Introduction
Endometrial carcinoma (EC) is one of the most common
malignancies in women, and a leading cause of cancer-associated
mortality worldwide (1). The most
frequently occurring subtype of endometrial cancer is endometrioid
endometrial carcinoma, which accounts for about 90% of all EC cases
(2). Surgery is the main treatment
strategy for patients with EC (3),
however the effectiveness of primary surgery is generally
unsatisfactory (3). Although the
incidence of EC is increasing, the molecular mechanism underlying
EC progression remains poorly understood (2). It is therefore necessary to explore the
mechanism and develop novel therapeutic strategies for EC
intervention.
Long non-coding RNAs (lncRNAs) are a large and
diverse class of transcribed RNA molecules usually with a length of
more than 200 nucleotides and limited protein-coding capability
(4). LncRNAs can regulate gene
expression via several different mechanisms, which include acting
as a molecular sponge for miRNAs to regulate the expression of its
target genes (5). Several studies
have demonstrated that lncRNAs are involved in a number of
biological processes, including tumor initiation, growth and
metastasis (6,7). The lncRNA gastric cancer associated
transcript 3 promotes colorectal cancer cell proliferation,
invasion and migration through competitive binding to miRNA-149
(8). LncRNA long intergenic
non-protein coding RNA 1510 promotes colorectal cancer cell growth
by modulating MET expression (9).
LncRNA FLVCR1-antisense RNA 1 acts as a miRNA-513c sponge to
regulate cancer cell proliferation, migration and invasion in
hepatocellular carcinoma (10).
LncRNA long intergenic non-protein coding RNA 958 accelerates
gliomagenesis through miRNA-203/cyclin-dependent kinase 2
regulation (11).
A recently study revealed that lncRNA colon cancer
associated transcript 1 (CCAT1) promotes cell proliferation,
migration and invasion in thyroid carcinoma via downregulation of
miRNA-143 (12). Other studies have
demonstrated that CCAT1 is involved in colorectal cancer (13), ovarian cancer (14), gastric cancer (15), myeloma (16) and breast cancer (17). The pathophysiological contribution of
CCAT1 to EC progression remains largely unknown. The aim of the
present study was to investigate the biological function of CCAT1
in EC. The current study demonstrated that CCAT1 was significantly
upregulated in EC tissue samples and cell lines. Knockdown of CCAT1
significantly decreased EC cell proliferation and migration.
Furthermore, the current study demonstrated that CCAT1 acts as a
molecular sponge of miRNA-181a-5p (miR-181a-5p) in EC cells and
inhibition of miR-181a-5p significantly reversed the effects of
CCAT1 knockdown. Taken together, the current study revealed that
the CCAT1/miR-181a-5p axis may serve a role in EC progression.
Materials and methods
Patient samples
The present study analyzed tissue samples from
patients diagnosed with endometrial cancer based on
histopathological evaluation. A total of 37 EC tissue and 21
matched adjacent normal tissue samples were collected from female
patients (mean age, 47.56±15.49 years) who had undergone surgical
resection at The First Affiliated Hospital of Kunming Medical
University between January 2010 and December 2016. None of the
patients received local or systemic treatment before the operation.
Endometrial cancer tissue and adjacent healthy tissue samples were
collected and immediately frozen in liquid nitrogen, and stored at
−80°C until further use. The current study was approved by the
Ethics Committee of The First Affiliated Hospital of Kunming
Medical University (Yunnan, China). Written informed consent was
obtained from all patients.
Cell culture and transfection
Human endometrial adenocarcinoma cell lines HEC-1-A
(ATCC® HTB-112™) and KLE (ATCC® CRL-1622™),
as well as the human endometrial stromal cell line T-HESC
(ATCC® CRL-4003™) were from American Type Culture
Collection (Manassas, VA, USA). Human endometrial adenocarcinoma
cell line Ishikawa (cat. no. 3111C0001CCC000686) was purchased from
China Infrastructure of Cell Line Resource (Beijing, China;
http://www.cellresource.cn/contact.aspx). All cell
lines were maintained in Dulbeco's modified Eagle medium (DMEM;
Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (FBS; HyClone; GE
healthcare, Chicago, IL, USA) and 1% penicillin-streptomycin
solution and maintained at 37°C in a 5% CO2-humidified
incubator. miR-181a-5p mimic (5′-AACAUUCAACGCUGUCGGUGAGU-3′),
miR-181a-5p inhibitor (5′-ACUCACCGACAGCGUUGAAUGUU-3′), negative
controls miRNA (5′-UCACAACCUCCUAGAAAGAGUAGA-3′), si-CCAT1
(5′-AGGGAAACAGGAGCAAUCAUCATTA-3′) and control siRNA
(5′-UUCUCCGAACGUGUCACGUTT-3′) were chemically synthesized by
Shanghai Integrated Biotech Solutions Co., Ltd. (Shanghai, China).
All cell transfection reactions were performed using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) with 100 nM miRNA or siRNA, according to the
manufacturer's protocol. Subsequent experiments were performed
following 48 h transfection.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from tissues or cultured
cells using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. Total
RNA (5 µg) was reverse transcribed into cDNA using M-MLV Reverse
Transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.) and
supplemented with Oligo(dT18) RT primers (Invitrogen; Thermo Fisher
Scientific, Inc.). qPCR was subsequently performed using SYBR
Premix Ex Taq (Takara Bio, Inc., Otsu, Japan) using a Bio-Rad CFX96
real-time PCR System (Bio-Rad Laboratories, Inc., Hercules, CA,
USA). The conditions of qPCR were as follows: 94°C for 15 min,
followed by 45 cycles of 94°C for 10 sec, 60°C for 30 sec, and 72°C
for 30 sec. Each sample assayed in triplicate in three independent
experiments. The mRNA levels were quantified using the
2−∆∆Cq method (18) and
normalized to internal reference gene U6. The sequence of the CCAT1
forward primer was 5′-TTTATGCTTGAGCCTTGA-3′ and reverse primer was
5′-CTTGCCTGAAATACTTGC-3′. The sequence of the miR-181a-5p forward
primer was 5′-GCCGAACATTCAACGCTGTCG-3′ and reverse primer was
5′-GTGCAGGGTCCGAGGT-3′. The sequence of the miR-181a-5p forward
primer was 5′-CTCGCTTCGGCAGCACA-3′ and reverse primer was
5′-AACGCTTCACGAATTTGCGT-3′. The sequence of U6 forward primer was
5′-CTCGCTTCGGCAGCACA-3′ and reverse primer was
5′-AACGCTTCACGAATTTGCGT-3′.
Cell proliferation assay
Cell counting kit-8 (CCK-8; Beyotime Institute of
Biotechnology, Haimen, China) was used to study cell proliferation.
KLE cells were seeded into 96-well plates at a density of
1×103 cells/well and cultured for 24, 48 or 72 h. Cells
were subsequently incubated with 10 µl of CCK-8 for 2 h at 37°C.
Following incubation with CCK-8, cell viability was determined by
measuring the absorbance at a wavelength of 450 nm.
Cell migration assay
Cell migration assays were performed using 8 µm
transwell inserts (EMD Millipore, Billerica, MA, USA). A total of
2×104 KLE cells were added to the top chamber containing
200 ml serum-free DMEM. DMEM supplemented with 10% FBS was added to
the lower chamber. After 48 h incubation, migrated cells in the
lower chamber were stained with 0.1% crystal violet for 1 h at
25°C. Using a light microscope at ×100 magnifications, five visual
fields were randomly selected to calculate the number of migrated
cells.
Western blot analysis
Total protein was extracted from KLE cells using
radioimmunoprecipitation lysis buffer (Beyotime Institute of
Biotechnology). Total protein was quantified using a bicinchoninic
acid assay kit and 40 µg protein/lane was separated via SDS-PAGE on
a 8–12% gel. The separated proteins were subsequently transferred
onto polyvinylidene fluoride membranes and blocked with 5% non-fat
milk for 1 h at 37°C. The membranes were incubated with primary
antibodies against GAPDH (1:5,000; cat. no. SAB2701826;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and c-MET (1:2,000;
cat. no. ab51067; Abcam, Cambridge, UK) overnight at 4°C. Following
primary incubation, the membranes were incubated with a goat
horseradish peroxidase-conjugated secondary antibody (1:5,000;
sc-2005; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) for 2 h
at room temperature. Protein bands were visualized using enhanced
chemiluminescence substrate (EMD Millipore).
Bioinformatics analysis
The target miRNAs of CCAT1 were predicted using
miRDB (http://mirdb.org/miRDB/index.html).
Luciferase reporter assay
A luciferase reporter assay was performed to
determine the direct binding of CCAT1 to miR-181a-5p. The wild type
(WT) and mutant (Mut) sequence of CCAT1 was directly synthesized by
Shanghai GenePharma Co., Ltd. (Shanghai, China) and then inserted
into pGL3 plasmid (Ambion; Thermo Fisher Scientific, Inc.). Cells
(2×104) were cultured in 24-well plates, and each well
was transfected with 0.2 µg WT or Mut pGL3-CCAT1 reporter plasmid,
and equal amounts of miR-181a-5p (50 nM) and NC mimics (50 nM)
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). After 24 h, Firefly and Renilla
luciferase activities were measured using the Dual-Luciferase
Reporter Assay Kit (Promega Corporation, Madison, WI, USA)
according to the manufacturer's protocols. Luciferase activities
were normalized to that of Renilla luciferase.
Statistical analysis
Data are presented as the mean ± standard deviation
of three independent experiments. All statistical analyses were
performed using SPSS software (version 20.0; IBM Corp., Armonk, NY,
USA). Student's t-test was performed for comparison analysis
between two groups. One-way analysis of variance followed by
Fisher's least significant difference post hoc test was performed
for multiple-group comparisons. Pearson's correlation coefficient
was used to measure the linear correlation between CCAT1 and
miR-181a-5p expression in EC tissues. P<0.05 was considered to
indicate a statistically significant difference.
Results
Expression patterns of CCAT1 and
miR-181a-5p in EC tissues
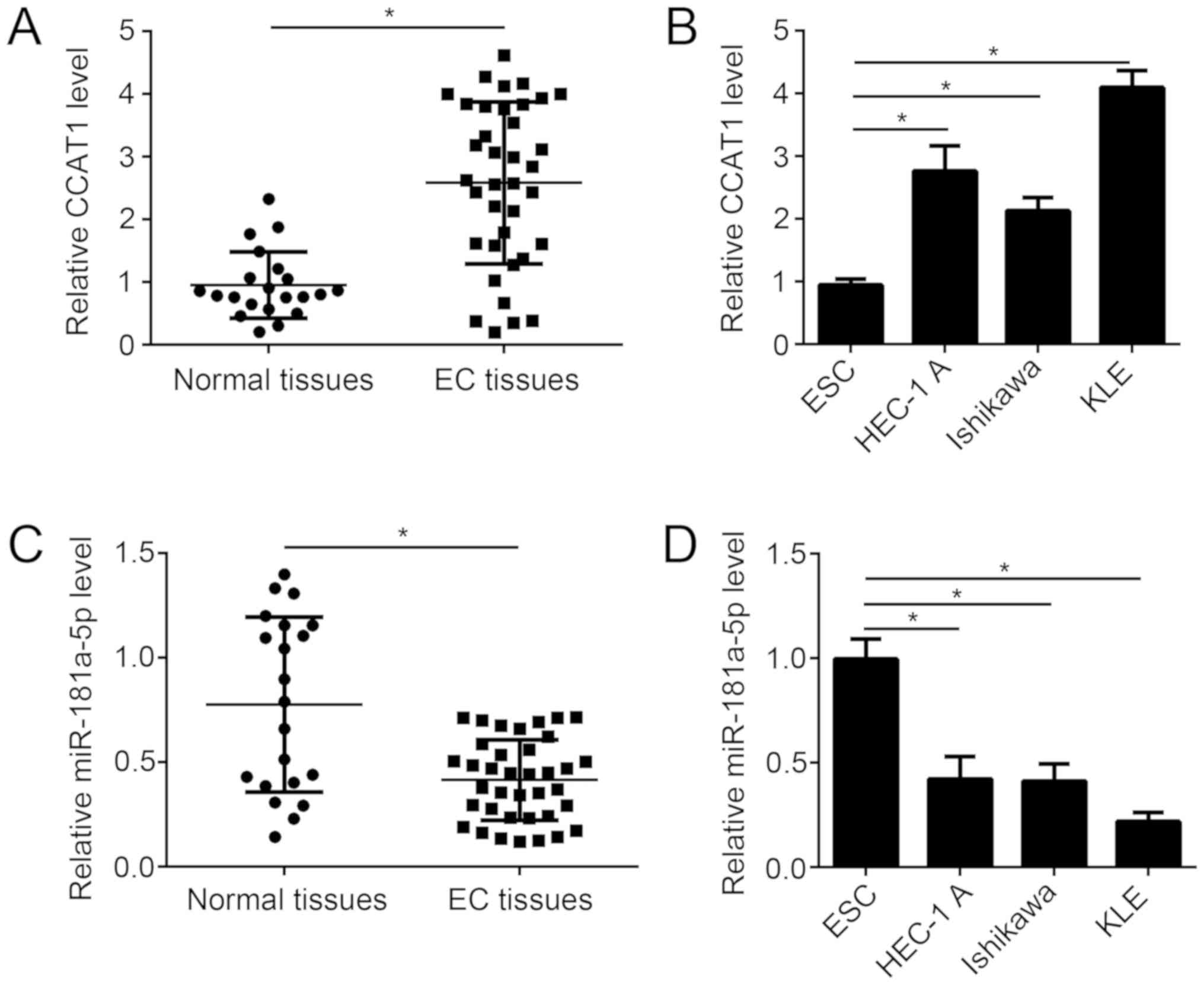
To investigate the role of CCAT1 in endometrial
cancer progression, CCAT1 expression was examined in tissue samples
from patients with endometrial cancer. The expression level of
CCAT1 was significantly increased in endometrial cancer tissue
samples compared with matched adjacent healthy tissue samples from
patients with endometrial cancer (Fig.
1A). Similarly, the expression level of CCAT1 significantly
increased in several endometrial cancer cell lines, compared with
normal human endometrial stromal cell line T-HESC (Fig. 1B). By contrast, miR-181a-5p
expression was significantly decreased in endometrial cancer tissue
samples compared with matched adjacent healthy tissue samples from
patients with endometrial cancer (Fig.
1C). Similarly, the expression level of miR-181a-5p
significantly decreased in several endometrial cancer cell lines,
compared with normal human endometrial stromal cell line T-HESC
(Fig. 1D). These results suggest
that CCAT1 may regulate miR-181a-5p expression in EC.
CCAT1 is a target of miR-181a-5p
To investigate the correlation between CCAT1 and
miR-181a-5p, bioinformatics analysis was performed using starBase
(starbase.sysu.edu.cn) to identify
potential targets of miR-181a-5p. starBase identified a potential
miR-181a-5p binding site in CCAT1 (Fig.
2A). Luciferase reporter gene assay was performed to confirm
the potential association between CCAT1 and miR-181a-5p. Following
transfection with miR-181a-5p mimic, the overexpression of
miR-181a-5p was confirmed by RT-qPCR (Fig. 2B). Overexpression of miR-181a-5p
significantly reduced the luciferase reporter activity of CCAT1-WT
compared with CCAT1-Mut in KLE cells (Fig. 2B). Following transfection with
siCCAT1, knockdown of CCAT1 expression was confirmed by RT-qPCR
(Fig. 2C). Knockdown of CCAT1
significantly increased miR-181a-5p expression in KLE cells
(Fig. 2D) By contrast,
overexpression of miR-181a-5p significantly reduced CCAT1
expression in KLE cells (Fig. 2E).
Furthermore, an inverse correlation was observed between
miR-181a-5p and CCAT1 expression in EC tissues (Fig. 2F). These results suggest that CCAT1
can act as a molecular sponge of miR-181a-5p in EC cells.
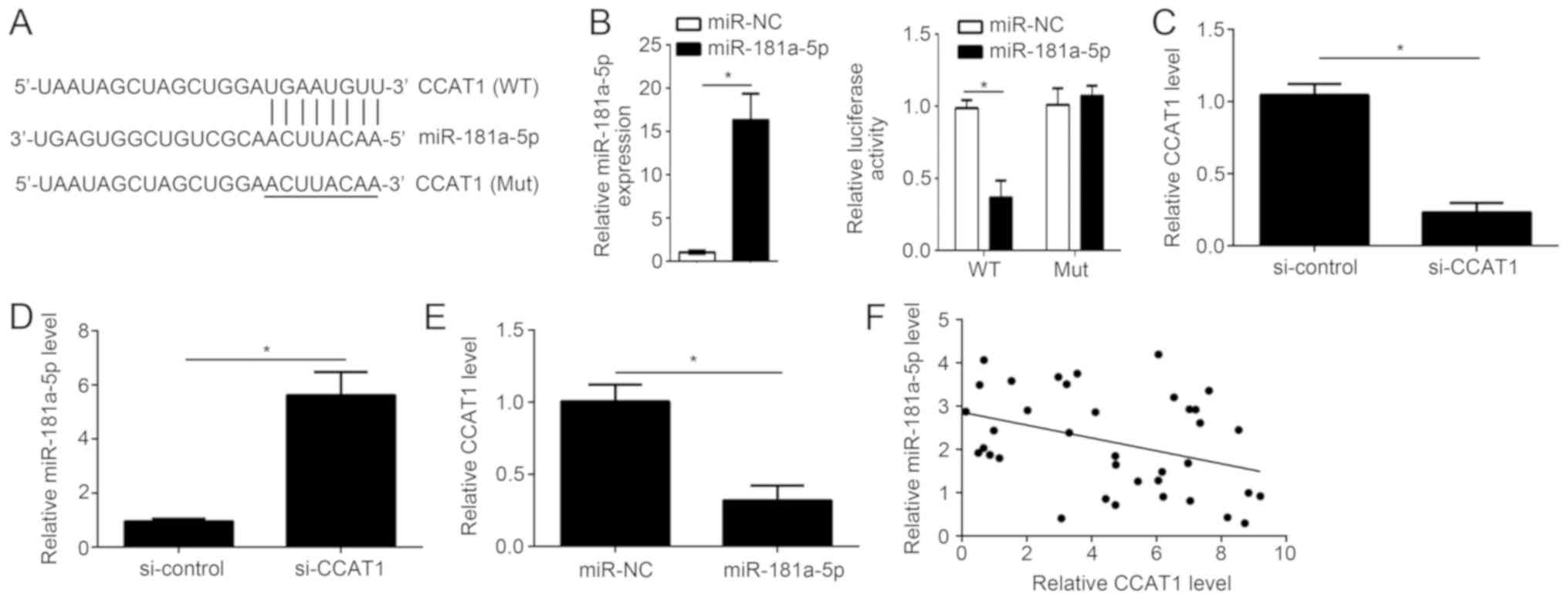 | Figure 2.CCAT1 is a target gene of
miR-181a-5p. (A) Bioinformatics analysis was used to identify
miR-181a-5p binding sites in CCAT1. (B) Following miR-181a-5p
overexpression, the expression level was detected by RT-qPCR and
the relative luciferase activity was measured. (C) Following
transfection with si-CCAT1 or control, the mRNA expression level of
CCAT1 was determined by RT-qPCR in human EC cell line, KLE. (D)
CCAT1 knockdown significantly enhanced miR-181a-5p expression in
KLE cells. (E) Overexpression of miR-181a-5p significantly reduced
the mRNA expression level of CCAT1 in KLE cells. (F) An inverse
correlation between CCAT1 and miR-181a-5p expression in EC tissue
samples was identified (r=−0.371; P=0.024). *P<0.05 vs. control
group. RT-qPCR, reverse transcription-quantitative polymerase chain
reaction; CCAT1, colon cancer-associated transcript 1; EC,
endometrial cancer; miR, microRNA; KLE, human endometrial stromal
cell line; miR-NC, endometrial cancer cell line KLE transfected
with scramble miR; miR-181a-5p, endometrial cancer cell line KLE
transfected with miR-181a-5p mimic; si-control, endometrial cancer
cell line KLE transfected with siRNA negative control; si-CCAT1,
endometrial cancer cell line KLE transfected with siRNA targeting
CCAT1; siRNA, small interfering RNA; CCAT1 (WT), wild-type 3′UTR of
CCAT1 cloned into luciferase reporter plasmid; CCAT1 (Mut), mutant
3′UTR of CCAT1 cloned into luciferase reporter plasmid. |
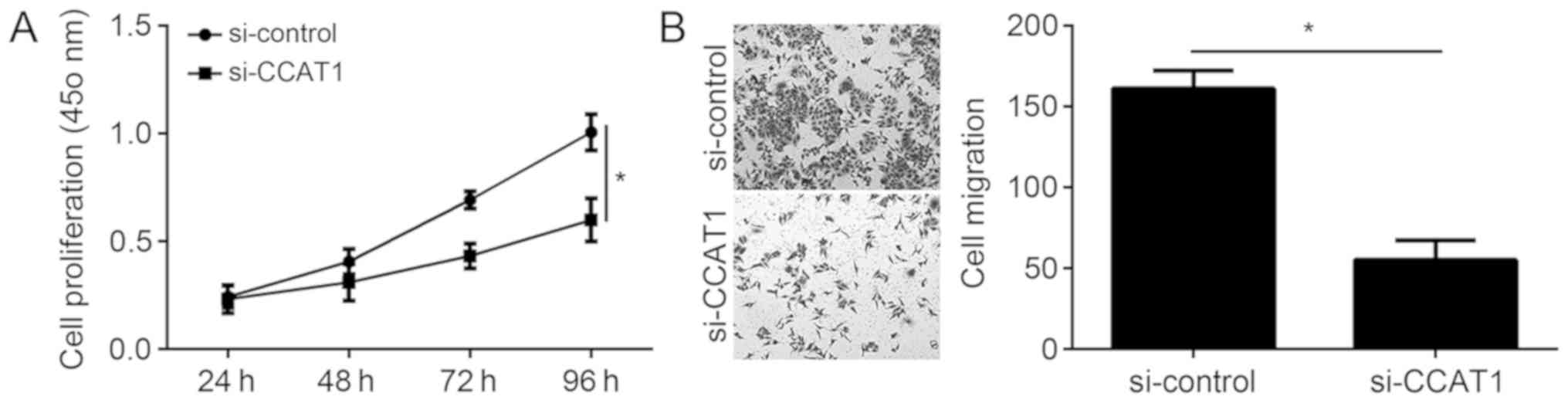
CCAT1 knockdown inhibits EC cell
proliferation and migration
The physiological functions of CCAT1 in EC were
further investigated. CCK-8 assays demonstrated that knockdown of
CCAT1 significantly inhibited the proliferation of KLE cells
(Fig. 3A). The association between
CCAT1 and migration was examined by Transwell migration assays. The
results from the migration assays demonstrated that knockdown of
CCAT1 significantly decreased the migration ability of KLE cells
(Fig. 3B).
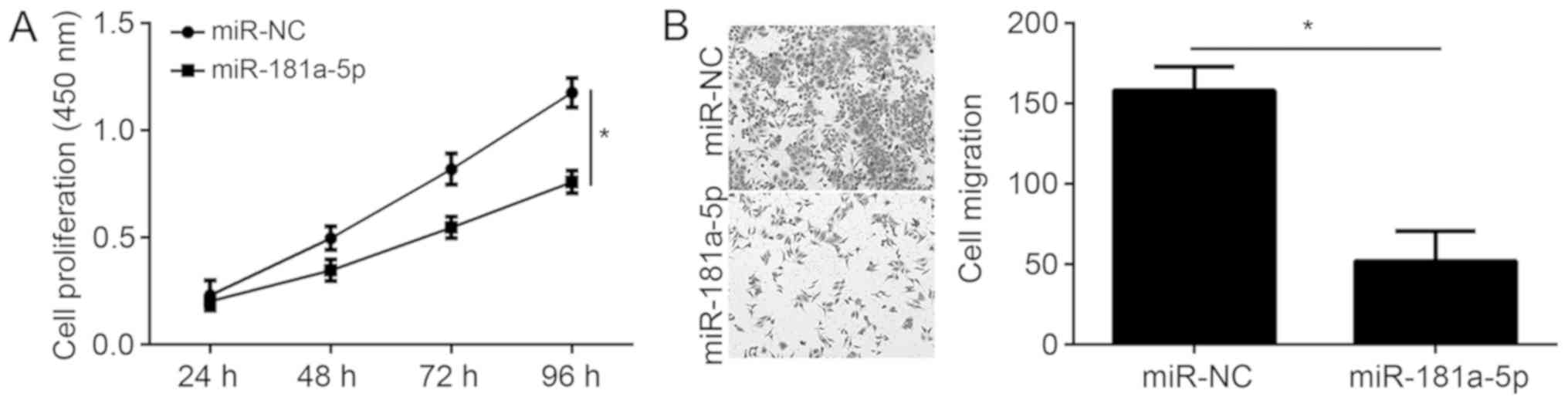
miR-181a-5p overexpression inhibits EC
cell proliferation and migration
The present study demonstrated that CCAT1 regulates
miR-181a-5p expression in EC cells. To determine the physiological
function of miR-181a-5p in EC, KLE cells were transfected with
miR-181a-5p mimic and subsequent CCK-8 and transwell assays were
performed. Overexpression of miR-181a-5p significantly reduced EC
cell proliferation and migration (Fig.
4A and B), suggesting that miR-181a-5p may function as a tumor
suppressor in EC.
miR-181a-5p inhibition significantly
reverses the effects of CCAT1 knockdown in EC cells
To determine whether CCAT1 functions in EC
progression through the inhibition of miR-181a-5p, rescue
experiments were performed in EC cells. Following transfection with
miR-181a-5p inhibitor alone, the relative expression level of
miR-181a-5p in KLE cells was confirmed by RT-qPCR (Fig. 5A). CCK-8 and Transwell migration
assays demonstrated that CCAT1 knockdown significantly inhibits the
proliferation and migration of KLE cells, however these effects
were reversed by the inhibition of miR-181a-5p (Fig. 5B and C). These results suggest that
CCAT1 may act as a sponge of miR-181a-5p to exert oncogenic roles
by regulating EC cell proliferation and migration.
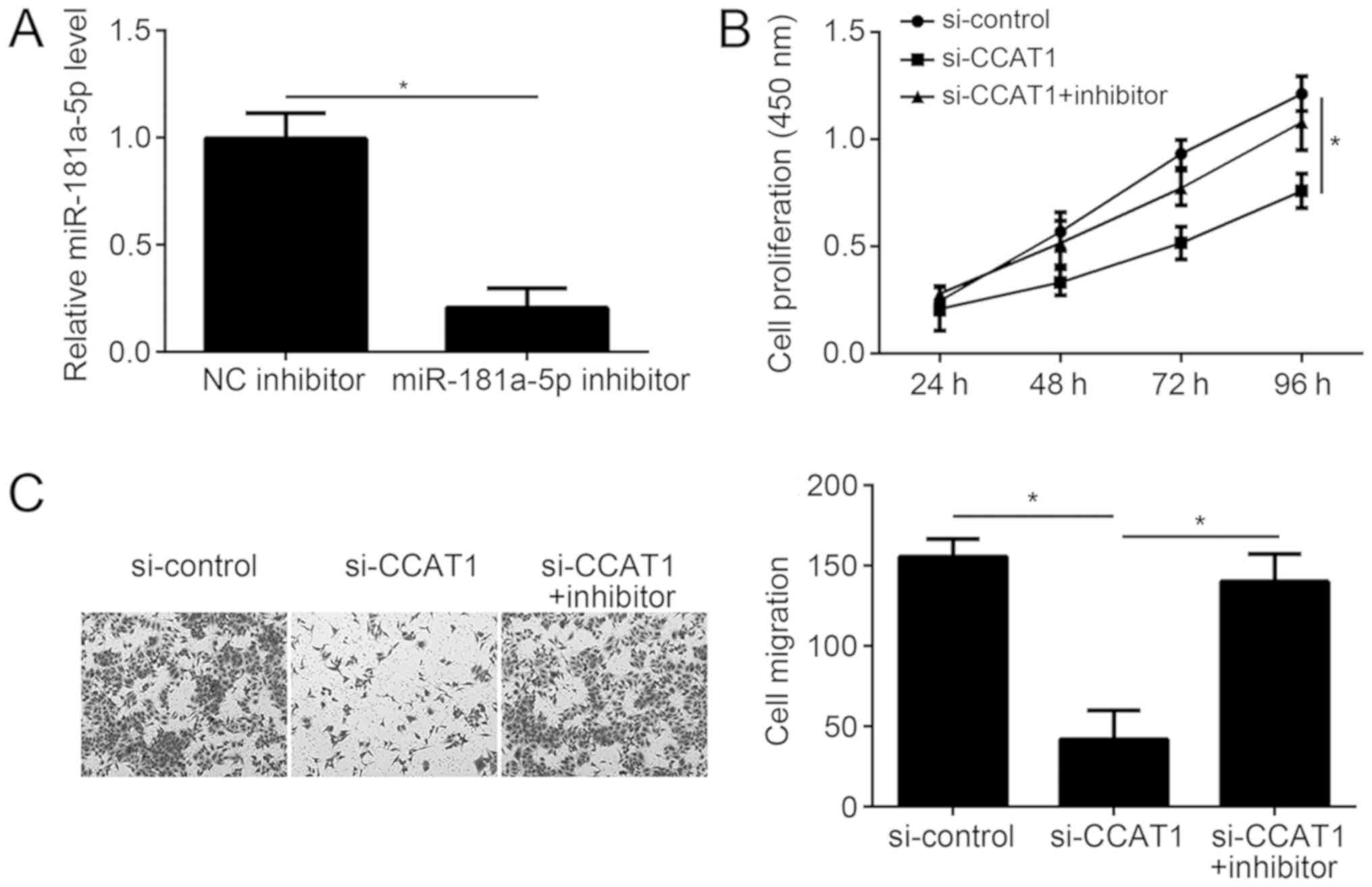 | Figure 5.miR-181a-5p inhibition significantly
reverses the effects of CCAT1 knockdown in EC cells. (A) Following
miR-181a-5p inhibition, the relative expression level of
miR-181a-5p was detected in KLE cells by reverse
transcription-quantification polymerase chain reaction. (B) The
CCK-8 assay was used to analyze the effect of CCAT1 knockdown and
miR-181a-5p inhibition on KLE cell proliferation. (C) The Transwell
migration assay was used to analyze the effect of CCAT1 knockdown
and miR-181a-5p inhibition on KLE cell migration. *P<0.05 as
indicated. CCAT1, colon cancer-associated transcript 1; EC,
endometrial cancer; miR, microRNA; KLE, human endometrial stromal
cell line; NC, negative control; NC inhibitor, endometrial cancer
cell line KLE transfected with negative control inhibitor;
miR-181a-5p inhibitor, endometrial cancer cell line KLE transfected
with miR-181a-5p inhibitor; si-control, endometrial cancer cell
line KLE transfected with siRNA negative control; si-CCAT1,
endometrial cancer cell line KLE transfected with siRNA targeting
CCAT1; siRNA, small interfering RNA; si-CCAT1 + inhibitor,
endometrial cancer cell line KLE transfected with siRNA targeting
CCAT1 and miR-181a-5p inhibitor. |
miR-181a-5p targets c-MET in KLE
cells
miR-181a-5p was previously reported to target c-MET
in liver cancer (19). Therefore,
the effect of miR-181a-5p on c-MET expression in KLE cells was
evaluated. Western blotting demonstrated that miR-181a-5p mimic
transfection suppressed c-MET expression in KLE cells (Fig. 6A). Furthermore, CCAT1 knockdown led
to decreased expression of c-MET (Fig.
6B), indicating that the CCAT1/miR-181a-5p axis may regulate
c-MET to exert roles in EC progression.
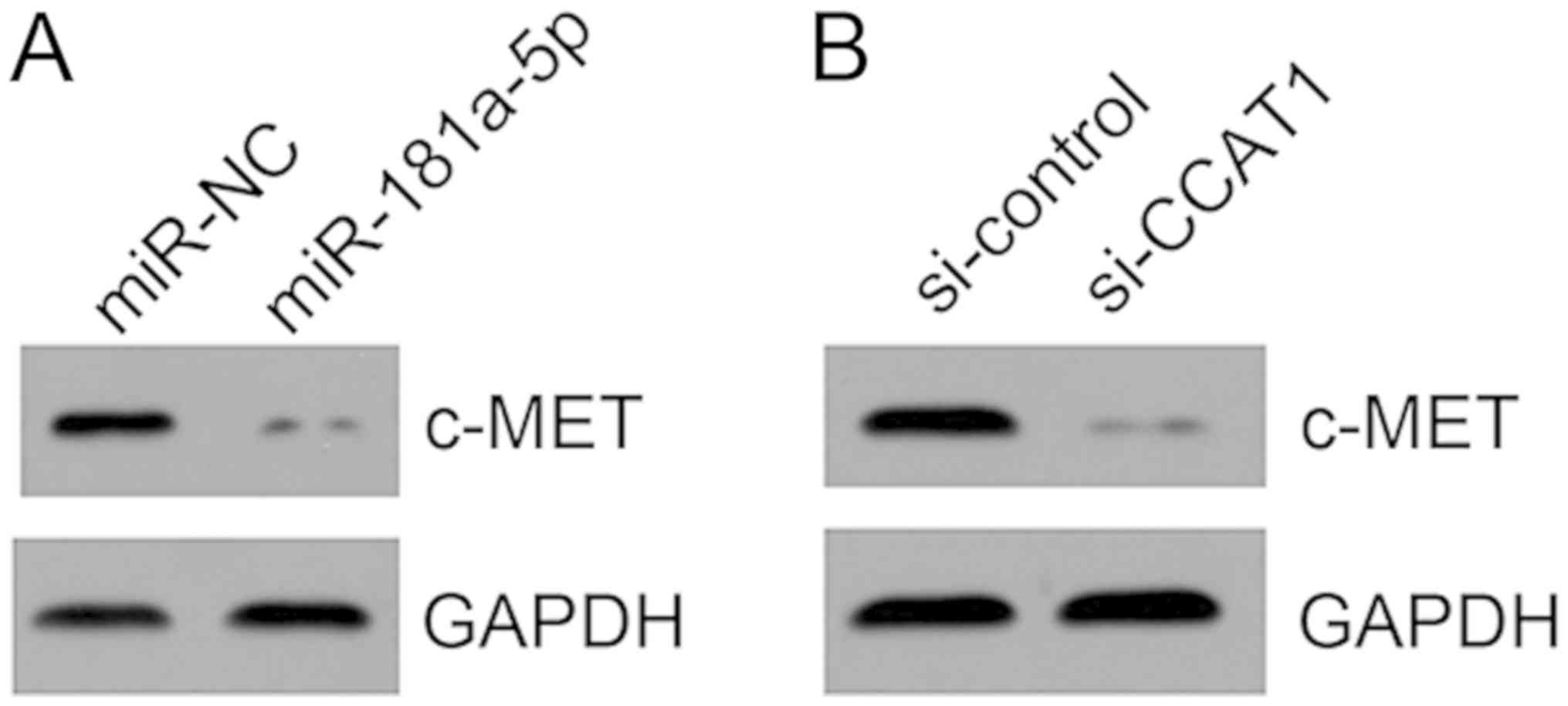 | Figure 6.miR-181a-5p targets c-MET in KLE
cells. (A) Following miR-181a-5p overexpression in KLE cells, the
protein expression level of c-MET was determined by western
blotting. (B) Following CCAT1 knockdown in KLE cells, the protein
expression level of c-MET was determined by western blotting.
CCAT1, colon cancer-associated transcript 1; miR, microRNA; KLE,
human endometrial stromal cell line; miR-NC, endometrial cancer
cell line KLE transfected with scramble miR; miR-181a-5p,
endometrial cancer cell line KLE transfected with miR-181a-5p
mimic; si-control, endometrial cancer cell line KLE transfected
with siRNA negative control; si-CCAT1, endometrial cancer cell line
KLE transfected with siRNA targeting CCAT1; siRNA, small
interfering RNA; c-MET, MET proto-oncogene, receptor tyrosine
kinase. |
Discussion
Endometrial cancer is one of the most common
malignancies in women, and a leading cause of cancer-associated
mortality worldwide (3).
Dysregulated expression of lncRNAs is closely associated with tumor
development and progression (20).
In addition, lncRNAs have been identified as promising biomarkers
for assessing cancer diagnosis and prognosis (21). Wang et al (22) reported that upregulation of lncRNA
HNF1A-antisense RNA 1 is associated with poor prognosis in bladder
cancer. Yang et al (23)
demonstrated that lncRNA small nucleolar RNA host gene 1 can
predict a poor prognosis and promote colon cancer tumorigenesis.
LncRNA BDNF-antisense RNA has functional role in human prostate
cancer and is associated with clinical outcomes (24). Xiao et al (25) revealed that lncRNA HOX transcript
antisense RNA can be used as a prognostic biomarker for the
proliferation and chemoresistance of colorectal cancer via
miRNA-203a-3p-mediated Wnt/β-catenin signaling pathway.
The pathophysiological function of lncRNAs in EC
requires further investigation. Previous studies have revealed that
CCAT1 may serve important roles in the progression and metastasis
of cancer. For example, Cao et al (26) indicated that CCAT1 promotes
metastasis and poor prognosis in epithelial ovarian cancer. Zhang
et al (27) demonstrated that
the CCAT1/miRNA-218/zinc finger protein X-linked axis regulates the
progression of laryngeal squamous cell cancer. In addition, the
CCAT1 promotes osteosarcoma proliferation and migration via
regulating the miRNA-148a/phosphoinositide-3-kinase interacting
protein 1 signal pathway (28).
Arunkumar et al (29)
revealed that CCAT1 is overexpressed in oral squamous cell
carcinoma and as CCAT1 overexpression may sponge miRNA15-5p and
let7b-5b, CCAT1 overexpression may predict poor prognosis. The role
of CCAT1 has not been defined in EC progression. The present study
demonstrated that CCAT1 expression was significantly upregulated in
endometrial cancer tissue samples compared with matched adjacent
healthy tissue samples from patients with endometrial cancer.
Similarly, CCAT1 expression was significantly upregulated in
several EC cell lines (KLE, Ishiwaka and HEC-1-A), compared with
normal human endometrial stromal cell line T-HESC. CCK-8 and
Transwell migration assays verified that CCAT1 knockdown
significantly decreased EC cell proliferation and migration,
suggesting that CCAT1 may contribute to EC progression.
Growing evidence has suggested that lncRNAs could
act as miRNA sponges to exert a physiological function (30). Xu et al (31) demonstrated that lncRNA
differentiation antagonizing non-protein coding RNA acts as a
sponge of miRNA-634 to regulate Ras-related protein Rab-1A
expression in glioma. Zhu et al (32) revealed that lncRNA taurine
up-regulated 1 acts as a sponge of miRNA-138-5p to regulate sirtuin
1 expression and cervical cancer progression. Chen et al
(16) revealed that CCAT1 can
promote multiple myeloma progression by acting as a molecular
sponge of miR-181a-5p and thereby regulating homeobox A1
expression. Additionally, previous studies have demonstrated that
lncRNA expression may be regulated by the binding of sponged miRNAs
(33,34). Consistent with previous findings, the
present study identified that CCAT1 acts as a molecular sponge of
miR-181a-5p to suppress miR-181a-5p expression in EC cells.
Previous studies have revealed that miR-181a-5p regulates the
progression of several types of cancer. Li et al (35) demonstrated that nuclear paraspeckle
assembly transcript 1 promotes proliferation and invasion via
targeting of miR-181a-5p in non-small cell lung cancer.
Additionally, miR-181a-5p promotes the proliferation and migration
of gastric cancer cells through regulation of protein tyrosine
phosphatase, non-receptor type 9 (36). Yang et al (37) indicated that miR-181a-5p promotes
proliferation and invasion, and inhibits apoptosis of cervical
cancer cells via targeting of inositol polyphosphate-5-phosphatase
A. The role of miR-181a-5p in EC has not been previously
investigated. The present study demonstrated that miR-181a-5p
expression was significantly downregulated in endometrial cancer
tissue samples compared with matched adjacent healthy tissue
samples from patients with endometrial cancer. Similarly,
miR-181a-5p expression was significantly downregulated in several
EC cell lines (KLE, Ishiwaka and HEC-1-A), compared with normal
human endometrial stromal cell line T-HESC. CCK-8 and Transwell
migration assays demonstrated that miR-181a-5p overexpression
inhibited EC cell proliferation and migration. Furthermore, rescue
experiments revealed that inhibition of miR-181a-5p significantly
reversed the effects of CCAT1 knockdown on EC cell proliferation
and migration. A previous study demonstrated that miR-181a-5p was
downregulated and suppressed motility, invasion and
branching-morphogenesis by directly targeting c-MET in liver cancer
(19). Additinally, c-MET signaling
is crucial in the development of EC (38). The present study revealed that either
miR-181a-5p overexpression or CCAT1 knockdown decreased c-MET
expression in EC cells, suggesting CCAT1 may be involved in EC
progression via c-MET signaling. Furthermore, miR-181a-5p may also
be sponged by other lncRNAs, including NEAT1 (35). CCAT1 may function with other lncRNAs
to sponge miR-181a-5p and regulate EC progression, however this
requires futher investigation.
In conclusion, the current study demonstrated that
CCAT1 may function as an oncogene in EC by acting as a molecular
sponge of miR-181a-5p. These results suggest a potential molecular
mechanism underlying tumorigenesis in EC and the CCAT1/miR-181a-5p
axis may be a promising target for EC therapy.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural
Science Foundation of China (grant no. 81560785).
Availability of data and materials
All datasets used and/or analyzed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contribution
JY and XH designed the study and analyzed the data.
XH wrote the manuscript. LJ, YG, QS, BL and YH performed the
experiments. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Ethics Committee of The First Affiliated Hospital of Kunming
Medical University (Yunnan, China). Written informed consent was
obtained from all participants.
Patient consent for publication
Patients have provided written informed consent for
publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Felix AS, Scott McMeekin D, Mutch D,
Walker JL, Creasman WT, Cohn DE, Ali S, Moore RG, Downs LS, Ioffe
OB, et al: Associations between etiologic factors and mortality
after endometrial cancer diagnosis: The NRG Oncology/Gynecologic
Oncology Group 210 trial. Gynecol Oncol. 139:70–76. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Morice P, Leary A, Creutzberg C,
Abu-Rustum N and Darai E: Endometrial cancer. Lancet.
387:1094–1108. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cui Z, An X, Li J, Liu Q and Liu W: LncRNA
MIR22HG negatively regulates miR-141-3p to enhance DAPK1 expression
and inhibits endometrial carcinoma cells proliferation. Biomed
Pharmacother. 104:223–228. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lin Z, Zhou Z, Guo H, He Y, Pang X, Zhang
X, Liu Y, Ao X, Li P, Wang J, et al: Long noncoding RNA gastric
cancer-related lncRNA1 mediates gastric malignancy through
miRNA-885-3p and cyclin-dependent kinase 4. Cell Death Dis.
9:6072018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhu P, Wang Y, Wu J, Huang G, Liu B, Ye B,
Du Y, Gao G, Tian Y, He L and Fan Z: LncBRM initiates YAP1
signalling activation to drive self-renewal of liver cancer stem
cells. Nat Commun. 7:136082016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou W, Wang L, Miao Y and Xing R: Novel
long noncoding RNA GACAT3 promotes colorectal cancer cell
proliferation, invasion and migration through miR-149. Onco Targets
Ther. 11:1543–1552. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cen C, Li J, Liu J, Yang M, Zhang T, Zuo
Y, Lin C and Li X: Long noncoding RNA LINC01510 promotes the growth
of colorectal cancer cells by modulating MET expression. Cancer
Cell Int. 18:452018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang K, Zhao Z, Yu J, Chen W, Xu Q and
Chen L: LncRNA FLVCR1-AS1 acts as miR-513c sponge to modulate
cancer cell proliferation, migration and invasion in hepatocellular
carcinoma. J Cell Biochem. 119:6045–6056. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guo E, Liang C, He X, Song G, Liu H, Lv Z,
Guan J, Yang D and Zheng J: Long Noncoding RNA LINC00958
Accelerates Gliomagenesis Through Regulating miR-203/CDK2. DNA Cell
Biol. 37:465–472. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang T, Zhai H, Yan R, Zhou Z, Gao L and
Wang L: lncRNA CCAT1 promotes cell proliferation, migration and
invasion by down-regulation of miR-143 in FTC-133 thyroid carcinoma
cell line. Braz J Med Biol Res. 51:e70462018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li Y, Jing F, Ding Y, He Q, Zhong Y and
Fan C: Long noncoding RNA CCAT1 polymorphisms are associated with
the risk of colorectal cancer. Cancer Genet. 222-223:13–19. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lai XJ and Cheng HF: LncRNA colon
cancer-associated transcript 1 (CCAT1) promotes proliferation and
metastasis of ovarian cancer via miR-1290. Eur Rev Med Pharmacol
Sci. 22:322–328. 2018.PubMed/NCBI
|
|
15
|
Li Y, Zhu G, Ma Y and Qu H: LncRNA CCAT1
contributes to the growth and invasion of gastric cancer via
targeting miR-219-1. J Cell Biochem. 2017.(Epub ahead of print).
View Article : Google Scholar
|
|
16
|
Chen L, Hu N, Wang C, Zhao H and Gu Y:
Long non-coding RNA CCAT1 promotes multiple myeloma progression by
acting as a molecular sponge of miR-181a-5p to modulate HOXA1
expression. Cell Cycle. 17:319–329. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lai Y, Chen Y, Lin Y and Ye L:
Down-regulation of LncRNA CCAT1 enhances radiosensitivity via
regulating miR-148b in breast cancer. Cell Biol Int. 42:227–236.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Korhan P, Erdal E and Atabey N:
MiR-181a-5p is downregulated in hepatocellular carcinoma and
suppresses motility, invasion and branching-morphogenesis by
directly targeting c-Met. Biochem Biophys Res Commun.
450:1304–1312. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang MH, Yang Y, Zhao Y, Wei HB, Ma YQ,
Yang CJ, Zhang XJ and Sun YL: LncRNA DQ786243 expression as a
biomarker for assessing prognosis in patients with gastric cancer.
Eur Rev Med Pharmacol Sci. 22:2304–2309. 2018.PubMed/NCBI
|
|
22
|
Wang YH, Liu YH, Ji YJ, Wei Q and Gao TB:
Upregulation of long non-coding RNA HNF1A-AS1 is associated with
poor prognosis in urothelial carcinoma of the bladder. Eur Rev Med
Pharmacol Sci. 22:2261–2265. 2018.PubMed/NCBI
|
|
23
|
Yang H, Wang S, Kang YJ, Wang C, Xu Y,
Zhang Y and Jiang Z: Long non-coding RNA SNHG1 predicts a poor
prognosis and promotes colon cancer tumorigenesis. Oncol Rep.
40:261–271. 2018.PubMed/NCBI
|
|
24
|
Li W, Dou Z, We S, Zhu Z, Pan D, Jia Z,
Liu H, Wang X and Yu G: Long noncoding RNA BDNF-AS is associated
with clinical outcomes and has functional role in human prostate
cancer. Biomed Pharmacother. 102:1105–1110. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xiao Z, Qu Z, Chen Z, Fang Z, Zhou K,
Huang Z, Guo X and Zhang Y: LncRNA HOTAIR is a prognostic biomarker
for the proliferation and chemoresistance of colorectal cancer via
MiR-203a-3p-mediated Wnt/β-catenin signaling pathway. Cell Physiol
Biochem. 46:1275–1285. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cao Y, Shi H, Ren F, Jia Y and Zhang R:
Long non-coding RNA CCAT1 promotes metastasis and poor prognosis in
epithelial ovarian cancer. Exp Cell Res. 359:185–194. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang Y and Hu H: Long non-coding RNA
CCAT1/miR-218/ZFX axis modulates the progression of laryngeal
squamous cell cancer. Tumour Biol.
39:10104283176994172017.PubMed/NCBI
|
|
28
|
Zhao J and Cheng L: Long non-coding RNA
CCAT1/miR-148a axis promotes osteosarcoma proliferation and
migration through regulating PIK3IP1. Acta Biochim Biophys Sin
(Shanghai). 49:503–512. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Arunkumar G, Murugan AK, Prasanna
Srinivasa Rao H, Subbiah S, Rajaraman R and Munirajan AK: Long
non-coding RNA CCAT1 is overexpressed in oral squamous cell
carcinomas and predicts poor prognosis. Biomed Rep. 6:455–462.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu D, Yu J, Gao G, Lu G, Zhang Y and Ma P:
LncRNA DANCR functions as a competing endogenous RNA to regulate
RAB1A expression by sponging miR-634 in glioma. Biosci Rep.
38(pii): BSR201716642018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhu J, Shi H, Liu H, Wang X and Li F: Long
non-coding RNA TUG1 promotes cervical cancer progression by
regulating the miR-138-5p-SIRT1 axis. Oncotarget. 8:65253–65264.
2017.PubMed/NCBI
|
|
33
|
Chen Z, Wu J, Huang W, Peng J, Ye J, Yang
L, Yuan Y, Chen C, Zhang C, Cai S, et al: Long non-coding RNA
RP11-789C1.1 suppresses epithelial to mesenchymal transition in
gastric cancer through the RP11-789C1.1/MiR-5003/E-cadherin axis.
Cell Physiol Biochem. 47:2432–2444. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Feng L, Shi L, Lu YF, Wang B, Tang T, Fu
WM, He W, Li G and Zhang JF: Linc-ROR promotes osteogenic
differentiation of mesenchymal stem cells by functioning as a
competing endogenous RNA for miR-138 and miR-145. Mol Ther Nucleic
Acids. 11:345–353. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li S, Yang J, Xia Y, Fan Q and Yang KP:
Long Noncoding RNA NEAT1 Promotes proliferation and invasion via
targeting miR-181a-5p in non-small cell lung cancer. Oncol Res.
26:289–296. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu Z, Sun F, Hong Y, Wang B, Tang T, Fu
WM, He W, Li G and Zhang JF: MEG2 is regulated by miR-181a-5p and
functions as a tumour suppressor gene to suppress the proliferation
and migration of gastric cancer cells. Mol Cancer. 16:1332017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yang M, Zhai X, Ge T, Yang C and Lou G:
MiR-181a-5p promotes proliferation and invasion and inhibits
apoptosis of cervical cancer cells via regulating inositol
polyphosphate-5-phosphatase A (INPP5A). Oncol Res. 26:703–712.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yang CH, Zhang XY, Zhou LN, Wan Y, Song
LL, Gu WL, Liu R, Ma YN, Meng HR, Tian YL and Zhang Y: LncRNA SNHG8
participates in the development of endometrial carcinoma through
regulating c-MET expression by miR-152. Eur Rev Med Pharmacol Sci.
22:1629–1637. 2018.PubMed/NCBI
|