Introduction
Glioma is the most common type of central nervous
system tumor in the clinical setting (1). Gliomas exhibiting a high pathological
grade indicate strong invasiveness. In most cases, the boundaries
between tumor and normal tissue are blurred and in high-grade
patients, can result in the incomplete surgical removal of diseased
tissues (1). In addition, high-grade
glioma is more resistant to radiation and chemotherapy (2,3).
microRNAs (miRs or miRNAs) are a class of endogenous
non-coding small RNAs, 18–24 nucleotides in length, that are
ubiquitous in viruses and eukaryote cells in highly conserved
sequences. It has been estimated that up to 50% of human
protein-coding genes are regulated by miRNAs (4). Each miRNA has the potential to regulate
hundreds of RNAs, and their expression is associated with tissue
status and disease subtype (5).
miRNAs have been demonstrated to serve an important role in the
development and prognosis of glioma. Lv et al (4) reported that low miR-320b expression in
glioma was associated with poor patient prognosis. The upregulation
of miR-320b enhanced apoptosis and attenuated the proliferation,
migration and invasive abilities of glioma cells. Santangelo et
al (6) collected serum samples
from patients with glioma and observed an increased expression of
miR-21, miR-222 and miR-124-3p in the serum exosomes of these
patients when compared with healthy participants. Patients with
high-grade glioma exhibited increased miR-21, miR-222 and
miR-124-3p expression when compared with the other patients with
low-grade glioma. Additionally, miR-211 was also indicated to be
reduced in glioma, and was closely associated with a decreased
patient survival time and considered to be an independent risk
factor for glioma prognosis (7). For
high-grade glioma, the upregulation of miR-126 may impair the
invasive ability of glioma cells by targeting KRAS, meaning miR-126
could be an effective therapeutic target (8). Therefore, these genes are considered to
be a potential biological target for glioma prognosis and
treatment.
Recently, miR-6852 has been demonstrated to be
associated with the development of tumors, including those in
gastric, colorectal and cervical cancer (9–11). The
association of miR-6852 with glioma remains undetermined.
Therefore, the present study aimed to assess miR-6852 expression in
glioma, and further determine the effect of miR-6852 in the
regulation of glioma progression.
Materials and methods
Glioma tissue collection
A total of 32 pairs of glioma tissues (14 males and
18 females; age range, 31–56 years old; median age, 47 years old)
and corresponding normal tissues were collected during biopsy from
Affiliated Hospital of Hebei University (Hebei, China) between
October 2014 and October 2016 for the current study. Normal tissues
were derived from the temporal lobes and saddle area 2 cm away from
tumor tissues. All glioma tissues were obtained from patients upon
first diagnosis of glioma. Among these patients, 18 cases were
high-grade and 14 cases were low-grade. All patients did not
recieve radiotherapy or chemotherapy prior to surgery. Patients
treated by radiotherapy or chemotherapy prior to surgery were
excluded. All participates had signed informed consent and the
present study has been approved by The Ethics Committee of
Affiliated Hospital of Hebei University.
Cell culture
Normal human astrocytes (NHAs; ScienCell Research
Laboratories, Inc.) were maintained in astrocyte medium (ScienCell
Research Laboratories, Inc.) according to manufacturers protocol.
In addition, human glioma cell lines (U251 and A172 cells),
purchased from the Type Culture Collection of the Chinese Academy
of Sciences were incubated at 37°C, 5% CO2 in DMEM
(Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS (Gibco;
Thermo Fisher Scientific, Inc.), 100 mg/ml streptomycin and 100
U/ml penicillin (Gibco; Thermo Fisher Scientific, Inc.).
Cell transfection
The mature miR-6852 mimics
(5′-CCCUGGGGUUCUGAGGACAUG-3′), miR-6852 inhibitors
(5′-CATGTCCTCAGAACCCCAGGG-3′) and negative control (NC;
5′-UCACAACCUCCUAGAAAGAGUAGA-3′) were synthesized and verified by
Shanghai GenePharma Co., Ltd. The coding sequence of lymphoid
enhancer binding factor 1 (LEF1) was constructed into the pcDNA3
vector (Invitrogen; Thermo Fisher Scientific, Inc.) to generate a
pcDNA3-LEF vector for the overexpression of LEF1. The pcDNA3 empty
vector was used for overexpressing control. A172 human glioma cells
were cultured under the aforementioned standard culture conditions
and prepared as a single cell suspension (1×106
cells/ml) with serum free DMEM. Cells were subsequently seeded into
six-well plates with 1 ml of cell suspension per well. A period of
1 day later, cells were grouped and transfected as follows: miR-NC
group (50 nM), cells were transfected with a miR-6852 mimics
negative control (50 nM); miR-6852 group (50 nM), cells were
transfected with miR-6852 mimics (50 nM); Inh-NC
(5′-GCGUAACUAAUACAUCGGAUUCGU-3′) group (50 nM), cells were
transfected with a miR-6852 inhibitor negative control;
Inh-miR-6852 group, cells were transfected with miR-6852
inhibitors; and miR-6852 + pcDNA3-LEF1 group, cells were
co-transfected with miR-6852 mimics and LEF1 overexpression
vectors. Transfection was performed using Lipofectamine 2000™
(Thermo Fisher Scientific, Inc.), according to manufacturer's
protocol. After 6 h, successfully transfected cells were confirmed
by qRT-PCR analysis and cultured in six-well plates using DMEM
containing 10% FBS for 2 days at 37°C and 5% CO2.
Cell Counting Kit (CCK)-8 assay
A total of 1×104 A172 cells of each
aforementioned group were inoculated in 96-well plates with 100 µl
DMEM, containing 10% FBS, and cultured at 37°C in 5% CO2
for 0, 24, 48 or 72 h. At each time point, cells were removed from
the incubator, and 10 µl CCK-8 solution (Beyotime Institute of
Biotechnology) was added into each well for subsequent incubation
for 4 h at 37°C. The optical density (OD) of each well was
monitored using an enzyme immunoassay analyzer (INC-STAR Corp.) at
450 nm.
Transwell experiments
A period of 2 days after transfection, A172 cells of
each aforementioned group were prepared as single cell suspensions
(1×105 cells/ml) with serum-free DMEM. Transwell
chambers were inserted into 24-well plates containing 600 µl of
DMEM (with 10% FBS) in the lower chamber. On the upper chamber, 100
µl of cell suspension was added with serum-free DMEM. All plates
were cultured for 48 h at 37°C, 5% CO2 in the incubator.
Non-migrated cells were scraped off and migrated cells were fixed
using 4% formaldehyde for 30 min at room temperature, stained using
0.5% crystal violet for 30 min at room temperature and counted
under a light microscope (magnification, ×200). The aforementioned
procedure was also used to detect cell invasion except Matrigel (BD
Biosciences) was spread onto the Transwell chamber.
Luciferase reporter gene assay
LEF1 was predicted as a potential miR-6852 target
using TargetScan7 (http://www.targetscan.org/vert_71/). The LEF1 3′-UTR
region was constructed into the pGL3 luciferase reporter vector
(Promega Corporation) to generate pGL3-LEF1-wild-type (WT)
reporter. The binding site for miR-6852 in LEF1 3′-UTR was mutated
to obtain the pGL3-LEF1-mutant reporter. For the luciferase
reporter assay, A172 cells in the miR-NC and miR-6852 group
underwent subsequent transfection with pGL3-LEF1-WT or
pGL3-LEF1-mutant respectively using Lipofectamine 2000™. Cells were
incubated at 37°C, 5% CO2 for 1 day. Luciferase activity
of each well was measured using a Dual-Luciferase Reporter System
(Promega Corporation) and normalized to Renilla luciferase
activity.
RNA extraction and reverse
transcription-quantitative (RT-q) PCR
Total RNA was extracted from glioma, normal tissues
and glioma cell lines using TRIzol reagent (Thermo Fisher
Scientific, Inc.). cDNA template synthesis was performed using the
Prime Script RT Reagent kit (Takara Bio, Inc.) according to the
manufacturer's protocol. The reaction was performed with incubation
at 42°C for 1 h, and the enzyme was subsequently inactivated by
incubation at 85°C for 5 min.. miR-6852 expression was determined
using SYBR Premix Ex Taq II (GeneCopoeia, Inc.) according to the
following conditions: 10 min of pre-denaturation at 95°C, followed
by 40 cycles of 10 sec denaturation at 95°C, 20 sec annealing at
60°C and 30 sec extension at 72°C. LEF1 mRNA expression was
measured using a SYBR Green PCR Master Mix (Applied Biosystems;
Thermo Fisher Scientific, Inc.) with the following procedures: 10
min pre-denaturation at 95°C, followed by 36 cycles of 10 sec
denaturation at 95°C, 20 sec annealing at 60°C and 34 sec extension
at 72°C. U6 and GAPDH were used as internal references for miR-6852
and LEF1 mRNA expression, respectively. Relative miR-6852 and LEF1
mRNA were processed using the 2-ΔΔCq method (12). The primer sequences were: miR-6852
forward, 5′-AACGAGACGACGACAGAC-3′ and reverse,
5′-CCCTGGGGTTCTGAGGACATG-3′; U6 forward,
5′-GTGCTCGCTTCGGCAGCACAT-3′ and reverse,
5′-TACCTTGCGAAGTGCTTAAAC-3′; LEF1 forward,
5′-AGAACACCCCGATGACGGA-3′ and reverse, 5′-GAGGGTCCCTTGTTGTAGAGG-3′;
and GAPDH forward, 5′-ACCCAGAAGACTGTGGATGG-3′ and reverse,
5′-TCTAGACGGCAGGTCAGGTC-3′.
Western blot analysis
Total proteins in each cell sample were extracted
using RIPA lysis buffer (Beyotime Institute of Biotechnology).
Proteins were then quantified using a bicinchoninic acid kit
(Pierce; Thermo Fisher Scientific, Inc.), then 40 µg protein/lane
were separated on 10% SDS-PAGE, then transferred to a
polyvinylidene difluoride membrane (PVDF) for 2 h. The PVDF
membrane was subsequently incubated with 5% skim milk for 1 h at
room temperature. Tris buffered saline with Tween 20 (TBST) was
used to wash the membrane. LEF1 primary antibody (1:1,000; cat. no.
ab137872, Abcam) was used to incubate the membrane for 12 h at 4°C.
Horseradish peroxidase-conjugated secondary antibodies (1:5,000;
cat. no. ab7090; Abcam) was then used to further incubate the
membrane at room temperature for 2 h. The membrane was washed with
TBST 3 times for 15 min each time. LEF1 protein expression was
monitored using the Pierce™ ECL Western Blotting Substrate (Thermo
Fisher Scientific, Inc.) and quantified using Quantity One software
v4.62 (Bio-Rad Laboratories Inc.). GAPDH was used as the internal
reference.
Statistical analysis
All data included in the current study was repeated
three times and is expressed as the mean ± standard deviation. A
Student's t-test was used to analyze the differences between two
groups. A one-way ANOVA followed by a Tukey's post-hoc test was
used for multiple comparisons. A Pearson's correlation coefficient
analysis was used to determine the correlation between miR-6852 and
LEF1 expression. SPSS 19.0 (IBM Corp.) was used for statistical
analysis. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-6852 expression is reduced in
glioma tissues and cells
Previous reports have indicated the
tumor-suppressive roles of miR-6852 in a variety of cancer types,
including gastric and colorectal cancer (9–11).
Therefore, it is reasonable to speculate that miR-6852 may be
associated with glioma. miR-6852 expression was assessed using
RT-qPCR in 32 pairs of glioma and corresponding normal tissue
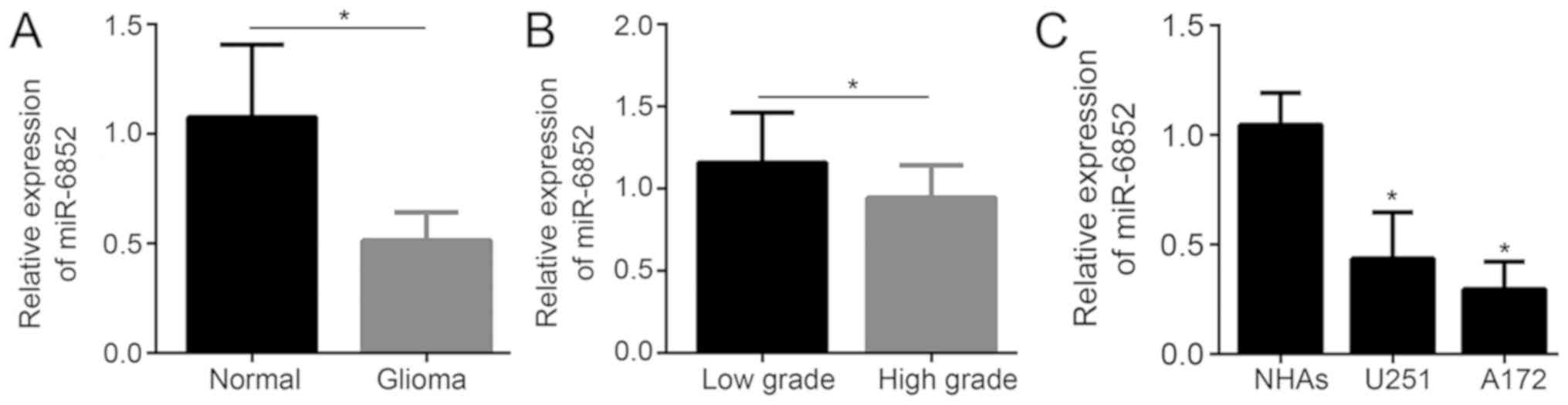
samples. As demonstrated in Fig. 1A,
when compared with normal tissues, glioma tissues exhibited
significantly decreased relative miR-6852 expression (P<0.05).
Tissues of high-grade glioma (n=18) exhibited significantly
decreased relative miR-6852 expression when compared with tissues
of low-grade glioma (n=14; P<0.05; Fig. 1B). miR-6852 expression was
significantly decreased in glioma cells (including U251 and A172
cells) when compared with expression in NHA cells (P<0.05;
Fig. 1C).
miR-6852 upregulation inhibits A172
cell proliferation, migration and invasion
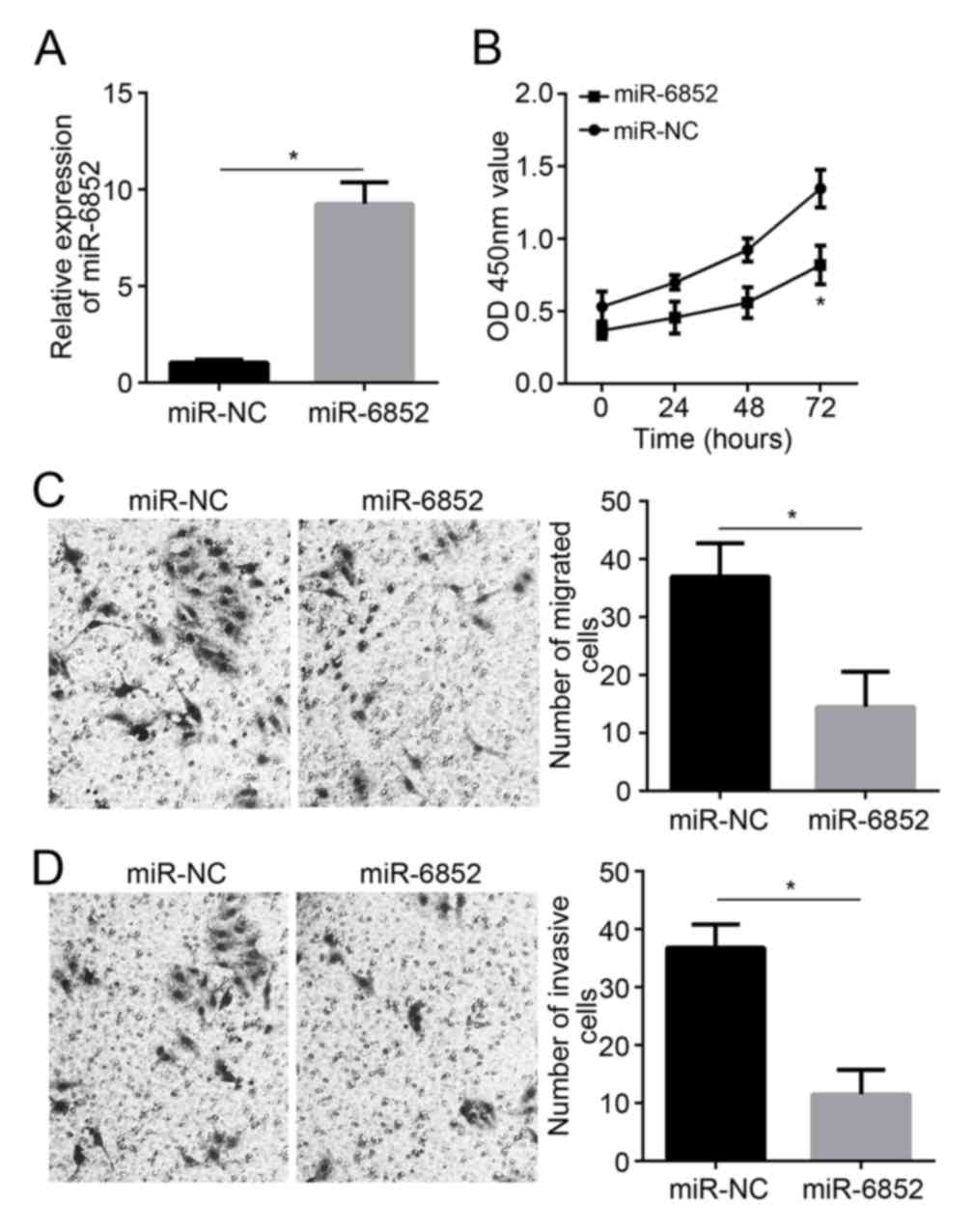
A172 miR-6852 expression significantly increased
subsequent to transfection with miR-6852 mimics (P<0.05;
Fig. 2A). A period of 72 h after
transfection, A172 cells in the miR-6852 group revealed a
significantly decreased OD450 value when compared with the miR-NC
group (P<0.05; Fig. 2B). The
results of the transwell assay revealed that A172 cells in the
miR-6852 group also exhibited a decreased number of migrated and
invasive cells when compared with those in the miR-NC group (all,
P<0.05; Fig. 2C and D).
miR-6852 downregulation promotes A172
cell proliferation, migration and invasion
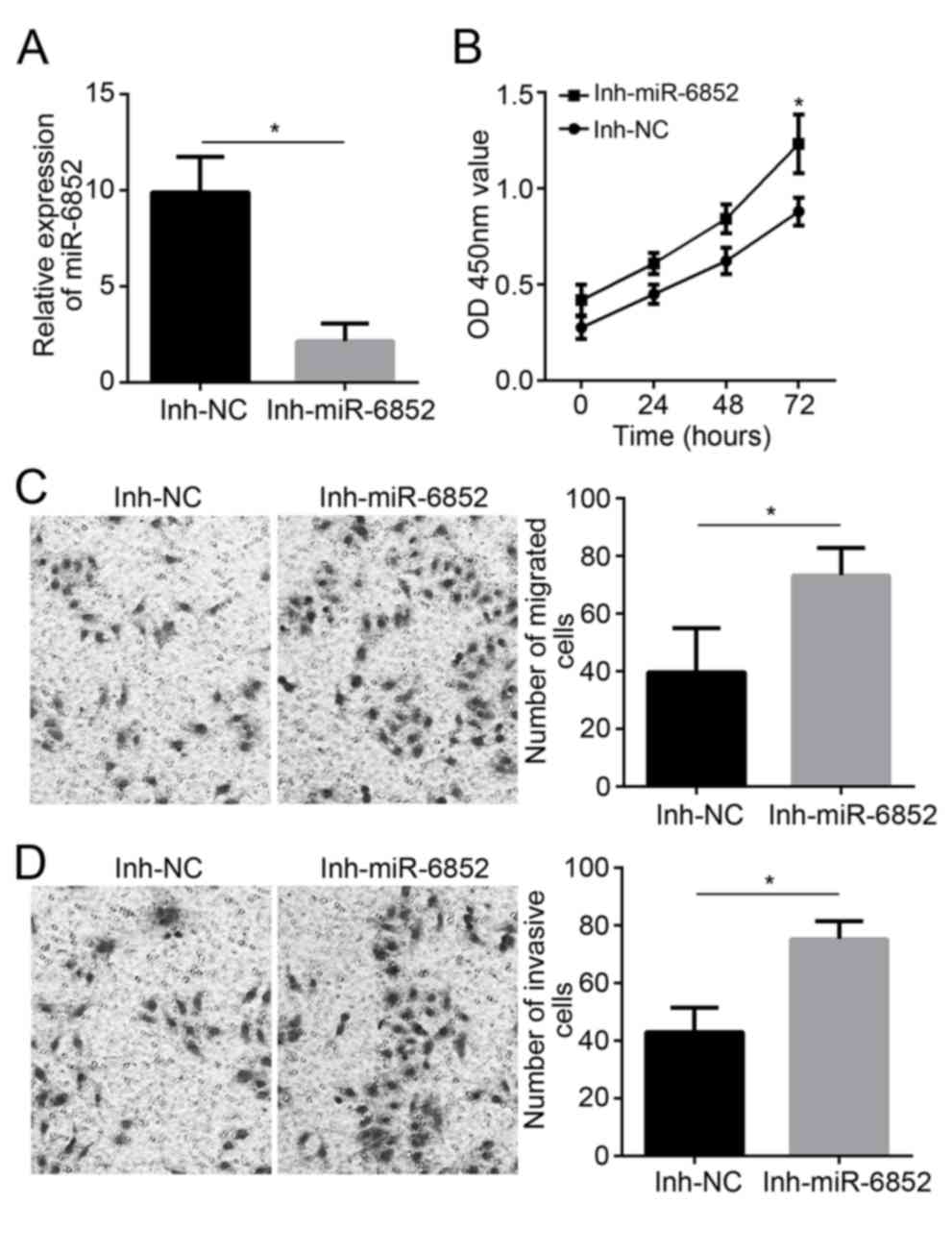
A172 cells transfected with miR-6852 inhibitors and
miR-6852 inhibitor negative controls. As presented in Fig. 3A, transfection with the Inh-miR-6852
inhibitor decreased relative miR-6852 expression in A172 cells
(P<0.05). This decreased miR-6852 expression significantly
enhanced A172 cell proliferation at 72 h subsequent to
transfection, as indicated by the higher OD450 value of A172 cells
in the Inh-miR-6852 group compared with the Inh-NC group
(P<0.05; Fig. 3B). In addition,
the number of migrated and invasive cells of the Inh-miR-6852 group
was significantly increased when compared with the number exhibited
in the Inh-NC group (P<0.05; Fig. 3C
and D).
LEF1 expression is inhibited by
miR-6852
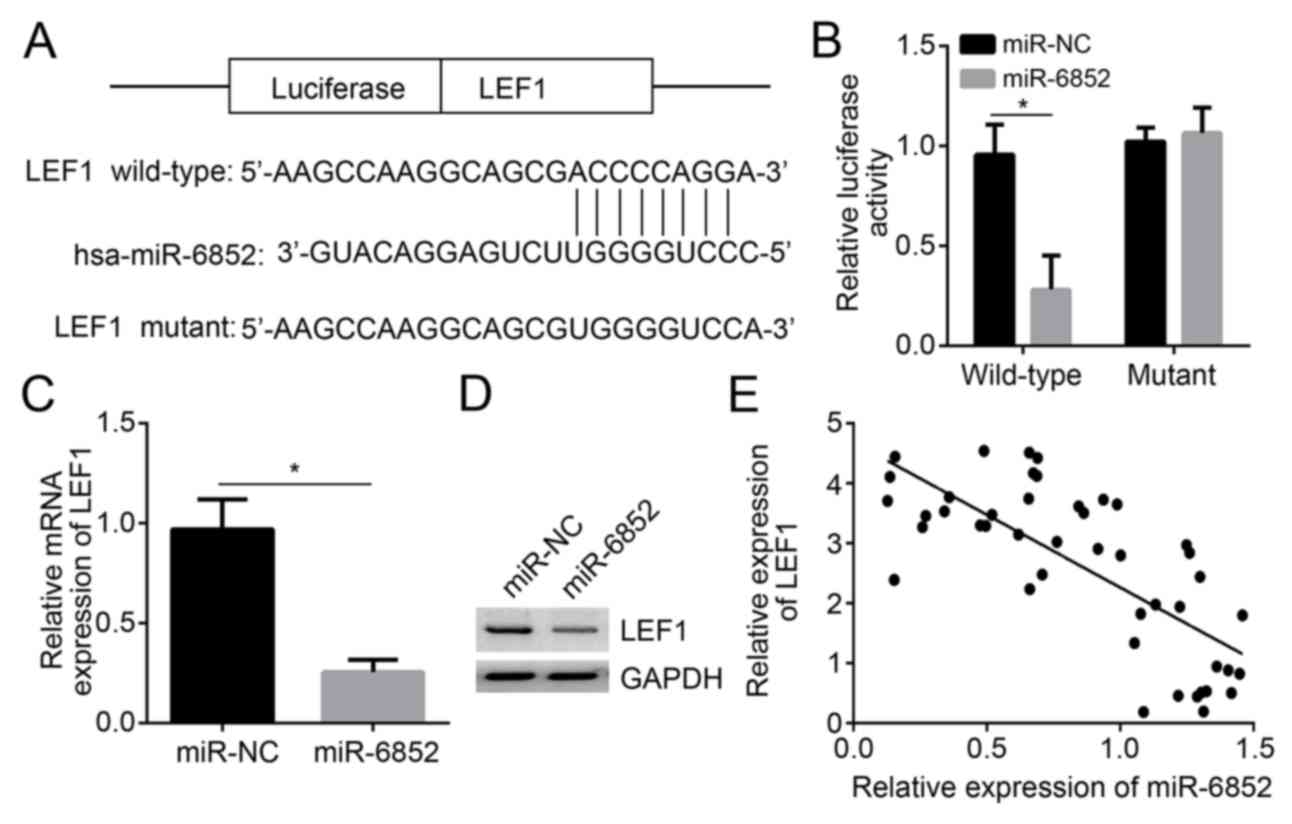
TargetScan predicted that miR-6852 has a direct
binding site with LEF1. Previous studies have also indicated that
miR-6852 targets forkhead box protein J1 (FOXJ1) and transcription
factor 7 (TCF7) in a variety of other types of cancer (9,10).
Therefore, in the present study, WT and mutant sequences of LEF1
were constructed (Fig. 4A).
Furthermore, FOXJ1 and TCF7 containing the binding sites for
miR-6852 were designed (data not shown). The luciferase reporter
gene assay indicated that insertion of LEF1 mutant-type sequences
did not influence the relative luciferase activity of A172 cells in
the miR-NC and miR-6852 group. However, LEF1 wild-type sequence
insertion significantly reduced the relative luciferase activity of
A172 cells in the miR-6852 group when compared with the miR-NC
group (P<0.05; Fig. 4B),
indicating that LEF1 expression was directly suppressed by
miR-6852. Furthermore, the FOXJ1 or TCF7 mutations did not
influence the relative luciferase activity in A172 cells (data not
shown), indicating that miR-6852 targets LEF1 in glioma. In
addition, A127 cell LEF1 mRNA expression in the miR-6852 group was
significantly deceased compared with the expression in the miR-NC
group (P<0.05; Fig. 4C). LEF1
protein level was also decreased by miR-6852 (Fig. 4D). Pearson's correlation analysis
demonstrated that LEF1 expression was negatively correlated with
miR-6852 expression in glioma tissues (Fig. 4E).
LEF1 overexpression reverses the
impaired proliferation, migration and invasion of A172 cells
induced by miR-6852
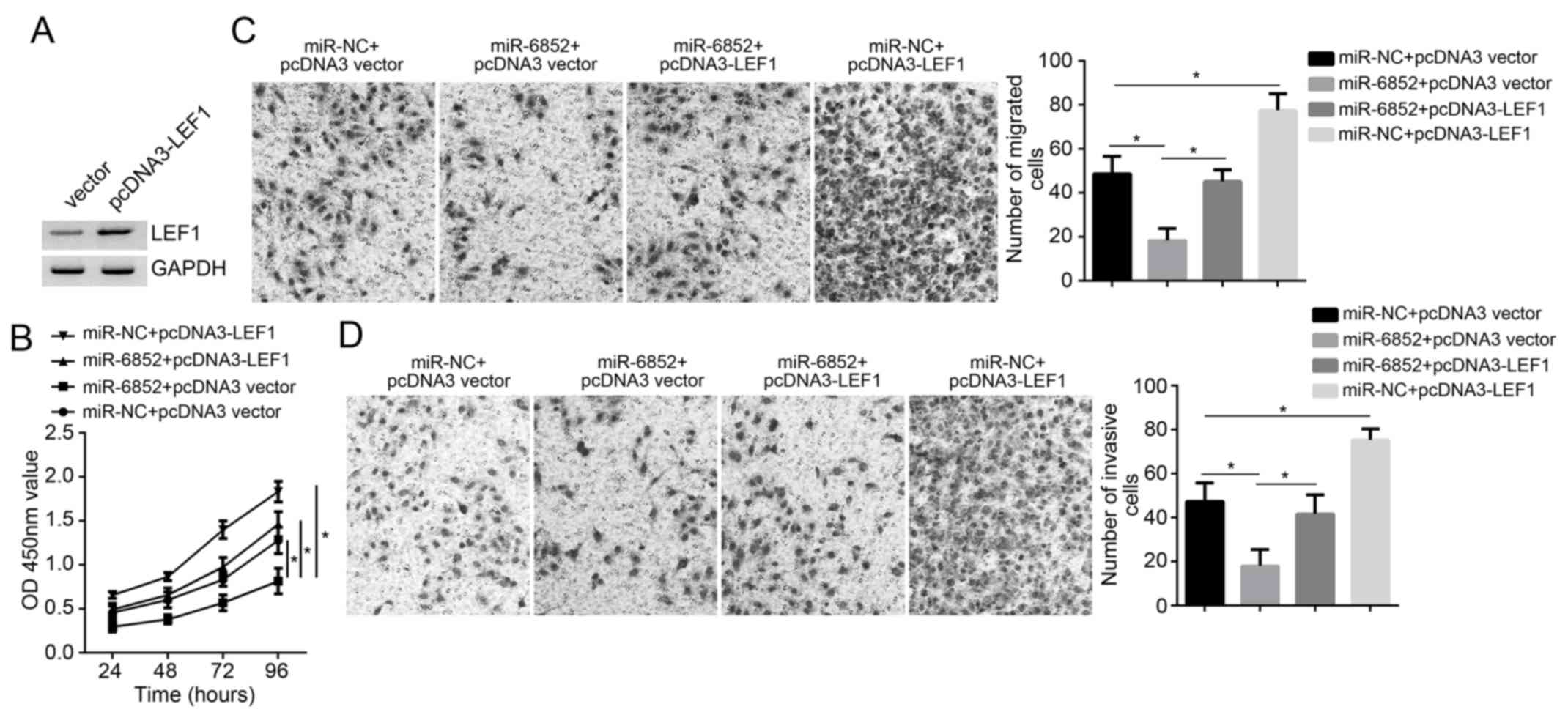
LEF1 protein expression in A172 cells was markedly
upregulated after transfection with pcDNA3-LEF1 overexpression
vectors (Fig. 5A). A172 cell
proliferation, migration and invasion in the miR-NC + pcDNA3 vector
group, miR-6852 + pcDNA3 vector group, miR-6852 + pcDNA3-LEF1 group
and miR-NC + pcDNA3-LEF1 group were also assessed. At 72 h, the
OD450 value of A172 cells in the miR-6852 + pcDNA3 vector group was
lower than that exhibited in the miR-NC + pcDNA3 vector group,
miR-NC + pcDNA3-LEF1 group and miR-6852 + pcDNA3-LEF1 group
(P<0.05; Fig. 5B). In addition,
the miR-6852 + pcDNA3 vector group exhibited a significant decline
in the number of migrated and invasive cells when compared with
those in the miR-6852 + pcDNA3-vector group (P<0.05; Fig. 5C and D). The results indicated that
LEF1 overexpression reversed the impaired proliferation, migration
and invasion of A172 cells induced by miR-6852.
Discussion
Glioma is one of the most deadly malignancies
worldwide (1). Currently, the
standard treatment for glioma is surgical resection and adjuvant
chemotherapy and radiotherapy (1).
However, due to treatment resistance and tumor recurrence, these
treatment strategies do not achieve the desired therapeutic effect
(13). The treatment of glioma is a
clinical problem that requires urgent attention.
The understanding of the molecular pathology of
glioma had led to the discovery of underlying mechanisms that are
associated with glioma development and progression. Previous
studies have demonstrated that in addition to a large number of
coding genes, various non-coding genes (particularly miRNAs) are
also associated with the regulation of glioma development (14,15). A
study of 148 clinical specimens revealed a significant decrease in
miR-424 expression in glioma tumor tissues compared with the
corresponding adjacent normal tissues (14). Results of an in vitro cell
assay indicated the enhanced apoptotic ability and decreased
invasive and migration abilities of glioma cells after miR-424
upregulation (14). Overexpressed
miR-130b was demonstrated to suppress glioma cell apoptosis by
selectively targeting CYLD, which is associated with poor prognosis
of patients with glioma (15).
Therefore, miR-130b was considered as an independent predictor of
poor prognosis in patients with glioma (15). In the current study, miR-6852
expression in glioma tissues and cells was decreased. High-grade
glioma tissues exhibited lower miR-6852 expression than low-grade
glioma tissues. Poudyal et al (11) revealed that the upregulation of
miRNA-6852 could arrest cervical cancer cells at the
G2/M phase and induce cell necrosis. Cui and Hong
(9) demonstrated that miR-6852
expression was reduced in colorectal cancer and was closely
associated with the metastasis and poor prognosis of patients.
In vitro, colorectal cancer cell invasion and proliferation
abilities and the proportion of cells in the S phase were
significantly reduced after miR-6852 overexpression. This research
also indicated that miR-6852 overexpression could suppress glioma
cell proliferation, migration and invasion, while miR-6852
downregulation had the opposite effect.
LEF1 exerts an important regulatory role in the
development of a variety of tissue types and its abnormal
expression and dysfunction could lead to developmental disorders of
tissues (16–18). LEF1 is highly expressed in numerous
animal embryonic tissues and its expression rapidly decreases as
the individual matures (16–18). However, previous research has
demonstrated that LEF1 is upregulated in multiple tumor types. Fang
et al (19) demonstrated that
LEF1 is a novel oncogene that promoted hepatocellular carcinoma
cell proliferation, invasion and migration abilities and
self-renewal ability. Increased LEF1 expression was associated with
advanced TNM stage, lymph node metastases and the reduced overall
survival of patients with colorectal cancer (20). Gao et al (21) revealed that, in glioblastoma
multiforme, LEF1 silencing suppressed U251 cell proliferation,
invasion and migration, indicating that LEF1 could be considered a
novel treatment target for glioblastoma multiforme. In glioma,
elevated LEF-1 expression was one of the hallmarks of malignant
glioma, and this could be used as an important marker for malignant
transformation (22). It has been
previously reported that LEF1 is associated with the signal
transduction of Wnt/β-catenin downstream molecules. Previous
studies have revealed that LEF1 binds to β-catenin to form a
β-catenin/LEF complex, thereby promoting the expression of Wnt
signaling pathway downstream molecules, including c-Myc and Cyclin
D1 (23,24). LEF1 tumor expression was also
mediated by miRNAs, including miR-218, miR-181a and miR-227
(23,25,26). To
the best of our knowledge, the current study revealed that LEF1 is
directly inhibited by miR-6852 for the first time, and the
overexpression of LEF1 reversed the impaired proliferation,
migration and invasion of A172 cells induced by miR-6852.
In conclusion, the current study indicated that
miR-6852 inhibited the proliferation, migration and invasion of
glioma cells by inhibiting the expression of LEF1. Therefore,
miR-6852 could be used as a novel biological target for the
treatment of glioma. Subsequent data on how miR-6852 affects glioma
should be obtained to provide a more reliable theoretical basis for
this targeted glioma therapy.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
JW and WD contributed to the conception and design
of the present study. In addition, WD analyzed and interpreted the
results and wrote the manuscript. JW, HL, KZ and SZ performed the
experiments. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by The Ethics
Committee of Affiliated Hospital of Hebei University. Written
informed consent was obtained from all enrolled patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lu B, Zhou Y, Su Z, Yan A and Ding P:
Effect of CCL2 siRNA on proliferation and apoptosis in the U251
human glioma cell line. Mol Med Rep. 16:3387–3394. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Latha K, Yan J, Yang Y, Gressot LV, Kong
L, Manyam G, Ezhilarasan R, Wang Q, Sulman EP, Xu J, et al:
Abstract 5606: Fibrinogen-like protein 2 drives malignant tumor
progression in glioma. Cancer Res. 77:56062017.
|
|
3
|
Ellert-Miklaszewska A, Ciechomska IA and
Kaminska B: Chapter e11 cannabinoid signaling in glioma cells and
therapeutic implications. Handbook Cannabis Relat Pathologies.
e111–e121. 2017. View Article : Google Scholar
|
|
4
|
Lv QL, Du H, Liu YL, Huang YT, Wang GH,
Zhang X, Chen SH and Zhou HH: Low expression of microRNA-320b
correlates with tumorigenesis and unfavorable prognosis in glioma.
Oncol Rep. 38:959–966. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kalogianni DP, Kalligosfyri PM, Kyriakou
IK and Christopoulos TK: Advances in microRNA analysis. Anal
Bioanal Chem. 410:695–713. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Santangelo A, Imbrucè P, Gardenghi B,
Belli L, Agushi R, Tamanini A, Munari S, Bossi AM, Scambi I, Benati
D, et al: A microRNA signature from serum exosomes of patients with
glioma as complementary diagnostic biomarker. J Neurooncol.
136:51–62. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang J, Lv J, Zhang F, Che H, Liao Q,
Huang W, Li S and Li Y: MicroRNA-211 expression is down-regulated
and associated with poor prognosis in human glioma. J Neurooncol.
133:553–559. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li Y, Li Y, Ge P and Ma C: MiR-126
Regulates the ERK pathway via targeting KRAS to inhibit the glioma
cell proliferation and invasion. Mol Neurobiol. 54:137–145. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cui BH and Hong X: miR-6852 serves as a
prognostic biomarker in colorectal cancer and inhibits tumor growth
and metastasis by targeting TCF7. Exp Ther Med. 16:879–885.
2018.PubMed/NCBI
|
|
10
|
Yu H, Zhang J, Wen Q, Dai Y, Zhang W, Li F
and Li J: MicroRNA-6852 suppresses cell proliferation and invasion
via targeting forkhead box J1 in gastric cancer. Exp Ther Med.
16:3249–3255. 2018.PubMed/NCBI
|
|
11
|
Poudyal D, Herman A, Adelsberger JW, Yang
J, Hu X, Chen Q, Bosche M, Sherman BT and Imamichi T: A novel
microRNA, hsa-miR-6852 differentially regulated by Interleukin-27
induces necrosis in cervical cancer cells by downregulating the
FoxM1 expression. Sci Rep. 8:9002018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lin L, Cai J and Jiang C: Recent advances
in targeted therapy for glioma. Curr Med Chem. 24:1365–1381. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jin C, Li M, Ouyang Y, Tan Z and Jiang Y:
MiR-424 functions as a tumor suppressor in glioma cells and is
down-regulated by DNA methylation. J Neurooncol. 133:247–255. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xiao ZQ, Yin TK, Li YX, Zhang JH and Gu
JJ: miR-130b regulates the proliferation, invasion and apoptosis of
glioma cells via targeting of CYLD. Oncol Rep. 38:167–174. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guo X, Zhang R, Liu J, Li M, Song C, Dovat
S, Li J and Ge Z: Characterization of LEF1 high expression and
novel mutations in adult acute lymphoblastic leukemia. PLoS One.
10:e01254292015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Menter T, Trivedi P, Ahmad R, Flora R,
Dirnhofer S, Tzankov A and Naresh KN: Diagnostic utility of
lymphoid enhancer binding factor 1 immunohistochemistry in small
B-cell lymphomas. Am J Clin Pathol. 147:292–300. 2017.PubMed/NCBI
|
|
18
|
Santiago L, Daniels G, Wang D, Deng FM and
Lee P: Wnt signaling pathway protein LEF1 in cancer, as a biomarker
for prognosis and a target for treatment. Am J Cancer Res.
7:1389–1406. 2017.PubMed/NCBI
|
|
19
|
Fang S, Lei LI, Liu M, Tsang J and Guan X:
Abstract 2434: LEF1 negatively regulates differentiation of HCC by
activation of Notch signaling pathway. Cancer Res. 77:24342017.
|
|
20
|
Wang WJ, Yao Y, Jiang LL, Hu TH, Ma JQ,
Ruan ZP, Tian T, Guo H, Wang SH and Nan KJ: Increased LEF1
expression and decreased Notch2 expression are strong predictors of
poor outcomes in colorectal cancer patients. Dis Markers.
35:395–405. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gao X, Mi Y, Ma Y and Jin W: LEF1
regulates glioblastoma cell proliferation, migration, invasion, and
cancer stem-like cell self-renewal. Tumor Biol. 35:11505–11511.
2014. View Article : Google Scholar
|
|
22
|
Pećina-Šlaus N, Kafka A, Tomas D, Marković
L, Okštajner PK, Sukser V and Krušlin B: Wnt signaling
transcription factors TCF-1 and LEF-1 are upregulated in malignant
astrocytic brain tumors. Histol Histopathol. 29:1557–1564.
2014.PubMed/NCBI
|
|
23
|
Liang J, Li X, Li Y, Wei J, Daniels G,
Zhong X, Wang J, Sfanos K, Melamed J, Zhao J and Lee P: LEF1
targeting EMT in prostate cancer invasion is mediated by miR-181a.
Am J Cancer Res. 5:1124–1132. 2015.PubMed/NCBI
|
|
24
|
Lyou Y, Habowski AN, Chen GT and Waterman
ML: Inhibition of nuclear Wnt signaling: Challenges of an elusive
target for cancer therapy. Br J Pharmacol. 174:4589–4599. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Qiu J, Hao Y, Huang S, Ma Y, Li X, Li D
and Mao Y: MiR-557 works as a tumor suppressor in human lung
cancers by negatively regulating LEF1 expression. Tumour Biol.
39:10104283177094672017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu Y, Yan W, Zhang W, Chen L, You G, Bao
Z, Wang Y, Wang H, Kang C and Jiang T: MiR-218 reverses high
invasiveness of glioblastoma cells by targeting the oncogenic
transcription factor LEF1. Oncol Rep. 28:1013–1021. 2012.
View Article : Google Scholar : PubMed/NCBI
|