Introduction
Gastric cancer (GC) is the fourth most common cancer
worldwide (1,2). GC originates from the mucosal
epithelium of the stomach and causes a significant number of
mortalities worldwide (1,2). GC is difficult to diagnose at an early
stage and thus the majority of patients with GC present with
advanced disease or tumor metastasis, making the disease difficult
to treat (3). In recent decades,
although great progress has been made in GC treatment, including
surgical resection combined with chemotherapy, the survival of
patients with advanced GC has not improved (1–3). It is
therefore necessary to assess the molecular mechanisms underlying
GC development and progression, which may help identify novel and
promising therapeutic targets for this disease.
The human genome produces coding and non-coding
RNAs, the latter of which is the predominant RNA species (4,5). Long
non-coding RNAs (lncRNAs), a class of non-coding RNAs that are
comprised of >200 nucleotides, often exhibit spatial and
temporal-specific expression patterns (4,5). It has
been widely reported that lncRNAs serve important roles in
regulating a variety of cellular biological processes, including
cell proliferation, apoptosis, migration, invasion and
tumourigenesis (6–9). Furthermore, a large number of lncRNAs
are dysregulated during the development or progression of certain
types of human cancer, including GC (10–14). For
instance, the lncRNA X-inactive specific transcript (XIST) is
significantly upregulated in GC cell lines and the knockdown of
XIST inhibits the growth of GC cells (15). Small Nucleolar RNA Host Gene 12
(SNHG12), which is associated with tumor size and metastasis, is
overexpressed in GC (16). The
inhibition of SNHG12 reduces GC cell growth, colony formation,
proliferation and invasion (16).
Distal-less homeobox 6 antisense 1 (DLX6-AS1), a
lncRNA localized at the 7q21.3 chromosomal region, has been
revealed to be frequently upregulated, serving an oncogenic role in
several types of common cancer, including lung adenocarcinoma
(17), renal cell carcinoma
(18) and hepatocellular carcinoma
(19). For instance, DLX6-AS1 levels
are significantly higher in lung adenocarcinoma tissues than in
adjacent healthy lung tissues and its overexpression is closely
associated with poor histological differentiation and advanced TNM
stage (17). However, to the best of
our knowledge, the expression and function of DLX6-AS1 in GC has
not been previously studied.
The present study therefore aimed to assess DLX6-AS1
expression in GC tissues and cell lines and to examine the
association between DLX6-AS1 expression and GC clinical
characteristics. Furthermore, the function of DLX6-AS1 in cell
proliferation, apoptosis, migration and invasion was assessed in GC
in vitro.
Materials and methods
Clinical tissue samples
The present study was approved by the Ethics
Committee of Xiangya Hospital (Changsha, China). A total of 62 GC
tissue and matched adjacent healthy tissue samples (3 cm from the
tumor edge) were collected from 62 primary patients with GC
admitted to Xiangya Hospital (Changsha, China) between March 2015
and June 2017 (Table I). Written
informed consent had been previously obtained. None of the patients
involved in the current study underwent chemotherapy or
radiotherapy prior to surgery. After surgical resection, tissues
were frozen using liquid nitrogen and stored at −80°C until further
use.
 | Table I.Association between DLX6-AS1
expression and the clinicopathological characteristics of patients
with gastric cancer. |
Table I.
Association between DLX6-AS1
expression and the clinicopathological characteristics of patients
with gastric cancer.
|
|
| DLX6-AS1
expression |
|
|---|
|
|
|
|
|
|---|
| Variables | Cases (n=62) | Low levels
(n=33) | High levels
(n=29) | P-value |
|---|
| Age (years) |
|
|
| 0.794 |
| ≤7 | 23 | 13 | 10 |
|
|
>60 | 39 | 20 | 19 |
|
| Sex |
|
|
| 0.302 |
|
Male | 37 | 22 | 15 |
|
|
Female | 25 | 11 | 14 |
|
| Tumor size
(cm) |
|
|
| 0.322 |
|
≤32 | 30 | 18 | 12 |
|
| >5
cm | 32 | 15 | 17 |
|
|
Differentiation |
|
|
| 0.068 |
| Well
and moderately | 38 | 24 | 14 |
|
|
Poor | 24 | 9 | 15 |
|
| Node
metastasis |
|
|
| 0.024a |
|
Present | 44 | 19 | 25 |
|
|
Absent | 18 | 14 | 4 |
|
| Distant
metastasis |
|
|
| 0.018a |
|
Present | 16 | 4 | 12 |
|
|
Absent | 46 | 29 | 17 |
|
| Clinical stage |
|
|
| 0.018a |
|
I–II | 23 | 17 | 6 |
|
|
III–IV | 39 | 16 | 23 |
|
Cell culture
Normal human gastric mucosa epithelial GES-1 cells
and GC cell lines (including HGC27, BGC823, SGC7901 and AGS cells)
were purchased from the Cell Bank of Chinese Academy of Sciences.
Cell lines were cultured in DMEM (Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (Thermo Fisher Scientific Inc.) at 37°C
in a humidified atmosphere with 5% CO2.
Cell transfection
AGS and BGC823 cells (1×106 cells/well)
were seeded in six-well plates and cultured at 37°C to ~80%
confluence. Cells were subsequently transfected with 100 nM
DLX6-AS1 small interfering RNA (siRNA; siDLX6-AS1) or 100 nM
negative control siRNA (siNC; both Shanghai GenePharma Co., Ltd.)
using Lipofectamine® 2000 (Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. Following 48-h
transfection, cells were used in further experimentation.
Reverse transcription-quantitative
(RT-q) PCR
Total RNA was extracted from tissues or cell lines
using TRIzol® reagent (Thermo Fisher Scientific, Inc.).
Total RNA (1 µg) was reverse transcribed into cDNA using the High
Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific,
Inc.), according to manufacturer's protocol. qPCR was performed
using SYBR Green Reverse Transcription PCR Master mix (Takara
Biotechnology Co., Ltd.) on an Applied Biosystems 7300 plus reverse
transcription PCR system (Thermo Fisher Scientific, Inc.). The
following primer pairs were used for the qPCR: DLX6-AS1 forward,
5′-AGTTTCTCTCTAGATTGCCTT-3′ and reverse,
5′-ATTGACATGTTAGTGCCCTT-3′; GAPDH forward,
5′-CTGGGCTACACTGAGCACC-3′ and reverse, 5′-AAGTGGTCGTTGAGGGCAATG-3′.
The following thermocycling conditions were used for the qPCR:
Initial denaturation at 95°C for 30 sec; 40 cycles of 95°C for 5
sec and 60°C for 30 sec. Expression levels were quantified using
the 2−∆∆Cq method and GAPDH was used as an internal
reference gene (20).
Cell counting kit-8 (CCK-8) assay
AGS and BGC823 cells (5×103 cells/well)
were plated into 96-well plates and incubated at 37°C for 0, 24, 48
or 72 h. CCK-8 reagent (10 µl; Beyotime Institute of Biotechnology)
was then added to each well. After incubation at 37°C for 2 h, the
optical density absorbance at 450 nm was measured using a
microplate reader.
Colony formation assay
Transfected AGS and BGC823 cells (1.5×102
cells/well) were seeded into six-well plates and cultured in DMEM
with 10% FBS at 37°C for 10 days. Cells were then washed with DPBS
(Thermo Fisher Scientific, Inc.) and stained with 0.1% crystal
violet for 5 min at room temperature. The number of colonies
consisting of >50 cells were counted under a light microscope
(magnification, ×40; Olympus Corporation).
Flow cytometry for cell cycle
analysis
Transfected AGS and BGC823 cells were washed with
PBS and fixed in 75% ethanol overnight at 4°C. Cells were
subsequently washed three times with PBS and stained with 500 µl
eBioscience™ propidium iodide staining solution (cat. no.
00-6990-50; Thermo Fisher Scientific, Inc.) at room temperature for
15 min. Cell cycle distribution was examined using a FACScan flow
cytometer (BD Biosciences) and BD Accuri C6 system software
(version 1.0; BD Biosciences).
Wound healing assay
Transfected AGS and BGC823 cells (1×106
cells/well) were seeded into six-well plates and cultured in DMEM
with 10% FBS at 37°C at ~24 h to ~90% confluence. Wounds were
created using a 200 µl pipette tip. Wound healing within the scrape
line was observed and imaged at 0 and 24 h using a light microscope
(magnification, ×400; Olympus Corporation).
Transwell cell invasion assay
A transwell assay was performed using a 24-well
transwell chamber (8 mm pore size; Corning, Inc.) pre-coated with
Matrigel Basement Membrane Matrix (BD Biosciences). Transfected
cells (3×103 cells/well) in 300 µl DMEM without serum
were added to the upper chamber, while 500 µl DMEM with 10% FBS was
added to the lower chamber. Cells were then incubated at 37°C for
24 h. Cells inside the insert were removed using a cotton-tipped
swab. Cells that had invaded were fixed with 4% paraformaldehyde
(Sigma-Aldrich; Merck KGaA) for 30 min at room temperature. Fixed
cells were subsequently stained with 0.5% crystal violet (Beyotime
Institute of Biotechnology) for 10 min at room temperature.
Invading cells were observed under a light microscope
(magnification, ×400; Olympus Corporation).
Western blot analysis
Transfected AGS and BGC823 cells were lysed using
RIPA buffer (Beyotime Institute of Biotechnology). Protein
concentration was determined using the Pierce BCA Protein Assay kit
(Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol. Protein samples (50 µg) were separated via SDS-PAGE on a
10% gel. The separated proteins were transferred onto
nitrocellulose membranes (EMD Millipore) and blocked with 5%
non-fat dry milk overnight at 4°C. The membranes were incubated
with primary antibodies against E-cadherin (1:500; ab133597),
N-cadherin (1:500; ab245117), Vimentin (1:200; ab16700) and GAPDH
(1:500; ab9485; all Abcam) for 3 h at room temperature. Following
primary incubation, membranes were incubated with horseradish
peroxidase-conjugated goat anti-rabbit second antibody (1:5,000;
ab6721; Abcam) for 1 h at room temperature. Protein bands were
visualized using the SuperSignal West Femto Maximum Sensitivity
substrate (Thermo Fisher Scientific, Inc.). Protein expression was
quantified using ImageJ software (version 1.46; National Institutes
of Health).
Statistical analysis
Data presented as the mean ± standard deviation.
SPSS version 22.0 for windows (IBM Corp.) was used for all
analyses. A Student's t-test was used for analysing the differences
between two groups. One-way ANOVA followed by a Tukey's post-hoc
test was used for the comparison of >2 groups. A Chi-square test
was used to analyse the association between DLX6-AS1 expression and
the clinicopathological characteristics of patients with GC.
Kaplan-Meier survival curves were analysed with a log-rank test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
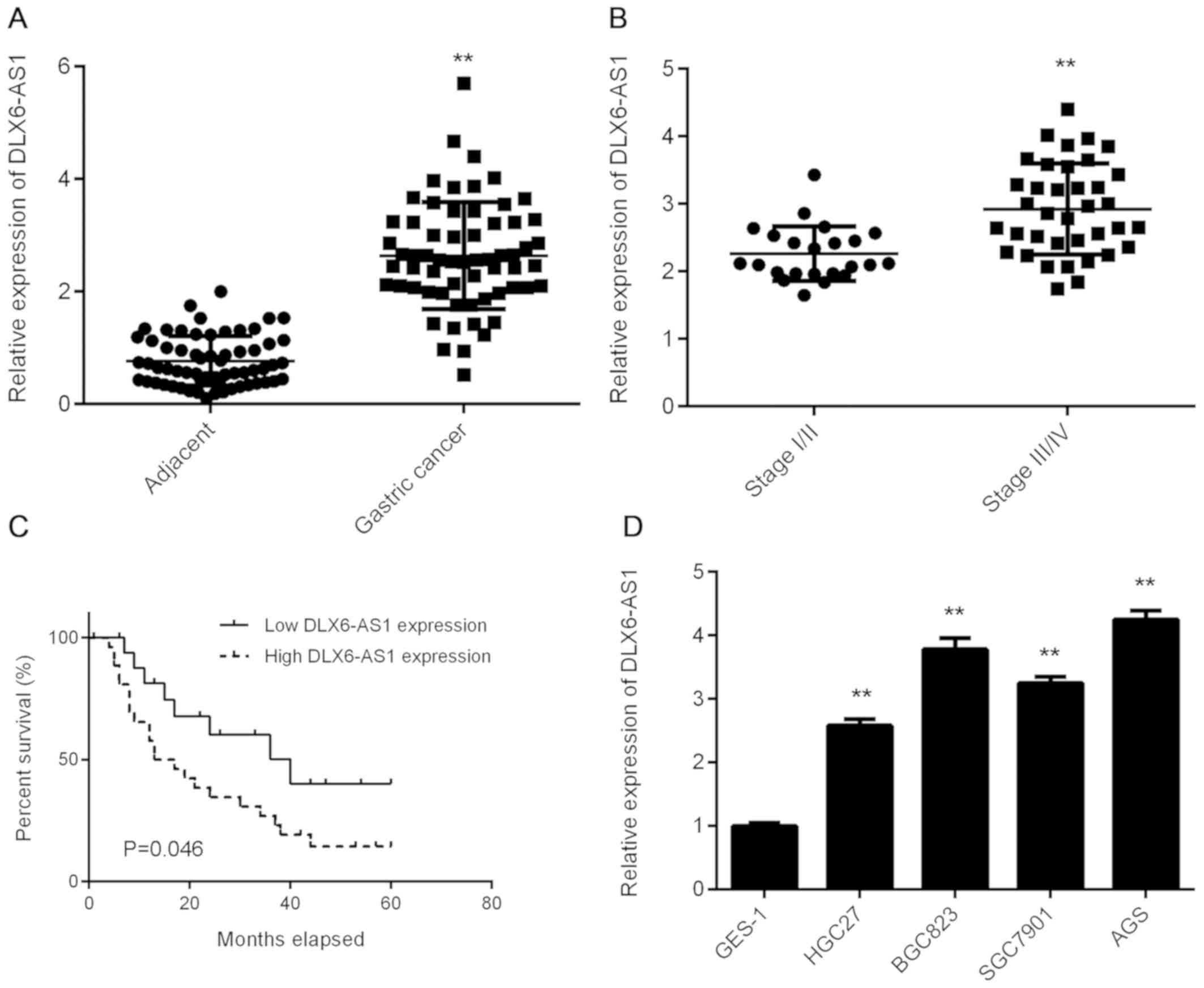
Upregulation of DLX6-AS1 in GC
To assess the biological function of DLX6-AS1 in GC,
RT-qPCR analysis was performed to determine its expression in a
total of 62 primary GC tissues and their matched adjacent normal
tissues. The data revealed that DLX6-AS1 expression was
significantly increased in GC tissues compared with adjacent normal
tissues (Fig. 1A). The expression of
DLX6-AS1 was also observed to be higher in advanced GC tissue
samples (III/IV) compared with early-stage samples (I/II; Fig. 1B), indicating that the upregulation
of DLX6-AS1 was associated with GC progression. To further clarify
whether DLX6-AS1 expression was associated with the
clinicopathological characteristics of patients with GC, patients
were divided into DLX6-AS1 low expression and high expression
groups using the median level of DLX6-AS1 as a cut-off (2.67). As
indicated in Table I, high DLX6-AS1
expression was not associated with sex, age, tumor size or
differentiation, but was significantly associated with metastasis
and advanced TNM stage. The results indicate that patients with GC
and a high DLX6-AS1 expression exhibit a lower percentage survival
than those with a low DLX6-AS1 expression (Fig. 1C). These results indicate that
DLX6-AS1 expression may be used as a predictive marker for GC
prognosis and as such, further study should be conducted to explore
the function of DLX6-AS1 in GC. The expression of DLX6-AS1 was
subsequently examined in a normal human gastric mucosa epithelial
cell line, GES-1 and several GC cell lines, including HGC27,
BGC823, SGC7901 and AGS. As presented in Fig. 1D, DLX6-AS1 expression was
significantly higher in GC cell lines compared with GES-1 cells.
AGS and BGC823 cell lines were subsequently selected to perform
in vitro experiments, as they exhibited the highest levels
of DLX6-AS1 expression.
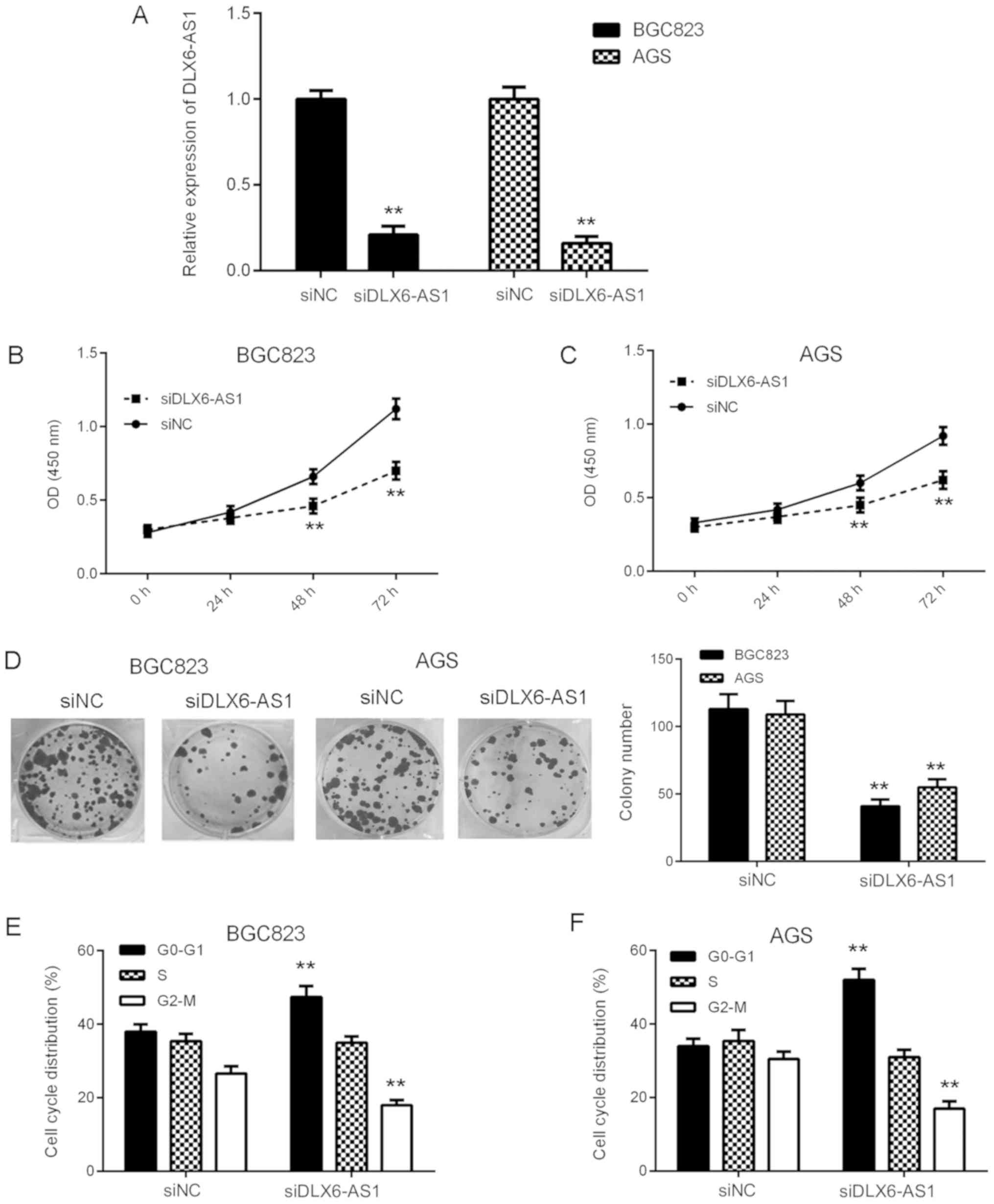
Inhibition of DLX6-AS1 suppresses GC
cell proliferation, colony formation and cell cycle
progression
To examine the function of DLX6-AS1 in GC in
vitro, AGS and BGC823 cells were transfected with DLX6-AS1
siRNA to downregulate its expression. At 48 h after transfection,
RT-qPCR was performed to assess DLX6-AS1 expression. As presented
in Fig. 2A, the expression of
DLX6-AS1 was significantly reduced in the siDLX6-AS1 group compared
with the siNC group. A CCK-8 assay was then performed in order to
assess cell proliferation. The results revealed that the
proliferation of AGS and BGC823 cells was significantly inhibited
at 48 and 72 h in the siDLX6-AS1 group compared with the siNC group
(Fig. 2B and C). These results
indicate that DLX6-AS1 may serve a growth-promoting role in GC. To
confirm this, a colony formation assay was performed, the results
of which indicated that the colony formation capacity of cells was
significantly reduced in the siDLX6-AS1 group compared with the
siNC group (Fig. 2D). Cell cycle
regulation serves a key role in cell proliferation (21). Therefore, flow cytometry was
performed to assess the function of DLX6-AS1 in the regulation of
cell cycle progression. As presented in Fig. 2E and F, the knockdown of DLX6-AS1
caused significant G1 and G2-M stage arrest compared with siNC
cells in AGS and BGC823 cells. The aforementioned data indicates
that DLX6-AS1 may promote cell proliferation, colony formation and
cell cycle progression in GC.
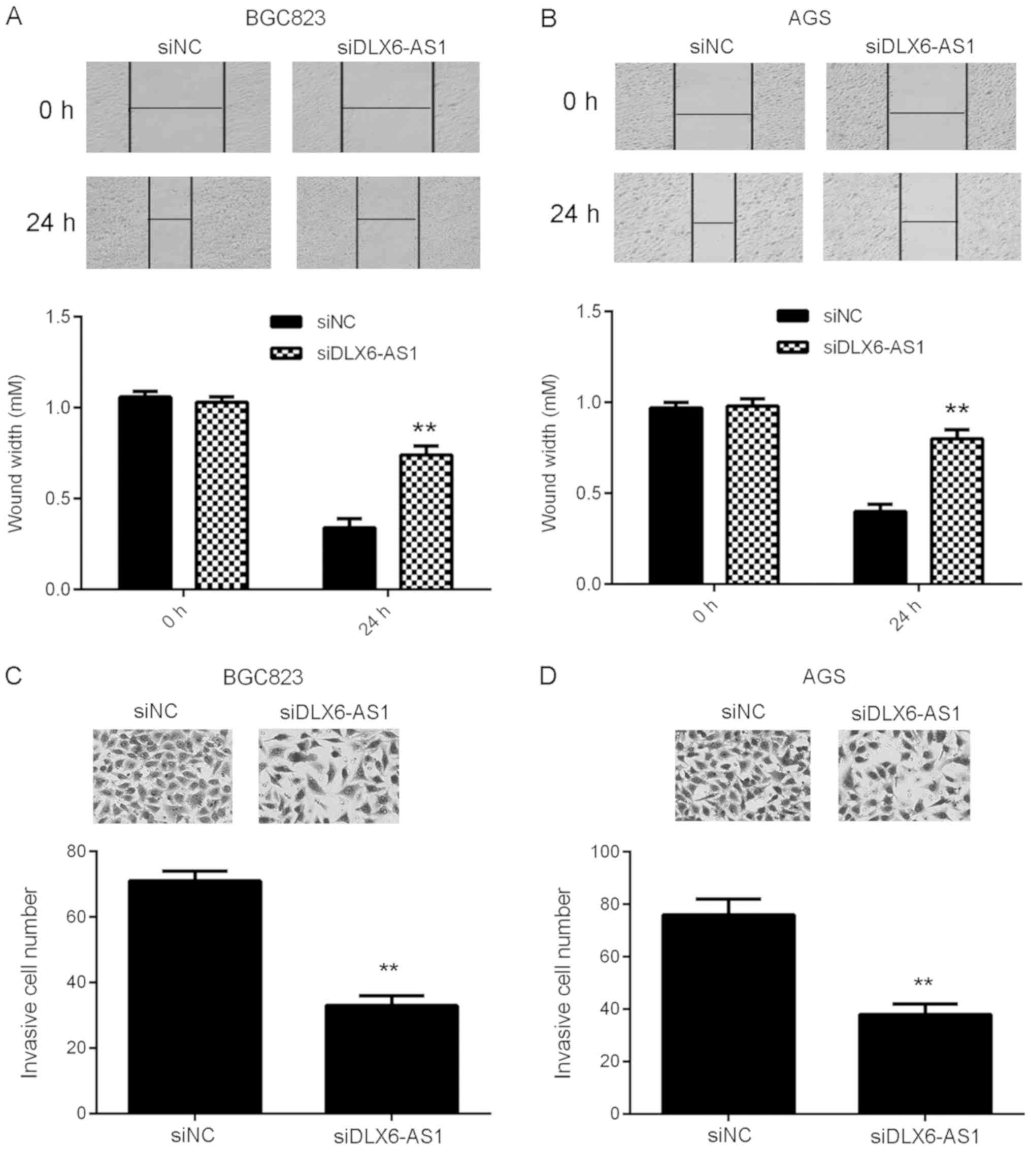
Silencing of DLX6-AS1 inhibits GC cell
migration, invasion and epithelial-mesenchymal transition
(EMT)
Tumor cell migration and invasion are essential for
tumor metastasis (22). Wound
healing and transwell assays were performed to assess the function
of DLX6-AS1 in GC cell migration and invasion. As presented in
Fig. 3A and B, wound healing assay
data revealed that compared with cells in the siNC group, the
migratory capacity of DLX6-AS1 inhibited AGS and BGC823 cells was
significantly reduced. Similarly, fewer invasive cells were
observed in the siDLX6-AS1 group than in the siNC group, indicating
that DLX6-AS1 silencing inhibits GC cell invasion (Fig. 3C and D). These data indicate that
DLX6-AS1 serves a promoting role in the migration and invasion of
GC cells, which may contribute to GC metastasis.
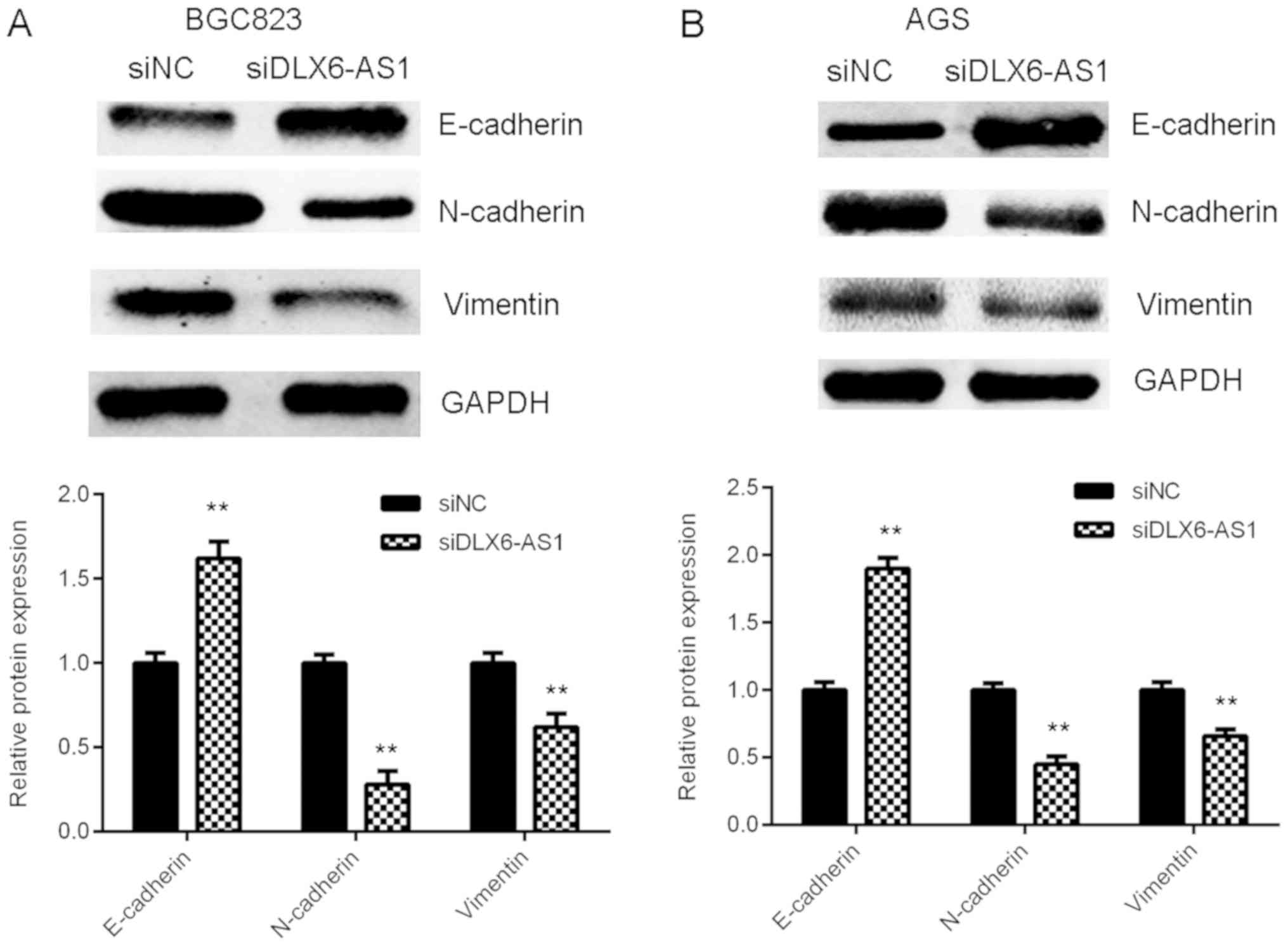
As EMT serves a key role during tumor cell migration
and invasion (23), the expression
of EMT markers in AGS and BGC823 cells were assessed with or
without DLX6-AS1 inhibition. The results of western blot analysis
demonstrated that DLX6-AS1 silencing expression caused the
significant upregulation of E-cadherin (an epithelial marker) and
the marked downregulation of N-cadherin and Vimentin (mesenchymal
markers) in AGS and BGC823 cells compared with the cells in the
siNC group (Fig. 4). The results
indicate that the knockdown of DLX6-AS1 suppresses GC cell
migration and invasion by inhibiting EMT.
Discussion
Although non-protein coding RNAs occupy >90% of
the human genome's transcriptional output (7), the molecular mechanisms of lncRNAs
underlying cancer development and progression remain largely
unknown. The current study revealed that the expression of DLX6-AS1
was significantly increased in GC tissues and cell lines and that
its upregulation was associated with advanced clinical stage, lymph
node metastasis, distant metastasis and poor prognosis in patients
with GC. The knockdown of DLX6-AS1 inhibited GC cell proliferation,
colony formation, cell cycle progression, migration, invasion and
EMT in vitro.
In recent years, a large number of lncRNAs have been
identified as key regulators during cancer development and
progression (24,25). The poor prognosis of patients with
advanced GC is primarily thought to be due to rapid tumor
metastasis (26). This therefore
reveals that furthering understanding into the underlying
mechanisms of GC may be beneficial for the development of novel and
effective therapies (27,28). Previous studies have reported that
certain lncRNAs, including Small nuclear RNA host gene 20 (SNHG20)
(29), terminal
differentiation-induced ncRNA (30),
nuclear paraspeckle assembly transcript 1 (NEAT1) (31) and XIST (15) are aberrantly expressed in GC and
regulate tumour growth and metastasis. For instance, NEAT1 is an
unfavourable prognostic factor in GC and serves a promoting role in
cell migration and invasion (31).
SNHG20 promotes GC progression via the downregulation of p21
expression and the upregulation of the glycogen synthase kinase 3
β/β-catenin signalling pathway (29). The results of the present study
indicated that DLX6-AS1 is significantly upregulated in GC tissues
and cell lines, compared with adjacent normal tissues and non-tumor
GES-1 cells. In addition, it was revealed that the expression of
DLX6-AS1 was higher in advanced GC tissue samples (III/IV) compared
with early-stage samples (I/II). It was also demonstrated that a
high DLX6-AS1 expression was significantly associated with lymph
node metastasis and advanced TNM stage. Patients with GC and a high
DLX6-AS1 expression exhibited decreased survival times compared
with those with a low DLX6-AS1 expression. These results indicate
that the upregulation of DLX6-AS1 may serve a key role in GC
progression.
To further assess the function of DLX6-AS1 in GC
growth and metastasis, loss-of-function assays were performed using
AGS and BGC823 cells by transfecting DLX6-AS1-specific siRNA. It
was demonstrated that silencing DLX6-AS1 expression caused a
significant reduction in GC cell proliferation, colony formation,
cell cycle progression, migration and invasion. Similarly, Zeng
et al (18) revealed that
DLX6-AS1 expression was also upregulated in renal cell carcinoma
cells, which was significantly associated with tumour progression.
In addition, Zhang et al (19) demonstrated that DLX6-AS1 promotes the
proliferation, invasion and migration of hepatocellular carcinoma
cells in vitro, as well as tumor growth in vivo. EMT,
characterized by the loss of an epithelial phenotype and the
acquisition of mesenchymal properties, serves a central role in
cell migration and invasion and is closely associated with tumor
metastasis (32). Many lncRNAs have
been suggested to regulate EMT in various cancers (33–35). For
instance, the overexpression of lncRNA colorectal neoplasia
differentially expressed facilitates EMT and is associated with a
poor prognosis of patients with intrahepatic cholangiocarcinoma
(33). However, to the best of our
knowledge, the detailed role of DLX6-AS1 in EMT regulation in GC
cells has not previously been studied. The results of the present
study revealed that the knockdown of DLX6-AS1 significantly
promotes E-cadherin protein expression and suppresses N-cadherin
and vimentin protein expression in GC cells, indicating that EMT
was inhibited. It is therefore reasonable to suggest that the
suppressive effects of DLX6-AS1 downregulation in GC cell invasion
and migration may occur via the inhibition of EMT.
In summary, the present study demonstrated that the
lncRNA, DLX6-AS1, is upregulated in GC and serves an oncogenic
role, indicating that DLX6-AS1 may serve as a novel therapeutic
target for GC treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All datasets used and/or generated during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
XF and YT collected clinical tissues, designed the
study and wrote the manuscript. XF, WG, SW and WK performed the
clinical and cell experiments.
Ethics approval and consent to
participate
The current study was approved by the Ethics
Committee of Xiangya Hospital (Changsha, China). All written
informed consents have been obtained.
Patient consent for publication
All participants provided written informed consent
for publication.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Röcken C: Molecular classification of
gastric cancer. Expert Rev Mol Diagn. 17:293–301. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Silva A, Bullock M and Calin G: The
clinical relevance of long non-coding RNAs in cancer. Cancers
(Basel). 7:2169–2182. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hung T and Chang HY: Long noncoding RNA in
genome regulation: Prospects and mechanisms. RNA Biol. 7:582–585.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wapinski O and Chang HY: Long noncoding
RNAs and human disease. Trends Cell Biol. 21:354–361. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Clark MB and Mattick JS: Long noncoding
RNAs in cell biology. Semin Cell Dev Biol. 22:366–376. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xiao Z, Qu Z, Chen Z, Fang Z, Zhou K,
Huang Z, Guo X and Zhang Y: LncRNA HOTAIR is a prognostic biomarker
for the proliferation and chemoresistance of colorectal cancer via
MiR-203a-3p-mediated Wnt/ß-catenin signaling pathway. Cell Physiol
Biochem. 46:1275–1285. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu K, Yao H, Wen Y, Zhao H, Zhou N, Lei S
and Xiong L: Functional role of a long non-coding RNA
LIFR-AS1/miR-29a/TNFAIP3 axis in colorectal cancer resistance to
pohotodynamic therapy. Biochim Biophys Acta Mol Basis Dis.
1864:2871–2880. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li PF, Chen SC, Xia T, Jiang XM, Shao YF,
Xiao BX and Guo JM: Non-coding RNAs and gastric cancer. World J
Gastroenterol. 20:5411–5419. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu R, Zhu X, Chen F, Huang C, Ai K, Wu H,
Zhang L and Zhao X: LncRNA XIST/miR-200c regulates the stemness
properties and tumourigenicity of human bladder cancer stem
cell-like cells. Cancer Cell Int. 18:412018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xiong W, Huang C, Deng H, Jian C, Zen C,
Ye K, Zhong Z, Zhao X and Zhu L: Oncogenic non-coding RNA NEAT1
promotes the prostate cancer cell growth through the SRC3/IGF1R/AKT
pathway. Int J Biochem Cell Biol. 94:125–132. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Peng Z, Liu C and Wu M: New insights into
long noncoding RNAs and their roles in glioma. Mol Cancer.
17:612018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Q, Chen B, Liu P and Yang J: XIST
promotes gastric cancer (GC) progression through TGF-beta1 via
targeting miR-185. J Cell Biochem. 119:2787–2796. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang H and Lu W: LncRNA SNHG12 regulates
gastric cancer progression by acting as a molecular sponge of
miR320. Mol Med Rep. 17:2743–2749. 2018.PubMed/NCBI
|
|
17
|
Li J, Li P, Zhao W, Yang R, Chen S, Bai Y,
Dun S, Chen X, Du Y, Wang Y, et al: Expression of long non-coding
RNA DLX6-AS1 in lung adenocarcinoma. Cancer Cell Int. 15:482015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zeng X, Hu Z, Ke X, Tang H, Wu B, Wei X
and Liu Z: Long noncoding RNA DLX6-AS1 promotes renal cell
carcinoma progression via miR-26a/PTEN axis. Cell Cycle.
16:2212–2219. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang L, He X, Jin T, Gang L and Jin Z:
Long non-coding RNA DLX6-AS1 aggravates hepatocellular carcinoma
carcinogenesis by modulating miR-203a/MMP-2 pathway. Biomed
Pharmacother. 96:884–891. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lim S and Kaldis P: Cdks, cyclins and
CKIs: Roles beyond cell cycle regulation. Development.
140:3079–3093. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mowers EE, Sharifi MN and Macleod KF:
Functions of autophagy in the tumor microenvironment and cancer
metastasis. FEBS J. 285:1751–1766. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Luo J, Chen J, Li H, Yang Y, Yun H, Yang S
and Mao X: LncRNA UCA1 promotes the invasion and EMT of bladder
cancer cells by regulating the miR-143/HMGB1 pathway. Oncol Lett.
14:5556–5562. 2017.PubMed/NCBI
|
|
24
|
Zhu H, Zheng T, Yu J, Zhou L and Wang L:
LncRNA XIST accelerates cervical cancer progression via
upregulating Fus through competitively binding with miR-200a.
Biomed Pharmacother. 105:789–797. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhou Y, Chen Y, Ding W, Hua Z, Wang L, Zhu
Y, Qian H and Dai T: LncRNA UCA1 impacts cell proliferation,
invasion, and migration of pancreatic cancer through regulating
miR-96/FOXO3. IUBMB Life. 70:276–290. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kahroba H, Hejazi MS and Samadi N:
Exosomes: From carcinogenesis and metastasis to diagnosis and
treatment of gastric cancer. Cell Mol Life Sci. Feb 8–2019.Doi:
10.1007/s00018-019-03035-2. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tan HY, Wang C, Liu G and Zhou X: Long
noncoding RNA NEAT1-modualted miR-506 regulates gastric cancer
development through targeting STAT3. J Cell Biochem. 120:4827–4836.
2018. View Article : Google Scholar
|
|
28
|
Ren K, Liu QQ, An ZF, Zhang DP and Chen
XH: MiR-144 functions as tumor suppressor by targeting PIM1 in
gastric cancer. Eur Rev Med Pharmacol Sci. 21:3028–3037.
2017.PubMed/NCBI
|
|
29
|
Liu J, Liu L, Wan JX and Song Y: Long
noncoding RNA SNHG20 promotes gastric cancer progression by
inhibiting p21 expression and regulating the GSK-3beta/beta-catenin
signaling pathway. Oncotarget. 8:80700–80708. 2017.PubMed/NCBI
|
|
30
|
Chen Z, Liu H, Yang H, Gao Y, Zhang G and
Hu J: The long noncoding RNA, TINCR, functions as a competing
endogenous RNA to regulate PDK1 expression by sponging miR-375 in
gastric cancer. Onco Targets Ther. 10:3353–3362. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fu JW, Kong Y and Sun X: Long noncoding
RNA NEAT1 is an unfavorable prognostic factor and regulates
migration and invasion in gastric cancer. J Cancer Res Clin Oncol.
142:1571–1579. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ye Y, Xiao Y, Wang W, Yearsley K, Gao JX,
Shetuni B and Barsky SH: ERalpha signaling through slug regulates
E-cadherin and EMT. Oncogene. 29:1451–1462. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xia XL, Xue D, Xiang TH, Xu HY, Song DK,
Cheng PG and Wang JQ: Overexpression of long non-coding RNA CRNDE
facilitates epithelial-mesenchymal transition and correlates with
poor prognosis in intrahepatic cholangiocarcinoma. Oncol Lett.
15:4105–4112. 2018.PubMed/NCBI
|
|
34
|
Li C, Wan L, Liu Z, Xu G, Wang S, Su Z,
Zhang Y, Zhang C, Liu X, Lei Z and Zhang HT: Long non-coding RNA
XIST promotes TGF-beta-induced epithelial-mesenchymal transition by
regulating miR-367/141-ZEB2 axis in non-small-cell lung cancer.
Cancer Lett. 418:185–195. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jia L, Tian Y, Chen Y and Zhang G: The
silencing of LncRNA-H19 decreases chemoresistance of human glioma
cells to temozolomide by suppressing epithelial-mesenchymal
transition via the Wnt/beta-Catenin pathway. Onco Targets Ther.
11:313–321. 2018. View Article : Google Scholar : PubMed/NCBI
|