Introduction
Cholangiocarcinomas originate from cholangiocytes of
small intrahepatic bile ducts or bile ductules, or of large hilar
or extrahepatic bile ducts (1,2).
Intrahepatic cholangiocarcinoma (ICC) is the second most common
type of primary liver cancer (3,4). As ICC
is the most deadly disease of the biliary tree due to its poor
prognosis (4), exploration of its
molecular mechanisms is urgently required for identification of
novel therapeutic targets and development of effective treatment
strategies.
Long non-coding RNAs (lncRNAs), a class of small
non-coding RNAs of >200 nucleotides in length, comprise ~80% of
non-coding RNAs and function through interaction with microRNAs
(miRs) or proteins (5,6). In recent decades, a large number of
lncRNAs have been identified, which have key roles in a variety of
physiological and pathological processes, including
differentiation, development, angiogenesis, cell proliferation,
apoptosis and motility, as well as carcinogenesis (6–10).
Furthermore, certain lncRNAs have been reported to be deregulated
and to have key roles in ICC (11–13). For
instance, upregulation of lncRNA colorectal neoplasia
differentially expressed correlates with poor prognosis in ICC and
promotes epithelial-mesenchymal transition (EMT) in ICC cells
(11). Zhang et al (12) reported that lncRNA glucosaminyl
(N-acetyl) transferase 2 (I blood group) promoted ICC cell
migration, invasion and EMT through inhibition of miR-152
expression. In addition, Lv et al (13) performed a correlation analysis
between lncRNA expression levels and clinicopathological
characteristics, which revealed that EMP1-008, ATF3-008 and
RCOR3-013 were significantly downregulated in metastatic ICC,
suggesting that their downregulation may participate in ICC
metastasis.
The lncRNA urothelial cancer-associated 1 (UCA1) is
frequently upregulated in various common types of human cancer, and
has a promoting role in tumour progression (14–16). For
instance, Zhou et al (17)
reported that UCA1 was upregulated in pancreatic cancer, promoted
pancreatic cancer cell proliferation, invasion, migration, and
inhibited cell apoptosis through the downregulation of miR-96 and
the upregulation of forkhead box O3. Furthermore, the expression of
UCA1 was increased in bladder cancer tissues when compared with
that in normal tissues, and UCA1 promoted the migration, invasion
and EMT of bladder cancer cells by inhibiting the expression of
miR-143 and miR-145, while increasing the expression of high
mobility group box 1, zinc finger E-box binding homeobox 1 and 2,
and fascin actin-bundling protein 1 (18,19). In
addition, UCA1 was upregulated in oral squamous cell carcinoma
(OSCC) tissues and cell lines, as well as in cisplatin-resistant
OSCC cells, promoted OSCC cell proliferation and induced cisplatin
resistance through inhibition of miR-184 expression (20). Recently, UCA1 was reported to
indicate an unfavourable prognosis in cholangiocarcinoma patients,
and to promote cancer cell growth, migration and invasion via
regulating the AKT/GSK-3β signalling pathway (21). However, the exact role of UCA1 in ICC
has remained to be elucidated.
Therefore, the present study aimed to determine the
expression and function of UCA1 in ICC. In addition, the regulatory
mechanisms of UCA1 underlying ICC progression were
investigated.
Materials and methods
Clinical tissue collection
The present study was approved by the Ethics
Committee of Hunan Province People's Hospital (Changsha, China).
ICC tissues and their paired adjacent non-tumour tissues were
collected from 66 ICC patients who received surgical resection at
Hunan Province People's Hospital (Changsha, China) between May 2011
and March 2013. These patients included 35 males and 31 females,
from 42–74 years old with mean of 61 years old. Written informed
consent was obtained from all of these patients. These patients did
not receive any treatment prior to surgery. All tissue samples were
stored at −80°C until use. Follow-up after surgical resection was
performed until the patients' death, which occurred mainly due to
cancer recurrence and metastasis.
Cell culture and transfection
The RBE, HCCC-9810 and LICCF human ICC cell lines
and a normal human intrahepatic biliary epithelial cell (HIBEC)
line were purchased from the Cell Bank of the Chinese Academy of
Sciences (Shanghai, China). Cells were cultured in Dulbecco's
modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) with 10% fetal bovine serum (FBS; Thermo Fisher
Scientific, Inc.) in a cell incubator containing 5% CO2
at 37°C. For cell transfection, HCCC-9810 cells were transfected
with 100 nM negative control (NC) small interfering (si)RNA (cat.
no. 4457287), UCA1 siRNA (cat. no. 4390771; both Thermo Fisher
Scientific, Inc.), a blank vector (cat. no. V0006) or a UCA1
expression plasmid (cat. no. P1104; both Yearthbio, Inc., Changsha,
China), or co-transfected with a UCA1 expression plasmid and
miR-122 mimics (cat. no. 4464066) or miR-NC mimics (cat. no.
4464058; both Thermo Fisher Scientific, Inc.) using Lipofectamine
2000™ (Thermo Fisher Scientific, Inc.). At 48 h after
transfection, the cells were used for the subsequent assays.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues or cells using
TRIzol reagent (Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Total RNA was reversely transcribed into
complementary DNA using SuperScript III Reverse Transcriptase
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. qPCR was performed to examine the UCA1 and miR-122
expression by using SYBR® Premix Ex Taq™
(Takara Bio Inc., Otsu, Japan) according to the manufacturer's
protocol. GAPDH was used as the internal reference for UCA1 and U6
was used as the internal reference for miR-122. The reaction
conditions were 95°C for 5 min, followed with 35 cycles at 95°C for
30 sec and 60°C for 30 sec. The relative expression levels of UCA1
and miR-122 were determined using the 2−ΔΔCq method
(22).
Luciferase reporter gene assay
The target miRNAs of UCA1 were predicted using
RNAhybrid 2.12 (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/).
The wild-type (WT) and mutant-type (MT) UCA1 luciferase reporter
plasmids were obtained from Yearthbio, Inc., which contain the UCA1
sequences with or without the miR-122 binding sites. To study the
targeting association between UCA1 and miR-122, Lipofectamine 2000
was used to co-transfect 9810 cells with WT or MT UCA1 luciferase
reporter plasmid and miR-122 or miR-NC mimics. After transfection
for 48 h, the luciferase activity was examined using the Dual
Luciferase Reporter Assay System (Promega Corp., Madison, WI, USA)
according to the manufacturer's protocol.
Cell proliferation assay
A Cell Counting Kit-8 (CCK-8; Dojindo Molecular
Technologies, Kumamoto, Japan) was used to assess cell
proliferation according to the manufacturer's protocol. The
transfected cells were seeded into 96-well plates (3×103
cells per well). After incubation at 37°C for 0, 24, 48 or 72 h, 10
µl CCK-8 solution was added to each well. The absorbance was
measured at 450 nm using a Bio-Tek Synergy HT Multi Detection
Microplate Reader (Bio-Tek, Taipei, Taiwan).
Cell invasion assay
A Transwell assay was used to study cell invasion.
After transfection for 48 h, cells (105 cells per well
in 200 µl serum-free DMEM) were seeded into the upper chambers of
Transwell plates (Corning Inc., Corning, NY, USA). DMEM with 10%
FBS was added to the lower chamber of the Transwell plates. After
24 h, non-invading cells were carefully removed from the upper
chamber with cotton swabs. The filters were then stained with 0.01%
crystal violet solution at room temperature for 10 min. Images of
invading cells were captured under an inverted microscope.
Statistical analysis
Values are expressed as the mean ± standard
deviation from three independent experiments. The differences
between two groups were analyzed using two-tailed Student's
t-tests. The differences among more than two groups were analysed
using one-way analysis of variance, followed by Tukey's post-hoc
test. Kaplan-Meier survival curves were generated and differences
were determined by using the log-rank test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Upregulation of UCA1 is associated
with ICC progression and poor prognosis
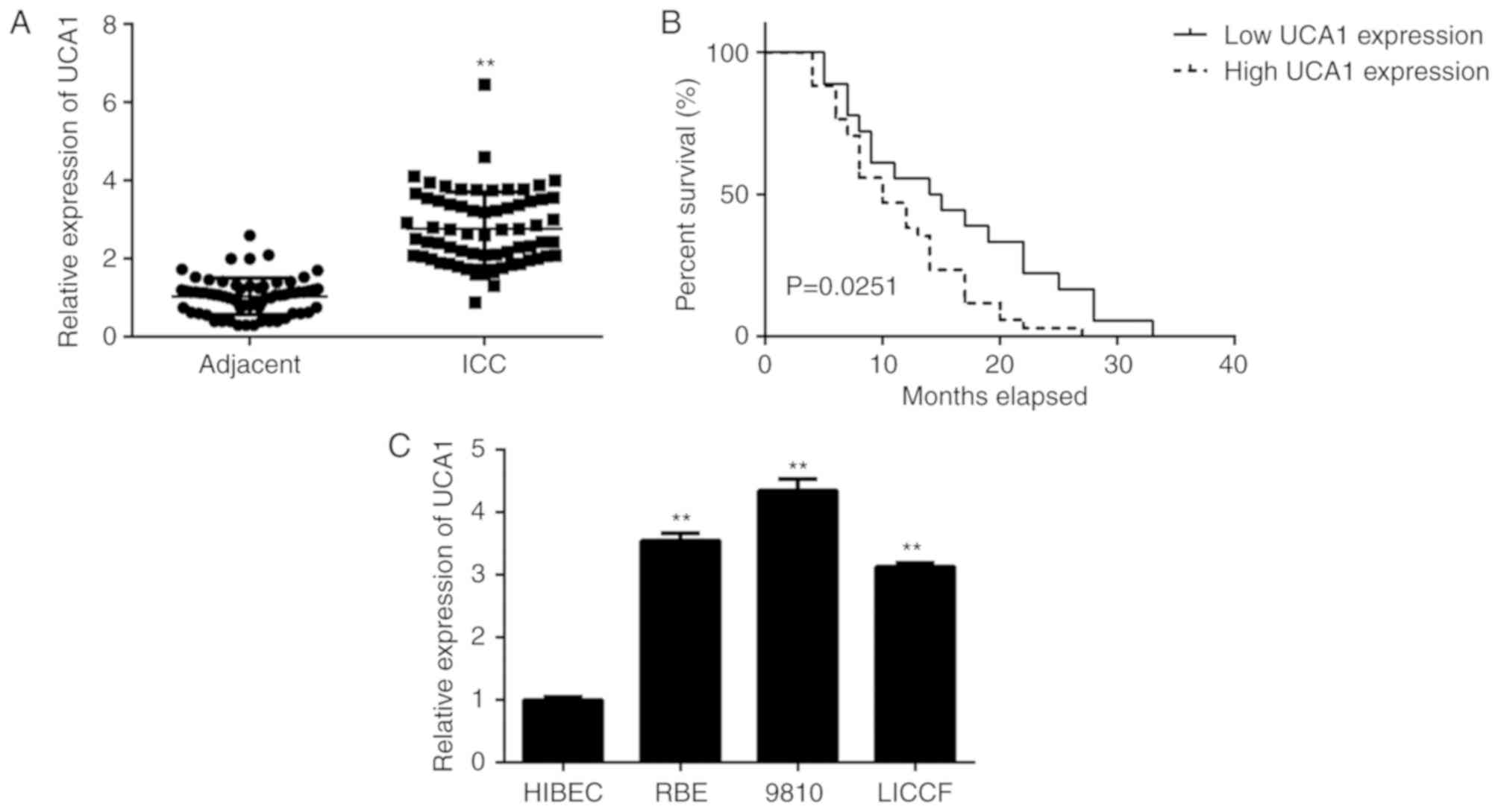
In the present study, RT-qPCR analysis indicated
that the expression levels of UCA1 were significantly higher in ICC
tissues when compared with those in adjacent non-tumour tissues
(Fig. 1A). The ICC patients were
then divided into a high and a low UCA1 expression group, based on
their mean expression level. As indicated in Table I, high expression of UCA1 was
significantly associated with clinical T-stage and lymph node
metastasis. Furthermore, those ICC patients with a high expression
of UCA1 had a significantly shorter survival when compared with
that of patients with low UCA1 expression (Fig. 1B). These results suggested that
upregulation of UCA1 is likely involved in ICC progression.
Consistent with the data obtained with the clinical tissues, the
expression of UCA1 was also higher in human ICC cell lines when
compared with that in HIBECs (Fig.
1C). As 9810 cells displayed the highest expression of UCA1
among the ICC cell lines, this cell line was used in the subsequent
in vitro experiments.
 | Table I.Association between UCA1 expression
and clinicopathological characteristics of patients with
intrahepatic cholangiocarcinoma. |
Table I.
Association between UCA1 expression
and clinicopathological characteristics of patients with
intrahepatic cholangiocarcinoma.
|
|
| UCA1 expression |
|
|---|
|
|
|
|
|
|---|
| Variables | Cases (n=66) | Low (n=32) | High (n=34) | P-value |
|---|
| Age (years) |
|
|
| 0.810 |
|
<55 | 30 | 14 | 16 |
|
| ≥55 | 36 | 18 | 18 |
|
| Sex |
|
|
| 0.460 |
| Male | 35 | 15 | 20 |
|
|
Female | 31 | 17 | 14 |
|
| Tumor focality |
|
|
| 0.477 |
|
Solitary | 57 | 29 | 28 |
|
|
Multiple | 9 | 3 | 6 |
|
| Histologic grade |
|
|
| 0.171 |
|
Well-moderate | 48 | 26 | 22 |
|
| Poor | 18 | 6 | 12 |
|
| Lymph node
metastasis |
|
|
| 0.034 |
|
Present | 14 | 3 | 11 |
|
|
Absent | 52 | 29 | 23 |
|
| Vascular
invasion |
|
|
| 0.083 |
|
Present | 26 | 9 | 17 |
|
|
Absent | 40 | 23 | 17 |
|
| HBV infection |
|
|
| 0.477 |
|
Present | 9 | 3 | 6 |
|
|
Absent | 57 | 29 | 28 |
|
| Clinical
T-stage |
|
|
| 0.027 |
|
T1/T2 | 38 | 23 | 15 |
|
|
T3/T4 | 28 | 9 | 19 |
|
Promoting effects of UCA1 on ICC cell
proliferation and invasion
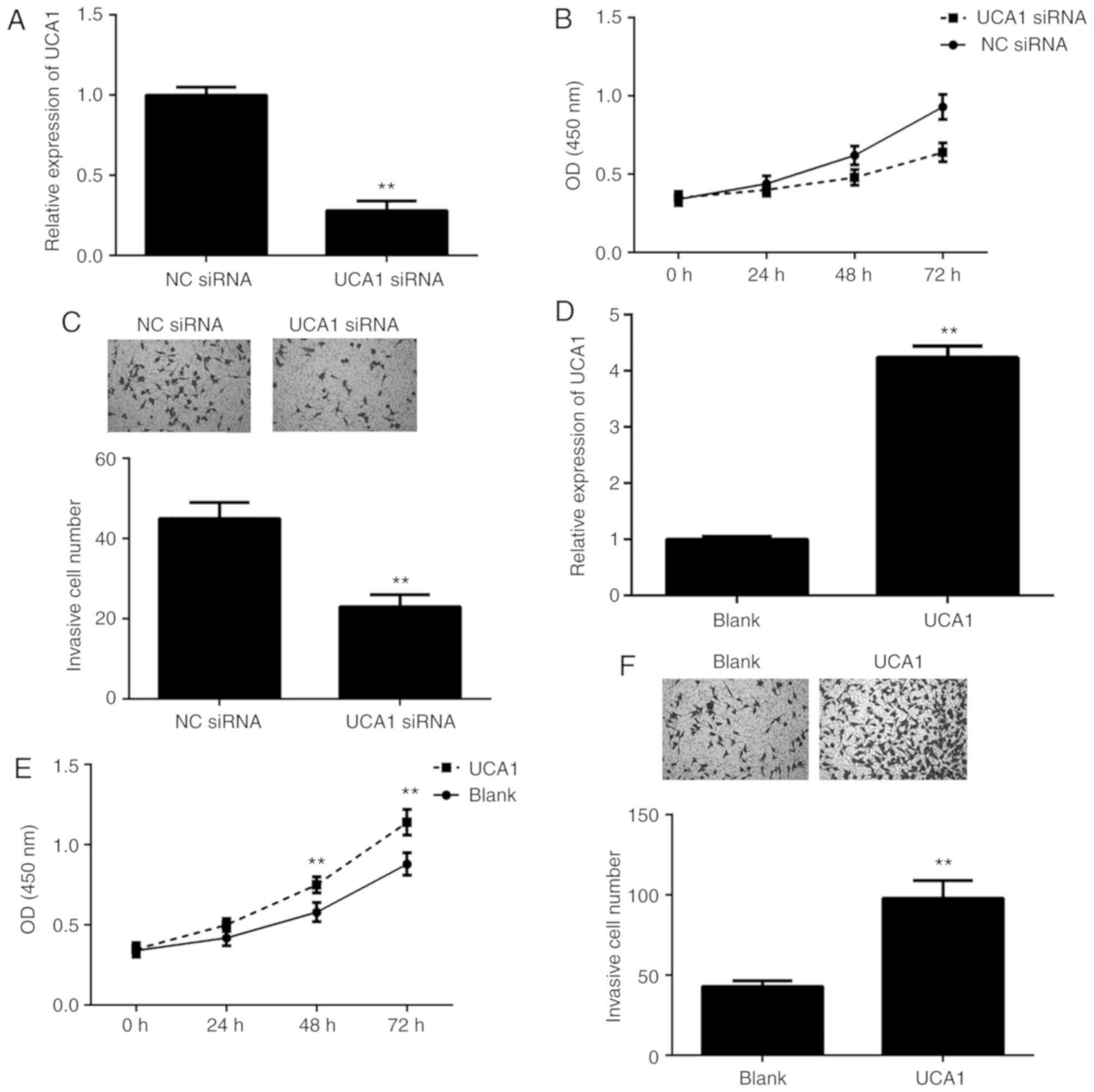
To further clarify the function of UCA1 in ICC, 9810
cells were transfected with UCA1 siRNA or NC siRNA. RT-qPCR
indicated that the expression of UCA1 was significantly
downregulated in the UCA1 siRNA group compared with that in the NC
siRNA group (Fig. 2A). A CCK-8 assay
and a Transwell assay were then performed to assess cell
proliferation and invasion. As presented in Fig. 2B and C, knockdown of UCA1 led to a
significant decrease in 9810 cell proliferation and invasion. To
further confirm these results, 9810 cells were transfected with
UCA1 plasmid or blank vector. After transfection, the expression
levels of UCA1 were significantly upregulated in the UCA1 group
when compared with those in the blank group (Fig. 2D). Furthermore, overexpression of
UCA1 markedly promoted the proliferation and invasion of 9810 cells
(Fig. 2E and F). These results
suggested that UCA1 promotes ICC cell proliferation and
invasion.
UCA1 directly targets miR-122 in ICC
cells
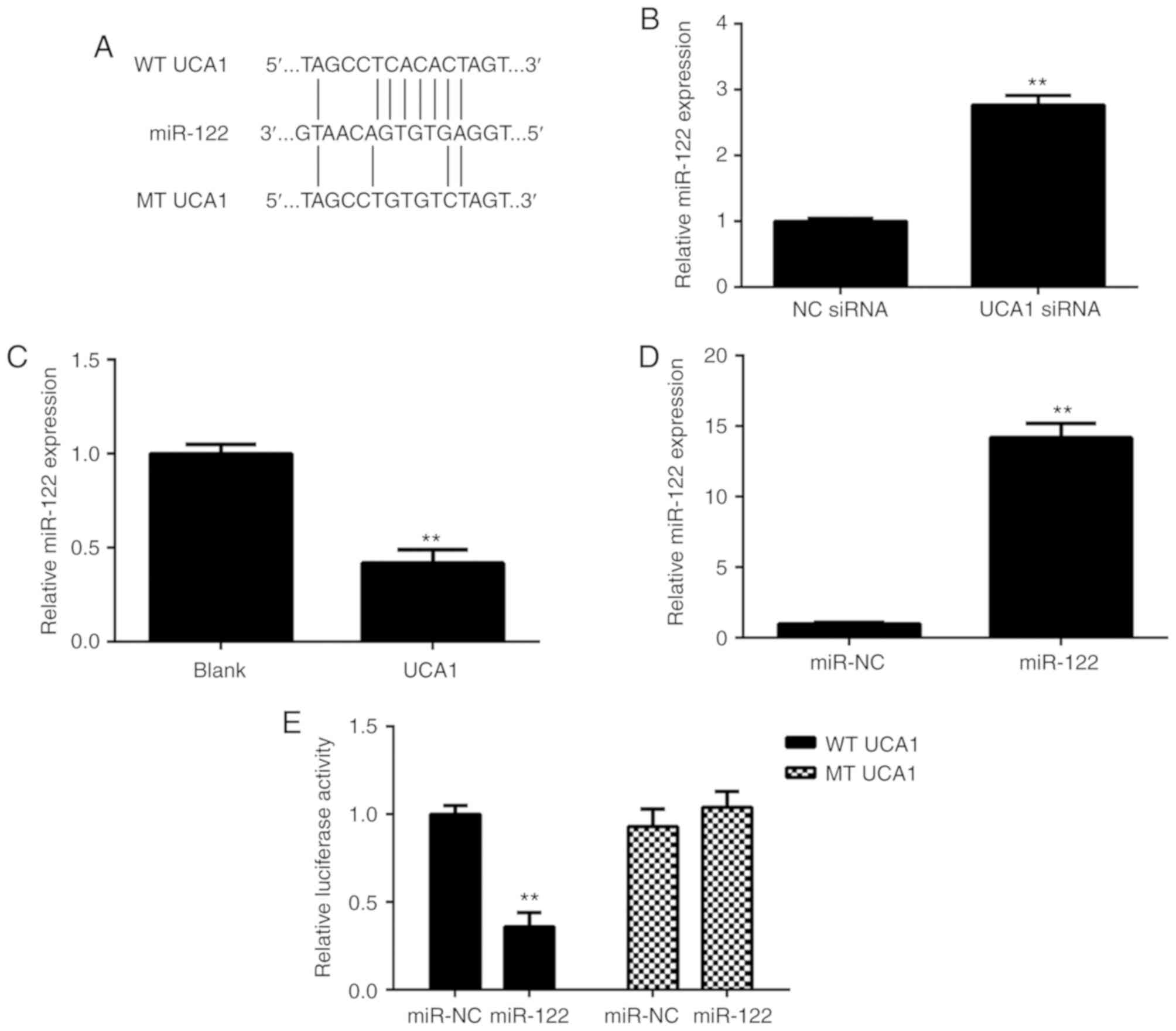
As lncRNAs generally act as ligands for pools of
target miRNAs, miRanda software (http://www.micro-RNA.org/) was used to predict
potential UCA1-miR interactions. As presented in Fig. 3A, miR-122 had a potential binding
site for UCA1. The effects of UCA1 upregulation or downregulation
on the miR-122 expression in 9810 cells were then studied. As
presented in Fig. 3B and C, ectopic
overexpression of UCA1 significantly reduced the expression of
miR-122 in 9810 cells, while knockdown of UCA1 markedly promoted
the miR-122 expression in 9810 cells. Next, 9810 cells were
transfected with the miR-NC or miR-122 mimics. After transfection,
RT-qPCR confirmed that the expression of miR-122 was significantly
increased in the miR-122 group compared with that in the miR-NC
group (Fig. 3D). These results
suggested that transfection with the miR-122 mimics effectively
upregulated miR-122 expression in 9810 cells. To verify the
predicted direct binding of miR-122 with UCA1, the WT or MT UCA1
luciferase reporter gene plasmids with or without the binding sites
with miR-122, respectively, were purchased (Fig. 3A). A luciferase reporter gene assay
was then performed in 9810 cells. As presented in Fig. 3E, transfection with the miR-122
mimics caused a significant decrease in the luciferase activity of
cells transfected with the WT UCA1 luciferase reporter plasmid;
however, the miR-122 mimics had no effect on the luciferase
activity of cells transfected with the MT UCA1 luciferase reporter
plasmid. Therefore, it was proven that UCA1 directly targets
miR-122 in ICC cells.
miR-122 functions as a downstream
effector in the UCA1-mediated ICC cell proliferation and
invasion
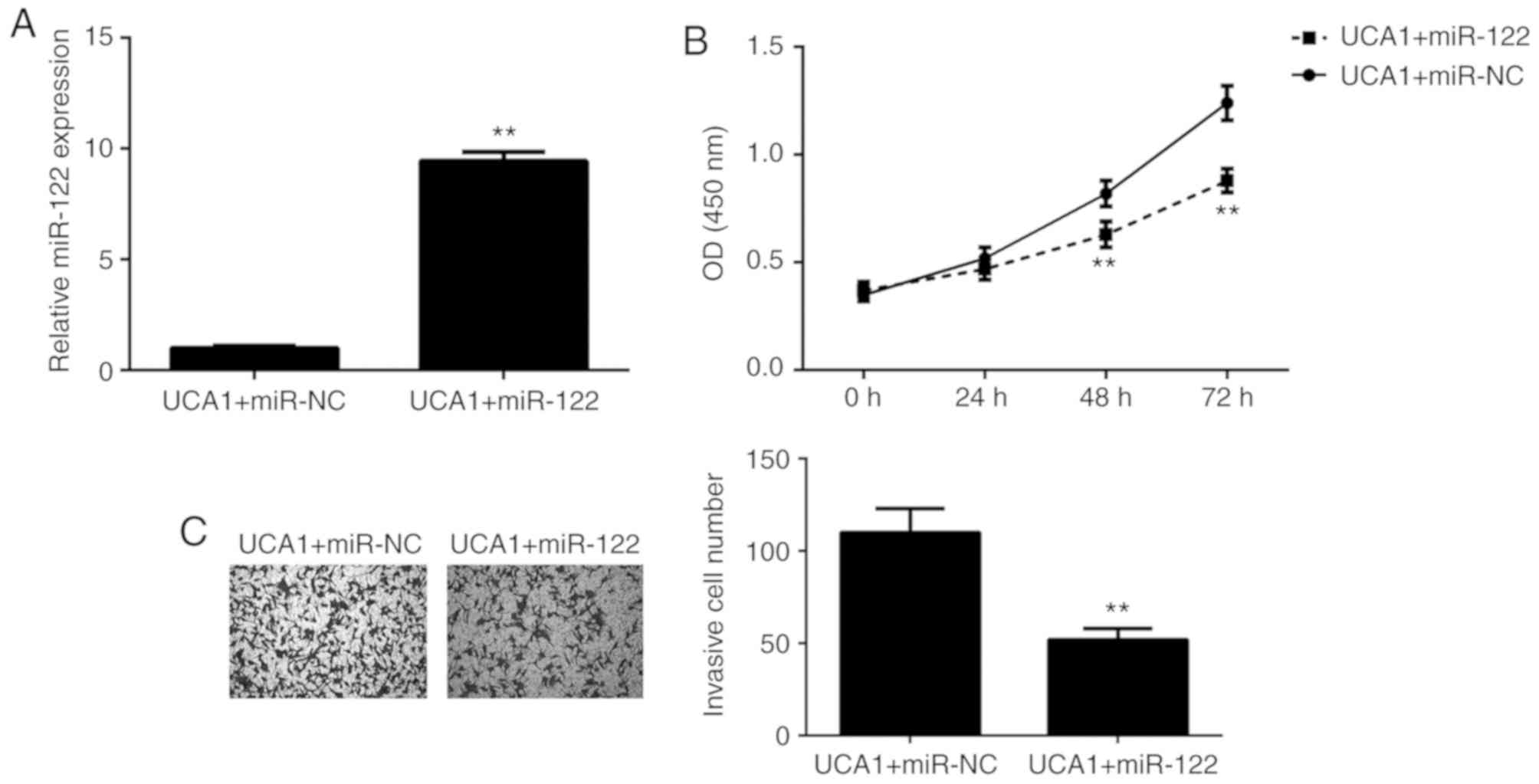
As UCA1 was proven to directly target miR-122 and
negatively regulate its expression in ICC cells, it was then
investigated whether miR-122 functions as a downstream effector in
the UCA1-mediated proliferation and invasion of ICC cells. 9810
cells were co-transfected with UCA1 plasmid and either miR-122
mimics or miR-NC mimics. After transfection, the miR-122 levels
were significantly increased in the UCA1+miR-122 group when
compared with those in the UCA1+miR-NC group (Fig. 4A). Next, a CCK-8 assay and a
Transwell assay were performed to assess cell proliferation and
invasion, respectively. As presented in Fig. 4B and C, respectively, the
proliferation and invasion of 9810 cells were significantly
downregulated in the UCA1+miR-122 group when compared with those in
the UCA1+miR-NC group. These results suggested that overexpression
of UCA1 promotes the proliferation and invasion of 9810 cells via
inhibition of miR-122 expression. Therefore, miR-122 functioned as
a downstream effector in the UCA1-mediated ICC cell proliferation
and invasion.
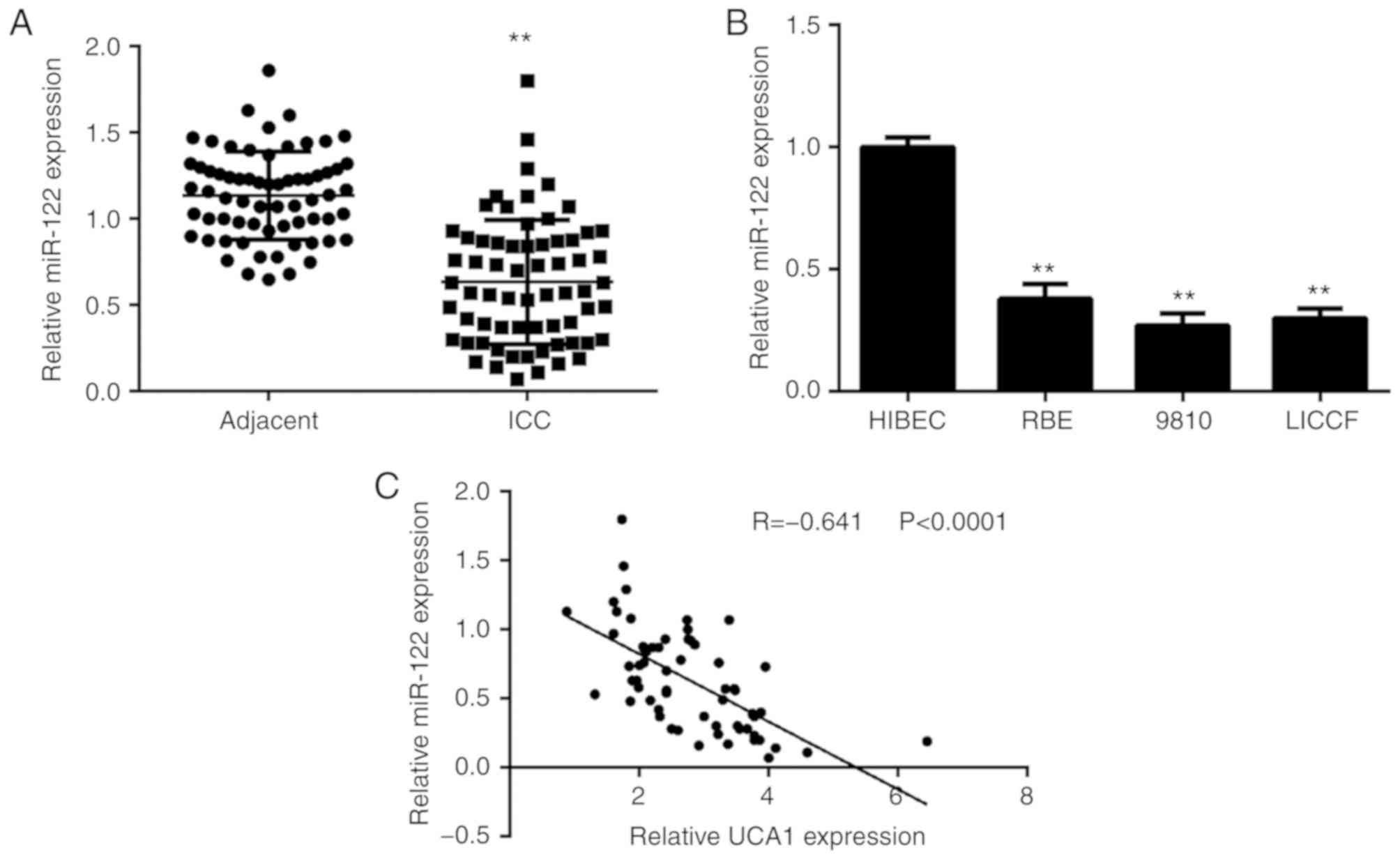
miR-122 is downregulated in ICC, with
an inverse correlation to the expression of UCA1
Finally, the expression levels of miR-122 were
determined in ICC tissues and cell lines. As presented in Fig. 5A, the expression of miR-122 was
significantly reduced in ICC tissues when compared with that in
adjacent non-tumour tissues. Consistently, the expression of
miR-122 was also downregulated in ICC cell lines when compared with
that in HIBECs (Fig. 5B). Of note,
an inverse correlation was observed between the UCA1 and miR-122
expression in ICC tissues (Fig. 5C).
Therefore, the reduced expression of miR-122 may at least in part
be due to the increased expression of UCA1 in ICC tissues.
Discussion
In the present study, UCA1 was observed to be
significantly upregulated in ICC tissues and cell lines, when
compared with that in the adjacent non-tumour tissues and HIBECs,
respectively. The increased expression of UCA1 was significantly
associated with lymph node metastasis and clinical T-stage in ICC.
Furthermore, the ICC patients with high expression of UCA1 had
shorter survival times when compared with those with low UCA1
expression. Knockdown of UCA1 caused a significant decrease in ICC
cell proliferation and invasion, and overexpression of UCA1
significantly promoted the proliferation and invasion of ICC cells.
Furthermore, UCA1 was confirmed to directly bind to miR-122, and
the expression of miR-122 was negatively regulated by UCA1 in ICC
cells. Of note, miR-122 mimics inhibited the promoting effects of
UCA1 on ICC cell proliferation and invasion. In addition, an
inverse correlation between miR-122 and UCA1 expression in ICC
tissues was observed.
Several studies have focused on the function of
lncRNAs in ICC. For instance, Wang et al (2) performed a lncRNA microarray using ICC
tissues and paired adjacent non-tumour tissues, and identified
2,773 lncRNAs that were significantly upregulated and 2,392 lncRNAs
that were downregulated in ICC tissues. They then observed a
positive correlation between 4 lncRNA-mRNA pairs in ICC tissues,
including RNA43085 and sulfatase 1, RNA47504 and lysine demethylase
8, RNA58630 and proprotein convertase subtilisin/kexin type 6, and
RNA40057 and cytochrome P450 family 2 subfamily D member 6 (CYP2D6)
(2). Furthermore, the ICC patients
with high CYP2D6 and RNA40057 expression had a better prognosis
(2). These results suggested that
certain lncRNAs may become promising diagnostic and prognostic
biomarkers for ICC. Ma et al (23) reported that the lncRNA
carbamoyl-phosphate synthase 1-intronic transcript 1 was
upregulated in ICC tissues, and its upregulation was associated
with poor liver function and shorter survival time of ICC patients.
In addition, Zeng et al (24)
indicated that the lncRNA taurine up-regulated 1 (TUG1) was
upregulated in ICC, which correlated with ICC progression and poor
prognosis, and inhibition of TUG1 expression reduced ICC cell
proliferation, migration and invasion in vitro and tumour
growth in vivo. However, the function of lncRNA UCA1 in ICC
has not been previously reported, to the best of our knowledge. The
results of the present study indicated that UCA1 was significantly
upregulated in ICC tissues and cell lines. Furthermore, high
expression of UCA1 was identified to be associated with clinical
T-stage and lymph node metastasis, as well as shorter survival
times of ICC patients. These results suggested that the increased
expression of UCA1 may contribute to the malignant progression of
ICC. To further study the role of UCA1 in ICC, two common ICC cell
lines were used to perform in vitro experiments. Knockdown
of UCA1 inhibited ICC cell proliferation and invasion, and
overexpression of UCA1 promoted the proliferation and invasion of
ICC cells. These results further suggested that the lncRNA UCA1 may
participate in ICC growth and metastasis.
As several cellular responses function through
interaction with miRs, the present study subsequently focused on
the downstream miRs of UCA1 in ICC. miRs, another type of
non-coding small RNA with 22–25 nucleotides, are important
regulators for gene expression, and have key roles during cancer
development and progression. A Bioinformatics analysis suggested
that miR-122 has a potential binding site with UCA1, which was
confirmed in the ICC cell lines by using a luciferase reporter gene
assay. A previous study reported that miR-122 is significantly
downregulated in ICC tissues (25).
In line with this, a downregulation of miR-122 in ICC tissues and
cell lines was also observed in the present study. Furthermore, an
inverse correlation between the UCA1 and miR-122 expression in ICC
tissues was observed. As ectopic overexpression of UCA1
significantly reduced the miR-122 expression and knockdown of UCA1
markedly promoted miR-122 expression in ICC cells, it was suggested
that the downregulation of miR-122 in ICC may be due to the
upregulation of UCA1. Furthermore, miR-122 mimics impaired the
effects of UCA1 upregulation on ICC cell proliferation and
invasion, suggesting that miR-122 functions as a downstream
effector in the UCA1-mediated ICC cell proliferation and
invasion.
In addition to ICC, the association between UCA1 and
miR-122 has been reported in glioma and breast cancer cells
(26). Sun et al (26) indicated that UCA1 targeted miR-122 to
promote glioma cell proliferation, migration and invasion. Zhou
et al (27) reported that
histocompatibility minor 13 regulated the UCA1-mediated invasion of
breast cancer cells through UCA1 decay and decreasing the
interaction between UCA1 and miR-122. Therefore, the present study
expands on the understanding of the function of the UCA1/miR-122
interaction in human cancers.
In conclusion, the present study demonstrates for
the first time that lncRNA UCA1 promotes the proliferation and
invasion of ICC cells through targeting miR-122 and thus suggests
that the UCA1/miR-22 interaction may be used as a potential
therapeutic target for the treatment of ICC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
WY collected clinical tissues. OL, PY and CG
performed the clinical analyses and cell experiments. OL and CP
designed the study and wrote the manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
Hunan Province People's Hospital (Changsha, China). Written
informed consent was obtained from all subjects.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sempoux C, Jibara G, Ward SC, Fan C, Qin
L, Roayaie S, Fiel MI, Schwartz M and Thung SN: Intrahepatic
cholangiocarcinoma: New insights in pathology. Semin Liver Dis.
31:49–60. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang J, Xie H, Ling Q, Lu D, Lv Z, Zhuang
R, Liu Z, Wei X, Zhou L, Xu X and Zheng S: Coding-noncoding gene
expression in intrahepatic cholangiocarcinoma. Transl Res.
168:107–121. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Andersen JB: Molecular pathogenesis of
intrahepatic cholangiocarcinoma. J Hepatobiliary Pancreat Sci.
22:101–113. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Deng G, Zhu L, Huang F, Nie W, Huang W, Xu
H, Zheng S, Yi Z and Wan T: SALL4 is a novel therapeutic target in
intrahepatic cholangiocarcinoma. Oncotarget. 6:27416–27426. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Peng Z, Liu C and Wu M: New insights into
long noncoding RNAs and their roles in glioma. Mol Cancer.
17:612018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Smolle MA and Pichler M: The role of long
non-coding RNAs in osteosarcoma. Noncoding RNA. 4(pii):
E72018.PubMed/NCBI
|
|
7
|
Xu S, Kong D, Chen Q, Ping Y and Pang D:
Oncogenic long noncoding RNA landscape in breast cancer. Mol
Cancer. 16:1292017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ruan X: Long noncoding RNA central of
glucose homeostasis. J Cell Biochem. 117:1061–1065. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wapinski O and Chang HY: Long noncoding
RNAs and human disease. Trends Cell Biol. 21:354–361. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Taylor DH, Chu ET, Spektor R and Soloway
PD: Long non-coding RNA regulation of reproduction and development.
Mol Reprod Dev. 82:932–956. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xia XL, Xue D, Xiang TH, Xu HY, Song DK,
Cheng PG and Wang JQ: Overexpression of long non-coding RNA CRNDE
facilitates epithelial-mesenchymal transition and correlates with
poor prognosis in intrahepatic cholangiocarcinoma. Oncol Lett.
15:4105–4112. 2018.PubMed/NCBI
|
|
12
|
Zhang S, Xiao J, Chai Y, Du YY, Liu Z,
Huang K, Zhou X and Zhou W: LncRNA-CCAT1 promotes migration,
invasion, and EMT in intrahepatic cholangiocarcinoma through
suppressing miR-152. Dig Dis Sci. 62:3050–3058. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lv L, Wei M, Lin P, Chen Z, Gong P, Quan Z
and Tang Z: Integrated mRNA and lncRNA expression profiling for
exploring metastatic biomarkers of human intrahepatic
cholangiocarcinoma. Am J Cancer Res. 7:688–699. 2017.PubMed/NCBI
|
|
14
|
Wei Y, Sun Q, Zhao L, Wu J, Chen X, Wang
Y, Zang W and Zhao G: LncRNA UCA1-miR-507-FOXM1 axis is involved in
cell proliferation, invasion and G0/G1 cell cycle arrest in
melanoma. Med Oncol. 33:882016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bian Z, Jin L, Zhang J, Yin Y, Quan C, Hu
Y, Feng Y, Liu H, Fei B, Mao Y, et al: LncRNA-UCA1 enhances cell
proliferation and 5-fluorouracil resistance in colorectal cancer by
inhibiting miR-204-5p. Sci Rep. 6:238922016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang HM, Lu JH, Chen WY and Gu AQ:
Upregulated lncRNA-UCA1 contributes to progression of lung cancer
and is closely related to clinical diagnosis as a predictive
biomarker in plasma. Int J Clin Exp Med. 8:11824–11830.
2015.PubMed/NCBI
|
|
17
|
Zhou Y, Chen Y, Ding W, Hua Z, Wang L, Zhu
Y, Qian H and Dai T: LncRNA UCA1 impacts cell proliferation,
invasion, and migration of pancreatic cancer through regulating
miR-96/FOXO3. IUBMB Life. 70:276–290. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Luo J, Chen J, Li H, Yang Y, Yun H, Yang S
and Mao X: LncRNA UCA1 promotes the invasion and EMT of bladder
cancer cells by regulating the miR-143/HMGB1 pathway. Oncol Lett.
14:5556–5562. 2017.PubMed/NCBI
|
|
19
|
Xue M, Pang H, Li X, Li H, Pan J and Chen
W: Long non-coding RNA urothelial cancer-associated 1 promotes
bladder cancer cell migration and invasion by way of the
hsa-miR-145-ZEB1/2-FSCN1 pathway. Cancer Sci. 107:18–27. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fang Z, Zhao J, Xie W, Sun Q, Wang H and
Qiao B: LncRNA UCA1 promotes proliferation and cisplatin resistance
of oral squamous cell carcinoma by sunppressing miR-184 expression.
Cancer Med. 6:2897–2908. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu Y, Yao Y, Leng K, Li Z, Qin W, Zhong X,
Kang P, Wan M, Jiang X and Cui Y: Long non-coding RNA UCA1
indicates an unfavorable prognosis and promotes tumorigenesis via
regulating AKT/GSK-3β signaling pathway in cholangiocarcinoma.
Oncotarget. 8:96203–96214. 2017.PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ma SL, Li AJ, Hu ZY, Shang FS and Wu MC:
Co-expression of the carbamoyl-phosphate synthase 1 gene and its
long non-coding RNA correlates with poor prognosis of patients with
intrahepatic cholangiocarcinoma. Mol Med Rep. 12:7915–7926. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zeng B, Ye H, Chen J, Cheng D, Cai C, Chen
G, Chen X, Xin H, Tang C and Zeng J: LncRNA TUG1 sponges miR-145 to
promote cancer progression and regulate glutamine metabolism via
Sirt3/GDH axis. Oncotarget. 8:113650–113661. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Karakatsanis A, Papaconstantinou I,
Gazouli M, Lyberopoulou A, Polymeneas G and Voros D: Expression of
microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c,
miR-221, miR-222, and miR-223 in patients with hepatocellular
carcinoma or intrahepatic cholangiocarcinoma and its prognostic
significance. Mol Carcinog. 52:297–303. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sun Y, Jin JG, Mi WY, Zhang SR, Meng Q and
Zhang ST: Long noncoding RNA UCA1 targets miR-122 to promote
proliferation, migration, and invasion of glioma cells. Oncol Res.
26:103–110. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhou Y, Meng X, Chen S, Li W, Li D, Singer
R and Gu W: IMP1 regulates UCA1-mediated cell invasion through
facilitating UCA1 decay and decreasing the sponge effect of UCA1
for miR-122-5p. Breast Cancer Res. 20:322018. View Article : Google Scholar : PubMed/NCBI
|