Introduction
Human T-cell leukemia virus type 1 (HTLV-1) is the
causative agent of adult T-cell leukemia/lymphoma (ATL) (1), HTLV-1-associated myelopathy/tropical
spastic paraparesis (HAM/TSP) (2),
and HTLV-1-associated uveitis (3).
Moreover, HTLV-1 virions are thought to be very poorly infectious
(4,5). It is still unclear whether the virions
are unusually unstable, are poorly released from the infected cell,
or inefficiently bind and penetrate into the host cell. After
HTLV-1 infection, some of the infected cells exhibit clonal
proliferation, albeit slowly, to achieve carrier conditions.
Additional genetic and epigenetic changes in these clonally
proliferating cells provide them with the selective advantage of
growth, which eventually leads to leukemia/lymphoma, including ATL
(6).
APOBEC3 (A3) enzymes have been reported to be potent
host-antiviral restriction factors that encode DNA-editing
deaminase enzymes. A3F and A3G are the most studied of the seven A3
genes (A3A, A3B, A3C, A3D/E, A3F, A3G, and A3H) encoded in the
human genome (7). The APOBEC3B
mutation signature is remarkably enriched in six types of cancers,
including those of the cervix, bladder, lung (adeno and squamous
cell), head and neck, and breast (8,9). There
are several reports that A3B level is increased in tumor virus
infections, such as those caused by HBV and HPV (10,11). On
the other hand, in HIV infection, whose causative agent is a
retrovirus, the levels of A3C, A3F, and A3G are increased, but that
of A3B is not (12). However,
APOBEC3 superfamily has been found to exert varying levels of
activity under different experimental conditions. We analyzed
APOBEC3 superfamily expression in HTLV-1 infection because HTLV-1
is a retrovirus and causative agent for malignancy.
Recently, we established a humanized mouse model of
HTLV-1 infection in the presence of specific adaptive immune
responses. These HTLV-1-infected humanized mice showed distinct
ATL-like features, including hepatosplenomegaly and the appearance
of flower cells (13). In this
report, we provide evidence of the pathogenesis of different
expressions of APOBEC superfamily in patients with HAM, who were
non-ATL patients, and HTLV-1-infected humanized mice. Moreover, we
discuss the role of APOBEC superfamily in the antiviral resistance
realized by the innate immunity of HTLV-1-infected humanized
mice.
Materials and methods
Patients
A total of 5 patients with HAM and 5 healthy donors
were included in the present study between April 2013 and December
2016 at Kansai Medical University Hospital (Hirakata, Japan). All
patients with HAM were of Japan. Another 5 healthy subjects were
included in the normal control group. The age and sex composition
were not significantly different (P>0.05; Table I). Human leukocytes (derived from
patients with HAM and normal donors) were separated from the
peripheral blood mononuclear cells (PBMCs) by density gradient
centrifugation using Ficoll Histopaque. The cells were maintained
in a RPMI-1640 medium supplemented with 10% fetal calf serum and 1%
penicillin/streptomycin. All procedures were approved by the Ethics
Committee of Kansai Medical University. Written informed consent
was obtained from all patients or their families.
 | Table I.Characteristics of the study sample
from human. |
Table I.
Characteristics of the study sample
from human.
| Patient | Status | Sex | Age (year) | Proviral DNA loads
per 100 PBMCs |
|---|
| Case 1 | Healthy donor | M | 47 | N.D. |
| Case 2 | Healthy donor | F | 63 | N.D. |
| Case 3 | Healthy donor | F | 67 | N.D. |
| Case 4 | Healthy donor | M | 70 | N.D. |
| Case 5 | Healthy donor | M | 54 | N.D. |
|
| Average/SD |
|
| 60.2±7.8 | N.D. |
|
| Case 6 | HAM patient | F | 69 | 11.88 |
| Case 7 | HAM patient | M | 63 | 7.89 |
| Case 8 | HAM patient | F | 44 | 8.4 |
| Case 9 | HAM patient | M | 66 | 13.8 |
| Case 10 | HAM patient | M | 62 | 9.79 |
|
| Average/SD |
|
| 60.8±8.0 | 10.4±2.5 |
HTLV-1-infected humanized mice
Female 4-week-old NOD/Shi-scid/IL-2Rγc null (NOG)
mice were purchased from the Central Institute of Experimental
Animals (Kawasaki, Japan). Mice were handled under sterile
conditions and maintained in germ-free isolators. The Animal Care
Committees of Kansai Medical University approved all animal
experiments. Five-week-old NOG mice were sublethally irradiated
with 250 cGy from a 137Cs source (Gammacell 40 exactor; Nordion
International). Within 24 h of irradiation, each mouse was injected
with 5×104 human CD133+ cells by intrabone
marrow injection. After 5–6 months, the HTLV-1-infected T-cell line
JEX28, HTLV-1 molecular clone pX1 MT-M was prepared by
electroporation into jurkat cells, was irradiated with 10 Gy from a
137Cs source irradiator. Then, the irradiated JEX28 cells
(2.5×106) or phosphate-buffered saline were
intraperitoneally inoculated (Table
II). All infections were performed in a bio-safety P2A
Laboratory level in accordance with the guidelines of Kansai
Medical University.
 | Table II.Characteristics of the study sample
from mouse. |
Table II.
Characteristics of the study sample
from mouse.
| Patient | Status | Week after
infection | Cell type | Proviral DNA loads
per 100 PBMCs |
|---|
| Case 11 | Uninfected | – | PBMC | – |
| Case 12 | Uninfected | – | PBMC | – |
| Case 13 | Uninfected | – | PBMC | – |
| Case 14 | Uninfected | – | PBMC | – |
| Case 15 | Uninfected | – | PBMC | – |
| Case 16 | HTLV-1
infected | 7 | PBMC,
splenocyte | 97.7 |
| Case 17 | HTLV-1
infected | 5 | PBMC,
splenocyte | 99.4 |
| Case 18 | HTLV-1
infected | 9 | PBMC,
splenocyte | 75.3 |
| Case 19 | HTLV-1
infected | 9 | PBMC,
splenocyte | 76.8 |
| Case 20 | HTLV-1
infected | 5 | PBMC,
splenocyte | 96.7 |
Preparation of cDNA samples
The RNeasy kit (Qiagen, Inc., Valencia, CA, USA) was
employed for the isolation of human total RNA. A concentration of
0.5 µg of total RNA was used for the synthesis of complementary DNA
(cDNA) samples by the reverse transcriptase ReverTra Ace (MMLV
Reverse Transcriptase RNase H-; Toyobo Co., Ltd., Osaka, Japan).
The RNA concentration was determined by using an IMPLEN
NanoPhotometer. Further, humanized mouse RNA was isolated by using
the ZR-Duet™ DNA/RNA MiniPrep kit, and humanized mouse cDNA samples
were synthesized using the same method as in human cDNA
synthesis.
RT-qPCR
The primers used for RT-PCR are listed in Table III. cDNA levels were quantified by
using the RT-PCR method (MyiQ Single color real-time PCR detection
system; Bio-Rad Laboratories, Inc., Hercules, CA, USA). Reactions
were performed in 96-well plates each of which containing 10 µl of
SYBR premix Ex Taq GC (Takara Bio, Inc., Otsu, Japan). The cycling
conditions used were as follows: initial denaturation at 95°C for
10 min, followed by 15 cycles at 95°C for 10 sec and 60°C for 30
sec, and 35 cycles at 95°C for 10 sec and 59°C for 30 sec. The
relative RNA expression levels were calculated by the MyiQ system
(Bio-Rad Laboratories, Inc.).
 | Table III.The primer sets for this study. |
Table III.
The primer sets for this study.
| Gene | Forward primer | Reverse primer | Probes |
|---|
| HTLV-1 pX |
5′-acaaagttaaccatgcttattatcagc-3′ |
5′-tctccaaacacgtagactgggt-3′ |
5′FAM-acaaagttaaccatgcttattatcagc-BHQ3′ |
| Human β-globin |
5′-tgaggagaagtctgccgttac-3′ |
5′-tggtctccttaaacctgtcttg-3′ |
5′FAM-tgaggagaagtctgccgttac-BHQ3′ |
|
| Gene | Forward
primer | Reverse
primer |
|
|
| APOBEC3A |
5′-gagaagggacaagcacatgg-3′ |
5′-tggatccatcaagtgtctgg-3′ |
|
| APOBEC3B |
5′-gaccctttggtccttcgac-3′ |
5′-gcacagccccaggagaag-3′ |
|
| APOBEC3C |
5′-agcgcttcagaaaagagtgg-3′ |
5′-aagtttcgttccgatcgttg-3′ |
|
| APOBEC3D |
5′-acccaaacgtcagtcgaatc-3′ |
5′-cacatttctgcgtggttctc-3′ |
|
| APOBEC3F |
5′-ccgtttggacgcaaagat-3′ |
5′-ccaggtgatctggaaacactt-3′ |
|
| APOBEC3G |
5′-ccgaggacccgaaggttac-3′ |
5′-tccaacagtgctgaaattcg-3′ |
|
| APOBEC3H |
5′-agctgtggccagaagcac-3′ |
5′-cggaatgtttcggctgtt-3′ |
|
| HTLV-1 tax |
5′-atcccgtggagactcctcaa-3′ |
5′-aacacgtagactgggtatcc-3′ |
|
| HPRT |
5′-ccttggtcaggcagtataatcca-3′ |
5′-ccaacaaagtctggcttatatccaa-3′ |
|
DNA isolation and HTLV-1 proviral load
measurement
Genomic DNA was extracted from splenocyte or
peripheral blood by using ZR-Duet™ DNA/RNA MiniPrep kit (Zymo
Research Corp., Irvine, CA, USA). Proviral loads (PVLs) were
measured by real-time PCR using the previously described protocol
with some modifications (TaqMan Fast Advanced Master Mix; Applied
Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) (MyiQ
or CFX96 real-time PCR system; Bio-Rad Laboratories, Inc.)
(13). The primers and probes
targeting HTLV-1 pX and human β-globin (HBB; as an internal
control) are listed in Table III.
PVLs were calculated by the following formula: [(copy number of
pX)/(copy number of HBB/2)] × 100.
Microarray hybridization and data
analyses
RNA was extracted from each sample and purified by
using an RNeasy mini kit (Qiagen, Inc.) following the
manufacturer's instructions. RNA labeling and hybridization were
performed using a commercial kit from Hokkaido System Science Co.,
Ltd., Sapporo, Japan. A 200-ng aliquot of total RNA was converted
to double-stranded cDNA, and Cyanie3-labeled cRNA was produced
using the Low Input Quick Amp Labeling kit (Agilent Technologies,
Inc., Santa Clara, CA, USA). The resulting labeled cRNA was
purified by using the RNeasy mini kit (Qiagen, Inc.) and fragmented
into strands in length in accordance with the protocols of the
Agilent Technologies, Inc. These strands were then hybridized to
GeneChip Array, and the hybridization data were analyzed using
SurePrint G3 Human GE 8×60k 1color (Agilent Technologies,
Inc.).
Flow cytometric analysis and cell
sorting
Mice with a body weight of below 17 g were
euthanized. Single-cell spleen suspensions were prepared as
described previously (14). To stain
surface markers, anti-human CD45-PerCP or APC-Cy7, CD3-fluorescein
isothiocyanate (FITC) or phycoerythrin (PE)-Cy7, CD4-PE, CD25-FITC
antibodies were used, along with mouse immunoglobulin G1 and FITC
as isotype controls (BD Biosciences, Franklin Lakes, NJ, USA). Flow
cytometric analysis was performed on a BD FACSCan for 3-color
staining and a BD FACSCant II (BD Biosciences) for 7-color
staining. The CellQuest and Diva software programs were used for
data acquisition (BD Biosciences), and the collected data were
analyzed by using FACS express 3 (De Novo Software, Piscataway, NJ,
USA). T cells expressing human CD4+, CD8−,
and CD25 were sorted from splenic mononuclear cells by using
FACSAria or FACSAria III (BD Biosciences).
Statistical analysis
Mann-Whitney U-test and Wilcoxon's signed-rank test
were used to determine the significance of differences and data
comparison. P<0.05 was considered to indicate a statistically
significant difference.
Results
Differences in APOBEC3 superfamily
expression
The APOBEC superfamily proteins has several and
important functions in human health and disease (15). We first analyzed the mRNA expression
of the APOBEC3 superfamily of in vivo samples from healthy
donors and patients with HAM. However, we found no significant
differences between these groups (Fig.
1A). HTLV-1 tax is known as a transactivator that can activate
many genes (16) and its expression
is suppressed in vivo (17).
Therefore, we hypothesized that the tax might influence the
expression of the APOBEC3 superfamily. As the HTLV-1 tax expression
increased after 24 h of ex vivo culture, we compared the
mRNA expression of the APOBEC3 superfamily in the samples of
patients with HAM before and after the ex vivo culture.
After culture, tax expression was apparently increased as well as
that of A3C, A3D, A3F, and A3G. On the other hand, there was a
tendency of decreasing A3B expression (Fig. 1B). As a negative control, we
conducted the same experiment by using samples from healthy donors.
We found that after 24 h of ex vivo culture, A3A expression
was decreased; a tendency of decrease was also exhibited by the
level of A3B expression. However, no changes were observed in the
expression of A3C, A3D, A3F, A3G, and A3H (Fig. 1C). There are reports that A3B is a
mutagen (8,9), whereas ATL is known as a malignant
disease that is caused by HTLV-1 infection. Therefore, A3B
expression might be increased in HTLV-1-infected individuals.
However, these results did not show the tendency. HTLV-1 infection
might not influence A3B expression because we used samples from the
patients with HAM in the first experiment, which is an inflammatory
disease not a malignant disease. Recently, we developed a humanized
mouse model similar to ATL (13).
For further research, we used PBMCs that were obtained from
uninfected humanized mouse and 4–6 weeks postinfected humanized
mouse and analyzed the mRNA expression of the APOBEC3 superfamily
in these humanized mouse models. Although no significant changes in
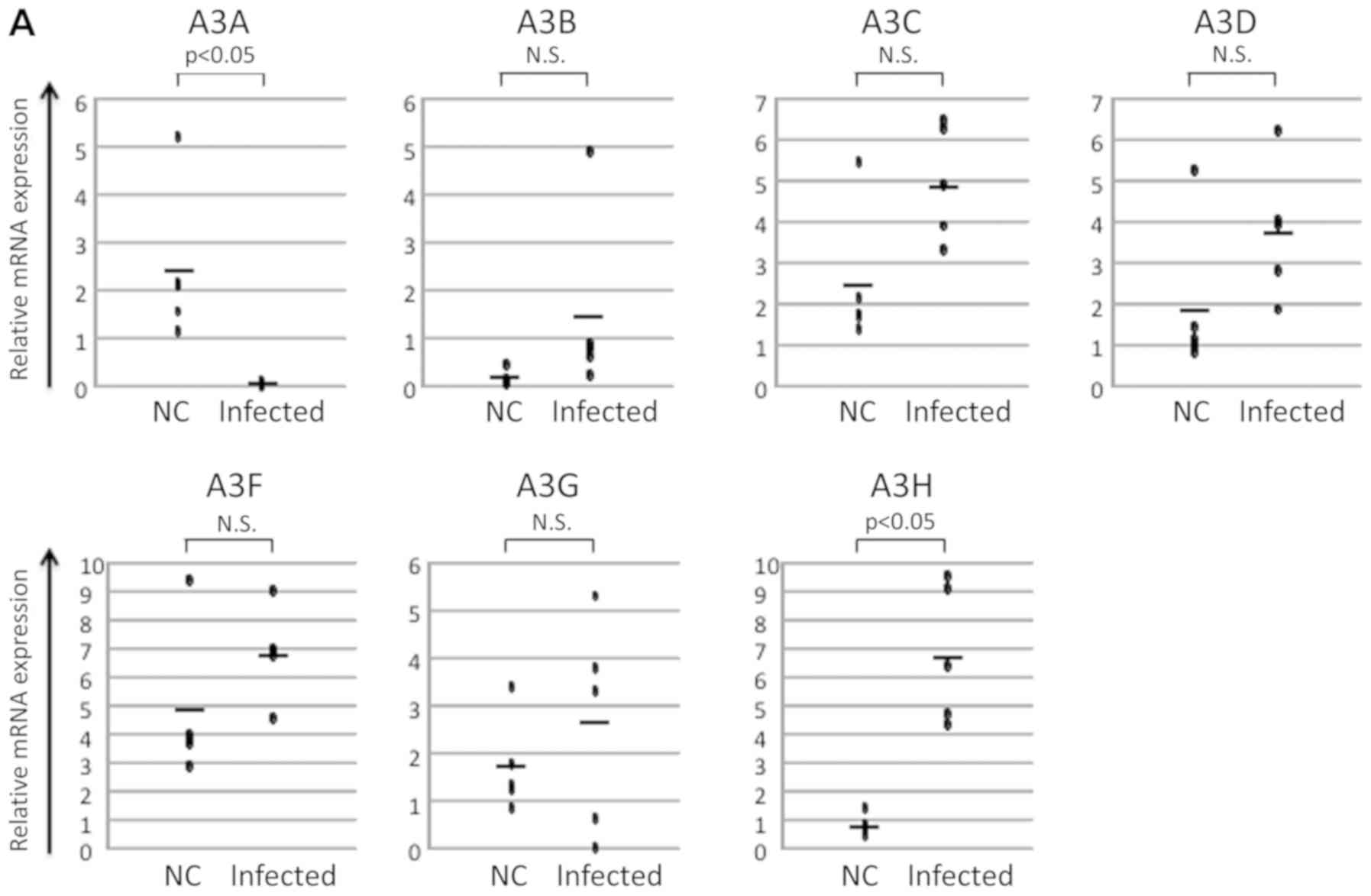
the expression of A3C, A3D, A3F, and A3G were observed, an increase
in the level of expression of A3B and a decrease in the level of
expression of A3A was noted. In contrast, the level of expression
of A3H was significantly increased (Fig.
2A). As it is difficult to extract sufficient qualities of
PBMCs from the animals in humanized mouse model, we used
splenocytes instead of PBMCs for ex vivo culture
experiments. After 24-h ex vivo culture, the levels of the
expression of tax and A3A apparently increased. There was a
tendency of decreasing A3B expression and increasing A3C, A3D, A3F,
A3G, and A3H expression (Fig. 2B).
Almost the data, except A3A, showed the same tendency with the
samples from the patients with HAM.
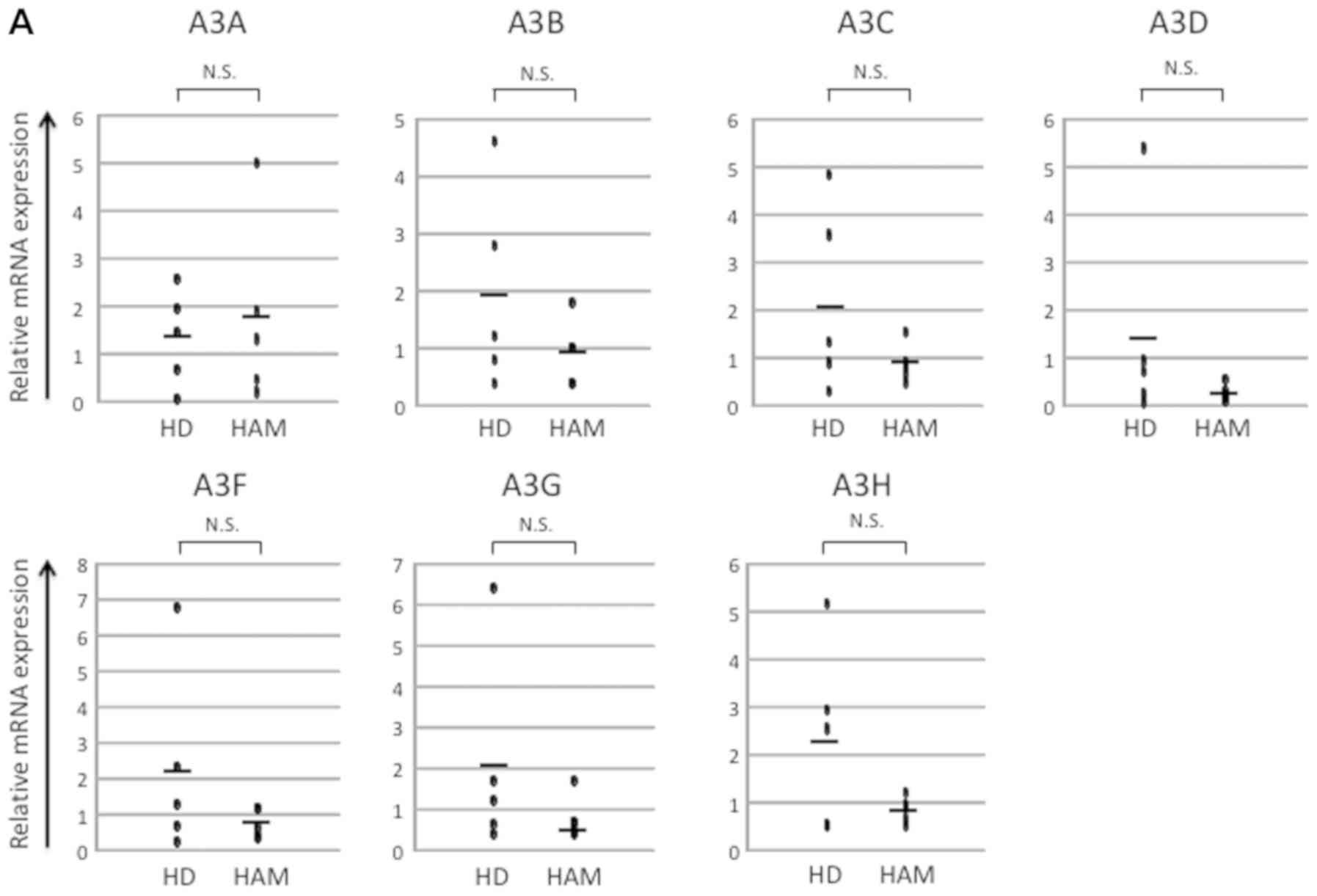 | Figure 1.Analysis of mRNA expression in human
samples. (A) mRNA expression of in vivo samples of healthy
donors and HAM patients. There was no significant difference in
APOBEC family expression; (B) mRNA expression of in vivo and
ex vivo samples of HAM patients. After 24 h of ex
vivo culture, tax expression was apparently increased
(P<0.05); the expression levels of A3C, A3D, A3F, and A3G were
also increased (P<0.05); on the other hand, there was a tendency
of decreasing A3B expression (0.05<P<0.1); (C) mRNA
expression of in vivo and ex vivo samples of healthy
donor. After 24 h of ex vivo culture, A3A was decreased
(P<0.05), A3B showed the tendency of decreasing
(0.05<P<0.1); however, no changes were observed in A3C, A3D,
A3F, A3G, and A3H expression. |
A3B expression increased in 18 weeks
post infection humanized mouse model
The levels of the expression of A3B did not change
in patients with HAM (Fig. 1A). In
contrast, it decreased after ex vivo culture in both the
samples from patients with HAM and infected humanized mouse model
(Figs. 1B and 2B). On the other hand, there was a tendency
of increasing A3B expression in the in vivo humanized mouse
samples (Fig. 2A). It was too short
to investigate the change in A3B expression, because we used the
ATL-like humanized mouse model samples with 4–6 weeks post
infection. Therefore, we analyzed the long-term humanized mouse
model 18 weeks after HTLV-1 infection. Initially, we checked the
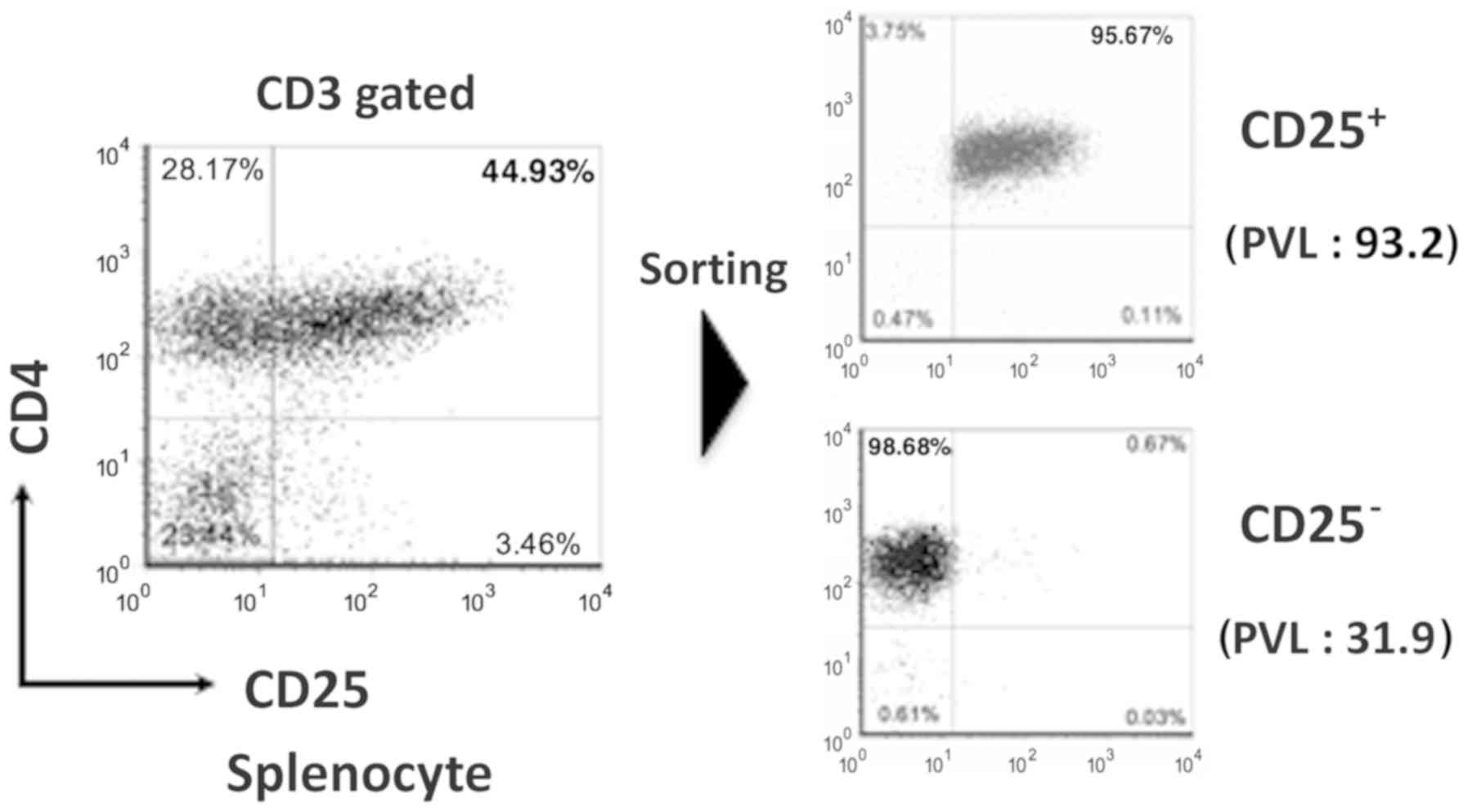
phenotype of the humanized mouse splenocytes by using FACS. In the
infected mice, the expression of CD4+ CD25+
cells was higher compared with that in the uninfected (Fig. 3). When the ATL-like humanized mouse
model was developed, CD4+ CD25+ cells was
quite increased (13). It is hard to
collect sufficient quantities of CD4+ CD25+
cells from uninfected control mice because this population is
exceedingly small. There is a report that CD4+
CD25− cells are also infected with HTLV-1 in
HTLV-1-infected individual (18).
Therefore, instead of CD4+ CD25+ cells, we
analyzed and compared CD4+ CD25− cells. Gene
expression array of CD4+ CD25− splenocytes
showed a 16.3-fold increase of CADM1 and a 12.6-fold increase of
APOBEC3B levels in the long-term humanized mouse model after HTLV-1
infection compared with the uninfected one (Table IV). In the humanized mouse model,
both CADM1 and APOBEC3B were increased in CD4+
CD25+ cells compared with those in CD4+
CD25− cells. On the other hand, APOBEC3 superfamily
expressions except APOBEC3B were not so increased in this humanized
mouse model (Table IV). These data
showed that APOBEC3B was increased in the ATL-like humanized mouse
model as well as such as CADM1, which are ATL indicators (19,20).
 | Table IV.Gene expression array of
CD4+ CD25−/CD4+ CD25+
splenocytes. |
Table IV.
Gene expression array of
CD4+ CD25−/CD4+ CD25+
splenocytes.
| Gene | CD4+
CD25− | CD4+
CD25+ |
|---|
| CADM1 | 16.3 | 42.3 |
| APOBEC3A |
0.9 |
1.0 |
| APOBEC3B | 12.6 | 34.2 |
| APOBEC3C |
2.2 |
1.8 |
| APOBEC3D |
1.0 |
1.2 |
| APOBEC3F |
2.0 |
2.0 |
| APOBEC3G |
2.4 |
1.7 |
| APOBEC3H |
3.2 |
3.0 |
Discussion
The host factor taking the role that is important to
the virus replication is targets to the development of a new virus
therapeutic drug (21). Since it has
been reported that APOBEC3G combines with HIV-1 vif and influences
infectivity of a virus in the past, new host-virus relations came
to be understood by through these molecules (22,23).
Previously it has been reported that multiple APOBEC3 enzymes may
limit the infectivity of HTLV-1 (24,25).
APOBEC3 genes are interferon-stimulated genes whose expression is
increased in response to various stimuli and exposure, including
the ligands of toll-like receptors TLR3, TLR4 and TLR7 (26,27).
Thus, the activation of innate immune responses that occurred after
infection with many different viruses could lead to increased
APOBEC3 activity. Therefore, APOBEC3 enzymes are involved in the
intrinsic/innate response to and the subsequent control of early
virus infection. Furthermore, there is increasing evidence that
APOBEC3 enzymes contribute to the adaptive immune response, more
specifically by affecting the generation of cytotoxic T lymphocytes
that recognize viral peptides and B-cell production of antiviral
antibodies. We expected that APOBEC superfamily expression was
increased in patients with HAM due to persistent viral infection.
Surprisingly, our results indicated that no significant differences
were observed in the APOBEC superfamily expression between the
PBMCs of healthy donors and patients with HAM (Fig. 1A). Similarly, Kataoka et al
reported that A3B was increased in both ATL patients and
asymptomatic HTLV-1 carriers (28).
On the other hand, Fan et al reported that A3G levels were
increased in patients with ATL (24). The results were contradictory because
we used samples from the patients with HAM.
A previous study has demonstrated that APOBEC3B is
enriched in the genomics of many human cancers. APOBEC enzymes
normally function in innate immune responses in according to viral
infection in the case of cancer developments (29). In ATL, it was reported that APOBEC3B
expression increases in comparison with healthy donor (28). In contrast, the expression of
APOBEC3B did not change significantly in HAM without tumor
characteristics (Fig. 1A). These
results suggest that A3B rises by malignant transformation not
infection itself. To solve this question, we should analyze the
patient sample in the process when ATL develops. It is, however,
quite difficult, because the onset rate of ATL is low
(approximately 5%), and extremely long periods of time are
necessary until the disease develops (30). The experimental system by an animal
model is needed to analyze a cause of the diseases of a virus. The
experimental limit is considered from individual's number and cost
side by an experimental animal by an ape. Thus, when it was seen
historically, a trial by the HTLV-1-infected animal model for which
mice, rats, rabbits were used had been performed, but there was
process, which didn't even come to clinical condition progress to
ATL (31). Recently, we developed a
humanized mouse model that showed ATL-like symptoms (13). Therefore, we analyzed changes of
APOBEC superfamily expression with HTLV-1 infected humanized mouse.
As expected, in the long-term humanized mouse model, A3B was
apparently increased after HTLV-1 infection (Table IV). In contrast, A3B increased only
slightly in the short-term model (Fig.
2A). We speculate that these different results might be caused
by the different lengths of the infection periods. Based on the
above consideration, we hypothesized that our animal model is ATL
pre-oncogenic, based on APOBEC3B, which induces mutations in
various human cancers (32) and that
this may lead to genomic instability and ultimately to
tumorigenesis. Further research is necessary in the future. In
fact, CADM1, known as an indicator of ATL, was apparently increased
in the long-term humanized mouse model after HTLV-1 infection
(Table IV). HTLV-1 infectivity to
the animal besides humans was very low and animal immunity systems
were so different from humans. The treatment development, which
also vaccinates with first as preclinical study is performed even
and it seems different to adaptation to humans. We tried using
HTLV-1-infected humanized mouse system. This study, which argued to
follow up a change with the passage of time using HTLV-1 infection
in humanized mouse models in vivo, was for the first
time.
To gain insights into the mechanisms underlying the
long latency and onset of HTLV-1-related diseases, we investigated
the interaction between Tax and APOBECs expression. We have already
reported when the cells were transferred to in vitro
conditions, tax expression in vivo resumed. Therefore, by
the change induced in Tax expression, we confirmed whether APOBECs
expression changed or not. In humanized mouse spleen, after 24 h of
ex vivo culture, tax expression apparently increased. The
levels of A3C, A3D, A3F, A3G, and A3H expression were also
increased. On the other hand, the expression of A3B was decreased
(Fig. 2B). Almost all the data
except those concerning A3A showed the same tendency as that in HAM
patient samples. These results suggest that tax expression does not
influence A3B expression directly. Therefore, other viral proteins,
such as HBZ, might likely be involved in APOBEC protein expression,
a subject we will push investigate further in the future.
Acknowledgements
The authors would like to thank Dr Taketani
(Department of Microbiology, Kansai Medical University) for kindly
providing critical revisions of the manuscript.
Funding
The present study was supported in part by a
Grant-in-Aid for Cancer Research from the Ministry of Education,
Culture, Sports, Science and Technology of Japan (MT: 26430126,
JIF: 24590562), and by the Japan Agency for Medical Research and
Development (AMED; www.amed.go.jp), the Ministry of Health Labour and
Welfare (www.mhlw.go.jp) (MT: 15fk0108030h0002,
NT: 16ek0109026h0003). The funders had no role in study design,
data collection and analysis, decision to publish, or preparation
of the manuscript.
Availability of data and materials
The analyses data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
JY was the guarantor for integrity of the study and
contributed to study concepts, experimental studies, data analysis
and manuscript preparation. MT contributed to study design and
prepared and revised the manuscript. NT contributed to manuscript
review, statistical analysis and clinical studies. YR and SL
contributed to established and maintained humanized mice. JIF
contributed to study design and provided intellectual content. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Kansai Medical University (Hirakata, Japan). All
patients provided informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Uchiyama T, Yodoi J, Sagawa K, Takatsuki K
and Uchino H: Adult T-cell leukemia: Clinical and hematologic
features of 16 cases. Blood. 50:481–492. 1977.PubMed/NCBI
|
|
2
|
Osame M, Usuku K, Izumo S, Ijichi N,
Amitani H, Igata A, Matsumoto M and Tara M: HTLV-1 associated
myelopathy, a new clinical entity. Lancet. 1:1031–1032. 1986.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nakao K, Ohba N and Matsumoto M:
Noninfectious anterior uveitis in patients infected with human
T-lymphotropic virus type I. Jpn J Ophthalmol. 33:472–481.
1989.PubMed/NCBI
|
|
4
|
Clapham P, Nagy K, Cheingsong-Popov R,
Exley M and Weiss RA: Productive infection and cell free
transmission of human T-cell leukemia virus in a non-lymphoid cell
line. Science. 222:1125–1127. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Derse D, Mikovits J, Polianova M, Felber
BK and Ruscetti F: Virions released from cells transfected with a
molecular clone of human T-cell leukemia virus type I give rise to
primary and secondary infections of T cells. J Virol. 69:1907–1912.
1995.PubMed/NCBI
|
|
6
|
Fialkow PJ: Clonal original of human
tumors. Annu Rev Med. 30:135–143. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Malim MH: APOBEC proteins and intrinsic
resistance to HIV-1 infection. Philos Trans R Soc Lond B Biol Sci.
364:675–687. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Roberts SA, Lawrence MS, Klimczak LJ,
Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL,
Saksena G, et al: An APOBEC cytidine deaminase mutagenesis pattern
is widespread in human cancers. Nat Genet. 45:970–976. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Burns MB, Temiz NA and Harris RS: Evidence
for APOBEC3B mutagenesis in multiple human cancers. Nat Genet.
45:977–983. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vartanian JP, Henry M, Marchio A, Suspène
R, Aynaud MM, Guétard D, Cervantes-Gonzalez M, Battiston C,
Mazzaferro V, Pineau P, et al: Massive APOBEC3 editing of hepatitis
B viral DNA in cirrhosis. PLoS Pathog. 6:e10009282010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vartanian JP, Guétard D, Henry M and
Wain-Hobson S: Evidence for editing of human papillomavirus DNA by
APOBEC3 in benign and precancerous lesions. Science. 320:230–233.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Albin JS and Harris RS: Interactions of
host APOBEC3 restriction factors with HIV-1 in vivo: Implications
for therapeutics. Expert Rev Mol Med. 12:e42010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tezuka K, Xun R, Tei M, Ueno T, Tanaka M,
Takenouchi N and Fujisawa J: An animal model of adult T-cell
leukemia: Humanized mice with HTLV-1-specific immunity. Blood.
123:346–355. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nie C, Sato K, Misawa N, Kitayama H,
Fujino H, Hiramatsu H, Heike T, Nakahata T, Tanaka Y, Ito M and
Koyanagi Y: Selective infection of CD4+ effector memory
T lymphocytes leads to preferential depletion of memory T
lymphocytes in R5 HIV-1-infected humanized NOD/SCID/IL-2Rgammanull
mice. Virology. 394:64–72. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Salter JD, Bennett RP and Smith HC: The
APOBEC protein family: United by structure, divergent in function.
Trends Biochem Sci. 41:578–594. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yoshida M, Suzuki T, Fujisawa J and Hirai
H: HTLV-1 oncoprotein tax and cellular transcription factors. Curr
Top Microbiol Immunol. 193:79–89. 1995.PubMed/NCBI
|
|
17
|
Furuta RA, Sugiura K, Kawakita S, Inada T,
Ikehara S, Matsuda T and Fujisawa J: Mouse model for the
equilibration interaction between the host immune system and human
T-cell leukemia virus type 1 gene expression. J Virol.
76:2703–2713. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yamano Y, Cohen CJ, Takenouchi N, Yao K,
Tomaru U, Li HC, Reiter Y and Jacobson S: Increased expression of
human T lymphocyte virus type I (HTLV–I) Tax11-19 peptide-human
histocompatibility leukocyte antigen A*201 complexes on
CD4+CD25+ T cells detected by
peptide-specific, major histocompatibility complex-restricted
antibodies in patients with HTLV-1-associated neurologic disease. J
Exp Med. 199:1367–1377. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sasaki H, Nishikata I, Shiraga T, Akamatsu
E, Fukami T, Hidaka T, Kubuki Y, Okayama A, Hamada K, Okabe H, et
al: Overexpression of a cell adhesion molecule, TSLC1, as a
possible molecular marker for acute-type adult T-cell leukemia.
Blood. 105:1204–1213. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Araki K, Harada K, Nakamoto K, Shiroma M
and Miyakuni T: Clinical significance of serum soluble IL-2R levels
in patients with adult T cell leukaemia (ATL) and HTLV-1 carriers.
Clin Exp Immunol. 119:259–263. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Henderson S and Fenton T: APOBEC3 genes:
Retroviral restriction factors to cancer drivers. Trends Mol Med.
21:274–284. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang Y, Kinlock BL, Shao Q, Turner TM and
Liu B: HIV-1 Vif inhibits G to A hypermutations catalyzed by
virus-encapsidated APOBEC3G to maintain HIV-1 infectivity.
Retrovirology. 11:892014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guerrero S, Libre C, Batisse J, Mercenne
G, Richer D, Laumond G, Decoville T, Moog C, Marquet R and Paillart
JC: Translational regulation of APOBEC3G mRNA by Vif requires its
5′UTR and contributes to restoring HIV-1 infectivity. Sci Rep.
6:395072016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fan J, Ma G, Nosaka K, Tanabe J, Satou Y,
Koito A, Wain-Hobson S, Vartanian JP and Matsuoka M: APOBEC3G
generates nonsense mutations in human T-cell leukemia virus type 1
proviral genomes in vivo. J Virol. 84:7278–7287. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ooms M, Krikoni A, Kress AK, Simon V and
Munk C: APOBEC3A, APOBEC3B, and APOBEC3H haplotype 2 restrict human
T-lymphotropic virus type 1. J Virol. 86:6097–6108. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lamb C and Arbuthnot P: Activating the
innate immune response to counter chronic hepatitis B virus
infection. Expert Opin Biol Ther. 16:1517–1527. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cardinaud S, Urrutia A, Rouers A, Coulon
PG, Kervevan J, Richetta C, Bet A, Maze EA, Larsen M, Iglesias MC,
et al: Triggering of TLR-3, −4, NOD2, and DC-SIGN reduces viral
replication and increases T-cell activation capacity of
HIV-infected human dendritic cells. Eur J Immunol. 47:818–829.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kataoka K, Nagata Y, Kitanaka A, Shiraishi
Y, Shimamura T, Yasunaga J, Totoki Y, Chiba K, Sato-Otsubo A, Nagae
G, et al: Integrated molecular analysis of adult T cell
leukemia/lymphoma. Nat Genet. 47:1304–1315. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vieira VC and Soares MA: The role of
cytidine deaminases on innate immune responses against human viral
infections. Biomed Res Int. 2013:6830952013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tajima K: The 4th nation-wide study of
adult T-cell leukemia/lymphoma (ATL) in Japan: Estimates of risk of
ATL and its geographical and clinical features. The T- and B-cell
malignancy study group. Int J Cancer. 45:237–243. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lairmore MD, Silverman L and Ratner L:
Animal models for human T-lymphotropic virus type 1 (HTLV-1)
infection and transformation. Oncogene. 24:6005–6015. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zou J, Wang C, Ma X, Wang E and Peng G:
APOBEC3B, a molecular driver of mutagenesis in human cancers. Cell
Biosci. 7:292017. View Article : Google Scholar : PubMed/NCBI
|