Introduction
Helicobacter pylori (Hp) is a globally
transmitted pathogen that has an infection rate of >90% in
developing countries (1). It is an
important pathogenic factor of chronic gastritis and gastroduodenal
ulcers, and is closely associated with gastric cancer or gastric
mucosa-associated malignant lymphoma (2,3). Studies
on the infection source and transmission route of Hp are becoming
more and more important.
The oral cavity is an ideal settling environment for
Hp as a facultative anaerobe (4). Hp
infection in the stomach is likely to cause oral diseases. As
children tend to have a high-sugar diet while lacking sufficient
knowledge to protect their teeth, they are easily affected by
pulpitis. The degradation of matrix proteins in pulpitis relies on
numerous different endogenous proteases, and proteolytic enzymes
are essential for the degradation of extracellular matrix.
According to their target molecules, proteolytic enzymes may be
divided into three types: Serine proteinases, cysteine proteinases
and matrix metalloproteinases (MMPs) (5). The role of MMPs in pulpitis has
attracted increasing attention in recent years. It was demonstrated
that the activation and expression of MMPs are involved in the
inflammatory process associated with pulpitis (6,7).
Upregulation of MMP9, a member of the MMP family, is also observed
in certain mucosal lesions that are closely associated with Hp
infection. However, it has remained elusive whether in pulpitis,
the levels of MMP9 are affected by the presence of Hp infection in
the stomach. In addition, upstream factors that regulate MMPs also
remain to be explored.
MicroRNAs (miRNA or miRs) are a class of non-coding
RNA molecules of 18–22 nucleotides in length, which regulate the
expression of their target proteins at the mRNA level (8). Altered expression of various miRNAs and
proteins in patients with hypertension suggests that miRNAs have
important roles in the regulation of proteins that are associated
with cardiovascular diseases (9–12). In
the present study, miRNA molecules associated with MMP9 were
screened using bioinformatics, and the association between miR-204
and MMP9 was identified. The expression of MMP9 was then assessed
in pediatric patients with pulpitis and Hp infection in the
stomach, and the association between MMP9 and miR-204 in this
context was investigated.
Materials and methods
Patients
A total of 26 pediatric patients with pulpitis who
received tooth extraction at the Children's Hospital of Nanjing
Medical University (Nanjing, China) between December 2014 and
August 2016 were also diagnosed with Hp infection in the stomach
and therefore included in the present study (HP+ group).
Furthermore, 19 contemporaneous pediatric patients with pulpitis
but without stomach infection of Hp were enrolled as a control
(HP− group). Pulp tissues, blood (serum) and saliva were
collected from all subjects. All three types of samples were tested
using the same protocol. The HP+ group comprised 16
males and 10 females (age range, 8–12 years; median age, 9.6 years)
and the HP− group comprised 11 males and 8 females (age
range, 7–12 years; median age, 9.2 years). None of the patients had
previously taken any non-steroidal drugs, proton pump inhibitors,
antibiotics or bismuth within two weeks prior to examination. All
procedures were approved by the Ethics Committee of Nanjing Medical
University (Nanjing, China). Written informed consent was obtained
from parents or guardians of all pediatric patients.
Reverse-transcription quantitative
polymerase chain reaction (RT-qPCR)
Pulp tissues (100 mg) were ground into powder in
liquid nitrogen and mixed with 1 ml TRIzol (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) for lysis. Total RNA was
extracted using the phenol chloroform method. The purity of the RNA
was determined by the absorbance at 260 vs. 280 nm using
ultraviolet spectrophotometry (Nanodrop ND2000; Thermo Fisher
Scientific, Inc.). Total RNA (1 µg) was reverse transcribed into
cDNA using the TIANScript II cDNA first strand synthesis kit (cat.
no. KR107; Tiangen, Beijing, China) and stored at −20°C. The PCR
reaction (25 µl) was composed of 12.5 µl SuperReal PreMix
(SYBR-Green; cat. no. FP204; Tiangen), 0.5 µl foward primer, 0.5 µl
reverese primer, 10.5 µl sterile water and 1 µl cDNA. The following
primer pairs were used: MMP9 forward, 5′-GCTGGCAGAGGAATACCTGTAC-3′
and reverse, 5′-CAGGGACAGTTGCTTCTGGA-3′; β-actin forward,
5′-CTGGAACGGTGAAGGTGACA-3′ and reverse,
5′-AAGGGACTTCCTGTAACAACGCA-3′. The reaction was performed in an iQ5
real-time PCR system (Bio-Rad Laboratories, Inc., Hercules, CA,
USA) using the following thermocycling conditions: Initial
denaturation at 95°C for 2 min; 40 cycles of 95°C for 15 sec, 60°C
for 30 sec and 72°C for 60 sec. The relative expression of MMP9
mRNA was quantified using the 2−ΔΔCq method and
normalized to the internal reference β-actin (13). Each sample was tested in
triplicate.
To assess the expression of miR-204, miRNA was
isolated using an miRcute miRNA separation kit (cat. no. FP401-02;
Tiangen), and cDNA was synthesized using a miRcute miRNA cDNA first
strand synthesis kit (cat. no. KR201; Tiangen). The level of
miR-204 was determined using a miRcute miRNA qPCR detection kit
(cat. no. FP401; Tiangen). The following primer pairs were used:
miR-204 forward, 5′-ACACTCCAGCTGGGTTCCCTTTGTCATCCTAT-3′ and
reverse, 5′-CTCAACTGGTGTCGTGGA-3′; U6 forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
The reaction was performed in an iQ5 real-time PCR system using the
following thermocycling conditions: Initial denaturation at 95°C
for 5 min; 40 cycles of 95°C for 15 sec, 62°C for 15 sec and 72°C
for 32 sec. The relative expression of miR-204 was quantified using
the 2−ΔΔCq method and normalized to the internal
reference U6. Each sample was tested in triplicate.
Western blot analysis
All types of samples were mixed with 100 µl
precooled radioimmunoprecipitation assay lysis buffer containing 1
mM phenylmethylsulfonyl fluoride for lysis of 15 min at 4°C. The
mixture was then centrifuged at 12,000 × g for 5 min at 4°C. The
supernatant was used to determine the protein concentration by
using a bicinchoninic acid protein concentration determination kit
(cat. no. RTP7102; Real-Times Biotechnology Co., Ltd., Beijing,
China). Protein samples were then mixed with SDS loading buffer
prior to denaturation in a boiling water bath for 5 min.
Subsequently, the samples (20 µg) were separated via SDS-PAGE on a
10% gel. The separated proteins were transferred onto
polyvinylidene difluoride membranes on ice (100 V, 2 h) and blocked
with 5% skimmed milk at room temperature for 1 h. The membranes
were then incubated with rabbit anti-human MMP9 polyclonal primary
antibody (1:1,000 dilution; cat. no. ab38898; Abcam, Cambridge, UK)
and rabbit anti-human β-actin primary antibody (1:5,000 dilution;
cat. no. ab6276; Abcam) at 4°C overnight. After extensive washing
with PBS containing Tween 20 (PBST) 3 times for 15 min each, the
membranes were incubated with horseradish peroxidase-conjugated
goat anti-rabbit polyclonal secondary antibody (1:3,000 dilution;
cat. no. ab6721; Abcam) for 1 h at room temperature prior to
washing with PBST 3 times of 15 min each. Protein bands were
visualized using an enhanced chemiluminescence detection kit (cat.
no. ab65623; Abcam) and protein expression was quantified using
Image lab v3.0 software (Bio-Rad Laboratories, Inc.) with β-actin
as the loading control.
ELISA
Blood samples were centrifuged at 1,000 × g for 10
min to obtain serum. Serum and saliva samples were tested using an
MMP9 ELISA kit (cat. no. 2219; BLKW Biotechnology, Beijing, China).
In microplates, standards (50 µl) and samples (10 µl sample liquid
and 40 µl diluent) were added into predefined wells. In the wells
for standards and samples, horseradish peroxidase-labelled
conjugates (100 µl) were added prior to sealing the plates for
incubation at 37°C for 1 h. After washing the plates for 5 times,
substrates A (50 µl) and B (50 µl) were added into each well. After
incubation at 37°C for 15 min, stop solution (50 µl) was added into
each well, and the absorbance of each well was measured at 450 nm
within 15 min.
Dual luciferase reporter assay
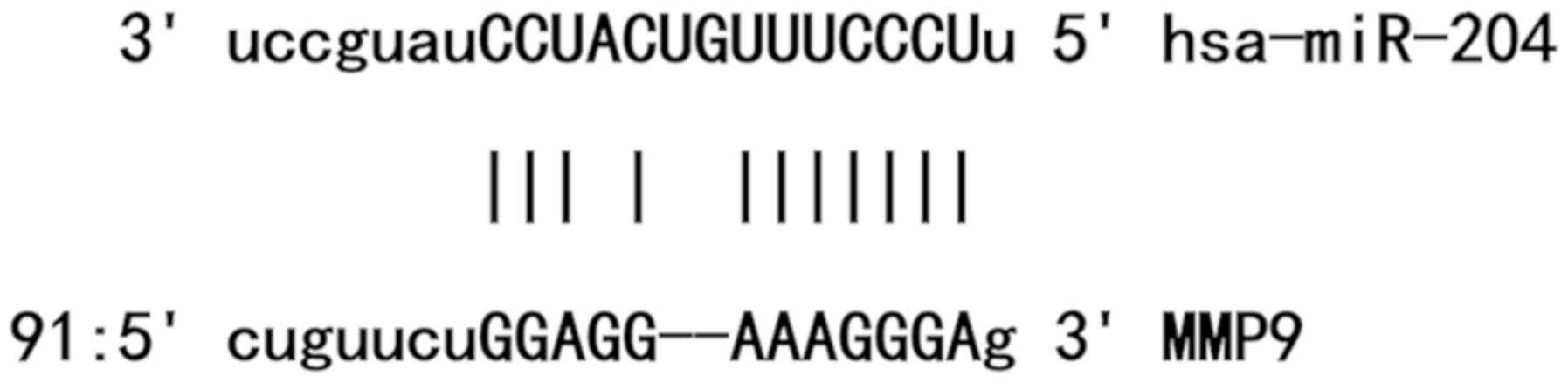
Bioinformatics prediction is a powerful tool for the
study of the functions of miRNAs. To understand the regulatory
mechanism of MMP9, miRanda (www.microrna.org/microrna/home.do), TargetScan
(www.targetscan.org), PiTa (genie.weizmann.ac.il/pubs/mir07/mir07_data.html),
RNAhybrid (bibiserv.techfak.uni-bielefeld.de/rnahybrid) and
PICTA (pictar.mdc-berlin.de) were used to
predict miRNA molecules that may regulate MMP9, and miR-204 was
identified as a potential regulator of MMP9 (Fig. 1). According to bioinformatics
analysis, wild-type (WT) and mutant 3′-untranslated region (UTR) of
the MMP9 mRNA containing the seed regions of miR-204 were
chemically synthesized in vitro (14) and cloned into the pMIR-REPORT
luciferase reporter plasmid (cat. no. AM5795; Ambion, Thermo Fisher
Scientific, Inc.) between the Spe-1 and HindIII
restriction sites. 293T cells were cultured in RPMI-1640 medium
(cat. no. SH30809.01; Thermo Fisher Scientific, Inc.) supplemented
with 10% fetal bovine serum (cat. no. SH30396.03; Thermo Fisher
Scientific, Inc.) at 37°C in a 5% CO2-humidified
incubator. 293T cells were co-transfected with plasmids (0.8 µg)
containing the WT or mutant sequences from the 3′-UTR of MMP9 mRNA
and agomiR-204 (100 nM; Sangon Biotech, Shanghai, China) using
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.).
293T cells in the negative control (NC) group were transfected with
agomiR-204 and empty pMIR-REPORT plasmid. Following 24-h
incubation, cells were lysed and luciferase activities were
measured using the Dual-Luciferase Reporter Assay system (Promega
Corporation, Madison, WI, USA), according to the manufacturer's
protocol. The luciferase activity was detected using a GloMax 20/20
luminometer (Promega Corporation). Firefly luciferase activity for
each group of cells was normalized to Renilla luciferase
activity as an internal reference.
Statistical analysis
The results were analyzed using SPSS 18.0
statistical software (SPSS, Inc., Chicago, IL, USA). Values are
expressed as the mean ± standard deviation. Data was analyzed using
one-way analysis of variance followed by a post hoc test. In case
of homogeneity of variance, the least significant difference and
Student-Newman-Keuls methods were used; in the case of
heterogeneity of variance, Tamhane's T2 or Dunnett's T3 method was
used. P<0.05 was considered to indicate a statistically
significant difference.
Results
Upregulation of MMP9 mRNA in pulp
tissues, serum and saliva from pediatric patients with pulpitis may
be associated with Hp infection in the stomach
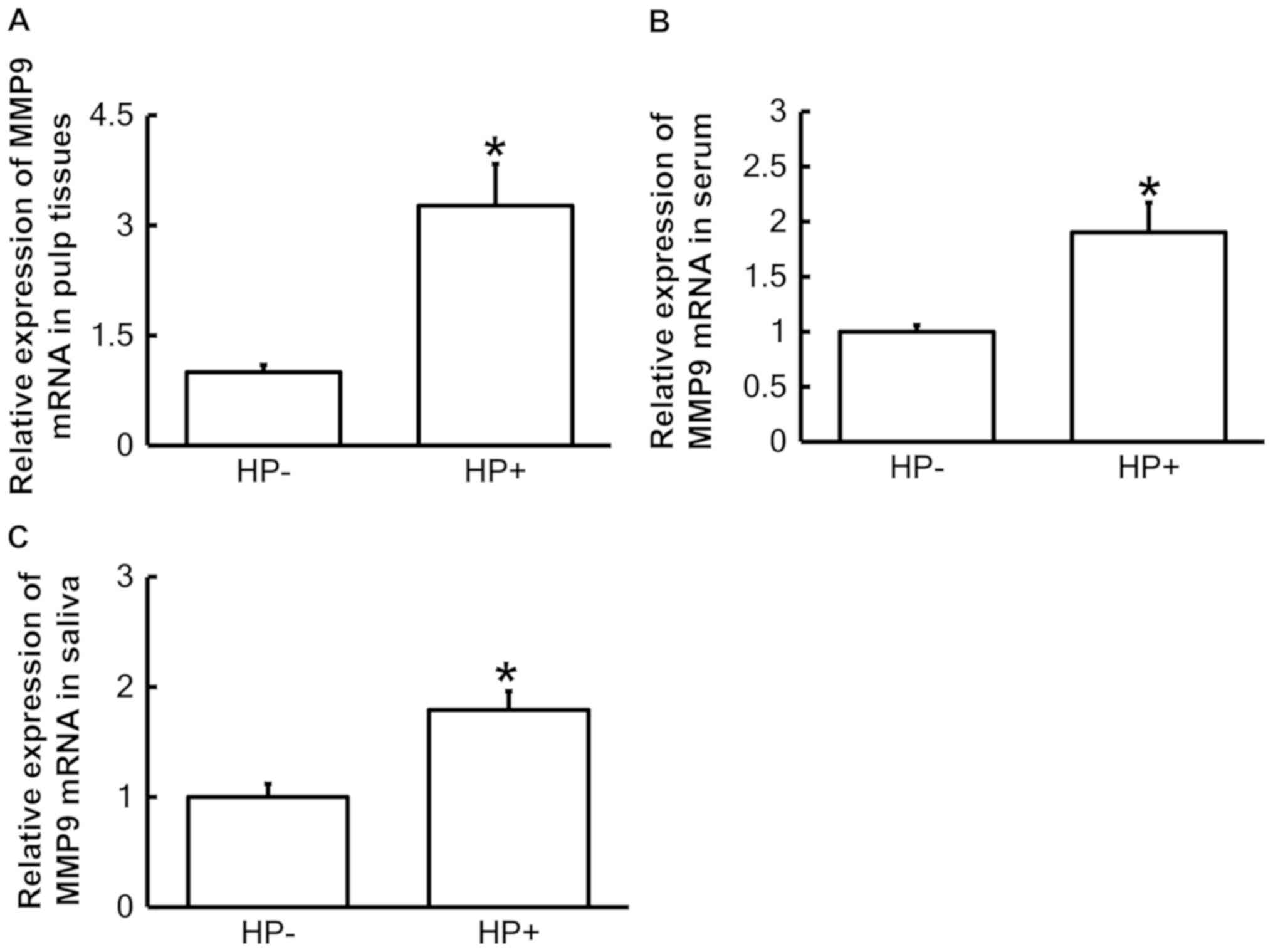
To determine the expression of MMP9 mRNA, RT-qPCR
was performed. The results indicated that the levels of MMP9 mRNA
in pulp tissues, serum and saliva from the pulpitis patients with
Hp infection in the stomach was significantly higher than that in
those without Hp infection (P<0.05; Fig. 2A-C). This result suggested that
upregulation of MMP9 mRNA in pulp tissues, serum and saliva from
pediatric pulpitis patients may be associated with Hp infection in
the stomach.
Increased protein expression of MMP9
in pulpitis pulp tissues is associated with stomach infection of
Hp
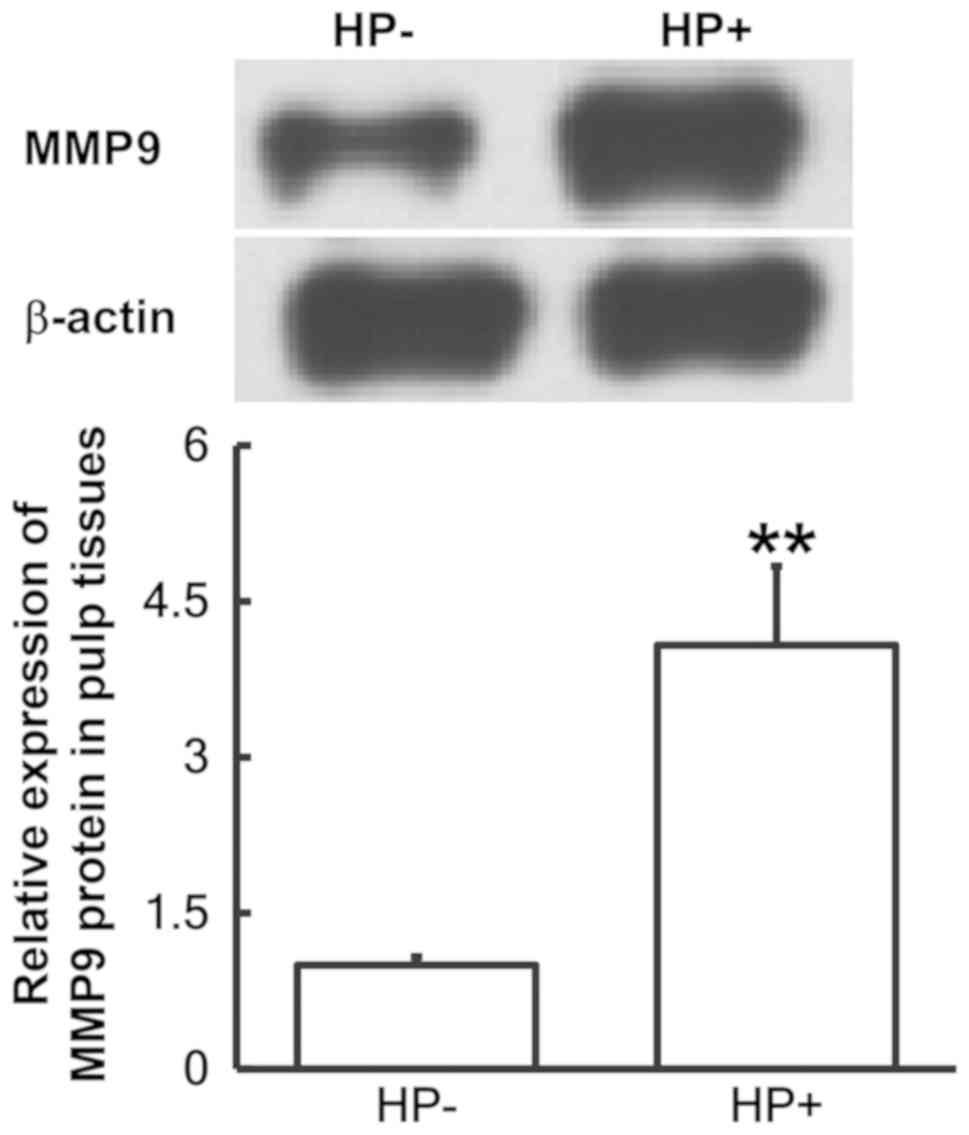
To determine the protein expression of MMP9 in pulp
tissues, western blot analysis was employed. The results indicated
that the protein levels of MMP9 in pulp tissues from pulpitis
patients with Hp infection in the stomach were significantly
elevated compared with those from pulpitis patients without Hp
infection (P<0.05; Fig. 3). This
result indicated that increased expression of MMP9 protein in pulp
tissues from pediatric pulpitis patients may be associated with
stomach infection of Hp.
Secretion of MMP9 protein into blood
and saliva from pulpitis patients is enhanced in the presence of Hp
infection in the stomach
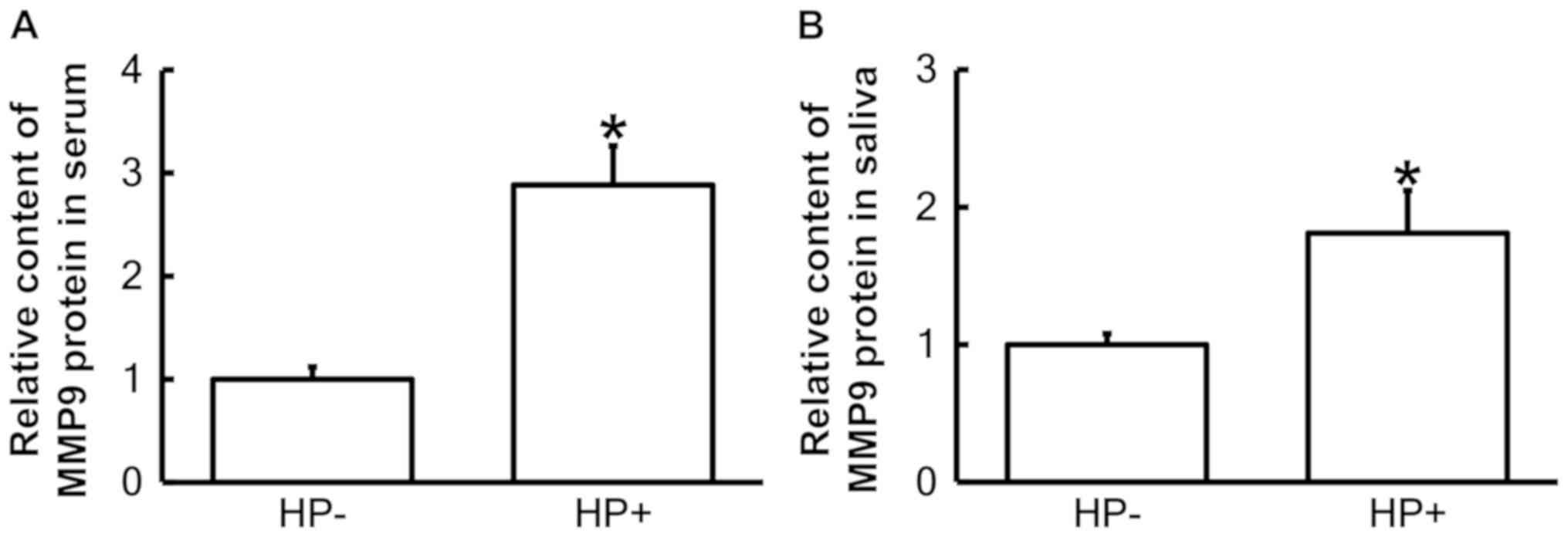
ELISA was performed to examine the contents of MMP9
protein in the serum and saliva. The results indicated that the
protein levels of MMP9 in serum and saliva from pediatric pulpitis
patients with Hp infection in the stomach were significantly higher
than those in pulpitis patients without Hp infection (P<0.05;
Fig. 4A and B). This result
suggested that the secretion of MMP9 protein into blood and saliva
is enhanced in the presence of Hp infection in the stomach.
miR-204 may have a regulatory role in
the effect of Hp infection in the stomach on pulpitis in pediatric
patients
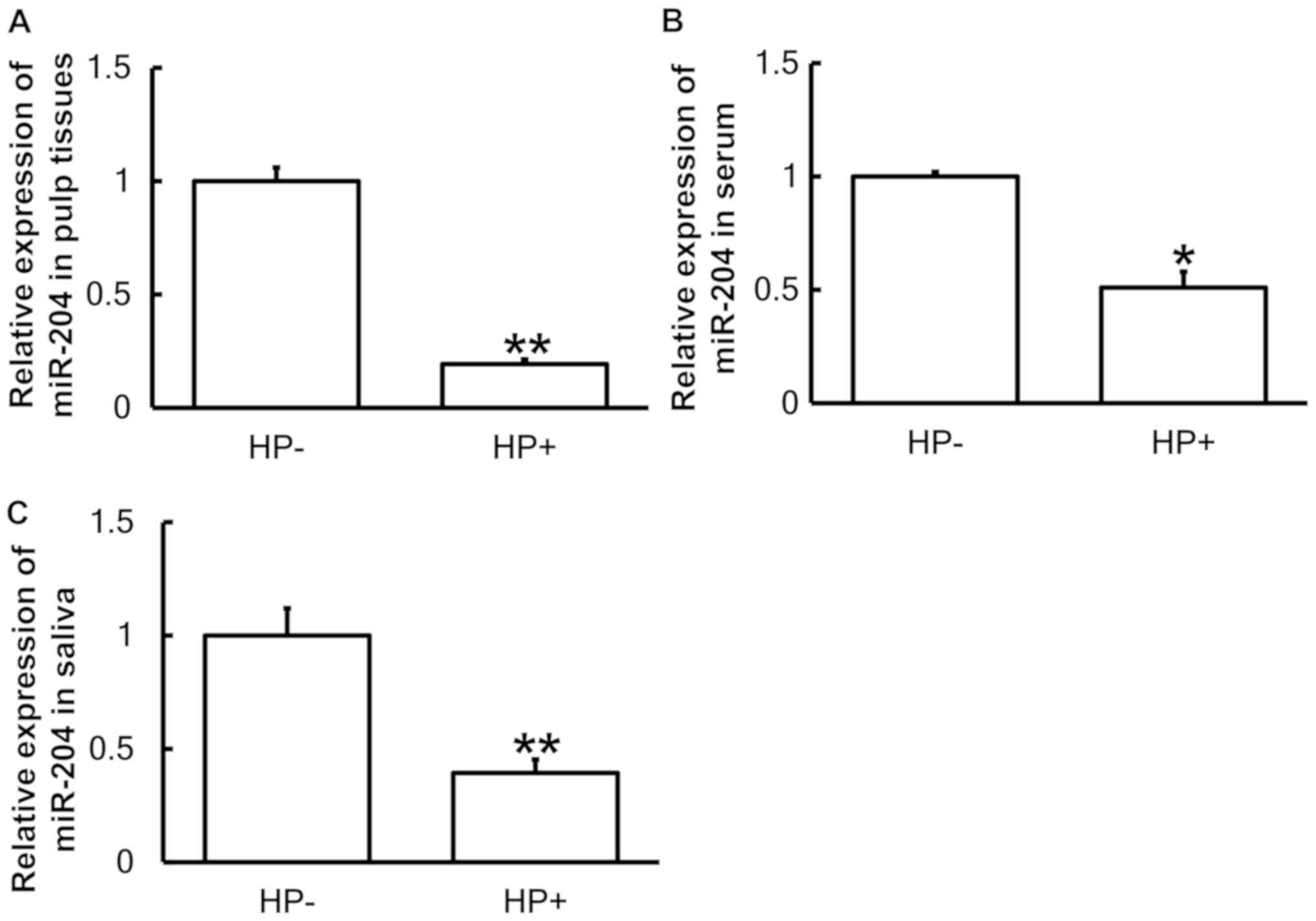
To determine the expression of miR-204 in all
samples, RT-qPCR was performed. The results indicated that the
levels of miR-204 in pulp tissues, serum and saliva from pediatric
patients with pulpitis and Hp infection in the stomach were
significantly reduced compared with those in pediatric pulpitis
patients without Hp infection (P<0.05; Fig. 5A-C). This result indicated that
miR-204 may have a regulatory role in the effect of Hp infection in
the stomach on pulpitis in pediatric patients.
miR-204 regulates the expression of
MMP9 by directly binding with the 3′-UTR of MMP9
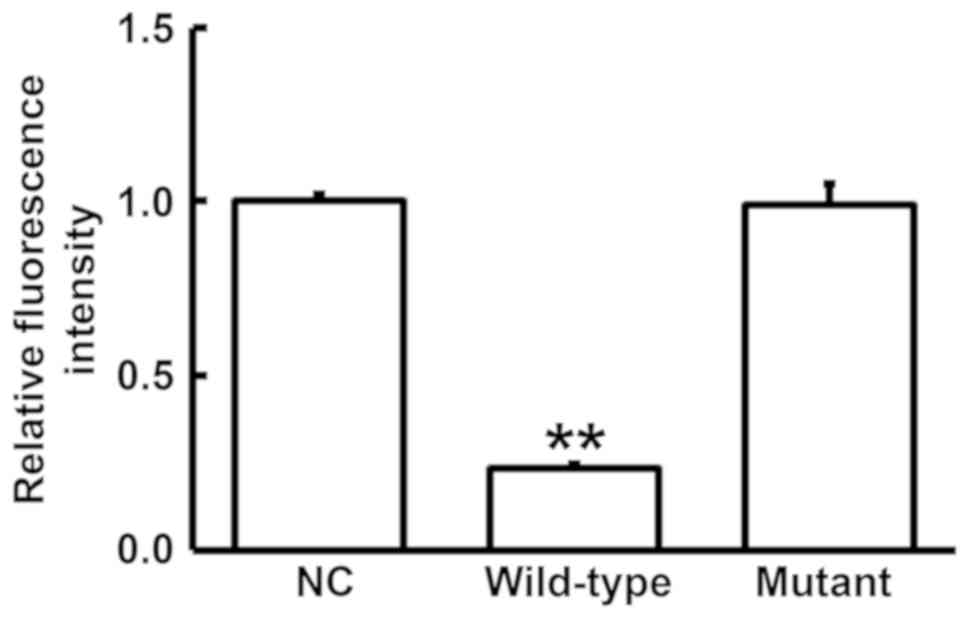
To test whether miR-204 directly targets MMP9, a
dual luciferase reporter assay was performed. The results indicated
that transfection with agomiR-204 and pMIR-REPORT in the WT group
resulted in significantly reduced fluorescence intensity compared
with that in NC group (P<0.05), while the fluorescence intensity
in mutant group was not significantly different from that in the
negative control group (P>0.05; Fig.
6). This result suggested that miR-204 regulates the expression
of MMP9 by directly binding with the 3′-UTR of MMP9.
Discussion
Hp was first isolated from gastric mucosa in
patients with gastritis in 1983 (15). Later, Hp was detected in the majority
of patients with active chronic gastritis, duodenal ulcer and
gastric ulcer, and is considered as an important pathogenic factor
for these diseases (16).
Sopata and Dancewicz (17) isolated MMP9 from neutrophile
granulocytes and discovered that it degrades collagen. Furthermore,
Murphy et al (18) revealed
that MMP9 decomposes type IV gelatin and type V collagen. MMP9 is
involved in various human diseases. Clinical studies have
demonstrated that MMP9 has important roles in renal (19), breast (20) and colorectal cancer (21,22).
Immunohistochemical study of MMP9 in the small mucosal type of
gastric cancer indicated that invasion into the lamina propria
epithelium is the first step in tumor invasion, and that MMP9 is
aberrantly expressed in most mucosal lesions (23).
Hp is associated with gastric diseases, which may
cause upregulation of MMP9. Previous studies on the association
between MMPs and periodontal disease have confirmed the implication
of MMPs (24,25). Tjaderhane et al (26) proposed that acid production by
bacteria activates MMP2 and MMP9 in the hosts, which then
participate in the degradation of carious dentin. Furthermore,
cytokines may also increase the expression of MMP2 and MMP9 in
human dental pulp cells (27). The
present study demonstrated that MMP9 levels in pulp tissues, serum
and saliva from pediatric patients with pulpitis and Hp infection
in the stomach were elevated, suggesting that Hp infection may
further aggravate pulpitis by increasing MMP9 levels in pulp
tissues, blood and saliva.
miRNAs are known to degrade their target mRNAs and
inhibit their translation (28). In
this way, miRNAs regulate the activity of multiple protein-coding
genes by up- or downregulating the translation of their mRNA, to
exert important roles in various diseases (8,29). Under
normal physiological conditions, the cleavage of MMP9 mRNA is
associated with the downregulation of its protein expression. The
bioinformatics study performed in the present study predicted that
miR-204 as an upstream miRNA that regulates MMP9. A previous study
reported that miR-204 affects the invasiveness of trophocytes by
regulating MMP9 (12). It was also
reported that miR-204 may be closely associated with inherited
retinal dystrophy (30). An et
al (31) discovered that
downregulation of miR-204 induces the proliferation and invasion of
human corneal epithelial cells. Regarding gastric disease, the role
of miR-204 was mostly elucidated in gastric cancer. Chang et
al (32) demonstrated that
miR-204 levels in Hp-negative gastric cancer tissues were
significantly higher than those in Hp-positive gastric cancer
tissues. Zhang et al (33)
reported that miR-204 inhibits the proliferation of gastric cancer
cells by downregulating the expression of ubiquitin
carboxyl-terminal hydrolase 47 and RAB22A. Zhou et al
(23) demonstrated that expression
of miR-204 in Hp-associated gastric carcinoma is significantly
decreased, and that SRY-box 4 is a direct target of miR-204. A
study by Zhang et al (34)
indicated that miR-204 downregulates sirtuin 1-induced epithelial
to mesenchymal transition, evasion of apoptosis and invasion of
gastric cancer cells. The above studies demonstrated that miR-204
is involved in mucosal cell invasion. The present study revealed
that expression of miR-204 was downregulated in pulp tissues from
pediatric patients with pulpitis and Hp infection in the stomach
compared with that in pediatric patients with pulpitis but without
Hp infection, suggesting that MMP9 is likely to be directly
regulated by miR-204. Downregulation of miR-204 expression weakened
the cleavage of MMP9 mRNA and increased the transcription and
translation of MMP9, finally aggravating the progression of
pulpitis induced by MMP9. In addition, the decrease of miR-204
levels in blood and saliva suggested that this miRNA may be
associated with oral infection by Hp. miR-204 is stable in serum
and saliva, and may be a potential marker for the diagnosis of oral
diseases in patients with Hp infection. Of note, sampling of saliva
is non-invasive and simple, and analysis of certain molecular
markers has high sensitivity and specificity, which therefore has
diagnostic prospects for Hp infection in the stomach (35). In conclusion, the present study
demonstrated that MMP9 expression in pulp tissues, blood and saliva
from pediatric patients with pulpitis and Hp infection in the
stomach was upregulated, while miR-204 expression was downregulated
compared with those in pediatric pulpitis patients without Hp
infection. miR-204 may affect inflammatory processes and other oral
diseases in pediatric patients with pulpitis and Hp infection via
MMP9, and may be a potential marker for the detection of Hp
infection in pediatric patients with pulpitis.
Acknowledgements
Not applicable.
Funding
The current study was supported by a grant from the
Jiangsu Provincial Preventive Medicine Research Foundation (grant
no. Y2013003).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SZ and JX contributed to the design of the study. SZ
performed the experiments. SZ and JX analyzed the data. SZ and JX
interpreted results and prepared the manuscript. The final version
of the manuscript has been read and approved by all authors.
Ethics approval and consent to
participate
All procedures performed were approved by the Ethics
Committee of Nanjing Medical University (Nanjing, China). Written
informed consent was obtained from all patients or their
families.
Patient consent for publication
Written informed consent for publication was
obtained from all patients or their parents, guardians or next of
kin.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Akopyanz N, Bukanov NO, Westblom TU and
Berg DE: PCR-based RFLP analysis of DNA sequence diversity in the
gastric pathogen Helicobacter pylori. Nucleic Acids Res.
20:6221–6225. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shiotani A, Cen P and Graham DY:
Eradication of gastric cancer is now both possible and practical.
Semin Cancer Biol. 23:492–501. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Villain P, Gonzalez P, Almonte M,
Franceschi S, Dillner J, Anttila A, Park JY, De Vuyst H and Herrero
R: European code against cancer 4th edition: Infections and cancer.
Cancer Epidemiol. 39 (Suppl 1):S120–S138. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen Y and Zhong LJ: The research progress
of the correlation between Helicobacter pylori and
periodontitis. Chin J Pract Stomatol. 4:752–754. 2011.(In
Chinese).
|
|
5
|
Birkedal-Hansen H, Moore WG, Bodden MK,
Windsor LJ, Birkedal-Hansen B, DeCarlo A and Engler JA: Matrix
metalloproteinases: A review. Crit Rev Oral Biol Med. 4:197–250.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wahlgren J, Salo T, Teronen O, Luoto H,
Sorsa T and Tjäderhane L: Matrix metalloproteinase-8 (MMP-8) in
pulpal and periapical inflammation and periapical root-canal
exudates. Int Endod J. 35:897–904. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gusman H, Santana RB and Zehnder M: Matrix
metalloproteinase levels and gelatinolytic activity in clinically
healthy and inflamed human dental pulps. Eur J Oral Sci.
110:353–357. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen K and Rajewsky N: The evolution of
gene regulation by transcription factors and microRNAs. Nat Rev
Genet. 8:93–103. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Greco S, Gorospe M and Martelli F:
Noncoding RNA in age-related cardiovascular diseases. J Mol Cell
Cardiol. 83:142–155. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bienertova-Vasku J, Novak J and Vasku A:
MicroRNAs in pulmonary arterial hypertension: Pathogenesis,
diagnosis and treatment. J Am Soc Hypertens. 9:221–234. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jamali Z, Asl Aminabadi N, Attaran R,
Pournagiazar F, Ghertasi Oskouei S and Ahmadpour F: MicroRNAs as
prognostic molecular signatures in human head and neck squamous
cell carcinoma: A systematic review and meta-analysis. Oral Oncol.
51:321–331. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yu Y, Wang L, Liu T and Guan H:
MicroRNA-204 suppresses trophoblast-like cell invasion by targeting
matrix metalloproteinase-9. Biochem Biophys Res Commun.
463:285–291. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fu X, Zeng L, Liu Z, Ke X, Lei L and Li G:
MicroRNA-206 regulates the secretion of inflammatory cytokines and
MMP9 expression by targeting TIMP3 in Mycobacterium
tuberculosis-infected THP-1 human macrophages. Biochem Biophys Res
Commun. 477:167–173. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Warren JR and Marshall B: Unidentified
curved bacilli on gastric epithelium in active chronic gastritis.
Lancet. 1:1273–1275. 1983.PubMed/NCBI
|
|
16
|
Marshall BJ and Warren JR: Unidentified
curved bacilli in the stomach of patients with gastritis and peptic
ulceration. Lancet. 1:1311–1315. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sopata I and Dancewicz AM: Presence of a
gelatin-specific proteinase and its latent form in human
leucocytes. Biochim Biophys Acta. 370:510–523. 1974. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Murphy G, Cawston TE, Galloway WA, Barnes
MJ, Bunning RA, Mercer E, Reynolds JJ and Burgeson RE:
Metalloproteinases from rabbit bone culture medium degrade types IV
and V collagens, laminin and fibronectin. Biochem J. 199:807–811.
1981. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bhuvarahamurthy V, Kristiansen GO,
Johannsen M, Loening SA, Schnorr D, Jung K and Staack A: In
situ gene expression and localization of metalloproteinases
MMP1, MMP2, MMP3, MMP9, and their inhibitors TIMP1 and TIMP2 in
human renal cell carcinoma. Oncol Rep. 15:1379–1384.
2006.PubMed/NCBI
|
|
20
|
Akkoc A, Inan S and Sonmez G: Matrix
metalloproteinase (MMP-2 and MMP-9) and steroid receptor
expressions in feline mammary tumors. Biotech Histochem.
87:312–319. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cavdar Z, Canda AE, Terzi C, Sarioglu S,
Fuzun M and Oktay G: Role of gelatinases (matrix metalloproteinases
2 and 9), vascular endothelial growth factor and endostatin on
clinicopathological behaviour of rectal cancer. Colorectal Dis.
13:154–160. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Park KS, Kim SJ, Kim KH and Kim JC:
Clinical characteristics of TIMP2, MMP2, and MMP9 gene
polymorphisms in colorectal cancer. J Gastroenterol Hepatol.
26:391–397. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhou X, Li L, Su J and Zhang G: Decreased
miR-204 in H. pylori-associated gastric cancer promotes cancer cell
proliferation and invasion by targeting SOX4. PLoS One.
9:e1014572014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
de Oliveira PA, de Pizzol-Junior JP,
Longhini R, Sasso-Cerri E and Cerri PS: Cimetidine reduces
interleukin-6, matrix metalloproteinases-1 and −9 immunoexpression
in the gingival mucosa of rat molars with induced periodontal
disease. J Periodontol. 88:100–111. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu N, Cao Y and Zhu G: Expression of
matrix metalloproteinases-2, −9 and reversion-inducing
cysteine-rich protein with Kazal motifs in gingiva in periodontal
health and disease. Arch Oral Biol. 75:62–67. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tjaderhane L, Larjava H, Sorsa T, Uitto
VJ, Larmas M and Salo T: The activation and function of host matrix
metalloproteinases in dentin matrix breakdown in caries lesions. J
Dent Res. 77:1622–1629. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
O'Boskey FJ Jr and Panagakos FS: Cytokines
stimulate matrix metalloproteinase production by human pulp cells
during long-term culture. J Endod. 24:7–10. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu K, Liu S, Zhang W, Jia B, Tan L, Jin Z
and Liu Y: miR-494 promotes cell proliferation, migration and
invasion and increased sorafenib resistance in hepatocellular
carcinoma by targeting PTEN. Oncol Rep. 34:1003–1010. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Conte I, Hadfield KD, Barbato S, Carrella
S, Pizzo M, Bhat RS, Carissimo A, Karali M, Porter LF, Urquhart J,
et al: MiR-204 is responsible for inherited retinal dystrophy
associated with ocular coloboma. Proc Natl Acad Sci USA.
112:E3236–E3245. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
An J, Chen X, Chen W, Liang R, Reinach PS,
Yan D and Tu L: MicroRNA expression profile and the Role of miR-204
in corneal wound healing. Invest Ophthalmol Vis Sci. 56:3673–3683.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chang H, Kim N, Park JH, Nam RH, Choi YJ,
Lee HS, Yoon H, Shin CM, Park YS, Kim JM and Lee DH: Different
microRNA expression levels in gastric cancer depending on
Helicobacter pylori infection. Gut Liver. 9:188–196. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang B, Yin Y, Hu Y, Zhang J, Bian Z,
Song M, Hua D and Huang Z: MicroRNA-204-5p inhibits gastric cancer
cell proliferation by downregulating USP47 and RAB22A. Med Oncol.
32:3312015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang L, Wang X and Chen P: MiR-204 down
regulates SIRT1 and reverts SIRT1-induced epithelial-mesenchymal
transition, anoikis resistance and invasion in gastric cancer
cells. BMC Cancer. 13:2902013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pickhardt PJ, Choi JR, Hwang I, Butler JA,
Puckett ML, Hildebrandt HA, Wong RK, Nugent PA, Mysliwiec PA and
Schindler WR: Computed tomographic virtual colonoscopy to screen
for colorectal neoplasia in asymptomatic adults. N Engl J Med.
349:2191–2200. 2003. View Article : Google Scholar : PubMed/NCBI
|