Introduction
Glioblastoma multiforme is the most aggressive
primary brain cancer in adults worldwide (1). Following meningioma, glioblastoma
multiforme is the second most common cancer of the central nervous
system, globally. Glioblastoma muliforme incidence is approximately
3/100,000 per year (1). After
diagnosis, the average survival time is approximately 12–15 months
and <5% of patients survive longer than 5 years, with survival
time being 3 months for patients who receive no medical treatment
(2). Therefore, it is important to
assess the mechanisms of cell viability and invasion ability in
glioblastoma multiforme.
Glial cell line-derived neurotrophic factor (GDNF)
promotes the longevity of dopamine neurons in the midbrain, spinal
motor neurons, peripheral sensory neurons and noradrenergic neurons
(3–6). A previous study demonstrated that GDNF
regulates neuronal phenotypes, the branching of neurites and
synaptic plasticity (7). The
opposite strand to the GDNF gene has been revealed to be associated
with the transcription of cis-antisense GDNF opposite strand
(GDNFOS) gene and four exons that are located in the GDNFOS gene
are spliced into various isoforms (8). GDNFOS1 and GDNFOS2 encode long
noncoding (lnc) RNAs, while GDNFOS3 encodes a protein of 105 amino
acids (8). GDNFOS1 has been reported
to overlap with GDNF transcription, whereas GDNFOS2 does not
(8). Lnc RNAs regulate the
homeostasis of a variety of cell and tissue types and may affect
signaling pathways and gene expression (9). Glioblastoma multiforme microglia
attraction has been demonstrated to be mediated by GDNF (10). GDNF has also been revealed to be
associated with the bone morphogenetic protein 4 (BMP4)-mediated
reversal of multi-drug resistance in glioblastoma multiforme
(11). The dysregulation of human
GDNF and GDNFOS has been revealed to be associated with the
pathogenesis of brain disease. For example, GDNFOS1 was
demonstrated to regulate middle temporal gyrus in Alzheimer's
disease (8). However, since the
GDNFOS gene was only discovered in 2011 (8), evidence of the association between
GDNFOS and a variety of cancer types is scarce and the role of
GDNFOS in glioblastoma is undetermined. The effects of GDNFOS1 on
GDNF expression in glioblastoma cells remain to be elucidated.
The current study aimed to assess the effects of
GDNFOS1 overexpression and interference on GDNF expression, cell
viability and invasion ability in U87 and U251 MG glioblastoma
cells.
Materials and methods
Cells and reagents
Lipofectamine 2000®, SYBR qPCR mix kit,
TRIzol reagent, SuperScript III reverse transcriptase, cDNA
synthesis kit and packaging mix were obtained from Invitrogen
(Thermo Fisher Scientific, Inc.). EcoRI/BamHI, T4 DNA
ligase and Opti-Minimum Essential Medium (Opti-MEM) were from
Thermo Fisher Scientific, Inc. PBS was supplied by Sigma-Aldrich
(Merck KGaA). CFX96 Touch™ Real-Time PCR Detection system was
supplied by (Bio-Rad Laboratories, Inc., and lysis buffer (cat. no.
P0013), protease and phosphatase inhibitors (cat. no. P1045),
enhanced chemiluminescence solution (cat. no. P0018A),
polyvinylidene fluoride (PVDF) membranes (cat. no. FFP24), BCA kit
(cat. no. P0012) and film (cat. no. FF057) were all supplied by
Beyotime Institute of Biotechnology. Primary antibody against GDNF
(cat. no. ab176564) and horse-radish peroxidase (HRP)-conjugated
goat anti-rabbit secondary antibody (cat. no. ab205718) were
supplied by Abcam. Transwell inserts (8-µm pore size; cat. no.
3422) were supplied by Corning, Inc.
Glioblastoma of unknown origin U87 (HTB-14; American
Type Culture Collection) and U251 MG cells (cat. no. 09063001;
European Collection of Authenticated Cell Cultures) were cultured
in DMEM (Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
(cat. no. SH30084.3; Hyclone; GE Healthcare) at 37°C in a
humidified atmosphere of 5% CO2. U87 cells
(2×105 cells/well; seeded into six-well plates) were
transduced with lentiviral particles at a multiplicity of infection
(MOI) of 5 (12). U251 cells
(2×105 cells/well; seeded into six-well plates) were
transduced at a MOI of 10 (13).
Cells were selected with 2 µg/ml blasticidin (Invitrogen; Thermo
Fisher Scientific, Inc.) and stable clones were subsequently
generated.
Construction of recombinant
plasmids
The lncRNA GDNFOS1 target sequence (600 bp; NCBI
accession number, JF824130.1) was synthesized. Specific primers
which included target sequences with enzyme excision sites were
designed and the sequences were as follows: Forward,
5′-AATTAAGGAAGCTAGAGCGCCGGGCTTTCC-3′ and reverse,
5′-TAAACCCAAGGCGCGGGCTAGCCACAAGATTTTTGCAC-3′. PCR was used to
amplify the target sequence, which was excised with restriction
enzymes NheI and AscI. Synthesized GDNFOS1 DNA was
amplified via PCR using KOD plus DNA polymerase (Toyobo Life
Science). The thermocycling conditions were as follows: 95°C for 30
sec, 60°C for 60 sec and 72°C for 60 sec (for 30 cycles). Vector
PDS159_pL6.3-CMV-GFPa1-IRES-MCS (Novobio Scientific, Inc.) was
excised with NheI and AscI and the products were
collected using a gel extraction kit (cat. no. AP-GX-50; Corning,
Inc.) according the manufacturer's protocol. The excised target
sequence and vector were ligated by T4 DNA ligase at 16°C for 4 h.
The product was transformed into DH5 α-cells (Beijing Transgen
Biotech Co., Ltd.). Positive clones were sequenced and used to
prepare the plasmids. A lncRNA GDNFOS1 overexpression vector with
cytomegalovirus promoter (Novobio Scientific, Inc.) was constructed
and named pL6.3-CMV-GFPa1-IRES-GDNFOS1. In addition, according to
the sequences of lncRNA GDNFOS1, three pairs of oligo sequences
were designed, respectively (Table
I). After annealing, double stranded DNA was inserted into the
lentivirus vector PDS019_pL_shRNA_F (BsmBI enzyme excision
site; Novobio Scientific, Inc.) to create the lncRNA GDNFOS1
interference vector. The sequence was confirmed via Chromas
software (Chromas v2.31; Technelysium Pty Ltd.). The interference
vectors were named pL-shRNA-GDNFOS1-9, pL-shRNA-GDNFOS1-49 and
pL-shRNA-GDNFOS1-248. Two vectors were combined and named
pL-shRNA-GDNFOS1-9+49, pL-shRNA-GDNFOS1-9+248, and
pL-shRNA-GDNFOS1-49+248. A scrambled vector was also used with a
sequence of 5′-TGAGACGAAGCTTCGTCTCGT-3′. The software utilized for
constructing the shRNA sequences was BLOCK-iT™ RNAi Designer
(Thermo Fisher Scientific, Inc.; rnaidesigner.thermofisher.com/rnaiexpress).
 | Table I.Oligonucleotide sequences used in
vector construction. |
Table I.
Oligonucleotide sequences used in
vector construction.
| Name | Sequences
(5′-3′) |
|---|
|
shRNA-GDNFOS1-9-F |
CACCGCTTTCCTCGCGCCTGTCGAACGAATTCGACAGGCGCGAGGAAAGC |
|
shRNA-GDNFOS1-9-R |
AAAAGCTTTCCTCGCGCCTGTCGAATTCGTTCGACAGGCGCGAGGAAAGC |
|
shRNA-GDNFOS1-49-F |
CACCGTGTCTCGCCCTCTCGCTTCTCGAAAGAAGCGAGAGGGCGAGACAC |
|
shRNA-GDNFOS1-49-R |
AAAAGTGTCTCGCCCTCTCGCTTCTTTCGAGAAGCGAGAGGGCGAGACAC |
|
shRNA-GDNFOS1-248-F |
CACCGAGTCACGGAAGAATAGAAGACGAATCTTCTATTCTTCCGTGACTC |
|
shRNA-GDNFOS1-248-R |
AAAAGAGTCACGGAAGAATAGAAGATTCGTCTTCTATTCTTCCGTGACTC |
Lentiviral packaging
A total of 3 µg recombinant lentiviral plasmid and 9
µg packaging mix were added into 1.5 ml Opti-MEM. In addition,
total of 1.5 ml Opti-MEM was mixed with 36 µl Lipofectamine 2000
and incubated for 5 min at room temperature. The diluted
Lipofectamine 2000 and plasmid solution were then mixed and
incubated for 5 min at room temperature. The mixture was added into
a culture dish with 293T cells (Novobio Scientific, Inc.) and
cultured at 37°C for 48 h. Cell supernatant was collected using a
pasteur pipette, centrifuged (1,500 × g for 10 min at room
temperature) and filtered. The virus solution was condensed using
centrifugation (50,000 × g for 2 h at 4°C) and re-suspended in
DMEM. The blank group refers to cells that were untransfected and
the negative control group refers to cells transfected with the
previously mentioned scrambled vector.
Reverse transcription-quantitative PCR
(RT-qPCR)
RT-qPCR was used to determine the efficiency of
interference and overexpression in U87 and U251 MG cells. Total RNA
was extracted using TRIzol reagent following manufacturer's
protocol and reverse transcription was performed using a cDNA
synthesis kit. Each reaction contained 0.5 µl Oligo dT primers (0.2
µg/µl; Table II) and 1 µl
SuperScript III reverse transcriptase (200 U/µl). Reverse
transcription was performed at 42°C. PCR was performed using a SYBR
qPCR mix kit. PCR conditions were as follows: Initial denaturation
at 95°C for 2 min; 40 cycles of 95°C for 10 sec, 60°C for 30 sec
and 70°C for 45 sec. A CFX96 Touch™ Real-Time PCR Detection system
was used during PCR. Gene expression was normalized to β-actin. The
2−ΔΔCq method was used to determine qPCR results
(14).
 | Table II.Primers used in quantitative PCR. |
Table II.
Primers used in quantitative PCR.
| Primers | Sequences
(5′-3′) |
|---|
| lncRNA
GDNFOS1-F |
AGTGGCGAGAAAAGGAGCTG |
| lncRNA
GDNFOS1-R |
GCACCTTGTGTTTGCCTGTT |
| GDNF-F |
AGTGACAAAGTAGGGCAGGC |
| GDNF-R |
CCACACCTTTTAGCGGAATGC |
| β-actin-F |
AGGGAAATCGTGCGTGAC |
| β-actin-R |
CGCTCATTGCCGATAGTG |
Western blot analysis
GDNF expression in U87 and U251 MG cells was
detected using western blot analysis. Cells were lysed in lysis
buffer with protease and phosphatase inhibitors at 4°C for 1 h. The
lysis mixture was centrifuged at 4°C at 10,000 × g for 10 min.
Supernatant with cellular proteins was collected and used in the
subsequent experiments. The protein concentration was measured
using a BCA kit and proteins were separated using SDS-PAGE (10%
gel; 40 µg protein/lane; 120 V). The separated proteins were
transferred to PVDF (100 V for 2 h). The membranes were blocked
with 5% non-fat milk at room temperature for 1 h and incubated with
primary antibodies against GDNF (1:300) and β-actin (cat. no.
ab8227; 1:1,000; Abcam) at 4°C overnight. After being washed with
Tris-buffered saline with Tween-20, membranes were incubated with
HRP-conjugated goat anti-rabbit secondary antibody (1:5,000) at
room temperature for 1 h. Membranes were subsequently incubated in
an enhanced chemiluminescence solution at room temperature for 10
min. Images were captured on film in a dark room. Experiments were
repeated three times. Blot images were quantified in greyscale
using ImageJ v1.42 (National Institutes of Health) and protein
expression was presented relative to the control group. The results
in control group were normalized using the mean results. However,
there was some variability in control samples.
Cell viability assay
U87 MG and U251 MG cell viability was detected using
a cell counting kit (CCK)-8 assay at 0, 24, 48 and 72 h after
GDNFOS1 overexpression or interference. CCK-8 reagent (Dojindo
Molecular Technologies, Inc.) was added into each well of 96-well
culture plates and incubated for 4 h. A microplate reader was used
to measure the absorbance at a wavelength of 490 nm. The reading of
each group was divided by the baseline reading at 0 h to determine
relative cell viability. Experiments were repeated three times.
Transwell invasion assay
Matrigel (1 g/l; 50 µl) was used to coat the
membrane of the upper transwell compartment, and incubated at 37°C
for 1 h. The upper compartment was filled with the U87 and U251 MG
cell suspension in DMEM (200 µl; 2×105 cells/ml) and the
lower compartment was filled with DMEM that contained 10% FBS (800
µl). Cells were subsequently incubated for 24 h at 37°C. The
microporous membrane was then fixed with 4% paraformaldehyde at
room temperature for 30 min. Crystal violet (1%) was used to stain
the lower side of membrane at room temperature for 10 min and was
subsequently washed with PBS twice. Cells were observed under a
light microscope (magnification, ×400; 6 random fields of view per
group) and cells that had invaded through the membrane were
counted. The average number of cells that transgressed through the
membrane was divided by that in the blank group to calculate the
relative cell invasion of U87 and U251 MG cells. Experiments were
repeated three times.
Statistical analysis
Statistical data were analyzed using GraphPad Prism
software (version 5.0; GraphPad Software, Inc.). The results are
presented as the mean ± standard error. Differences between three
and more groups were compared using a one-way ANOVA followed by the
Bonferroni post-hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Confirmation of GDNFOS1 overexpression
and interference in U87 and U251 MG cells
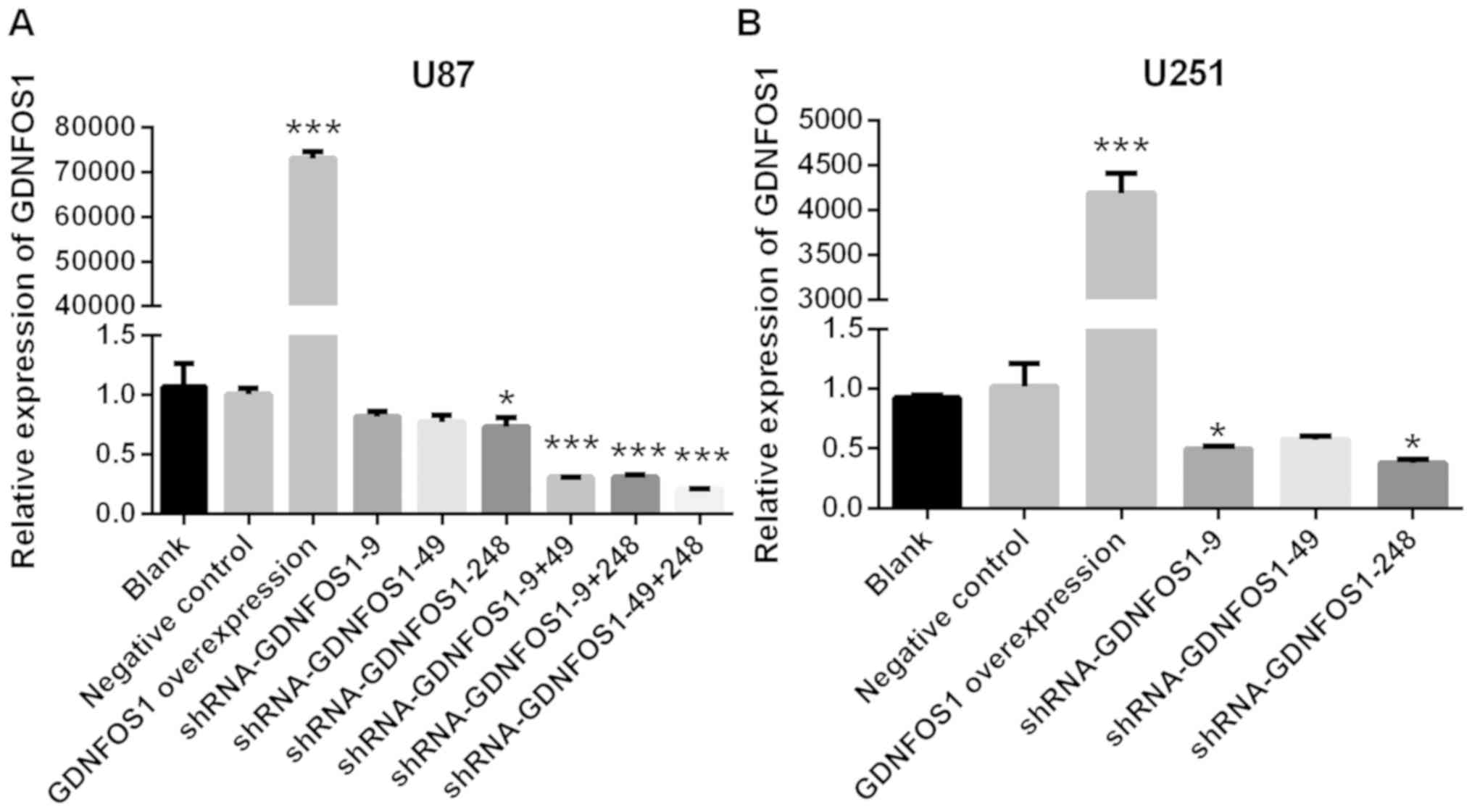
Compared with the negative control group, GDNFOS1
mRNA expression in U87 MG cells was increased significantly in the
GDNFOS1 overexpression group (P<0.001; Fig. 1A). In addition, GDNFOS1 mRNA
expression in U87 MG cells was significantly decreased in the
shRNA-GDNFOS1-248 group when compared with the negative control
group (P<0.05; Fig. 1A). A
combination of interference vectors caused higher GDNFOS1
expression inhibition in U87 MG cells. GDNFOS1 mRNA expression was
significantly decreased in the shRNA-GDNFOS1-9 + shRNA-GDNFOS1-49
combination, shRNA-GDNFOS1-9 + shRNA-GDNFOS1-248 combination and
shRNA-GDNFOS1-49 + shRNA-GDNFOS1-248 combination, when compared
with the negative control group (all P<0.001; Fig. 1A). The shRNA-GDNFOS1-49 +
shRNA-GDNFOS1-248 combination exhibited the highest inhibition of
GDNFOS1 expression in U87 MG cells. The shRNA-GDNFOS1-49 +
shRNA-GDNFOS1-248 combination was therefore used in the subsequent
experiments. Similarly, when compared with the negative control
group, GDNFOS1 mRNA expression in U251 MG cells was significantly
higher in the GDNFOS1 overexpression group (P<0.001; Fig. 1B) and significantly lower in the
shRNA-GDNFOS1-9 and shRNA-GDNFOS1-248 groups (P<0.05; Fig. 1B). ShRNA-GDNFOS1-248 exhibited the
highest interference ability in U251 cells and was therefore used
in the subsequent experiments.
Protein expression of GDNF in U87 and
U251 MG cells is significantly increased in the GDNFOS1
overexpression group and decreased in the GDNFOS1 interference
group
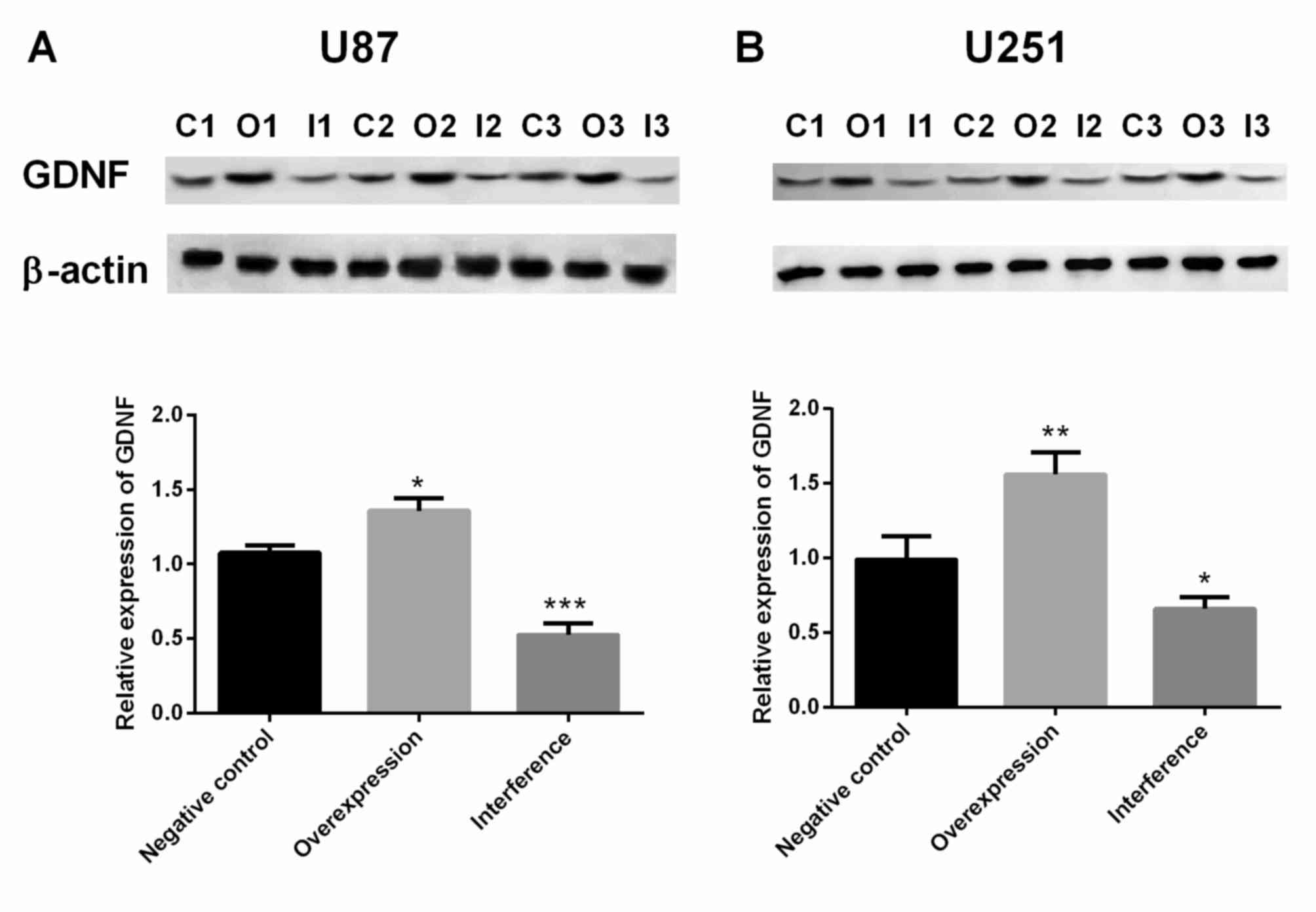
When compared with the control group, GDNF protein
expression in U87 MG cells was significantly increased in the
GDNFOS1 overexpression group (P<0.05; Fig. 2A) and significantly decreased in the
GDNFOS1 interference group (P<0.001; Fig. 2A). In addition, the protein
expression of GDNF in U251 MG cells was significantly increased in
the GDNFOS1 overexpression group (P<0.01; Fig. 2B) and significantly decreased in the
GDNFOS1 interference group (P<0.05; Fig. 2B).
Viability of U87 MG and U251 cells is
significantly increased in the GDNFOS1 overexpression group and
decreased in the GDNFOS1 interference group
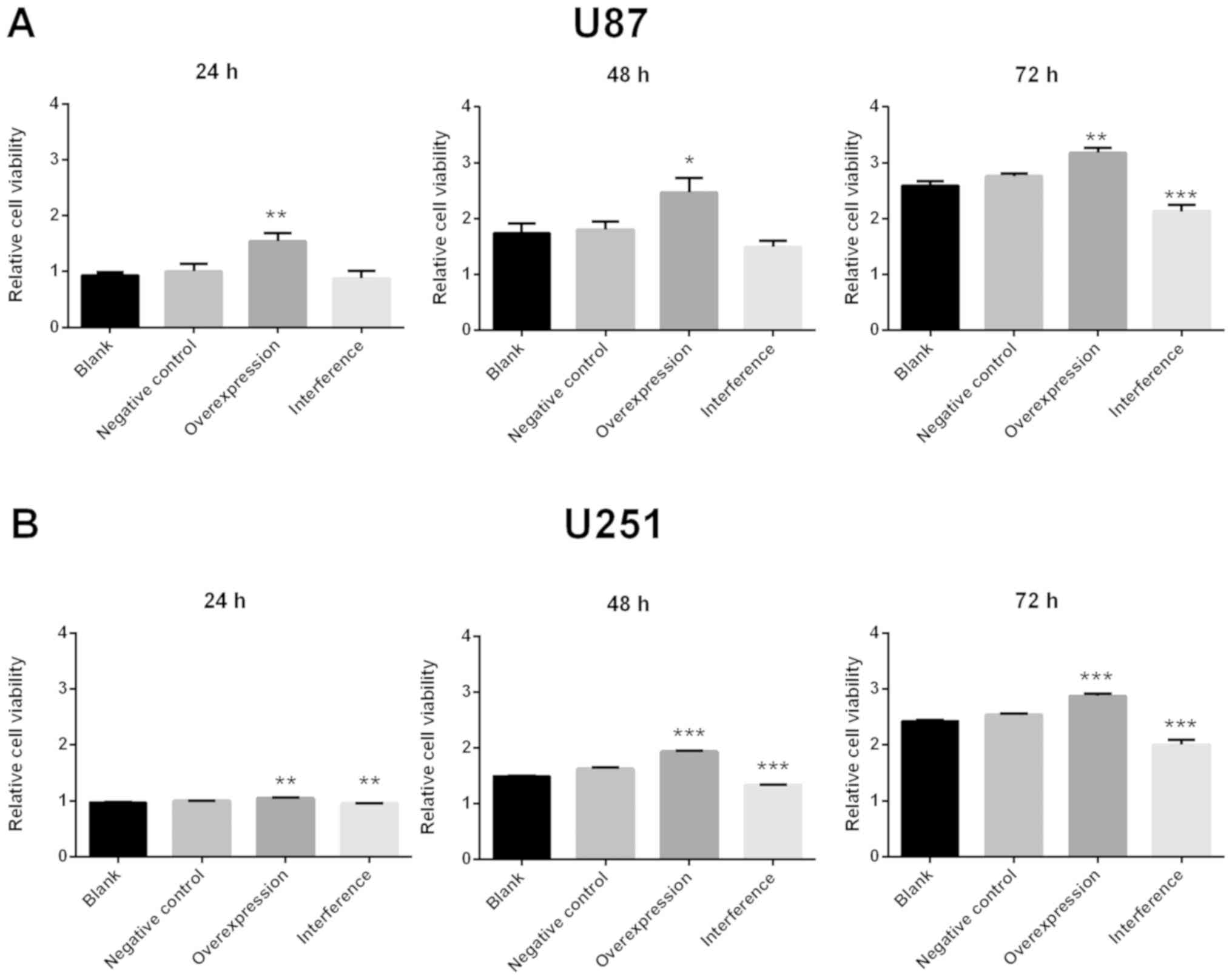
When compared with the negative control group, U87
MG cell viability increased significantly in the GDNFOS1
overexpression group at 24, 48 and 72 h (P<0.01, P<0.05 and
P<0.01, respectively; Fig. 3A).
Meanwhile, U87 MG cell viability decreased significantly in the
GDNFOS1 interference group at 72 h when compared with the negative
control group (P<0.001; Fig. 3A).
Furthermore, U251 MG cell viability significantly increased in the
GDNFOS1 overexpression group at 24, 48 and 72 h (P<0.01;
P<0.001; P<0.001, respectively; Fig. 3B) and significantly decreased in the
GDNFOS1 interference group at 24, 48 and 72 h (P<0.01;
P<0.001; P<0.001, respectively; Fig. 3B).
Invasion ability is significantly
increased in the GDNFOS1 overexpression group and decreased in the
GDNFOS1 interference group
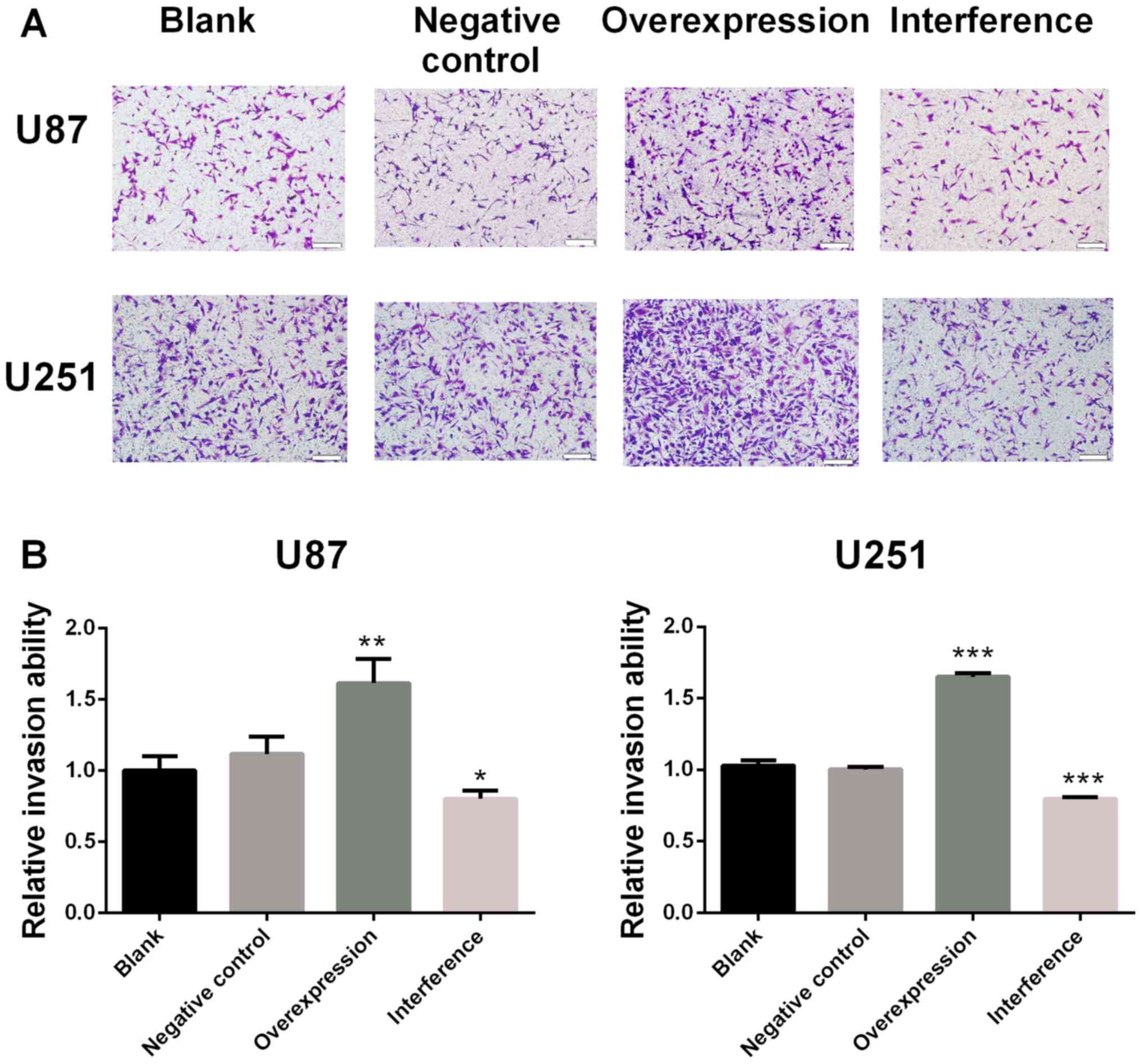
The relative invasion ability of U87 MG cells was
significantly increased in the GDNFOS1 overexpression group
(P<0.01; Fig. 4A) and
significantly decreased in the GDNFOS1 interference group when
compared with the negative control group (P<0.05; Fig. 4A). Similarly, when compared with the
negative control group, the relative invasive ability of U251 MG
cells was significantly increased in the GDNFOS1 overexpression
group (P<0.001; Fig. 4B) and
significantly decreased in the GDNFOS1 interference group
(P<0.001; Fig. 4B).
Discussion
In current study, the results demonstrated that GDNF
expression, cell viability and invasion ability of glioblastoma
cells significantly increased with GDNFOS1 overexpression and
decreased with GDNFOS1 interference.
GDNF has previously been reported to promote the
survival of peripheral motor and sensory neurons, GABAergic and
cholinergic neurons in the forebrain and pancreatic β-cells
(15–17). Furthermore, GDNF has been
demonstrated to affect the enteric nervous system (18–24). The
ability of enteric neural progenitors to develop into the enteric
nervous system has been revealed to increase with GDNF exposure
(18–21). The loss of enteric neurons in
diabetic rats has been demonstrated to be associated with decreased
GDNF expression (22). In diabetic
rats, the regeneration of lost enteric neurons, which is induced by
high frequency electroacupuncture, has been revealed to be
associated with the GDNF and PI3K/AKT signaling pathways (23). The markers of synaptic vesicles in
enteric neurons were revealed to be induced by GDNF (24). In addition, GDNF has also been
demonstrated to be associated with the physiology and
pathophysiology of glioblastoma multiforme. Glioblastoma
multiforme-induced attraction of microglia has been revealed to be
mediated by GDNF (10). In a
previous study, the modulation of GDNF was associated with the
BMP4-mediated reversion of multi-drug resistance in glioblastoma
multiforme (11). In rat C6
glioblastoma cells, the expression of GDNF has been indicated to be
promoted by calcium discharge from the endoplasmic reticulum via
mitogen-activated protein kinase-independent and dependent pathways
(25). Fibroblast growth factor
(FGF)-2 has been demonstrated to stimulate GDNF mRNA expression in
rat C6 glioblastoma cells (26).
After treatment with tumor necrosis factor-α or interleukin-1 for
24 h, U87 MG glioblastoma cells were stimulated to release GDNF.
The production of GDNF from U87 MG glioblastoma cells was also
indicated to be affected by FGF-1, 2 and 9, prostaglandins
(PGA2, PGE2 and PGI2),
dexamethasone and vitamin D3 (27).
GDNFOS1 belongs to the lncRNAs and the opposite
strand of the GDNF gene is used to transcribe the cis-antisense
GDNFOS gene (8). Unlike small
noncoding RNAs including microRNAs and small interfering RNAs, long
noncoding RNAs are intergenic noncoding RNAs (28). They are located between coding genes
rather than antisense to them or within introns (29). Kidney, testis and ovary display the
highest mRNA expression of GDNFOS1 (8). In the brain, higher expression GDNFOS1
was exhibited in the nucleus accumbens and cerebellum compared with
other brain regions (8). The
expression of the GDNF isoform Ex1_4L is higher than GDNFOS1
expression in the human brain (8).
The first exon of GDNFOS1 has been revealed to be composed of 136
nucleotides that are reversely complementary to the 5′-untranslated
region of the GDNF isoform Ex1_4L/S (30). Exon 2-short and exon 3-short are
spliced from exon 1 of GDNFOS1 via intra-exonal splicing, whereas
exon 4-short is spliced from exon 1 of GDNFOS1 via alternative
poly-adenylation (8).
In the present study, the results revealed that GDNF
expression, cell viability and invasion ability of glioblastoma
cells significantly increased with GDNFOS1 overexpression and
decreased with GDNFOS1 interference. Studies that assess the
association between GDNFOS1 and glioblastoma multiforme are
required. To the best of our knowledge, for the first time, the
present study revealed that GDNFOS1 is associated with the
regulation of viability and invasion in glioblastoma cell lines U87
and U251. The increased cell viability of glioblastoma cells
observed in the present study, may be associated with increased
GDNF expression due to the fact GDNFOS1 overlaps with GDNF mRNA.
The molecular mechanisms underlying how GDNFOS1 regulated the
expression of GDNF requires further investigation. The cross-link
regulation of precursor N-cadherin and fibroblast growth factor
receptor 1 by GDNF has been demonstrated to increase the viability
of U251 glioblastoma cells (31).
GDNF has also been revealed to enhance the viability of human
cumulus cells by downregulating miR-145-5p (32). In a previous study, GDNF reduced
apoptosis in dopaminergic neurons in vitro (33) and prevented ethanol-induced apoptosis
(34). In addition, GDNF has been
revealed to serve an important role in the aggressive behavior and
invasion of cancer. The perineural invasion of pancreatic cancer
cells has been reported to be induced by the secretion of GDNF from
macrophages (35). The GDNF
receptor, which is released by nerves, enhanced cancer cell
perineural invasion via GDNF-RET signaling (36). An antibody against the GDNF receptor
was revealed to provide a targeted therapeutic opportunity for
patients with breast cancer (37).
In addition, GDNFOS1 may stimulate the viability and invasion
ability of glioblastoma cells through other molecular mechanisms
not involving GDNF. Further studies are required to elucidate the
underlying biological mechanisms behind this interaction.
In the current study, the viability of glioblastoma
cells when GDNFOS1 was overexpressed or interfered was detected,
but this was not tested in normal cell lines. Migration ability was
also not determined in the present study. In previous studies
however, the changes are often the same between tumor invasion and
migration in both tumor cells and tumors (38–42).
Future experiments should address the limitations of the present
study.
In conclusion, the current study demonstrated that
GDNF expression, cell viability and invasion ability of
glioblastoma cells significantly increased with GDNFOS1
overexpression and decreased with GDNFOS1 interference.
Acknowledgements
Not applicable.
Funding
The present study was supported by National Natural
Science Foundation of China (grant no. 81473506; Dr Yihong Fan),
Zhejiang Provincial Construction Foundation of China (grant no.
WKJ-ZJ-1531; Dr Bin Lv), Zhejiang Provincial Natural Science
Foundation of China (grant no. LY17H290009; Dr Yihon Fan) and
Zhejiang Science and Technology Program of TCM (grant nos.
2016ZB047 and 2017ZA056; Dr Yihong Fan; and grant no. 2018ZB046; Dr
Yi Xu).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
SW and YF conceived and designed the current study.
SW, YX and LZ performed the experiments. LC and BL analyzed the
data. SW wrote the paper. All authors have read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bleeker FE, Molenaar RJ and Leenstra S:
Recent advances in the molecular understanding of glioblastoma. J
Neurooncol. 108:11–27. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gallego O: Nonsurgical treatment of
recurrent glioblastoma. Curr Oncol. 22:e273–e281. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Henderson CE, Phillips HS, Pollock RA,
Davies AM, Lemeulle C, Armanini M, Simmons L, Moffet B, Vandlen RA,
Simpson LC, et al: GDNF: A potent survival factor for motoneurons
present in peripheral nerve and muscle. Science. 266:1062–1064.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lin LF, Doherty DH, Lile JD, Bektesh S and
Collins F: GDNF: A glial cell line-derived neurotrophic factor for
midbrain dopaminergic neurons. Science. 260:1130–1132. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Arenas E, Trupp M, Akerud P and Ibáñez CF:
GDNF prevents degeneration and promotes the phenotype of brain
noradrenergic neurons in vivo. Neuron. 15:1465–1473. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Trok K, Hoffer B and Olson L: Glial cell
line-derived neurotrophic factor enhances survival and growth of
prenatal and postnatal spinal cord transplants. Neuroscience.
71:231–241. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Airaksinen MS and Saarma M: The GDNF
family: Signalling, biological functions and therapeutic value. Nat
Rev Neurosci. 3:383–394. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Airavaara M, Pletnikova O, Doyle ME, Zhang
YE, Troncoso JC and Liu QR: Identification of novel GDNF isoforms
and cis-antisense GDNFOS gene and their regulation in human middle
temporal gyrus of Alzheimer disease. J Biol Chem. 286:45093–45102.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shi C, Zhang L and Qin C: Long non-coding
RNAs in brain development, synaptic biology, and Alzheimer's
disease. Brain Res Bull. 132:160–169. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ku MC, Wolf SA, Respondek D, Matyash V,
Pohlmann A, Waiczies S, Waiczies H, Niendorf T, Synowitz M, Glass R
and Kettenmann H: GDNF mediates glioblastoma-induced microglia
attraction but not astrogliosis. Acta Neuropathol. 125:609–620.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu B, Chen Q, Tian D, Wu L, Dong H, Wang
J, Ji B, Zhu X, Cai Q, Wang L and Zhang S: BMP4 reverses multidrug
resistance through modulation of BCL-2 and GDNF in glioblastoma.
Brain Res. 1507:115–124. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Feng SY, Dong CG, Wu WK, Wang XJ, Qiao J
and Shao JF: Lentiviral expression of anti-microRNAs targeting
miR-27a inhibits proliferation and invasiveness of U87 glioma
cells. Mol Med Rep. 6:275–281. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li G, Zhang H, Liu Y, Kong L, Guo Q and
Jin F: Effect of temozolomide on livin and caspase-3 in U251 glioma
stem cells. Exp Ther Med. 9:744–750. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Trupp M, Rydén M, Jörnvall H, Funakoshi H,
Timmusk T, Arenas E and Ibáñez CF: Peripheral expression and
biological activities of GDNF, a new neurotrophic factor for avian
and mammalian peripheral neurons. J Cell Biol. 130:137–148. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Williams LR, Inouye G, Cummins V and
Pelleymounter MA: Glial cell line-derived neurotrophic factor
sustains axotomized basal forebrain cholinergic neurons in vivo:
Dose-response comparison to nerve growth factor and brain-derived
neurotrophic factor. J Pharmacol Exp Ther. 277:1140–1151.
1996.PubMed/NCBI
|
|
17
|
Mwangi S, Anitha M, Mallikarjun C, Ding X,
Hara M, Parsadanian A, Larsen CP, Thule P, Sitaraman SV, Anania F
and Srinivasan S: Glial cell line-derived neurotrophic factor
increases beta-cell mass and improves glucose tolerance.
Gastroenterology. 134:727–737. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
McKeown SJ, Mohsenipour M, Bergner AJ,
Young HM and Stamp LA: Exposure to GDNF enhances the ability of
enteric neural progenitors to generate an enteric nervous system.
Stem Cell Reports. 8:476–488. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Worley DS, Pisano JM, Choi ED, Walus L,
Hession CA, Cate RL, Sanicola M and Birren SJ: Developmental
regulation of GDNF response and receptor expression in the enteric
nervous system. Development. 127:4383–4393. 2000.PubMed/NCBI
|
|
20
|
Schiltz CA, Benjamin J and Epstein ML:
Expression of the GDNF receptors ret and GFRalpha1 in the
developing avian enteric nervous system. J Comp Neurol.
414:193–211. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chalazonitis A, Rothman TP, Chen J and
Gershon MD: Age-dependent differences in the effects of GDNF and
NT-3 on the development of neurons and glia from neural
crest-derived precursors immunoselected from the fetal rat gut:
Expression of GFRalpha-1 in vitro and in vivo. Dev Biol.
204:385–406. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Du F, Wang L, Qian W and Liu S: Loss of
enteric neurons accompanied by decreased expression of GDNF and
PI3K/Akt pathway in diabetic rats. Neurogastroenterol Motil.
21:1229–e114. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Du F and Liu S: Electroacupuncture with
high frequency at acupoint ST-36 induces regeneration of lost
enteric neurons in diabetic rats via GDNF and PI3K/AKT signal
pathway. Am J Physiol Regul Integr Comp Physiol. 309:R109–R118.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bottner M, Harde J, Barrenschee M, Hellwig
I, Vogel I, Ebsen M and Wedel T: GDNF induces synaptic vesicle
markers in enteric neurons. Neurosci Res. 77:128–136. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Oh-hashi K, Kaneyama M, Hirata Y and
Kiuchi K: ER calcium discharge stimulates GDNF gene expression
through MAPK-dependent and -independent pathways in rat C6
glioblastoma cells. Neurosci Lett. 405:100–105. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Suter-Crazzolara C and Unsicker K: GDNF
mRNA levels are induced by FGF-2 in rat C6 glioblastoma cells.
Brain Res Mol Brain Res. 41:175–182. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Verity AN, Wyatt TL, Lee W, Hajos B,
Baecker PA, Eglen RM and Johnson RM: Differential regulation of
glial cell line-derived neurotrophic factor (GDNF) expression in
human neuroblastoma and glioblastoma cell lines. J Neurosci Res.
55:187–197. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Akella A, Bhattarai S and Dharap A: Long
noncoding RNAs in the pathophysiology of ischemic stroke.
Neuromolecular Med; 2019, View Article : Google Scholar
|
|
29
|
Bakhtiarizadeh MR and Salami SA:
Identification and expression analysis of long noncoding RNAs in
fat-tail of sheep breeds. G3 (Bethesda). 9:1263–1276.
2019.PubMed/NCBI
|
|
30
|
Zhang Y, Liu XS, Liu QR and Wei L:
Genome-wide in silico identification and analysis of cis natural
antisense transcripts (cis-NATs) in ten species. Nucleic Acids Res.
34:3465–3475. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tang CX, Gu YX, Liu XF, Tong SY, Ayanlaja
AA, Gao Y, Ji GQ, Xiong Y, Huang LY and Gao DS: Cross-link
regulation of precursor N-cadherin and FGFR1 by GDNF increases
U251MG cell viability. Oncol Rep. 40:443–453. 2018.PubMed/NCBI
|
|
32
|
Cui L, Fang L, Mao X, Chang HM, Leung PCK
and Ye Y: GDNF-induced downregulation of miR-145-5p enhances human
oocyte maturation and cumulus cell viability. J Clin Endocrinol
Metab. 103:2510–2521. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Clarkson ED, Zawada WM and Freed CR: GDNF
reduces apoptosis in dopaminergic neurons in vitro. Neuroreport.
7:145–149. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
McAlhany RE Jr, West JR and Miranda RC:
Glial-derived neurotrophic factor (GDNF) prevents ethanol-induced
apoptosis and JUN kinase phosphorylation. Brain Res Dev Brain Res.
119:209–216. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cavel O, Shomron O, Shabtay A, Vital J,
Trejo-Leider L, Weizman N, Krelin Y, Fong Y, Wong RJ, Amit M and
Gil Z: Endoneurial macrophages induce perineural invasion of
pancreatic cancer cells by secretion of GDNF and activation of RET
tyrosine kinase receptor. Cancer Res. 72:5733–5743. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
He S, Chen CH, Chernichenko N, He S, Bakst
RL, Barajas F, Deborde S, Allen PJ, Vakiani E, Yu Z and Wong RJ:
GFRα1 released by nerves enhances cancer cell perineural invasion
through GDNF-RET signaling. Proc Natl Acad Sci USA.
111:E2008–E2017. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bhakta S, Crocker LM, Chen Y, Hazen M,
Schutten MM, Li D, Kuijl C, Ohri R, Zhong F, Poon KA, et al: An
Anti-GDNF family receptor alpha 1 (GFRA1) antibody-drug conjugate
for the treatment of hormone receptor-positive breast cancer. Mol
Cancer Ther. 17:638–649. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang Y, Li Z, Zhang H, Jin H, Sun L, Dong
H, Xu M, Zhao P, Zhang B, Wang J, et al: HIF-1α and HIF-2α
correlate with migration and invasion in gastric cancer. Cancer
Biol Ther. 10:376–382. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gao J, Yu SR, Yuan Y, Zhang LL, Lu JW,
Feng JF and Hu SN: MicroRNA-590-5p functions as a tumor suppressor
in breast cancer conferring inhibitory effects on cell migration,
invasion and epithelial-mesenchymal transition by downregulating
the Wnt-β-catenin signaling pathway. J Cell Physiol. 234:1827–1841.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wu D, Liu J, Chen J, He H, Ma H and Lv X:
miR-449a suppresses tumor growth, migration and invasion in
non-small cell lung cancer by targeting a HMGB1-mediated NF-κB
signaling pathway. Oncol Res. 27:227–235. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mansoori B, Mohammadi A, Ghasabi M,
Shirjang S, Dehghan R, Montazeri V, Holmskov U, Kazemi T, Duijf P,
Gjerstorff M and Baradaran B: miR-142-3p as tumor suppressor miRNA
in the regulation of tumorigenicity, invasion and migration of
human breast cancer by targeting Bach-1 expression. J Cell Physiol.
234:9816–9825. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sano M, Ijichi H, Takahashi R, Miyabayashi
K, Fujiwara H, Yamada T, Kato H, Nakatsuka T, Tanaka Y, Tateishi K,
et al: Blocking CXCLs-CXCR2 axis in tumor-stromal interactions
contributes to survival in a mouse model of pancreatic ductal
adenocarcinoma through reduced cell invasion/migration and a shift
of immune-inflammatory microenvironment. Oncogenesis. 8:82019.
View Article : Google Scholar : PubMed/NCBI
|