Introduction
Ellagic acid (EA4) is a
naturally-occurring phenolic compound (structure shown in Fig. 1) found in certain oak species
(1), medicinal mushroom Phellinus
linteus (2), and macrophyte
Myriophyllum spicatum (3). It
is also richly contained in some human food sources (4–12). High
levels of EA are found in Longan (also known as
Dimocarpus longan), Litchi (Litchi chinensis),
walnuts, pecans, cranberries, raspberries, strawberries, grapes,
and peach (4–12). EA has been reported to have a number
of biological activities, including antioxidant and
antiproliferative properties as observed in some of the in
vitro and animal models (10,13–16). As
with other polyphenol antioxidants, it has been suggested that EA
may have a chemoprotective effect in cellular models by inhibiting
reactive chemical carcinogens [e.g., nitrosamines (17,18) and
polycyclic aromatic hydrocarbons (19)] from covalently modifying DNA
(17–19). It is noteworthy that in recent years,
EA has been controversially marketed as a dietary supplement with a
number of assumed benefits against cancer, heart disease, as well
as other medical issues, and these claims have received warnings
from the US Food and Drug Administration (FDA) (20).
Cyclooxygenase I and II (COX I and II) are key
enzymes that catalyze the metabolism of arachidonic acid (AA),
resulting in the formation of important biological mediators
including prostaglandins (PGs), prostacyclins, thromboxanes, and
others (21–24). Since these mediators affect many
pathological and physiological processes, COX enzymes have become
important targets in pharmacology and toxicology. Pharmacological
modulation of the COX enzyme activity has become an effective
approach in treating many medical conditions (25–28).
We have recently shown that certain natural
phenolics, such as quercetin and myricetin, can activate the
catalytic activity of COX I and II in enzymatic assays by
functioning as reducing co-substrates for these enzymes (29). This phenomenon was further confirmed
when they were tested in cultured cells (29) and animal models (30). Notably, these compounds are effective
in activating COX enzyme activity for PG biosynthesis in intact
cells with effective concentrations in the nM range (29). Additional mechanistic studies showed
that some of the flavonoids can bind inside the peroxidase active
site of the enzymes and directly interact with protoporphorin IX
with FeIV inside (P+FeIV) to
facilitate the electron transfer from these reducing compounds to
the Fe ion (31).
Based on our earlier three dimensional
(3D)-QSAR/CoMFA models for COX I and II that were derived from
experimental study of representative flavonoids (29), we predicted that EA may share the COX
enzyme-modulating activity. In the present study, we aimed to
experimentally examine the ability of EA to modulate PG production
using cultured cells and intact animals. The possible mechanism for
its modulating effect was explored using computational modeling
approach by studying their binding interaction with the COX-1 and
COX-2 enzymes.
Materials and methods
Chemicals and reagents
EA (purity >99%), galangin, AA,
lipopolysaccharide (LPS; from Escherichia coli, serotype
055:b5), and Dulbecco's modified Eagle's medium (DMEM) were
purchased from Sigma-Aldrich (St. Louis, ΜO, USA). The anti-COX I
and anti-COX II antibodies were obtained from Abcam (Cambridge,
UK), and the anti-GAPDH antibody was obtained from Cell Signaling
Technology (Danvers, MA, USA). Fetal bovine serum (FBS) was
obtained from Gibco-Thermo Fisher Scientific, Inc. (Waltham, MA,
USA), and the enzymatic immunoassay (EIA) kit for detecting
PGE2 was obtained from Cayman Chemical (Ann Arbor, MI,
USA).
Cell culture experiments and compound
screening
The murine macrophage RAW264.7 cell line was
obtained from the American Type Culture Collection (Manassas, VA,
USA), and maintained in DMEM containing L-glutamine, glucose and
sodium bicarbonate supplemented with 10% FBS at 37°C under 5%
CO2. In the experiments that were designed to determine
the effect of these phenolic compounds on the formation of
PGE2 in cultured RAW264.7 cells, the cells were first
stimulated with 1 µg/ml LPS for 2 h to induce the expression of COX
enzymes. Then the medium was removed and replaced with 300 µl
serum-free DMEM with or without different concentrations (0.01,
0.1, 1, 10, and 100 µM) of a phenolic compound of interest. After
an additional 2-h incubation, the culture media were collected for
measurement of PGE2 level by using an EIA kit obtained
from Cayman Chemical.
In vivo animal experiments
All the procedures involving the use of live animals
as described in this study were approved by the Institutional
Animal Care and Use Committee of the Southern University of Science
and Technology (approval number: SUSTC-G-2014009), and the
guidelines for humane treatment of animals accepted by the National
Institutes of Health (NIH) of the USA were followed. The male
Sprague-Dawley rats (4 to 5-week-old, specific pathogen-free) were
obtained from Guangdong Medical Laboratory Animal Center
(Guangdong, China), and they were maintained in our institute's
central animal facility. After arrival, the animals were allowed to
acclimatize for one week prior to being used for experimentation.
The animals were housed under constant conditions of temperature
(20±1°C) and 12-h light/dark cycle, and had free access to food and
water.
Male rats were divided into the following two
groups: The control group (receiving vehicle treatment only) and
the EA group (treated with 6 mg/kg body weight EA, dissolved in 1.5
ml of 1% methyl cellulose). Blood samples were collected through
tail bleeding at different time points (0, 3, 6, 12, and 24 h)
following administration, and stored in small vials containing
heparin. Plasma was prepared from the collected blood by
centrifugation at 1,000 × g for 10 min at 37°C. The plasma level of
PGE2 was determined using an EIA kit (Cayman Chemical)
according to the manufacturer's instructions.
Molecular docking analysis of the binding
interaction of EA with COX II. Energy minimization and molecular
docking were performed on a Dell PowerEdge R730 Server with the
Discovery Studio modeling software (version 2007; Accelrys, San
Diego, CA, USA).
Protein processing
Since the X-ray structure of sheep COX I protein
[PDB code: 1q4g (32)] and mouse COX
II protein [(DB code: 3nt1 (33)] in
complex with P+FeIV are available, we used
these structures as templates for computational docking analysis.
All small molecules except P+FeIV that are
non-covalently attached to the COX protein were removed, and then
the amino acid residues in the protein structure were re-numbered
according to the correct known sequences. The Clean Protein module
in Discovery Studio was used to complete the side chains for amino
acid residues, correct bonding and bond orders, and add hydrogens
back. Notably, P+FeIV in the sheep COX I
protein [PDB code: 1q4g (32)] and
mouse COX II structure (PDB code: 3nt1 (33)] are already contained in the structure
as complex, and the ion atom is set as FeIV. Lastly, the
Prepare Protein module in Discovery Studio was used for protein
preparation along with the CHARMm force field.
Ligand processing
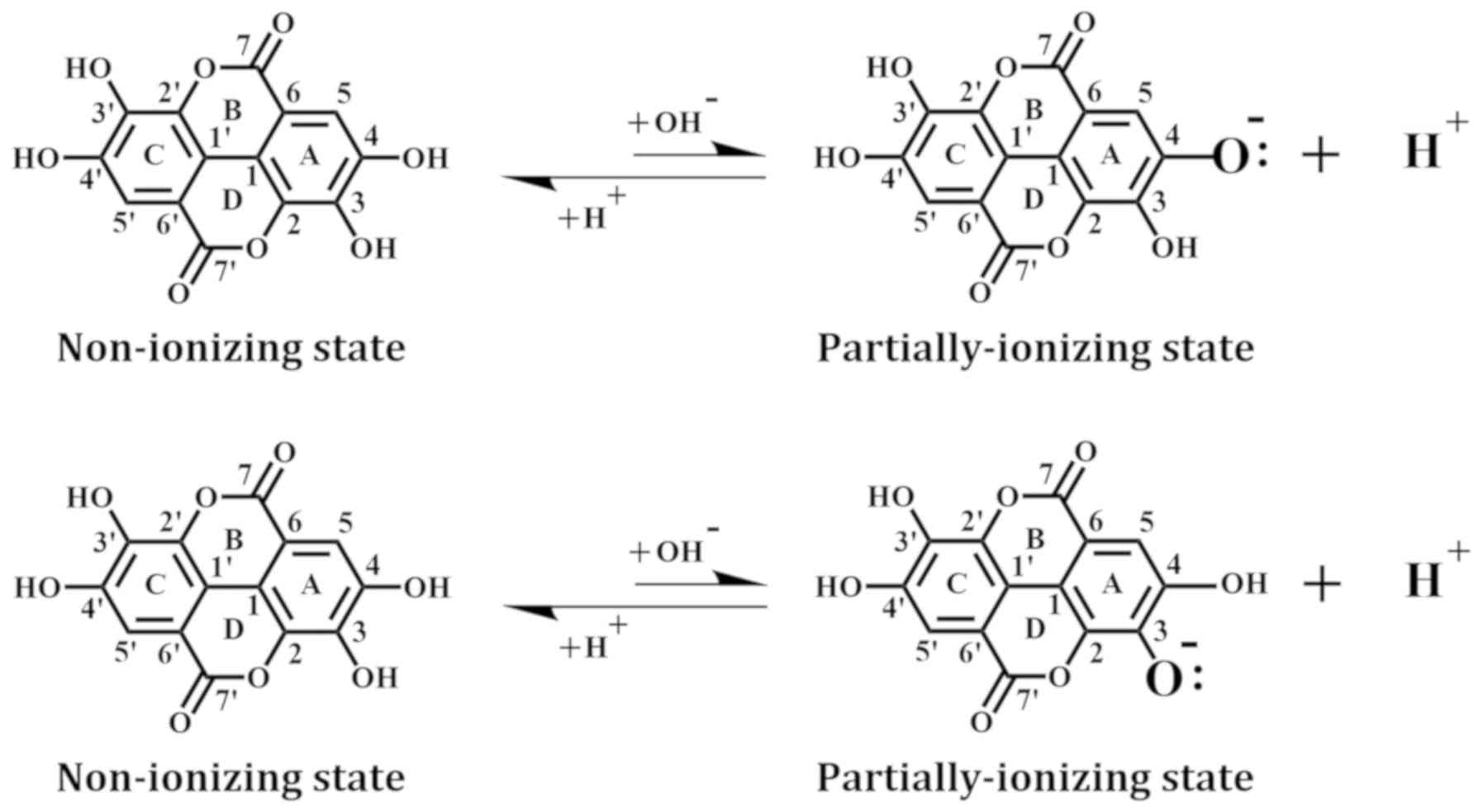
The structure of EA was downloaded from the Protein
Data Bank and minimized with the CHARMm force field. In addition,
we used the Prepare Ligands module to generate EA in a non-ionizing
state and two partially-ionizing states. The non-ionizing state has
all hydrogens of the four phenolic hydroxyl groups retained,
whereas the ionizing states each have one proton removed (i.e.,
deprotonation) from a different hydroxyl group in EA, which include
the C-4-OH in the A-ring (equivalent to the C-4′-OH in the
B-ring) and the C-3-OH in the A-ring (equivalent to
the C-3′-OH in the B-ring) (Fig.
1).
Flexible docking
For flexible docking, we used the Find Sites from
Receptor Cavities module to identify the binding site in the
prepared 1q4g COX I and 3nt1 COX II structures. According to our
earlier study, the target site is the peroxidase active site in
these two COX proteins (31). We
selected all amino acid residues within a 5-Å reach of the target
site and allow them to have flexible side chains. The SBD Site
Sphere is centered at the target site and then expanded to a
13-radius size. Under the Flexible Docking mode with conformation
method set to BEST, the Simulated Annealing docking method was then
applied to dock EA into the target sites of COX I and COX II.
Notably, two flexible docking modes were separately executed for
COX I and II, corresponding to the two different ionizing states of
EA. The whole structure of each COX protein was further minimized
with the CHARMm force field.
Calculation of binding energy
The Calculate Binding Energies module in Discovery
Studio is used to find the complexes with the lowest binding energy
values. According to Discovery Studio, the free energy for the
binding interaction between a protein and its ligand is estimated
according to the free energies of the complex, the protein, and the
ligand. These free energy values are separately calculated using
the CHARMm force field and the generalized Born with smooth
switching (GBSW) method (34). In
this approach, a van der Waals-based surface with a smooth
dielectric boundary is used in the calculation of the
self-electrostatic solvation energy. The ligand conformational
entropy is also considered during the free binding energy
calculation. The following equation is used to calculate the
binding energy (ΔGbinding) between EA and the COX
I or COX II protein: ΔGbinding =
Gcomplex - (GCOX +
Gligand), where Gcomplex is the
absolute free energy of the complex, GCOX is the
absolute free energy of the COX protein, and
Gligand is the absolute free energy of the ligand
(35,36). The ΔGbinding value
is used to reflect the relative interaction affinity between the
COX enzyme and EA.
Statistical analysis. Data were determined as mean ±
SD of triple determinations.
Results
Effect of EA on PGE2
production in vitro and in vivo
In vitro studies
To determine whether EA can modulate PG production
in cultured RAW264.7 cells, the cells were first stimulated with 1
µg/ml LPS for 2 h to induce the expression of COX proteins as well
as PG production (Fig. 2). We found
that LPS pretreatment mostly induced COX II expression in these
cells as confirmed by western blotting (Fig. 2A), which is in agreement with an
earlier report (21). The increased
expression of COX II also correlated with increased production of
PGE2, a representative PG selected for testing in this
study (Fig. 2B).
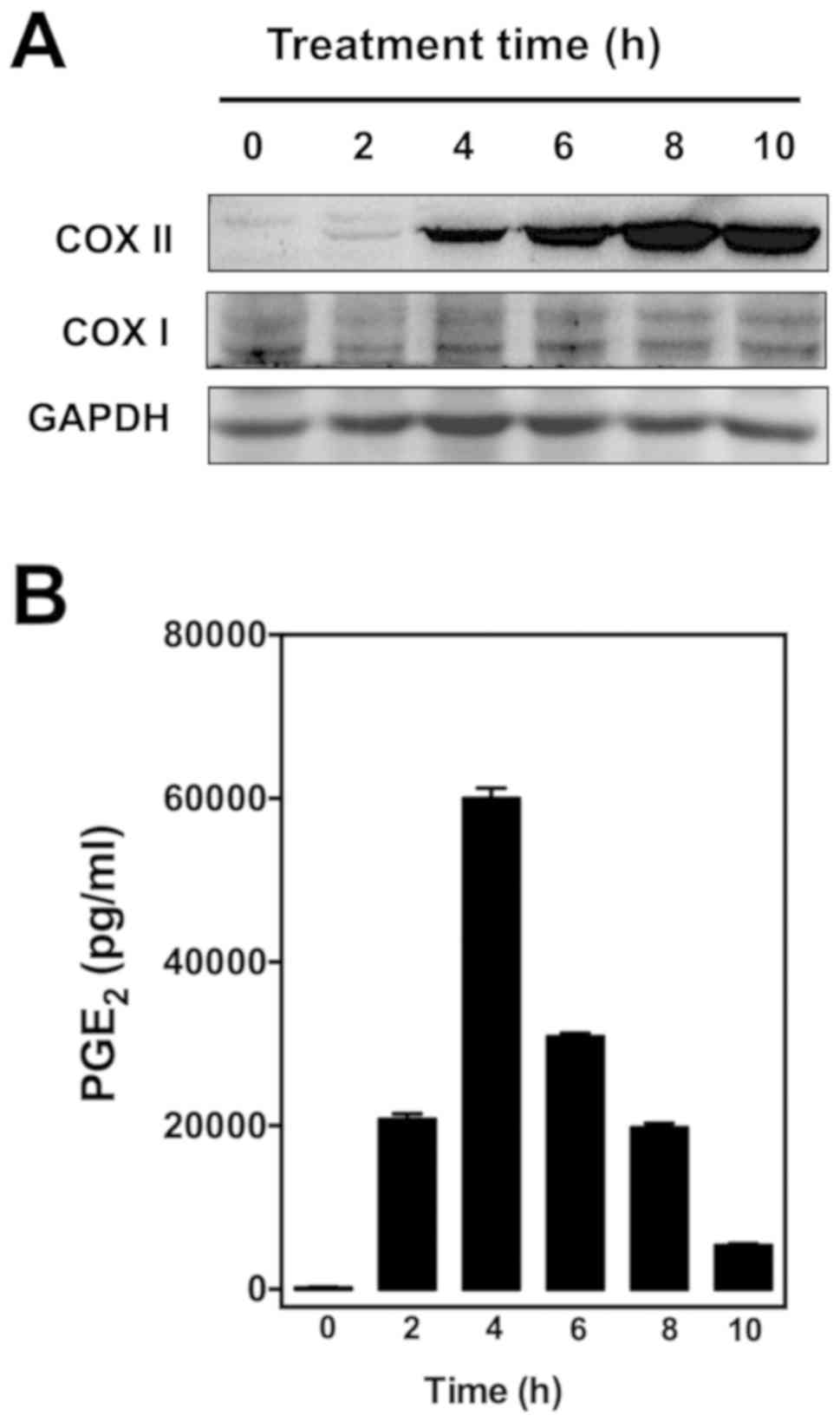 | Figure 2.Effect of different length of
treatment with LPS on (A) COX I/II protein levels in cultured
RAW265.7 cells and (B) PGE2 levels in cell culture media. After
treatment with 1 µg/ml LPS for 0, 2, 4, 6, 8, and 10 h, RAW264.7
cells were incubated with serum-free medium for an additional 2 h,
and the supernatants were collected for measurement of
PGE2 levels by using an enzymatic immunoassay kit.
Western blot analysis of cell lysates was performed with antibodies
COX I or II, coupled with a secondary antibody conjugated with
horseradish peroxidase. LPS, lipopolysaccharide; COX I and COX II,
cyclooxygenase I and II, respectively. |
Using LPS-pretreated RAW264.7 cells as an in
vitro model, we then tested the modulating effect of EA on
PGE2 production. Following LPS pretreatment, the medium
was removed and replaced with 300 µl serum-free DMEM with or
without different concentrations (0.01, 0.1, 1, 10, and 100 µM) of
EA. After an additional 2-h incubation, the culture media were
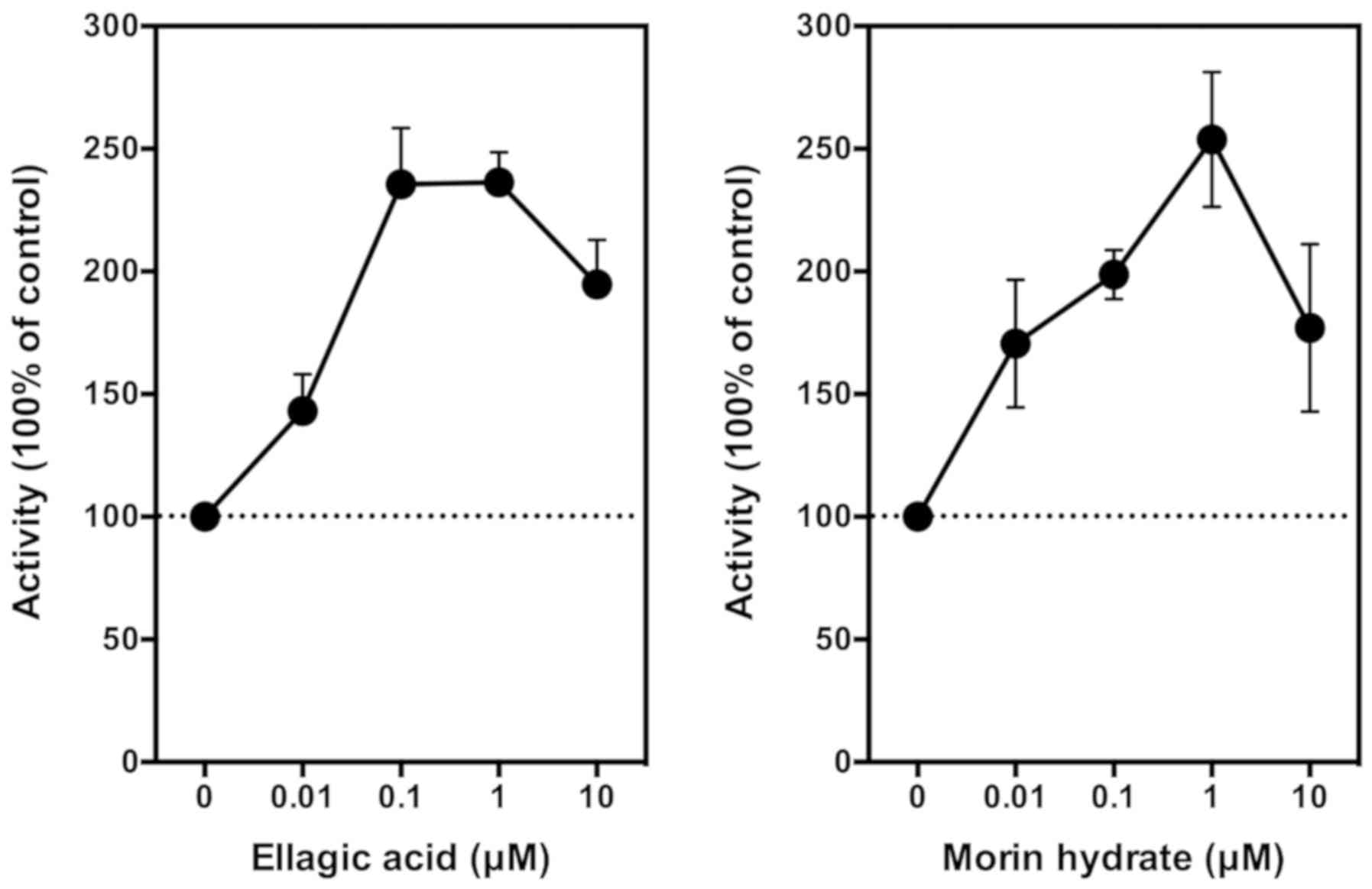
collected for measurement of PGE2. We found that EA at
10 nM showed a weak stimulatory effect on PGE2
production, and this stimulation reached a plateau when the
concentration of EA reached 100–1,000 nM. The maximal stimulation
of COX-mediated PGE2 production by EA was approximately
140% above the control in these cells (Fig. 3, left panel). Notably, when EA
concentration further increased to 10 µM, PGE2
production is slightly reduced. It is noteworthy that this
phenomenon was also observed in our earlier in vitro and
in vivo studies with other reducing co-substrates such as
quercetin and myricetin (29,30). For
comparison, we also included for testing the effect of morin (an
analog of quercetin) on PGE2 production. We found that
morin modulates the production of PGE2 in a similar
manner as EA (Fig. 3, right
panel).
In this study, we confirmed that galangin (a known
competitive inhibitor of the COX peroxidase active site) (37) does not have a significant stimulatory
effect on PGE2 production when it is added alone to
LPS-pretreated RAW264.7 cells in culture. However, when it is added
along with EA, it can inhibit EA-stimulated PGE2
production in a concentration-dependent manner, with an
IC50 value of <1 µM (Fig.
4, left panel). Notably, when galangin was added alone to the
LPS-pretreated RAW264.7 cells, it also inhibited the baseline
production of PGE2 in a similar manner (Fig. 4, right panel).
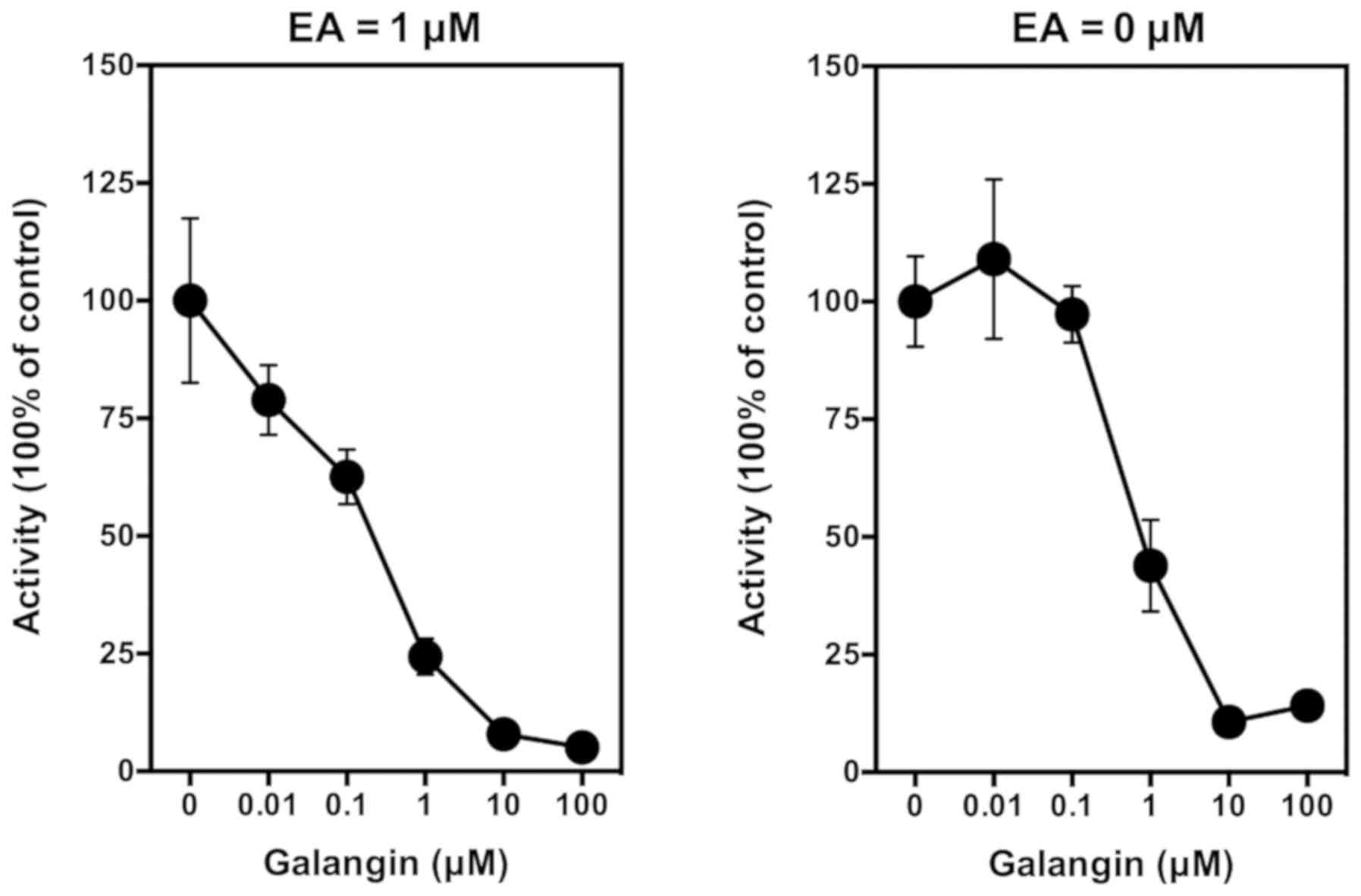 | Figure 4.Effect of galangin on ethyl
gallate-stimulated PGE2 release from LPS-pretreated
RAW264.7 cells. Cells were pretreated with 1 µg/ml LPS for 2 h to
induce COX II expression, and then the culture media were removed
and replaced with 300 µl serum-free medium containing 1 µM ethyl
gallate plus different concentrations (0.01, 0.1, 1 and 10 µM) of
galangin for another 2 h. The levels of PGE2 were
measured using an enzymatic immunoassay kit (Cayman Chemical, Ann
Arbor, MI, USA). Each point was the mean ± SD of triple
determinations. LPS, lipopolysaccharide; COX I and COX II,
cyclooxygenase I and II, respectively; EA, ellagic acid. |
In vivo studies
In the present study, we also determined the effect
of EA on the plasma levels of PGE2 by using normal male
Sprague-Dawley rats as an in vivo model. The reason for use
of this animal model is because it was successfully used earlier to
study the effect of other representative phenolic compounds on
plasma and tissue levels of several PG products (30). We found that administration (oral
route or injection) of these phenolic compounds to normal male rats
can significantly increase the tissue and blood levels of PG
products in vivo (30).
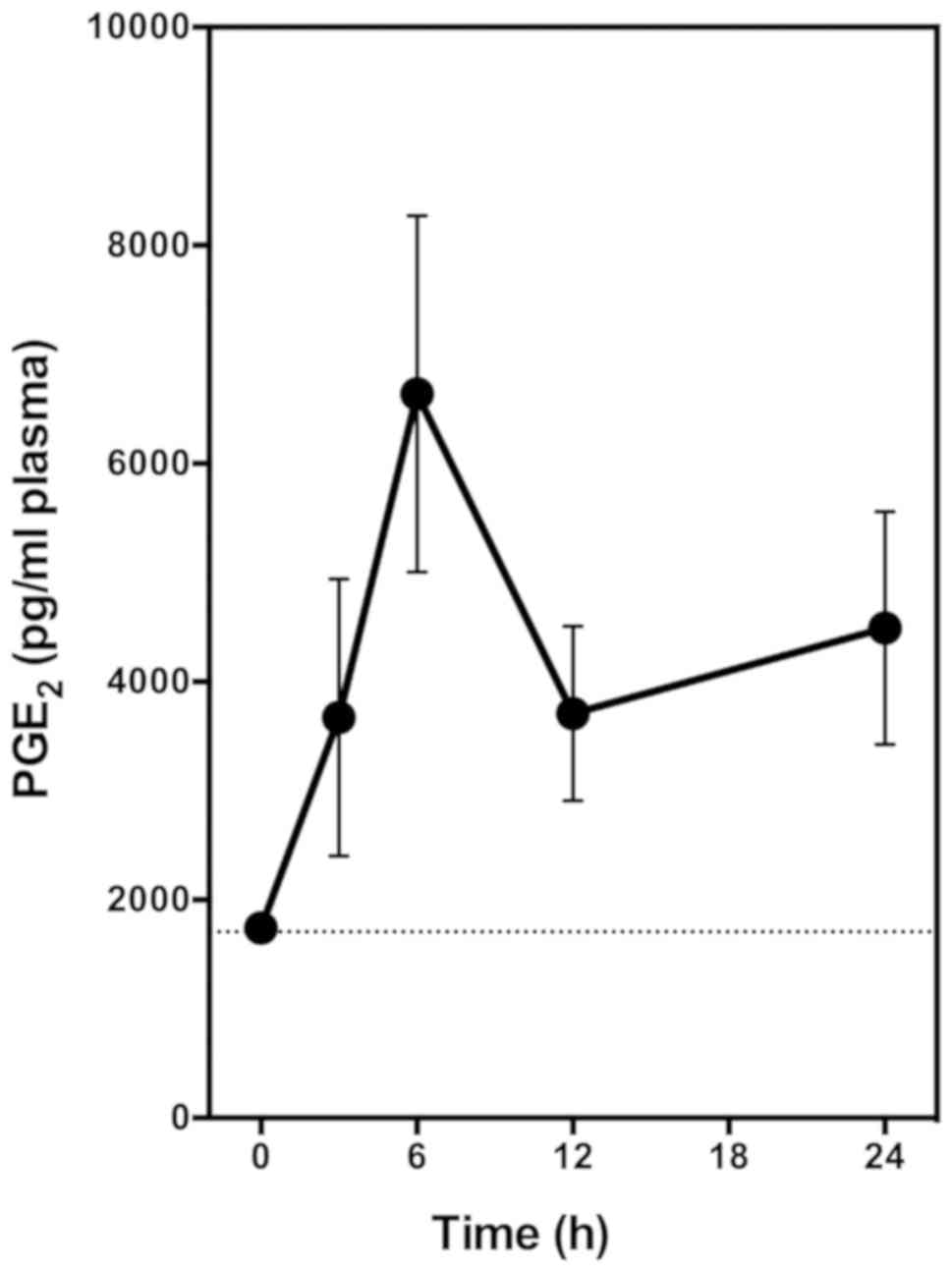
In this experiment, the animals receive a single
oral dose of EA alone (at 6 mg/kg body weight). Blood samples were
collected through tail bleeding at different time points, and
plasma samples were prepared for PGE2 measurement. We
found that oral administration of EA alone markedly increased the
plasma level of PGE2 in a time-dependent manner
(Fig. 5). Plasma PGE2
level started to increase significantly at 3 h after
administration, and peaked at approximately 6 h after
administration, with a maximal increase of the plasma
PGE2 level by approximately 3.5-fold (Fig. 5). This observation is very similar to
the observations made in our earlier study with two other COX
activators (quercetin and myricetin) (30).
In summary, in vitro experiments using
LPS-pretreated RAW264.7 cells and in vivo experiments using
rats both showed that EA is an activator of COX-mediated production
of PGE2. This effect is abrogated by the co-presence of
galangin, an inhibitor of the peroxidase activity of COX,
presumably by blocking the effect of the reduction of
co-substrates.
Computational docking analysis of EA binding inside
the peroxidase active sites of COX I and II
We employed sheep COX I [PDB code: 1q4g (32)] and mouse COX II [PDB code: 3nt1
(33)] proteins as templates to
model the docking interaction between EA and COX I/II. The 3D
structural models of these two proteins were prepared using
Discovery Studio. Using these structural models, we docked EA in
three different ionizing states (one non-ionizing state vs. two
partially-ionizing states) into the peroxidase active sites of COX
I/II (Figs. 6 and 7). The results are summarized below.
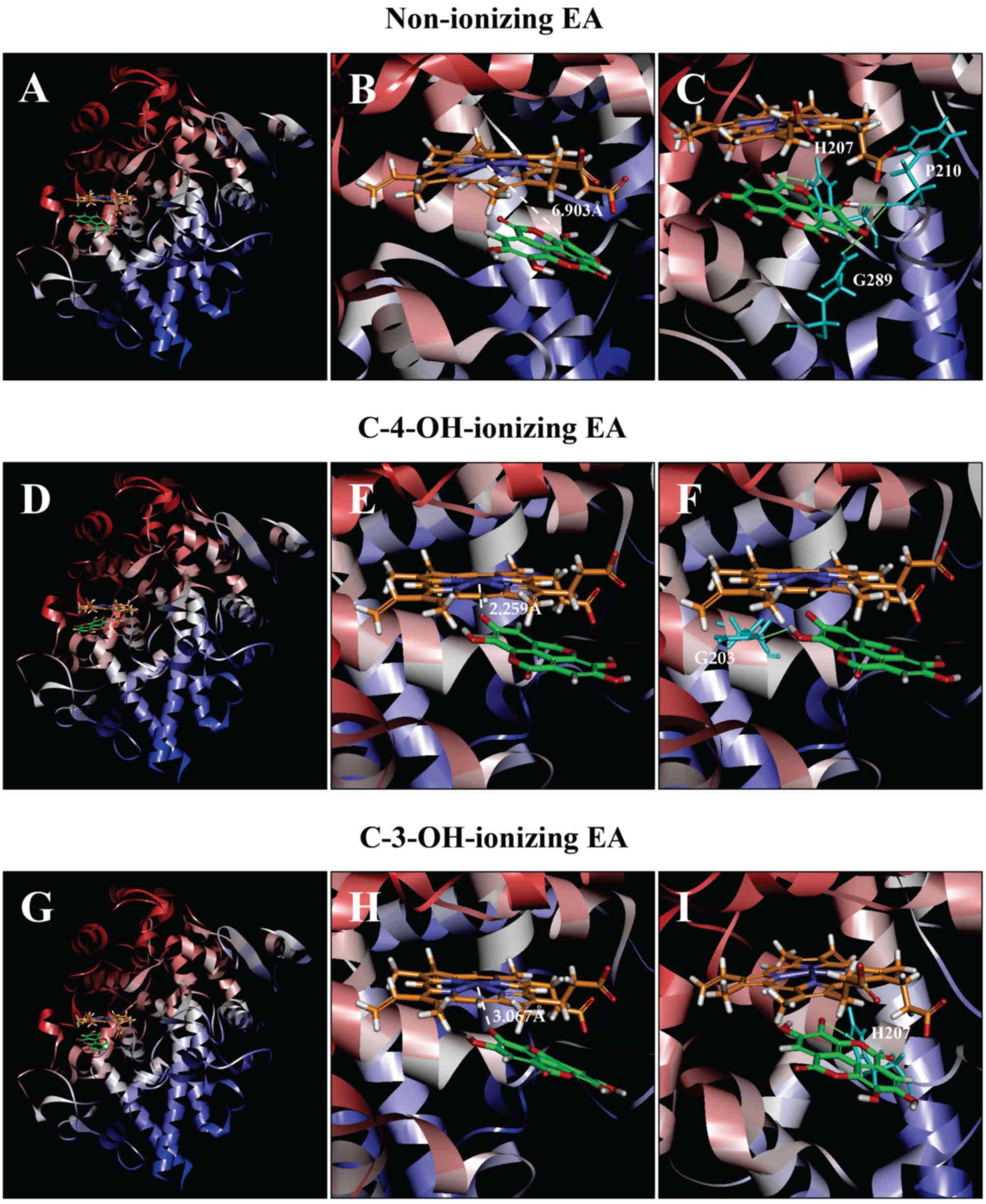 | Figure 6.Molecular docking analysis of the
binding interaction of COX I with non-ionizing EA (A-C),
C4-OH-ionizing EA (D-F), and C-3-OH-ionizing EA (G-I). (A, D and G)
The dominant docking result for non-ionizing EA (A), C4-OH-ionizing
EA (D) and C-3-OH-ionizing EA (G) inside the peroxidase active site
of COX I. The protein structure is shown in a flat ribbon format.
In P+FeIV, carbon is colored in orange,
nitrogen in blue, oxygen in red, hydrogen in white, and iron in
navy blue. In EA, carbon is colored in green, oxygen in red, and
hydrogen in white. (B, E and H) The same structure as in respective
panels (A, D and G) with a white dash line added to indicate the
distance between Fe4+ ion and O in one of EA's OHs. (C,
F and I) Suggested potential hydrogen bonds (green dash lines)
between the amino acid residues and the non-ionizing EA (C),
C-4-OH-ionizing EA (F), and C-3-OH-ionizing EA (I). The amino acid
residues are colored in light blue. EA, ellagic acid; COX I and COX
II, cyclooxygenase I and II, respectively. |
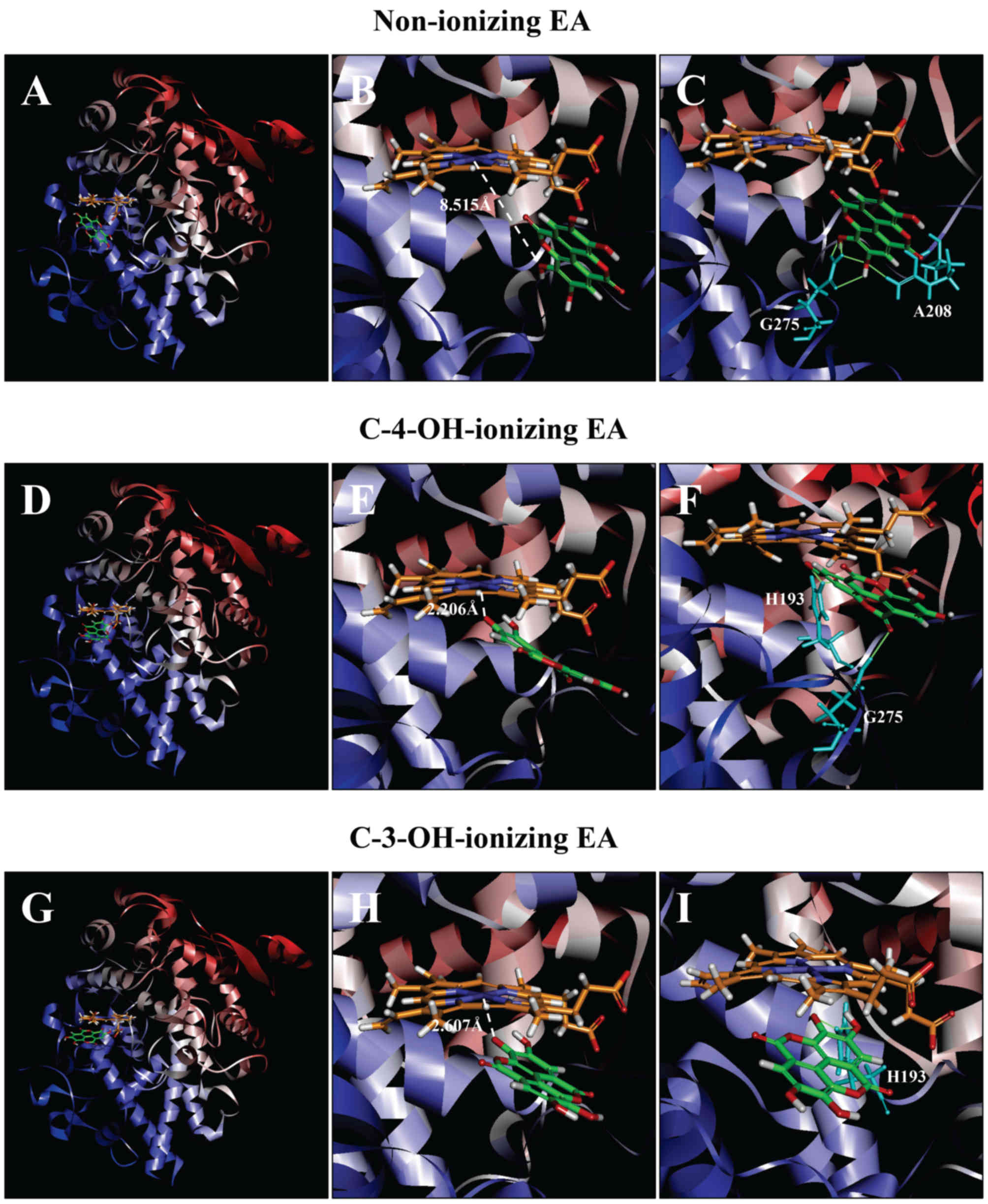 | Figure 7.Molecular docking analysis of the
binding interaction of COX II with non-ionizing EA (A-C),
C4-OH-ionizing EA (D-F), and C-3-OH-ionizing EA (G-I). (A, D and G)
The dominant docking result for non-ionizing EA (A), C4-OH-ionizing
EA (D) and C-3-OH-ionizing EA (G) inside the peroxidase active site
of COX II. The protein structure is shown in a flat ribbon format.
In P+FeIV, carbon is colored in orange,
nitrogen in blue, oxygen in red, hydrogen in white, and iron in
navy blue. In EA, carbon is colored in green, oxygen in red, and
hydrogen in white. (B, E and H) The same structure as in respective
panels (A, D and G) with a white dash line added to indicate the
distance between Fe4+ ion and O in one of EA's OHs. (C,
F and I) Suggested potential hydrogen bonds (green dash lines)
between the amino acid residues and the non-ionizing EA (C),
C-4-OH-ionizing EA (F), and C-3-OH-ionizing EA (I). The amino acid
residues are colored in light blue. EA, ellagic acid; COX I and COX
II, cyclooxygenase I and II, respectively. |
COX I
Docking analysis of EA in a non-ionizing state
suggests that it can bind inside the peroxidase active site in two
possible binding modes: One with its A-ring structure inside
the peroxidase site facing P+FeIV, and the
other one with its B-ring structure inside the peroxidase
site. Based on binding energy ΔGbinding values
(Table I), it is predicted that the
binding mode with its B-ring inside is the dominant binding
pose (ΔGbinding of −3.180 kcal/mol). However, in
this pose, all hydroxyl groups of EA are not too close to the Fe
ion of P+FeIV (Fig. 6A and B), suggesting that this binding
pose is an inactive pose, and would not be able to transfer its
electrons to the Fe ion of P+FeIV for
reduction. For poses ranked 2–10, all hydroxyl groups of EA are
even slightly farther away from the Fe ion of
P+FeIV than the dominant binding pose.
 | Table I.Computed binding energy values
(ΔGbinding, kcal/mol) for the molecular docking
analysis of the best binding poses between EA (partially-ionized
vs. non-ionized) and COX I/II proteins. |
Table I.
Computed binding energy values
(ΔGbinding, kcal/mol) for the molecular docking
analysis of the best binding poses between EA (partially-ionized
vs. non-ionized) and COX I/II proteins.
|
| Binding energy
value ΔGbinding (kcal/mol) |
|---|
|
|
|
|---|
| Type of
protein | No ionization | C-4-OH
ionization | C-3-OH
ionization |
|---|
| COX I protein | −3.180 | −78.952 | −15.470 |
| COX II protein | −3.061 | −76.923 | −47.614 |
Potential hydrogen bonds between COX I and EA in its
dominant binding pose as suggested by the Receptor-Ligand Hydrogen
Bonds module are shown in Fig. 6C,
and they involve five amino acid residues: two with His207 (1.314
and 2.289 Å), two with Phe210 (1.928 and 1.949 Å), and one with
Gln289 (2.219 Å).
It is estimated that under physiological conditions,
a small fraction of the hydroxyl groups in EA's A-ring would
undergo ionization (deprotonation), i.e., removal of a proton.
Results from our recent study (38)
suggest that the binding interaction of a reducing substrate (such
as quercetin) under partial ionization is dramatically enhanced in
comparison with the non-ionizing state. Therefore, we also
performed docking analysis using the partially-ionizing EA.
Predicted by the Discovery Studio, C-4-OH has a higher tendency to
deprotonate than C-3-OH under physiological conditions. In the
present study, we chose to determine the docking conformation when
deprotonation occurs only with one hydroxyl group at any given
moment, because simultaneous deprotonation of multiple protons in
the same molecule is considered nearly impossible to occur under
physiological pH conditions.
We found that when each of the hydroxyl groups in
A-ring is individually deprotonated, the dominant poses
(based on ΔGbinding values; Table I) all have its A-ring inside
(Fig. 6D and G). Under C-4-OH
deprotonation (Fig. 6E;
ΔGbinding of −78.7952 kcal/mol), the distance
between Fe and O− is 2.259 Å. Under C-3-OH deprotonation
(Fig. 6H;
ΔGbinding of −15.470 kcal/mol), the distance
between the Fe ion and O− is 3.067 Å.
Potential hydrogen bonds between COX I and EA in two
ionizing states as suggested by the receptor-ligand hydrogen bonds
are shown in Fig. 6F and I. Under
C-4-OH deprotonation, EA in its dominant binding pose only forms
one hydrogen bond with Gln203 (2.297 Å), and under C-3-OH
deprotonation, it forms two hydrogen bonds with His207 (1.494 and
1.786 Å).
COX II
As predicted according to the binding energy
ΔGbinding value (ΔGbinding of
−3.061 kcal/mol), the dominant binding pose of EA in a non-ionizing
state has its B-ring inside COX II's peroxidase site
(Fig. 7A). All hydroxyl groups in
this pose are very far away from the Fe ion of
P+FeIV (shortest distance at 8.515 Å;
Fig. 7B). For poses ranked 2–10, the
hydroxyl groups of EA are even slightly farther away from the Fe
ion of P+FeIV than the dominant pose.
We also analyzed the docking conformations when
deprotonation occurs individually with EA's C-4-OH and C-3-OH
hydroxyl groups (data shown in Fig. 7D
and E, respectively). We found that when C-4-OH is
deprotonated, the dominant pose has its A-ring closer to the
Fe ion of P+FeIV (ΔGbinding
of −76.923 kcal/mol), particularly the O− ion in EAs
C-4-OH (distance of 2.206 Å; Fig.
7G). Under C-3-OH deprotonation, the dominant pose (Fig. 7H) also has its A-ring inside
(ΔGbinding of −47.614 kcal/mol), with the
distance of 2.607 Å between Fe ion and O- ion in EA's C-3-OH. The
suggested hydrogen bonds in the dominant poses in three different
states are shown in Fig. 7C, F and
I.
Discussion
The results from both in vitro and in
vivo experiments in this study showed to the best of our
knowledge, for the first time, that EA is an activator of the COX
enzyme-catalyzed production of PGE2. Mechanistically, EA
likely exerts this effect through activating the peroxidase active
site of COX enzymes by serving as a reducing co-substrate for the
Fe ion of P+FeIV. The effect of EA is very
similar to the effect of some naturally-occurring flavonoids, such
as quercetin and myricetin, that were reported earlier by our
group, which can activate the catalytic activity of the COX I and
II enzymes by functioning as reducing co-substrates (29).
Our earlier study revealed that the hydroxyl groups
of the B-ring of quercetin play a critical role in
re-activating the COX I/II catalytic activity (29,31).
Recently, we have further shown that galangin, a flavonoid that has
the same A/C-ring structure as quercetin but does not have
any hydroxyl group in its B-ring, can function as a COX
inhibitor, by competitively blocking the binding of those
flavonoids that can serve as reducing co-substrates for the COX
enzymes (37). In this study, we
confirmed that galangin does not have a stimulatory effect on
PGE2 production when it is added to LPS-pretreated
RAW264.7 cells in culture, but it can inhibit EA-stimulated
PGE2 production in a concentration-dependent manner,
with an IC50 value of <1 µM (Fig. 4, left panel). Notably, when galangin
is added alone to the LPS-pretreated RAW264.7 cells in culture, it
also inhibits the baseline production of PGE2 in a
similar manner (Fig. 4, right
panel). This phenomenon was also observed in our recent study
(37), which likely is due to the
presence of other reducing substrates either indigenously produced
by the cells or contained in the cell culture medium, and these
compounds can support the basal COX activity as detected in
cultured cells. In support of this explanation, we observed earlier
that when galangin is tested in the in vitro biochemical
enzyme assays involving COX I and II proteins where no other
unknown chemicals are introduced, it does not have any meaningful
stimulatory or inhibitory effect (37). The observed modulating effect of
galangin on EA mirrors the effect of galangin on quercetin-induced
PG production as observed in our recent study (37), providing support for the concept that
EA has a similar mechanism of action as quercetin.
Computational docking analysis provides insight into
the mechanism of the COX-activating action of EA at the molecular
level. Comparison of EA in both non-ionizing and partially-ionizing
states indicates that ionization of C-4-OH and C-3-OH shortens the
distance between Fe4+ and the respective O- (from 6.903
to 2.259 and 3.067 Å, respectively) and increases the binding
infinity (from −3.180 to −78.952 and −15.470 kcal/mol,
respectively). These data suggest that deprotonation would
facilitate the transfer of electron from EA to
P+FeIV for peroxidase reduction. In addition,
when deprotonation of C-4-OH and C-3-OH is compared, the former
shows a shorter distance and higher binding infinity than the
latter, suggesting that C-4-OH can more readily transfer its
electron to P+FeIV than C-3-OH.
When a partially-ionizing EA is bound inside the
peroxidase active site, its estimated shortest distance is 2.259 Å,
and this interaction distance is expected to enable a facile
transfer of an electron from its hydroxyl group to
P+FeIV. Notably, while the best binding poses
of ionizing EA in COX I and II proteins are very different, the
distances between the Fe ion and oxygen ion are very similar, as
are their overall binding energy values. This observation provides
additional support for the suggestion that ionic interaction
between Fe ion and the respective O− is the dominant
force that determines the binding energy level and binding
affinity.
It appears that the number of suggested hydrogen
bonds does not correlate with the overall binding energy values.
The reasons for this apparent discrepancy might be: First, the
strong ionic interaction between ionized EA (which contains a
negatively-charged O− ion) and the positively-charged Fe
ion plays a more important role than hydrogen bonds (39). This suggestion is consistent with the
fact that hydrogen bonds are far weaker than the ionic
interactions. Second, some of the suggested hydrogen bonds may be
of negligible significance in strength due to their rather long
bond distance.
Earlier studies have shown that EA is richly present
in Longan and Litchi (Lychee) at high
concentrations (4–9,14).
Longan and Litchi are members of the soapberry family
(Sapindaceae). These plants are grown extensively in China and
South East Asia, as well as in Australia, Florida (USA), southern
Europe, and southern Africa (40,41). In
traditional Chinese medicine, Longan and Litchi are
fruits with health beneficial functions, but they are also best
known for their ‘hot’ properties when overdosed, i.e.,
overingestion of Longan and Litchi is known to
promote inflammatory-type responses. In recent years, there are
increasing reports in Southeast Asia regions of Litchi-associated
acute encephalitis syndrome among children (42–45).
However, the mechanism for some of their beneficial as well as
their pro-inflammatory effects is poorly understood at present. The
results of the present study showed that EA, a natural phenolic
compound richly contained in Longan and Litchi, can
stimulate the catalytic activity of the COX I and II enzymes in
vitro by functioning as a reducing co-substrate for these
enzymes. This unique effect may help partially account for some of
the beneficial as well as pro-inflammatory effects of Litchi
and Longan.
Lastly, it is noteworthy that most of the
pharmacological design and strategies aim to inhibit the COX I/II
enzymes, because of their well-known roles in some of the
pathogenic processes. However, it is of note that abnormally-low
levels of COX I activity are also associated with some serious
pathogenic conditions, such as gastrointestinal ulceration and
bleeding and cardiovascular diseases (28,46–48).
Thus, too low basal levels of the COX activity (particularly COX I)
are not beneficial for optimal health. Our finding that some of the
natural phenolics can be used in the body as reducing co-substrates
of COX enzymes to support their normal catalytic activity for
biosynthesis of PG-related mediators may offer a new mechanistic
explanation for some of their health beneficial functions in the
body.
In summary, both in vitro and in vivo
experiments showed that EA is an activator of PGE2
production. Mechanistically, it is suggested that EA can activate
the peroxidase active site of COX enzymes by serving as a reducing
co-substrate for the reduction of P+FeIV in
the catalytic site. The effect of EA is abrogated by the
co-presence of galangin, which is known to bind to COX's peroxidase
active site and thereby blocks the effect of the reducing
co-substrates.
Acknowledgements
Not applicable
Funding
The present study was supported by research grants
from the National Natural Science Foundation of China (NSFC nos.
81473224 and 81630096), Shenzhen City Basic Science Project (no.
JCYJ20140714151402768), and Shenzhen Peacock Plan (no.
KQTD2016053117035204).
Availability of data and materials
All data generated or analyzed during this study
are included in this published article.
Authors' contributions
HRW conducted the cell culture and animal
experiments, analyzed the data, and prepared part of the initial
draft of the manuscript; HCS performed the computational analysis,
analyzed the data, and prepared part of the initial draft of the
manuscript; BTZ had the initial ideas and designed all the
experiments, analyzed the data, and prepared and finalized the
manuscript.
Ethics approval and consent to
participate
All procedures involving the use of live animals as
described in the present study were approved by the Institutional
Animal Care and Use Committee of the Southern University of Science
and Technology (approval number: SUSTC-G-2014009), and the
guidelines for the humane treatment of animals accepted by the
National Institutes of Health (USA) were followed.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests..
Glossary
Abbreviations
Abbreviations:
|
EA
|
ellagic acid
|
|
COX I and COX II
|
cyclooxygenase I and II
|
|
AA
|
arachidonic acid
|
|
PG
|
prostaglandin
|
|
P+FeIV
|
protoporphorin IX with
FeIV inside
|
References
|
1
|
Mämmelä P, Savolainen H, Lindroos L,
Kangas J and Vartiainen T: Analysis of oak tannins by liquid
chromatography-electrospray ionisation mass spectrometry. J
Chromatogr A. 891:75–83. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nierenstein M: The formation of ellagic
acid from galloyl-glycine by penicillium. Biochem J. 9:240–244.
1915. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nakai S: Myriophyllum
spicatum-released allelopathic polyphenols inhibiting growth of
blue-green algae Microcystis aeruginosa. Water Res.
34:3026–3032. 2000. View Article : Google Scholar
|
|
4
|
Prasad KN, Yang B, Yang S, Chen Y, Zhao M,
Ashraf M and Jiang Y: Identification of phenolic compounds and
appraisal of antioxidant and antityrosinase activities from litchi
(Litchi sinensis Sonn.) seeds. Food Chem. 116:1–7. 2009.
View Article : Google Scholar
|
|
5
|
Estela de Rezende Q, Patto de Abreu CM,
Kelly da Silva O, Vinicius de Oliveira R and Fráguas RM: Bioactive
phytochemicals and antioxidant activity in fresh and dried lychee
fractions. Rev Ciênc Agron. 46:163–169. 2015. View Article : Google Scholar
|
|
6
|
Soong YY and Barlow PJ: Isolation and
structure elucidation of phenolic compounds from longan
(Dimocarpus longan Lour.) seed by high-performance liquid
chromatography-electrospray ionization mass spectrometry. J
Chromatogr A. 1085:270–277. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zheng G, Xu L, Wu P, Xie H, Jiang Y, Chen
F and Wei X: Polyphenols from longan seeds and their
radical-scavenging activity. Food Chem. 116:433–436. 2009.
View Article : Google Scholar
|
|
8
|
Rangkadilok N, Sitthimonchai S,
Worasuttayangkurn L, Mahidol C, Ruchirawat M and Satayavivad J:
Evaluation of free radical scavenging and antityrosinase activities
of standardized longan fruit extract. Food Chem Toxicol.
45:328–336. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tseng HC, Wu WT, Huang HS and Wu MC:
Antimicrobial activities of various fractions of longan
(Dimocarpus longan Lour. Fen Ke) seed extract. Int J Food
Sci Nutr. 65:589–593. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vattem DA and Shetty K: Biological
function of ellagic acid: A review. J Food Biochem. 29:234–266.
2005. View Article : Google Scholar
|
|
11
|
Usta C, Ozdemir S, Schiariti M and Puddu
PE: The pharmacological use of ellagic acid-rich pomegranate fruit.
Int J Food Sci Nutr. 64:907–913. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Infante R, Contador L, Rubio P, Aros D and
Peña-Neira Á: Postharvest sensory and phenolic characterization of
‘Elegant Lady’ and ‘Carson’ peaches. Chil J Agric Res. 71:445–451.
2011. View Article : Google Scholar
|
|
13
|
Seeram NP, Adams LS, Henning SM, Niu Y,
Zhang Y, Nair MG and Heber D: In vitro antiproliferative, apoptotic
and antioxidant activities of punicalagin, ellagic acid and a total
pomegranate tannin extract are enhanced in combination with other
polyphenols as found in pomegranate juice. J Nutr Biochem.
16:360–367. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Vattem DA and Shetty K: Biological
function of ellagic acid: A Review. J Food Biochem. 29:234–266.
2005. View Article : Google Scholar
|
|
15
|
Emanuele S, Lauricella M, Calvaruso G,
D'Anneo A and Giuliano M: Litchi chinensis as a functional
food and a source of antitumor compounds: An overview and a
description of biochemical pathways. Nutrients. 9:E9922017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Narayanan BA, Geoffroy O, Willingham MC,
Re GG and Nixon DW: p53/p21(WAF1/CIP1) expression and its possible
role in G1 arrest and apoptosis in ellagic acid treated cancer
cells. Cancer Lett. 136:215–221. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mandal S, Shivapurkar NM, Galati AJ and
Stoner GD: Inhibition of N-nitrosobenzylmethylamine metabolism and
DNA binding in cultured rat esophagus by ellagic acid.
Carcinogenesis. 9:1313–1316. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mandal S and Stoner GD: Inhibition of
N-nitrosobenzylmethylamine-induced esophageal tumorigenesis in rats
by ellagic acid. Carcinogenesis. 11:55–61. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Teel RW, Babcock MS, Dixit R and Stoner
GD: Ellagic acid toxicity and interaction with benzo[a]pyrene and
benzo[a]pyrene 7,8-dihydrodiol in human bronchial epithelial cells.
Cell Biol Toxicol. 2:53–62. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Casewatch, . https://www.casewatch.net/fdawarning/prod/2008/best_on_earth.shtmlDecember
2–2018
|
|
21
|
Marnett LJ: Cyclooxygenase mechanisms.
Curr Opin Chem Biol. 4:545–552. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Williams CS, Mann M and DuBois RN: The
role of cyclooxygenases in inflammation, cancer, and development.
Oncogene. 18:7908–7916. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fitzpatrick FA: Cyclooxygenase enzymes:
Regulation and function. Curr Pharm Des. 10:577–588. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mitchell JA and Kirkby NS: Eicosanoids,
prostacyclin and cyclooxygenase in the cardiovascular system. Br J
Pharmacol. Feb 21–2018.(Epub ahead of print).
|
|
25
|
Duggan KC, Walters MJ, Musee J, Harp JM,
Kiefer JR, Oates JA and Marnett LJ: Molecular basis for
cyclooxygenase inhibition by the non-steroidal anti-inflammatory
drug naproxen. J Biol Chem. 285:34950–34959. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Blobaum AL and Marnett LJ: Structural and
functional basis of cyclooxygenase inhibition. J Med Chem.
50:1425–1441. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Marnett LJ, Rowlinson SW, Goodwin DC,
Kalgutkar AS and Lanzo CA: Arachidonic acid oxygenation by COX-1
and COX-2. Mechanisms of catalysis and inhibition. J Biol Chem.
274:22903–22906. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kurumbail RG, Kiefer JR and Marnett LJ:
Cyclooxygenase enzymes: Catalysis and inhibition. Curr Opin Struct
Biol. 11:752–760. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bai HW and Zhu BT: Strong activation of
cyclooxygenase I and II catalytic activity by dietary
bioflavonoids. J Lipid Res. 49:2557–2570. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bai HW and Zhu BT: Myricetin and quercetin
are naturally occurring co-substrates of cyclooxygenases in vivo.
Prostaglandins Leukot Essent Fatty Acids. 82:45–50. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang P, Bai HW and Zhu BT: Structural
basis for certain naturally occurring bioflavonoids to function as
reducing co-substrates of cyclooxygenase I and II. PloS One 2010,.
5:e123162010. View Article : Google Scholar
|
|
32
|
Gupta K, Selinsky BS, Kaub CJ, Katz AK and
Loll PJ: The 2.0 Å resolution crystal structure of prostaglandin
H2 synthase-1: Structural insights into an unusual
peroxidase. J Mol Biol. 335:503–518. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Duggan KC, Walters MJ, Musee J, Harp JM,
Kiefer JR, Oates JA and Marnett LM: Molecular basis for
cyclooxygenase inhibition by the non-steroidal anti-inflammatory
drug naproxen. J Biol Chem. 285:34950–34959. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Im W, Lee MS and Brooks CL III:
Generalized born model with a simple smoothing function. J Comput
Chem. 24:1691–1702. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Uciechowska U, Schemies J, Scharfe M,
Lawson M, Wichapong K, Jung M and Sippl W: Binding free energy
calculations and biological testing of novel thiobarbiturates as
inhibitors of the human NAD+ dependent histone
deacetylase Sirt2. Med Chem Comm. 3:167–173. 2012. View Article : Google Scholar
|
|
36
|
Pouplana R, Lozano JJ and Ruiz J:
Molecular modelling of the differential interaction between several
non-steroidal anti-inflammatory drugs and human prostaglandin
endoperoxide H synthase-2 (h-PGHS-2). J Mol Graph Model.
20:329–343. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhu BT, Bai HW, Rao S and Sui HC: Galangin
inhibits cyclooxygenase by blocking the function of the reducing
cosubstrate at the peroxidase site. FASEB J. submitted. 2018.
|
|
38
|
Sui HC and Zhu BT: Catalytic mechanism of
the peroxidase activity of human cyclooxygenase and the role of
phenol as a reducing co-substrate. Sci Rep. submitted. 2018.
|
|
39
|
Anslyn EV and Dougherty DA: Modern
Physical Organic Chemistry. University Science; Sausalito, CA:
2004
|
|
40
|
Chen H: The production and uses of litchis
in China. http://ir4.rutgers.edu/GMUS/presentation%20pdf/day1Chen.pdfApril.
2018
|
|
41
|
Menzel CM and Waite GK: Litchi and Longan.
Botany, Production and Uses. CABI Publishing; Wallingford, UK:
2005
|
|
42
|
Spencer PS and Palmer VS: The enigma of
litchi toxicity: An emerging health concern in southern Asia.
Lancet Glob Health. 5:e383–e384. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Paireau J, Tuan NH, Lefrançois R,
Buckwalter MR, Nghia ND, Hien NT, Lortholary O, Poirée S,
Manuguerra JC, Gessain A, et al: Litchi-associated acute
encephalitis in children, Northern Vietnam, 2004–2009. Emerg Infect
Dis. 18:1817–1824. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Shrivastava A, Kumar A, Thomas JD,
Laserson KF, Bhushan G, Carter MD, Chhabra M, Mittal V, Khare S,
Sejvar JJ, et al: Association of acute toxic encephalopathy with
litchi consumption in an outbreak in Muzaffarpur, India, 2014: A
case-control study. Lancet Glob Health. 5:e458–e466. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Islam MS, Sharif AR, Sazzad HMS, Khan
AKMD, Hasan M, Akter S, Rahman M, Luby SP, Heffelfinger JD and
Gurley ES: Outbreak of sudden death with acute encephalitis
syndrome among children associated with exposure to lychee orchards
in Northern Bangladesh, 2012. Am J Trop Med Hyg. 97:949–957. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Krumholz HM, Ross JS, Presler AH and
Egilman DS: What have we learnt from Vioxx? BMJ. 334:120–123. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
McGettigan P and Henry D: Cardiovascular
risk and inhibition of cyclooxygenase: A systematic review of the
observational studies of selective and nonselective inhibitors of
cyclooxygenase 2. JAMA. 296:1633–1644. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
White WB: Cardiovascular risk,
hypertension, and NSAIDs. Curr Rheumatol Rep. 9:36–43. 2007.
View Article : Google Scholar : PubMed/NCBI
|