon AR, Takahashi S, Severgnini M,
Fanburg BL and Cochran BH: Role of the JAK-STAT pathway in
PDGF-stimulated proliferation of human airway smooth muscle cells.
Am J Physiol Lung Cell Mol Physiol. 282:L1296–L1304. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Shi S, Jin L, Zhang S, Li H, Zhang B and
Sun M: MicroRNA-590-5p represses proliferation of human fetal
airway smooth muscle cells by targeting signal transducer and
activator of transcription 3. Arch Med Sci. 14:1093–1101. 2018.
View Article : Google Scholar : PubMed/NCBI
|