Introduction
Long non-coding RNAs (LncRNAs) are a novel class of
non-coding RNA, which are longer than 200 nucleotides. Increasing
evidence have suggested that LncRNAs serve a crucial role in
various pathological and physiological processes, including cell
proliferation, differentiation and metabolism. Furthermore, LncRNAs
participate in various key regulatory processes, which are not
limited to X-chromosome silencing, genomic imprinting, chromatin
modification, transcriptional activation, transcriptional
interference and intranuclear transport (1).
Ischemia/reperfusion (I/R) is a pathological process
characterized by a reduction of blood supply to tissues, followed
by the subsequent restoration of perfusion and concomitant
re-oxygenation (2). Through a
multitude of mechanisms, the re-establishment of blood flow causes
secondary damage to the ischemic tissue, producing an I/R injury.
I/R injury predominantly contributes to the morbidity and mortality
of coronary artery disease. Myocardial infarction, and the
subsequent acute loss of viable myocardium, is the leading cause of
mortality in industrialized countries, and has been reported to
account for >400,000 fatalities a year in the United States
(3). Even if a patient survives the
acute phase of myocardial infarction, the subsequent injury to
cardiac function accompanied by myocardial remodeling will notably
lower the quality of life.
The pathogenesis of I/R injury is complex, and a
combination of several mechanisms act either independently or
together (3). LncRNAs are known to
have various functions binding proteins or microRNAs (miRs).
Numerous studies have demonstrated that LncRNAs may function as
competing endogenous RNAs that can crosstalk with mRNAs by
competitively binding to their common miRs (4–6). Among
these LncRNAs, Nuclear Enriched Abundant Transcript 1 (NEAT1) is a
critical structural component of paraspeckles and is essential for
paraspeckle formation (7). NEAT1 has
two isoforms that share the same transcription start site. With its
structural characteristics, NEAT1 has been reported to contribute
to the progression of various of types of cancer including breast
and prostate cancer. However, there are relatively few studies,
which have examined the association between NEAT1 and ischemic
injury.
In the present study, a hypoxia/reoxygenation (H/R)
cell model was established to explore the biological effect of
NEAT1 and miR-520a. The expression level of NEAT1 in the
H/R-induced cardiomyocyte injury model was evaluated. Subsequently,
a loss of function study was used to investigate the role of NEAT1
in myocardial injury. Furthermore, the underlying molecular
mechanism of NEAT1 in myocardial injury was explored. It was
identified that NEAT1 could be a potential therapeutic biomarker
for the diagnosis and treatment of ischemia heart disease. A novel
explanation for the mechanism of myocardial injury was
suggested.
Materials and methods
Cell culture and low oxygen
treatments
The H9c2 cardiomyocyte cell line was purchased from
American Type Culture Collection (Manassas, VA, USA). Cells were
cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented
with 12% fetal bovine serum (FBS) and 1% antibiotics (streptomycin
and penicillin) in a humidified incubator atmosphere containing 5%
CO2 at 37°C. To establish the H/R model, H9c2 cells were
cultured in a humid atmosphere containing 5% CO2 and 95%
N2 for 12 h followed by incubation under normal
conditions (5% CO2) for 0, 3, 6 and 12 h.
I/R model
A total of 40 Sprague Dawley male rats (age, 8
weeks; weight, 230–250 g) were randomly divided into five groups
(n=8/group): Sham, 0 min post-I/R, 30 min post-I/R, 60 min post-I/R
and 120 min post-I/R. Rats had access to food and water ad
libitum and were maintained under controlled conditions
(temperature, 22–25°C; 50–60% humidity; 12-h light-dark cycle, with
lights on at 0700 h). Rats were anesthetized with an
intraperitoneal injection of pentobarbital (40 mg/kg; Sinopharm
Chemical Reagent Co., Ltd., Shanghai, China) and artificially
ventilated. Following this, the left anterior descending coronary
arteries (LADs) were exposed and 6–0 silk sutures were used to
ligate the LADs. Cyanosis in the anterior ventricular walls was
used to confirm ligation. Ischemia was induced using small vinyl
tubes that were threaded through the ligatures. Following 30 min of
ischemia, tubes were translocated and coronary circulation was
restored for 0, 30, 60 or 120 min, as previously described
(8). Following reperfusion, the
blood vessel connecting the heart was severed. The hearts were then
removed. Subsequently, the infarct area of myocardium tissues and
blood samples were collected for further analysis. Sham control
animals were subjected to the entire surgical procedure without LAD
ligation. All procedures were performed following the Declaration
of Helsinki of the World Medical Association. The research protocol
was approved by the Ethics Committee of The First Affiliated
Hospital of Jiaxing University (Jiaxing, China).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from H9c2 cells and heart
tissue from I/R rats using TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the
manufacturer's protocol. RNA concentration was detected using a
NanoDrop Spectrophotometer (Thermo Fisher Scientific, Inc.). Total
RNA was reverse transcribed into cDNA using QuantiTect Reverse
Transcription kit (Qiagen China Co., Ltd., Shanghai, China) using
the following thermocycling conditions: 50°C for 60 min followed by
70°C for 15 min. The relative expression levels of lncRNAs and miRs
were detected using the SYBR Green qPCR Master mix (Applied
Biosystems; Thermo Fisher Scientific, Inc.) and a 7500 Fast
Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. The following
primer pairs were used for the qPCR: NEAT1 forward,
5′-GCTCTGGGACCTTCGTGACTCT-3′ and reverse,
5′-CTGCCTTGGCTTGGAAATGTAA-3′; GAPDH forward,
5′-CCTCGTCCCGTAGACAAAATG-3′ and reverse,
5′-TCTCCACTTTGCCACTGCAA-3′; miR-520a forward,
5′-ACACTCCAGCTGGGAAAGTGCTTCCC-3′ and reverse,
5′-CTCAACTGGTGTCGTGGA-3′; and U6 forward, 5′-CTCGCTTCGGCAGCACA-3′
and reverse, 5′-AACGCTTCACGAATTTGCGT-3′. The following
thermocycling conditions were used for the qPCR: Initial
denaturation at 50°C for 2 min, 95°C for 10 min; 40 cycles of 95°C
for 15 sec and 60°C for 60 sec. lncRNA and miR levels were
quantified using the 2−ΔΔCq method and normalized to
GAPDH and U6, respectively (9).
Adenovirus construction
The short hairpin (sh)RNA sequence targeting NEAT1
was 5′-GCCAUCAGCUUUGAAUAAAUU-3′, the shRNA control sequence was
5′-CCATGACTTCGGATCGGGTCG-3′, the sequence of miR-520a was
5′-AAAGUGCUUCCCUUUGGACUGU-3′ and the control scramble sequence of
miR-520a was 5′-UUCUCCGAACGUGUCACGUTT-3′ were synthesized by
Guangzhou RiboBio Co., Ltd. (Guangzhou, China). Adenovirus-mediated
overexpression of NEAT1 was performed using the following primer
pairs: NEAT1 forward, 5′-CTTCCTCCCTTTAACTTATCCATTCAC-3′ and
reverse, 5′-CTCTTCCTCCACCATTACCAACAATAC-3′; and for control
forward, 5′-ACTTTATATGCCGCTACCTACTAT-3′ and reverse,
5′-CCTATACTGTGGCTATGGCAGTACT-3′. These primers were synthesized by
Guangzhou RiboBio Co., Ltd. (Guangzhou, China). The adenovirus
construction system including the pHB-U6-CMVMCS-PGK-ZsGreen-pur,
pSPAX2 and pMD2G vectors were kindly provided by Tianjin Medical
University (Tianjin, China). The shRNA sequences were cloned into
the pHB-U6-CMVMCS-PGK-ZsGreen-puro vector followed by the
transduction of 293T cells with the packaging plasmid pSPAX2 and
the envelope plasmid pMD2G using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). Following 6-h
transfection, the medium was replaced with DMEM supplemented with
10% FBS. Following a 72-h incubation, the 293T lentiviral
supernatant (Ad-shNEAT1, Ad-shctrl, Ad-miR-520 and Ad-ctrl) was
harvested following 72-h for centrifugation at 1,000 × g for 2 h at
4°C to remove cell debris.
To determine the viral titer, 293T cells were seeded
into 96-well plates at a density of 5×103 cells/well.
Lentiviral supernatant was serially diluted three times and 10 µl
of serially diluted lentiviral vector was added to each
corresponding well, respectively. DMEM culture medium was added to
a total volume of 100 µl. Following 48-h incubation, the medium was
replaced with 100 µl complete DMEM medium supplemented with 1.5
µg/ml puromycin. Following 24-h incubation, the medium was replaced
with complete medium without puromycin. Fluorescence was observed
under a fluorescent microscope(Zeiss GmbH, Jena, Germany). Viral
titer was calculated using the following formula: viral titer
(TU/ml)= cell number × fluorescence percentage × MOI × virus
dilution multiplier × 103.
Transfection with shRN lentiviral
plasmid
In the in vitro study, H9c2 cells were seeded
into six-well plates and incubated for 24 h. Following incubation,
the adenovirus solution was used to transfect cells at a
multiplicity of infection (MOI) of 0. 1 for 24 h. Transfection
efficiency was examined following 24-h transfection. Cells were
observed under a fluorescence microscope (magnification, ×100,
Zeiss GmbH). The ratio of green (positive) cells compared with the
total number of cells indicated the infection efficiency.
In the in vivo study, 3 and 7 days prior to
the establishment of the I/R model, rats received a tail vein
injection with the adenovirus at a MOI of 0.6. The efficiency of
adenovirus knockdown and gene overexpression was evaluated using
RT-qPCR.
Luciferase assay
Bioinformatics analysis was performed using
TargetScanHuman (www.targetscan.org/vert_72) to predict NEAT1 as a
target gene of miR-520a and the luciferase assay was performed as
previously described (10). The
3′-untranslated region (UTR) of NEAT1, harboring either the
wild-type or mutant miR-520a binding site was cloned into the
psiCHECK-2 vector (Promega Corporation, Madison, WI, USA)
immediately downstream of the stop codon of the luciferase gene to
generate the psiCHECK-PDK1-3′-UTR luciferase reporter plasmid.
Plasmid DNA and miR-520a mimics or control miRs (Shanghai
GenePharma Co., Ltd., Shanghai, China) were co-transfected into
293T cells using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) for 72 h. Luciferase activities were
measured with a Dual-Glo Luciferase Assay System (Promega
Corporation). Firefly luciferase activity was normalized to
Renilla luciferase activity and all experiments were
performed in triplicate.
H/R model
H9c2 cells were cultured in a modular incubator
(Model 3131; Thermo Fisher Scientific, Inc.) containing 1%
O2, 94% N2 and 5% CO2. Following
hypoxia for 24 h, the cells were exposed to 95% air, 5%
CO2 and 37°C for 12 h. Cells in the normal control group
were cultured with 5% CO2 at 37°C.
Flow cytometry assay
Cells were trypsinized and re-suspended in PBS. Cell
apoptosis was analyzed using the Annexin- V-FITC Apoptosis
Detection kit (cat. no. C1062S; Beyotime Institute of
Biotechnology, Haimen, China). Briefly, following 48-h
transfection, 1×105 H9c2 cells were harvested with
trypsin, washed with PBS and resuspended in 100 µl binding buffer.
Cells were subsequently stained with 5 µl Annexin V-FITC in the
dark for 10 min at room temperature, followed by incubation with 5
µl PI solution for 5 min at room temperature. Cell apoptosis was
detected using a FACSVerse flow cytometer (BD Biosciences, San
Jose, CA, USA). Data were analyzed using FlowJo software (version
7.6.5; BD Biosciences).
Western blotting
Total protein was extracted from cells using
radioimmunoprecipitation assay buffer (Tianjin Bioco Biochemical
Co., Ltd., Tianjin, China) containing 0.1 mM proteinase inhibitor.
Total protein was quantified and 40 mg protein/lane was separated
via SDS-PAGE on 10% gels. The separated proteins were transferred
onto polyvinylidene fluoride membranes (EMD Millipore Corporation,
Billerica, MA, USA) and blocked for 2 h at room temperature with 5%
skimmed milk. The membranes were incubated with primary antibodies
against B-cell lymphoma-2 (Bcl-2, 1:200; cat. no. ab196495),
Bcl-2-associated × protein (Bax, 1:200; cat. no. ab182734), cleaved
Caspase-3 (1:200; cat. no. ab214430), Caspase-3 (1:200; cat. no.
ab90437), GAPDH (1:200; cat. no. ab181602; all Abcam, Cambridge,
MA, USA) overnight at 4°C. Membranes were washed three times in
Tris-buffered saline with Tween 20. Following primary incubation,
membranes were incubated with horseradish peroxidase-conjugated
secondary antibody (1:1,000; cat. no. A0208; Beyotime Institute of
Biotechnology) for 2 h at room temperature. Protein bands were
visualized using an enhanced chemiluminescence reagent (Tianjin
Bioco Biochemical Co., Ltd.). Protein expression was quantified
using ImageJ software (version 1.48; National Institutes of Health,
Bethesda, MD, USA) with GAPDH as an internal control.
Rescue experiments
To further confirm the association between miR-520a
and NEAT1, a rescue experiment was performed using
adenovirus-mediated overexpression of miR-520a and NEAT1. H9c2
cells were infected with miR-520a and NEAT1 overexpression
adenoviruses for 48 h. Following 48-h infection, cells were
subjected to H/R treatment. Flow cytometry and Terminal
deoxynucleotidyl-transferase-mediated dUTP nick end labeling
(TUNEL) staining were performed to evaluate apoptosis.
TUNEL staining
Heart specimens were embedded in paraffin and cut
into 5-µm-thick sections. Following deparaffinization and
rehydration, the paraffin sections were washed in PBS three times.
Subsequently, the sections were incubated with Proteinase K working
solution (10 µg/ml in 10 mM Tris/HCl, pH 7.5–8.0) at 37°C for 20
min and then treated with TUNEL working solution (Roche
Diagnostics, Basel, Switzerland) according to the manufacturer's
instructions. Following this, sections were counter stained with
4′, 6-diamidino-2-phenylindole for 5 min at 37°C.
Statistical analysis
All data were expressed as the mean ± standard
deviation, and P<0.05 was considered to indicate a statistically
significant difference. A two-tailed unpaired Student's t-test or
one-way analysis of variance (ANOVA) was used to compare two or
more than three groups, respectively. ANOVA with Scheffe's post hoc
test was used to evaluate the statistical significance of
differences between groups. Pearson correlation analysis was used
to examine the association between miR520a and NEAT1.
Results
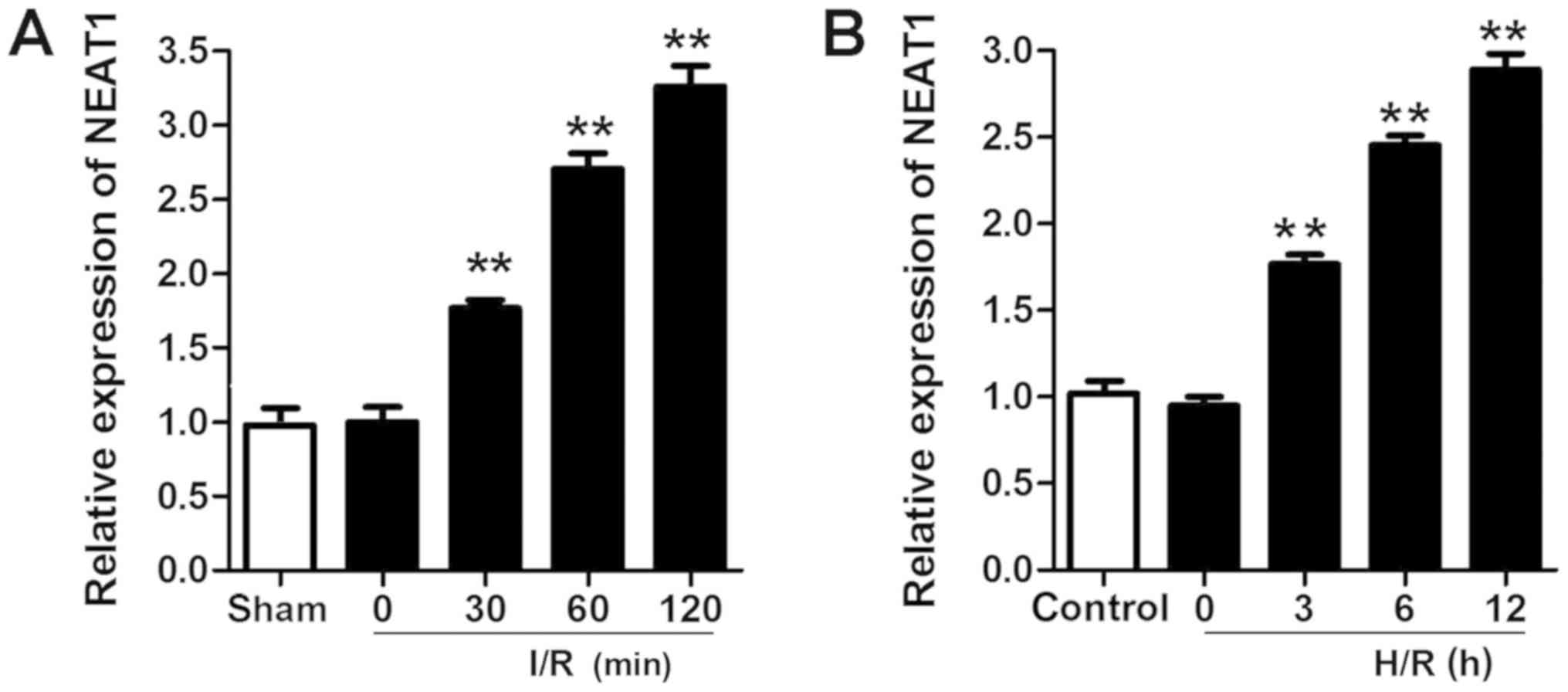
NEAT1 is upregulated in the myocardium
and cardiomyocytes subjected to I/R or H/R injury
A rat I/R injury model and a H/R-induced
cardiomyocyte injury model were established. RT-qPCR was performed
to evaluate the expression of NEAT1. NEAT1 was significantly
upregulated in the myocardium and cardiomyocytes subjected to I/R
and H/R injury (Fig. 1).
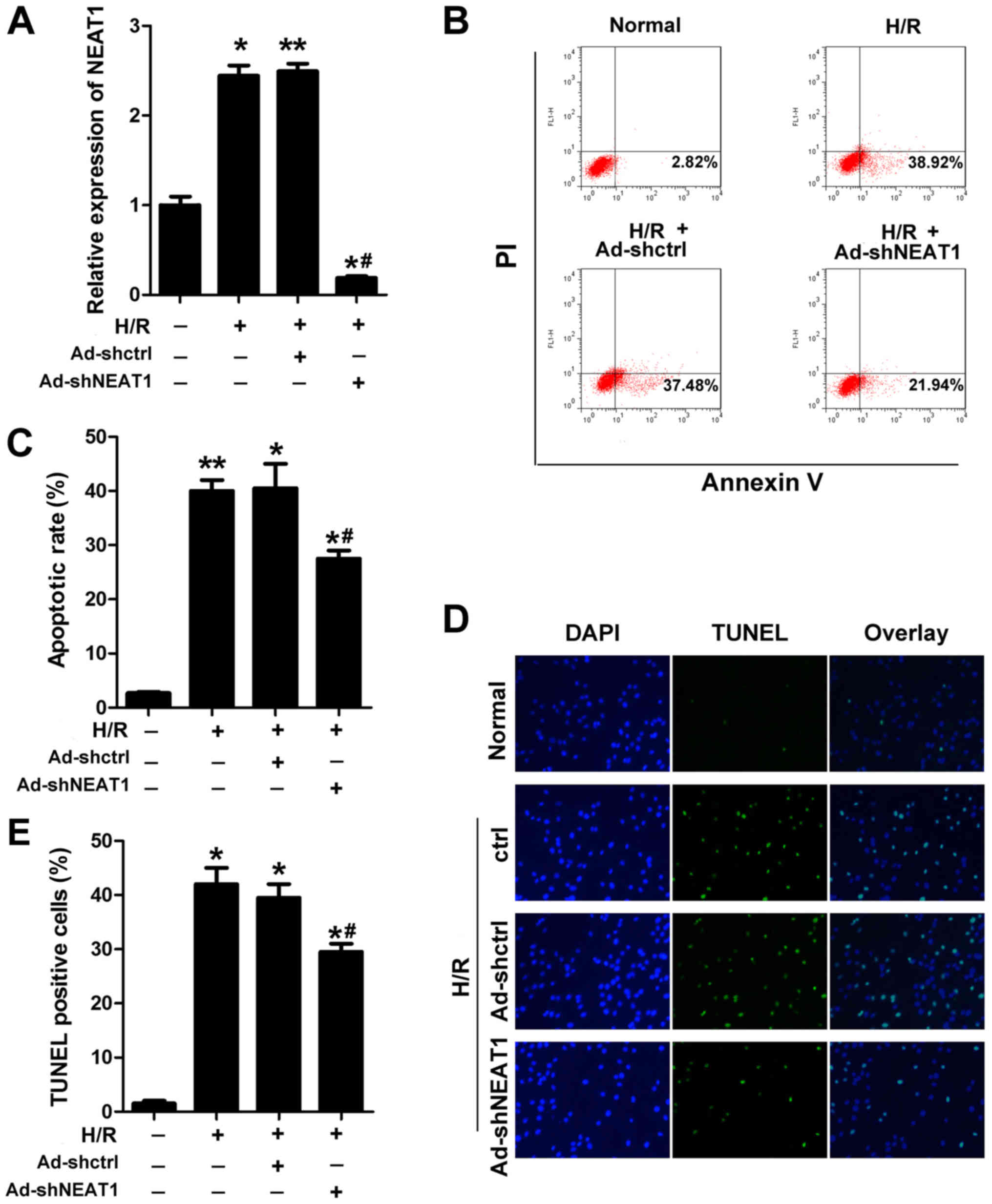
NEAT1 knockdown inhibits cardiomyocyte
apoptosis induced by H/R
The cardiomyocyte H/R model was established to
investigate the effect of NEAT1. Annexin V-FITC/PI staining with
flow cytometry and TUNEL staining were performed to evaluate the
apoptosis of cardiomyocytes. Ad-shNEAT1 was used to knock down the
expression of NEAT1, which was effective (Fig. 2A). NEAT1 knockdown with the
application of Ad-shNEAT1 significantly reduced the rate of
apoptosis of cardiomyocytes subjected to H/R (Fig. 2B-E).
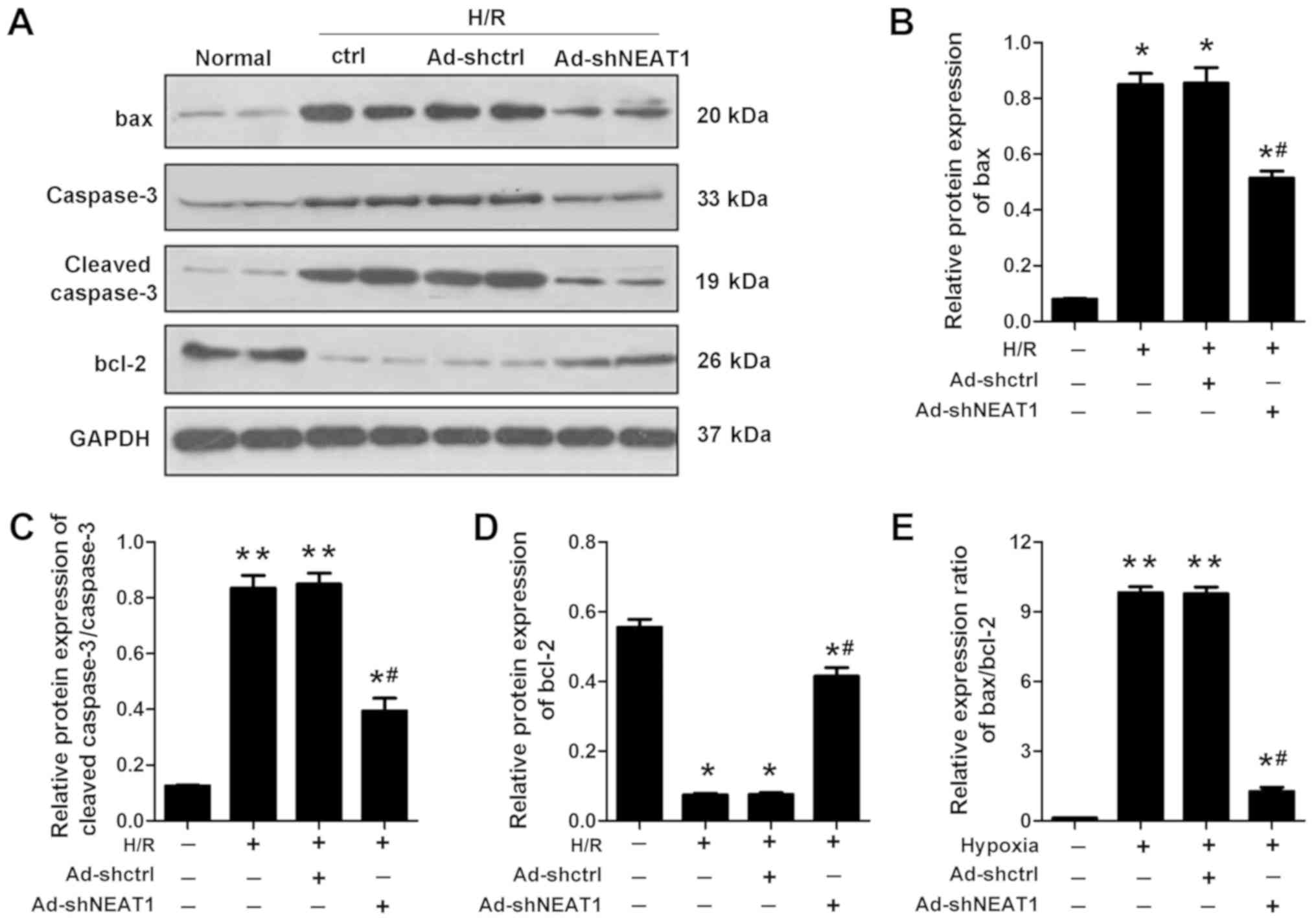
NEAT1 knockdown regulates the
expression of apoptotic proteins
To further determine that NEAT1 regulates the
apoptosis of cardiomyocytes, the relative protein expression levels
of apoptotic proteins Bax, Bcl-2 and cleaved-caspase3 were
assessed. H/R significantly elevated the expression levels of Bax,
caspase3 and cleaved caspase-3, but inhibited the expression level
of Bcl-2 compared with the control group. The knockdown of NEAT1
significantly reduced the expression levels of Bax, caspase3 and
cleaved-caspase3 that were elevated by H/R. Furthermore, knockdown
of NEAT1 significantly increased the expression levels of Bcl-2
that were inhibited by H/R (Fig.
3).
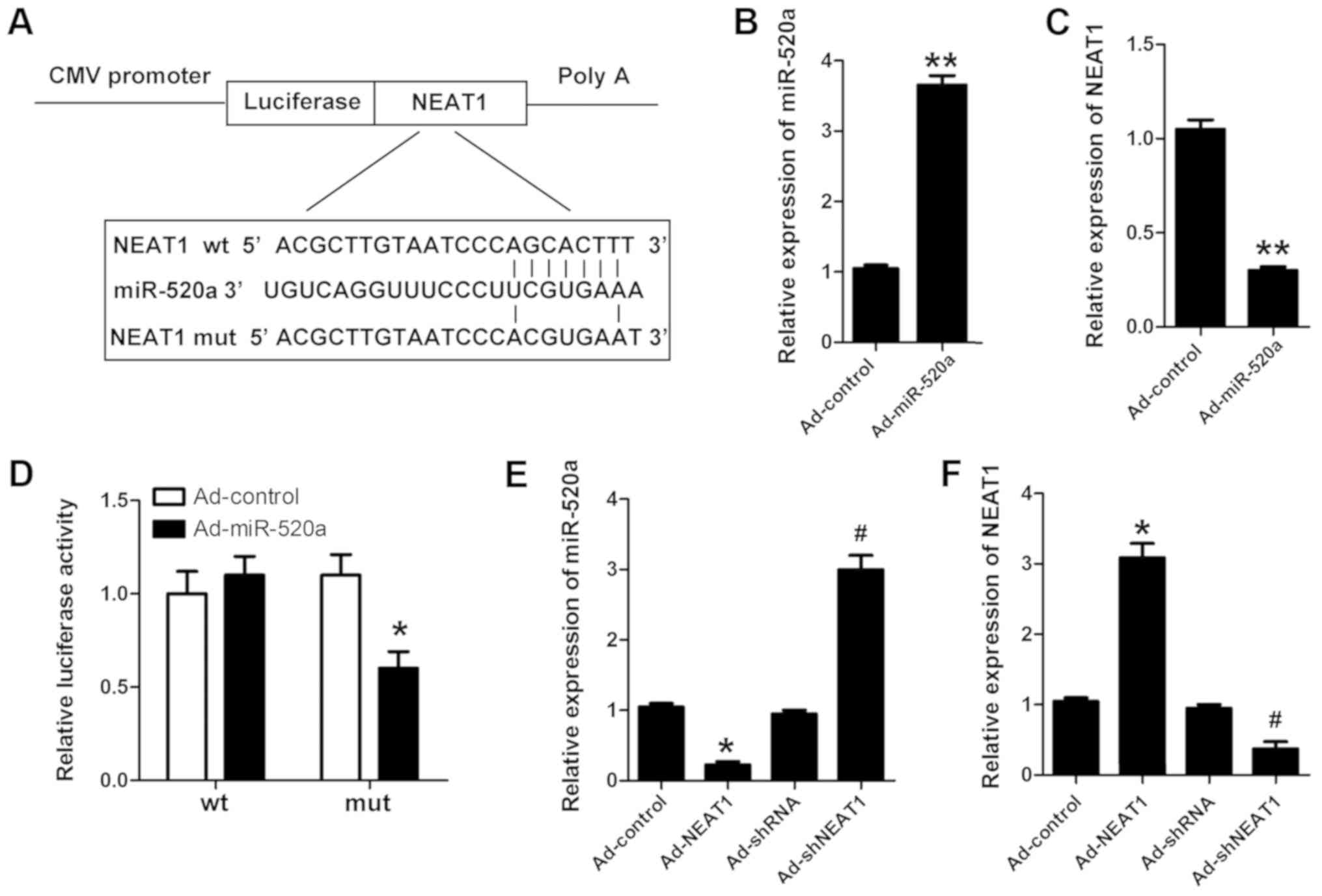
NEAT1 is the target of miR-520a
LncRNA participates in cell processes through
various mechanisms, including sequestering targeting miRs.
Bioinformatics analysis was used to predict NEAT1 as a target gene
of miR-520a (Fig. 4A). Luciferase
activity was assessed to determine whether NEAT1 binds to miR-520a.
miR-520a was successfully overexpressed in cardiomyocytes
transfected with Ad-miR-520a (Fig.
4B). The expression of NEAT1 was reduced in cardiomyocytes
transfected with Ad-miR-520a (Fig.
4C). Furthermore, miR-520a was indicated to directly target
NEAT1 (Fig. 4D-F).
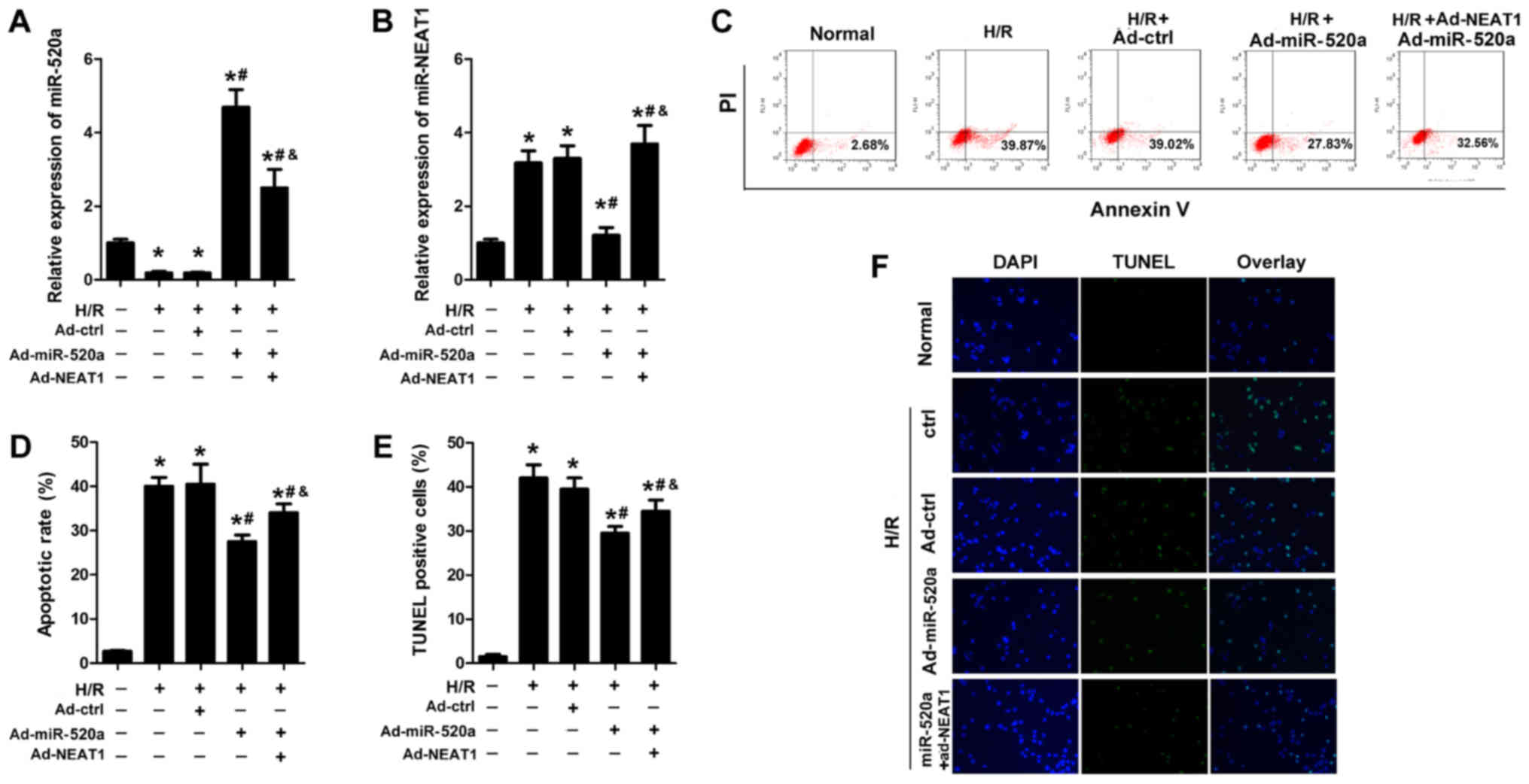
miR-520a overexpression inhibits
cardiomyocyte apoptosis induced by H/R, and NEAT1 reverses this
effect
The effect of miR-520a on cardiomyocyte apoptosis
was assessed. Ad-miR-520a and Ad-NEAT1 were used to elevate
miR-520a and NEAT1 expression, respectively (Fig. 5A and B). Flow cytometry and TUNEL
assay results indicated that miR-520a overexpression resulted in a
similar effect as knockdown of NEAT1 and significantly reduced the
apoptosis rate of cardiomyocytes subjected to H/R (Fig. 5C-F). NEAT1 overexpression
significantly reversed the effects of miR-520a on cardiomyocyte
apoptosis (Fig. 5C-F).
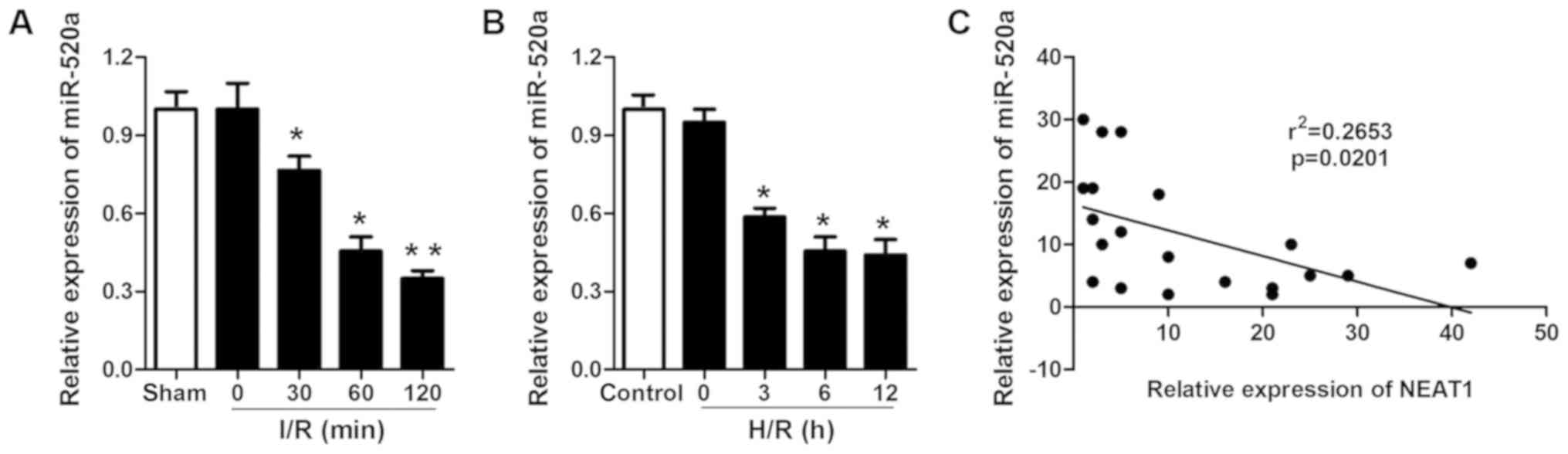
miR-520a is downregulated in I/R
myocardium and H/R cardiomyocytes and is negatively correlated with
the expression of NEAT1
As it was demonstrated that miR-520a possesses a
protective role in H/R-induced cardiomyocytes, the miR-520a
expression levels in ischemic myocardium and cardiomyocytes
subjected to I/R and H/R were evaluated. miR-520a was significantly
downregulated in ischemic myocardium and cardiomyocytes subjected
to I/R and H/R (Fig. 6A and B).
Pearson correlation analysis was performed to examine the
association between NEAT1 and miR-520a. The results indicated that
the relative expression of NEAT1 was negatively correlated with the
relative expression of miR-520a (Fig.
6C).
Discussion
Ischemic injury is a severe cardiovascular disease,
a serious threat to human life which can lead to congestive heart
failure and malignant arrhythmia (11). Thus, in order to provide valuable
benefits to current therapies it is necessary to distinguish novel,
highly sensitive and specific biomarkers for the early diagnosis of
acute myocardial infarction (AMI) and to identify possible
regulatory targets of AMI.
Apoptosis is characterized by cell shrinkage,
chromosomal DNA and nuclear fragmentation (12,13). It
is a type of programmed cell death that contributes to cardiac
diseases, including myocardial infarction, heart failure and I/R
injury. Cardiomyocyte apoptosis was notably increased in the
ischemic hearts in recent studies and subsequently lead to
continuous cardiomyocyte loss (14).
Several studies have demonstrated that inhibiting apoptosis of
cardiomyocytes protected against I/R injury and improved cardiac
dysfunction significantly (15,16).
Furthermore, it has been demonstrated that LncRNAs may regulate
cell apoptosis through altering the pro-apoptotic or anti-apoptotic
protein expression. However, how LncRNAs regulate the process of
cell apoptosis remains unclear. The major signaling pathways
mediating apoptosis involve the death receptor, mitochondrial and
endoplasmic reticulum-stress signaling pathways.
Mitochondrial cell death is regulated by
pro-apoptotic proteins, such as Bax, and anti-apoptotic proteins,
such as Bcl-2. Intracellular stress, including oxidative stress,
serum deprivation and DNA damage, contributes to mitochondrial
membrane permeability changes and subsequent release of several
pro-apoptotic factors such as cytochrome c,
apoptosis-inducing factor, endonuclease G and second
mitochondria-derived activator of caspases (Smac/Diablo). These
pro-apoptotic factors result in the initiation of apoptosis.
In the present study, NEAT1 was upregulated in the
myocardium and cardiomyocytes following I/R and H/R, respectively.
Further study demonstrated that the knockdown of NEAT1 inhibited
the apoptosis of cardiomyocytes. Additionally, NEAT1 knockdown
elevated the expression levels of Bax and cleaved caspase3, and
reduced the expression level of Bcl-2.
LncRNAs regulate molecules through multiple
mechanisms, including epigenetic regulation, genomic imprinting,
RNA stability, RNA alternative splice and miR regulation (1,17–20).
miRs are small, non-coding RNAs that regulate the expression of
protein-encoding genes at post-transcriptional level. Accumulating
evidence has reported the key roles of miRs in the process of
cardiac disease, including hypoxia-induced myocardial injury. In
the present study, the use of bioinformatics analysis predicted
that human NEAT1 could bind to miR-520a. Luciferase activity was
also assessed, which confirmed this prediction. Following this, it
was investigated whether miR-520a was involved in I/R injury in the
present study. The results indicated that miR-520a overexpression
also inhibited apoptosis induced by H/R. The results revealed that
infection of Ad-control alone did not significantly alter the
expression of miR-520a or NEAT1, nor did it significantly impact
cell apoptosis compared with the control group. For this reason,
co-infection of Ad-miR-520a and Ad-control as an additional control
in the rescue experiment was not performed.
In conclusion, the present study indicated the role
of NEAT1 and miR-520a in H/R and I/R injury. Knockdown of NEAT1 and
overexpression of miR-520a protected cardiomyocytes from
H/R-induced cell apoptosis. Furthermore, it was revealed that NEAT1
can directly bind to miR-520a. However, miRs participate in various
physical and pathological processes by regulating the level of
target genes. miRs may participate in the regulation of signal
pathways involved in cell apoptosis which include the PI3K/AKT
signaling pathway. Cardiomyocyte apoptosis was induced by altering
the expression levels of bax, bcl-2 and cyto-c, which are relevant
to the mitochondria. However, the mitochondrial membrane potential
was not examined and this may be considered a potential limitation
of the current study.
Taken together, the present findings may aid in the
design of an adenovirus vector for clinical use to knockdown NEAT1
expression, which may be protective in ischemia. Further studies
concerning the target of miR-520a and downstream genes are
required.
Acknowledgements
Not applicable.
Funding
This study was supported by grants from the 2015
Zhejiang Province Medical and Health Science and Technology
Research Fund (grant no. 2015KYB387), 2015 Zhejiang Province
Traditional Chinese Medicine Scientific Research Fund (grant no.
2015ZA203), 2016 Zhejiang Province Traditional Chinese Medicine
Scientific Research Fund (grant no. 2016ZA191), 2016 Zhejiang
Province Medical and Health Science and Technology Research Fund
(grant no. 2016KYB287), 2016 Jiaxing Science and Technology Project
Fund (grant no. 2016AY23038), 2017 Jiaxing Science and Technology
Project Fund (grant no. 2017AY33021), Jiaxing Cardiovascular Key
Discipline Fund (grant no. 2014-F07), Jiaxing Key Innovation Team
Fund (grant no. 2015-CX1) and the Class General Financial Grant
from the China Postdoctoral Science Foundation (grant no.
2017M611587).
Availability of data and materials
All datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
HW and GT performed the experiments and data
collection. WS, MH and HP performed the data analysis. CZ prepared
the manuscript. GQ contributed to the design of the study design
and reviewed the manuscript. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li J, Kim K, Jeong SY, Chiu J, Xiong B,
Petukhov PA, Dai X, Li X, Andrews RK, Du X, et al: Platelet protein
disulfide isomerase promotes glycoprotein ibα -mediated
platelet-neutrophil interactions under thromboinflammatory
conditions. Circulation. 139:1300–1319. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rahnavard M, Hassanpour M, Ahmadi M,
Heidarzadeh M, Amini H, Javanmard MZ, Nouri M, Rahbarghazi R and
Safaie N: Curcumin ameliorated myocardial infarction by inhibition
of cardiotoxicity in the rat model. J Cell Biochem.
18–Feb;2019.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Karreth FA, Reschke M, Ruocco A, Ng C,
Chapuy B, Léopold V, Sjoberg M, Keane TM, Verma A, Ala U, et al:
The BRAF pseudogene functions as a competitive endogenous RNA and
induces lymphoma in vivo. Cell. 161:319–332. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cremer S, Michalik KM, Fischer A,
Pfisterer L, Jaé N, Winter C, Boon RA, Muhly-Reinholz M, John D,
Uchida S, et al: Hematopoietic deficiency of the long non-coding
RNA MALAT1 promotes atherosclerosis and plaque inflammation.
Circulation. 139:1320–1334. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shen L, Wang Q, Liu R, Chen Z, Zhang X,
Zhou P and Wang Z: LncRNA lnc-RI regulates homologous recombination
repair of DNA double-strand breaks by stabilizing RAD51 mRNA as a
competitive endogenous RNA. Nucleic Acids Res. 46:717–729. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lin Y, Schmidt BF, Bruchez MP and McManus
CJ: Structural analyses of NEAT1 lncRNAs suggest long-range RNA
interactions that may contribute to paraspeckle architecture.
Nucleic Acids Res. 46:3742–3752. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yu L, Sun Y, Cheng L, Jin Z, Yang Y, Zhai
M, Pei H, Wang X, Zhang H, Meng Q, et al: Melatonin
receptor-mediated protection against myocardial
ischemia/reperfusion injury: Role of SIRT1. J Pineal Res.
2:228–238. 2014. View Article : Google Scholar
|
|
9
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2 (-Delta Delta C (T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Csaba G and Madarasz B: Ultrastructural
changes elicited in the Tetrahymena by primary exposure
(imprinting) and reexposure to hormone. Z MikroskAnatForsch.
99:884–890. 1985.
|
|
11
|
Heusch G and Gersh BJ: The pathophysiology
of acute myocardial infarction and strategies of protection beyond
reperfusion: A continual challenge. Eur Heart J. 38:774–784.
2017.PubMed/NCBI
|
|
12
|
Nawaz W, Khan FU, Khan MZ, Gang W, Yang M,
Liao X, Zhang L, Ihsan AU, Khan A, Han L and Zhou X:
Exo-organoplasty interventions: A brief review of past, present and
future directions for advance heart failure management. Biomed
Pharmacother. 88:162–172. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
White HD and Chew DP: Acute myocardial
infarction. Lancet. 372:570–584. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shi B, Ma M, Zheng Y, Pan Y and Lin X:
mTOR and Beclin1: Two key autophagy-related molecules and their
roles in myocardial ischemia/reperfusion injury. J Cell Physiol.
234:12562–12568. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhong CB, Chen X, Zhou XY and Wang XB: The
role of peroxisome proliferator-activated receptor γ in mediating
cardioprotection against ischemia/reperfusion injury. J Cardiovasc
Pharmacol Ther. 23:46–56. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
DeBerge M, Yeap XY, Dehn S, Zhang S,
Grigoryeva L, Misener S, Procissi D, Zhou X, Lee DC, Muller WA, et
al: MerTK cleavage on resident cardiac macrophages compromises
repair after myocardial ischemia reperfusion injury. Circ Res.
121:930–940. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Klattenhoff CA, Scheuermann J, C Surface
LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey
L, Haas S, et al: Braveheart, a long noncoding RNA required for
cardiovascular lineage commitment. Cell. 152:570–583. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guttman M, Donaghey J, Carey BW, Garber M,
Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al:
lincRNAs act in the circuitry controlling pluripotency and
differentiation. Nature. 477:295–300. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yildirim E, Kirby JE, Brown DE, Mercier
FE, Sadreyev RI, Scadden DT and Lee JT: Xist RNA is a potent
suppressor of hematologic cancer in mice. Cell. 152:727–742. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gartler SM and Riggs AD: Mammalian
X-chromosome inactivation. Annu Rev Genet. 17:155–190. 1983.
View Article : Google Scholar : PubMed/NCBI
|