Introduction
Approximately 103 distinct bacterial
species, the total number of which is >10-fold that of
eukaryotic cells, colonize in the human gastrointestinal tract
(1,2). The commensal intestinal microbiota
fulfills important roles in digestion and absorption of nutrients,
the composition of the gut mucosal barrier, host metabolism and the
innate immune system (3–7). A healthy gut microbiota maintains
homeostasis within the host, which may be crucial to the normal
operation of other vital organs, including the liver (8–11) and
the brain (12–15). Accumulating evidence suggests that
long-term dietary habits may result in various gut microbiota,
comprising different bacterial ecosystems, which are known as
enterotypes (16–18). A Western-style diet, high in total
and saturated fat, is relevant to the prevalence of numerous
illnesses and conditions, including obesity, type 2 diabetes,
inflammatory bowel disease (IBD) and digestive tract cancers,
probably due to an imbalance in gut microbial populations. A study
by Kim et al (19), which
revealed that a high-fat diet (HFD) induced low-grade inflammation,
demonstrated the pro-inflammatory effects of energy-dense diets on
vagal gutbrain communication. David et al (20) reported that increases in the growth
of the sulfite-reducing pathobiont Bilophila wadsworthia in
the presence of a diet rich in animal fats supported an association
between dietary fat, bile acids and the outgrowth of microbes,
which was able to trigger IBD. Schulz et al (21) demonstrated that an HFD accelerated
tumor progression in the small intestine of K-rasG12Dint
mice, which was independent of the occurrence of obesity. Ou et
al (22) reported that the
effects of dietary fat on the occurrence of colorectal cancer (CRC)
may be indirectly mediated by secondary bile acids, which are
produced via the enterobacterial 7α-dehydroxylation of primary bile
acids and are considered to be potential carcinogens in the
etiology of CRC. However, a direct association between diet, the
intestinal microbiota and the pathogenesis of CRC has yet to be
conclusively demonstrated, even in animal models. Of note, a number
of epidemiological studies have demonstrated that excessive intake
of animal fat is associated with an increased risk of colon cancer,
which may promote the expansion of sulfate-reducing bacteria that
are thereby able to produce genotoxic agents, including hydrogen
sulfide.
Probiotics consist of live beneficial bacterial
species for promoting health of the host, typically lactobacilli
and bifidobacteria, and these directly improve the composition of
the colonic microbiota (23). A
study by Hu et al (24) that
employed a high cholesterol diet-fed rat model suggested that two
Lactobacillus strains exerted a positive effect on lipid
metabolism. Lactobacillus delbrueckii and Bifidobacterium
animalis var. lactis administered alone were indicated
to ameliorate colonic preneoplastic lesions in mice, although their
administration in combination did not prove to be effective
(25). Bertkova et al
(26) demonstrated that probiotic
Lactobacillus plantarum together with bioactive compounds
was able to suppress colon carcinogenesis in
N,N-dimethylhydrazine-induced rats. In China, capsules containing
live, combined Lactobacillus, Bifidobacterium and
Enterococcus have been widely used as probiotics in clinical
settings, including the treatment of non-alcoholic fatty liver
disease (27), irritable bowel
syndrome (28) and even
gastrointestinal tumors (29,30), and
these probably work via reducing the inflammatory response and
forming a biological protective barrier.
Various functional effects of different probiotic
strains on the gut microbiota and relevant diseases have been
clearly demonstrated. However, certain studies have identified that
gut microbiota undergo a less pronounced response to oral
probiotics, partly due to inter-individual variation in microbiota
composition elicited by dietary habits, in addition to the genetic
background, age, other environmental factors, or the application of
different analytical methods. In this regard, the concept of three
enterotypes was raised by Arumugam et al (16), namely Bacteroides, Prevotella
and Ruminococcus. In reality, it was difficult to categorize
human beings as a particular ‘enterotype’, since dietary factors
may have an impact on gut microbial populations (31,32). A
previous study by our group demonstrated a different composition of
stool bacterial genera when comparing between individuals on either
a high- or a low-fat diet (33).
Since diet may shape the composition and function of the gut
microbiota, in turn influencing host health, one promising
therapeutic strategy aimed at improving health would be to alter
the gut microbiome using dietary intervention.
Although the positive effects of probiotics and
dietary intervention on IBD and CRC have been broadly demonstrated,
comparative studies to identify the efficacy of probiotic
supplementation and dietary intervention as prophylactic tools
previously under HFD conditions require further investigation ahead
of the occurrence of gastrointestinal diseases. Therefore, the
present study aimed to assess whether probiotic treatment and
dietary intervention exert any impact on the gut microbiota in
human subjects on an HFD.
Subjects and methods
Subjects, diets and experimental
design
A total of 36 healthy volunteers (age, 45–65 years;
males/females, 16/20) from Zhouzhuang Town (Jiangyin City, China),
who were consuming an HFD with dietary fat accounted for >40% of
total energy (34), were enrolled
mainly according to the results of questionnaire and physical
examination during the community health survey. All experiments of
the present study were approved by the Ethics Committee of Shanghai
Tenth People's Hospital (Shanghai, China) and Jiangyin People's
Hospital (Jiangyin, China). Written informed consent was obtained
from all of the participants prior to their enrolment.
Subsequently, these volunteers were randomly assigned to four
groups: i) The HFD group, where the HFD was maintained due to their
constant habits to establish a control group (n=9); ii) the dietary
intervention (DI) group, where the HFD was replaced by a low-fat
diet (LFD) in which dietary fat accounted for <40% of total
energy (n=9); iii) an HFD + Probiotic group, where the HFD was
supplemented with a daily dose of 2 g live combined
Lactobacillus acidophilus [≥1.0×107
colony-forming units (Cfu)/g], Bifidobacterium longum
(≥1.0×107 Cfu/g) and Enterococcus faecalis
(≥1.0×107 Cfu/g) powder (Shanghai Xinyi Pharmaceutical
Co., Ltd.) administered orally in water according to the
manufacturer's recommendation (n=9); and iv) the DI + Probiotic
group, which received a combination of the LFD and the
above-mentioned probiotic microorganisms (n=9). No significant
differences in daily intake of dietary fiber, or the percentages of
calories derived from fat, were identified among the four groups
prior to the experiment (Fig. 1).
The volunteers also had a similar gender ratio, were of a similar
age and body mass index, yielded similar valid sequencing reads for
fecal microbiota and had similar gut microbiota compositions at the
start of the experiment. These details were presented in
Supplementary Table SI. Probiotic
treatment and dietary intervention were continued for 4 months
(from Jan 1st, 2015 to Apr 30th, 2015), which hopefully allowed
sufficient duration for intestinal flora change. All of the
volunteers completed the study under strict quality control with
dietary changes and medication compliance monitored throughout the
study.
Stool collection
A freshly voided stool sample was collected from
each subject at the end of the experiment (Apr 30th, 2015), and was
snap-frozen in liquid nitrogen. All samples were finally
transferred to a deep freezer at −80°C until the microbiota was
analyzed.
Microbiota analysis
DNA extraction
Metagenomic DNA was extracted from each fecal sample
using a MicroElute Genomic DNA kit (cat. no. D3096-01; Omega;
BioTek, Inc.) according to the manufacturer's protocol. The
MicroElute Genomic DNA kit extraction controls were included
through the DNA extraction and PCR steps as a negative control.
Total DNA was eluted in 50 µl elution buffer using a method
modified from the manufacturer's protocol (Qiagen; GE Healthcare)
and stored at −20°C prior to performing qPCR.
PCR amplification and 16S rDNA
sequencing
The primers 319F and 806R were used to amplify the
double hypervariable V3 and V4 regions of the bacterial 16S rRNA.
PCR procedures were as follows: Initial denaturation (30 sec at
98°C), followed by 35 cycles of amplification including
denaturation (10 sec at 98°C), annealing (30 sec at 54°C) and
extension (45 sec at 72°C), and then final extension (10 min at
72°C). The PCR products for each example were normalized using an
AxyPrep® TM Mag PCR Normalizer (Axygen Biosciences),
followed by purification with AMPure XT beads (Beckman Coulter
Genomics), and quantification using the Illumina Library
Quantification kit (Kapa Biosciences). Pyrosequencing was performed
using a MiSeq System (Illumina, Inc.).
Bioinformatics analysis
After filtering the raw data, the high-quality
sequences were identified, which were subsequently clustered into
operational taxonomic units (OTUs) using the CD-hit-est-based
clustering method (35). The numbers
of the sequences and OTUs for each sample were calculated by using
the software PyNAST (http:/qiime.org/pynast/). Subsequently, analysis of
the α-diversity (i.e., the mean species diversity in sites or
habitats at a local scale), including the Observed species,
Shannon, Simpson and Chao1 indexes, was performed. A Venn diagram
of the common and unique OTUs among the four groups was constructed
using online software (http://bioinfogp.cnb.csic.es/tools/venny/). Unweighted
Unifrac distance metrics analysis and principal coordinate analysis
(PCoA) were also performed in terms of the matrix of distance to
assess the β-diversity (i.e., the ratio between regional and local
species diversity). The relative abundance (%) of bacteria at the
phylum and species taxa levels in each sample were calculated by
using RDP-derived taxonomic communities. A heatmap on species
information was constructed using Heml software (version 1.0.3.3)
(36).
Statistical analysis
Student's t-test and Kruskal-Wallis one-way analysis
of variance with Bonferroni's correction control were performed
using SPSS version 19.0 (IBM Corp.). P<0.05 was considered to
indicate a statistically significant difference.
Results
Effects of dietary intervention and
oral probiotics on the gut microbial diversity indices in an HFD
population
A total of 2,203,533 pyrosequencing reads for the
fecal microbiota were analyzed with a mean of 63,573, 57,697,
63,598 and 59,968 reads for the HFD, DI, HFD + Probiotic and DI +
Probiotic group, respectively. According to the indices of Observed
species, Shannon and Chao1 for stool (Fig. S1), but not according to the Simpson
index, the αdiversity of the fecal microbiota of the DI + Probiotic
group was higher compared with that of the DI group or the HFD
group (P<0.05). However, there was no significant difference
between the HFD and the DI group, or between the HFD + Probiotic
and the DI + Probiotic group. β-diversity analysis revealed that
the fecal samples were distributed diffusely and apparent
non-overlapping clusters among groups were also formed (Fig. 2).
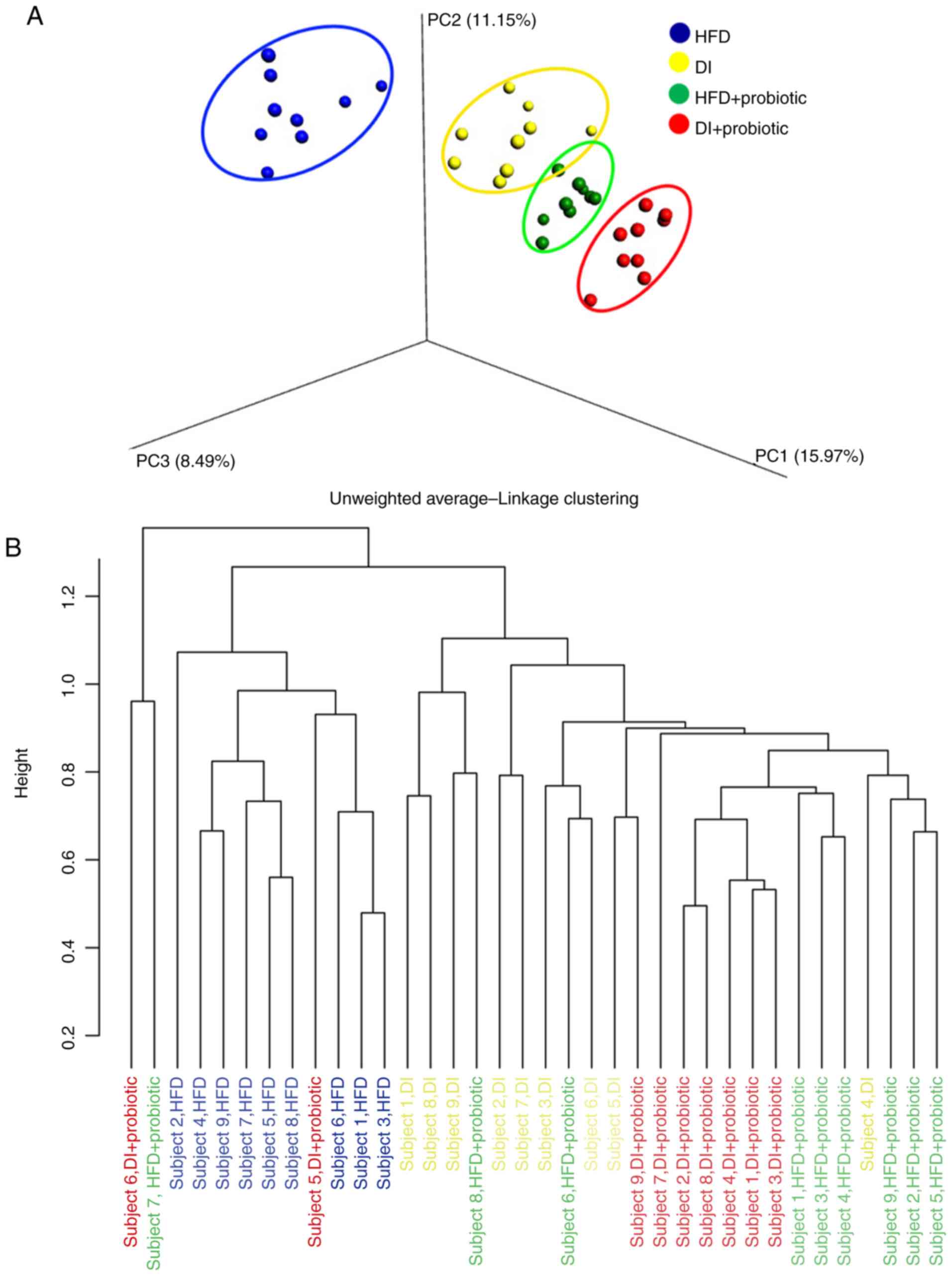 | Figure 2.Clustering of samples based on fecal
microbiota communities. (A) Principal coordinates analysis of
samples from the HFD, DI, HFD + Probiotic and DI + Probiotic
groups. (B) Unweighted pairgroup method with arithmetic mean of
samples from the HFD, DI, HFD + Probiotic and DI + probiotic
groups. Probiotics included Bifidobacterium longum,
Lactobacillus acidophilus and Enterococcus faecalis.
PC1, PC2 and PC3, three eigenvalues calculated by distance matrix
of fecal samples, represent the top three principal coordinate
components explaining as much of the variability in the data as
possible. HFD, high-fat diet; DI, dietary intervention with low-fat
diet. |
Effects of dietary intervention and
oral probiotics on the gut microbiota composition of an HFD
population
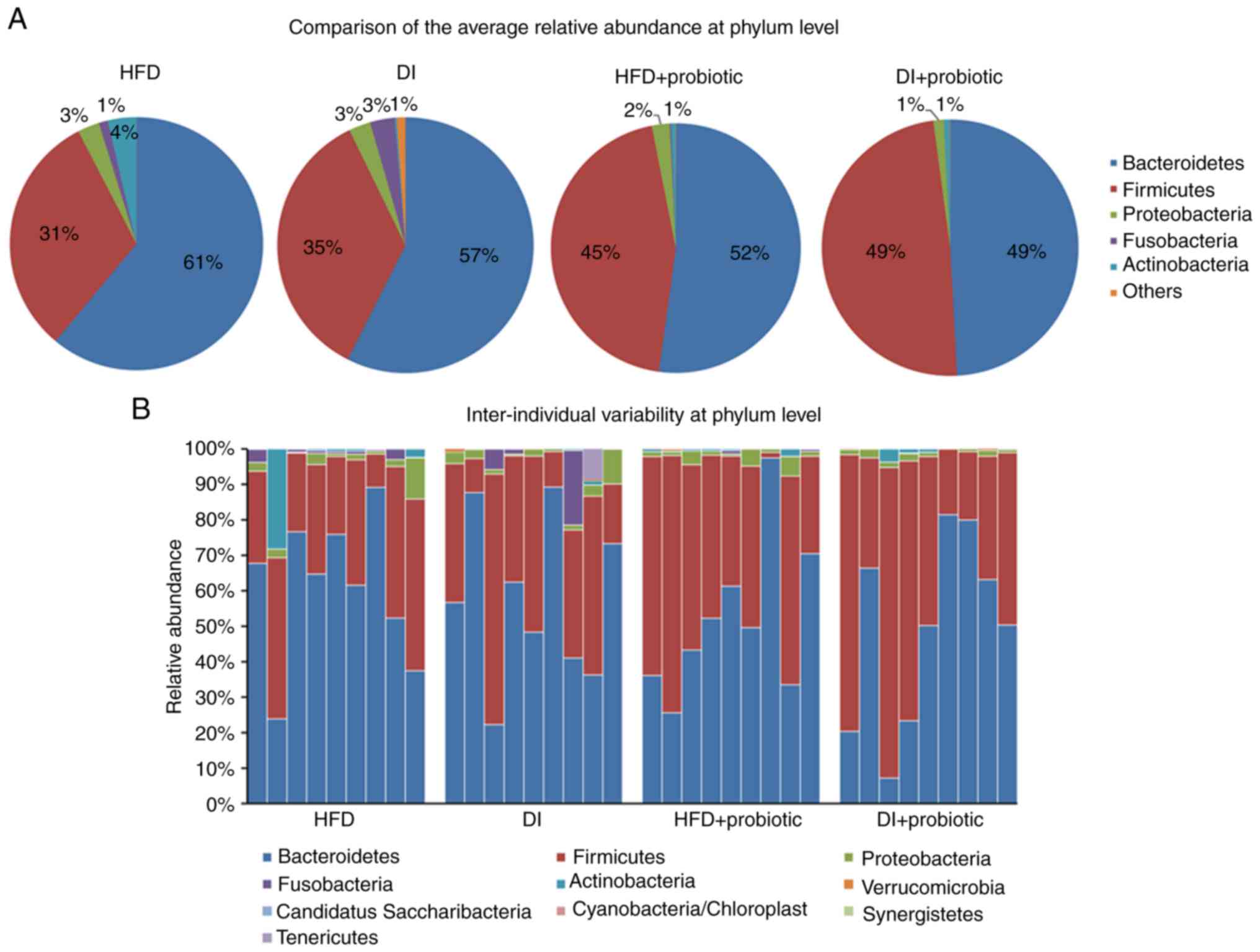
The three most abundant phyla in the fecal
microbionta, including Firmicutes, Bacteroidetes and
Proteobacteria, were not significantly different among the
groups at 4 months (Fig. 3A). The
relative abundance of the other phyla (mainly including
Actinobacteria and Fusobacteria) was low in all four
groups. There was no apparent inter-individual variability at the
phylum level within each group (Fig.
3B).
Subsequently, the differences in shared species
among groups were analyzed. Overall, there were 467 species
detected in stool samples (323, 385, 411 and 418 species in the
HFD, DI, HFD + Probiotic and DI + Probiotic group, respectively).
When comparing 100% (9/9), 89% (8/9), 78% (7/9), 67% (6/9), 56%
(5/9), 44% (4/9) and 11% (1/9) of the populations in each group
with each other, the number of species shared among all groups was
5, 9, 22, 43, 74, 109 and 270, respectively. These results
suggested that the number of bacterial species shared between the
groups is dependent on how many subjects in each group were used
for the comparison between the groups (Fig. S2).
Subsequently, the differences in species composition
of the fecal microbiota were analyzed. In total, 30 species were
different among the four groups, and the majority of these were
unclassified with the exception of two distinct species,
Prevotella copri and Bacteroides ovatus. It was
revealed that 27 species were altered in subjects receiving dietary
intervention and probiotic treatment compared with the other three
groups. The relative abundance of 5, 9 and 25 species in the DI,
HFD + Probiotic and DI + Probiotic groups, respectively, was
significantly different compared with the HFD group (P<0.05;
Fig. 4). The relative abundance of
17 species, predominantly belonging to the Ruminococcaceae
and Lachnospiraceae families of the order
Clostridiales and the phylum Firmicutes, was
increased in subjects from the DI + Probiotic group compared with
the HFD group, whereas 8 species were decreased in the DI +
Probiotic group, particularly two Bacteroides species with
relatively higher abundance (B. ovatus and one unclassified)
belonging to the Bacteroidaceae family of the order
Bacteroidales and the phylum Bacteroidetes. Other
reduced unclassified species associated with probiotic
supplementation and dietary intervention belonged to various
different bacterial families: Flavonifractor, Clostridium XVIII,
Veillonella, Anaerostipes and Fusobacterium.
Discussion
In the present study, healthy human subjects who
were consuming an HFD were selected as research subjects rather
than rodents, e.g. mice or rats, whose confounding factors,
including genetic background, age, sex and diet, may be well
controlled, and also in preference to human subjects who had
already incurred disease entities, including obesity, type 2
diabetes and intestinal disorders. Furthermore, under clinical
conditions, not all individuals on an HFD develop obesity,
hyperlipemia, IBD and intestinal tumors, as other factors are
involved, including genetic background, dietary pattern, energy
expenditure, metabolic capability and intestinal barrier state.
Although a large number of reviews have described how probiotics
are regarded as a gut microbiota-targeted therapy to treat
HFD-induced obesity, type-2 diabetes and gastrointestinal diseases
(23,37–39), the
role of combined oral probiotics and dietary intervention in the
modulation of the gut microbiota of healthy populations consuming
an HFD remained to be fully elucidated. The results of the present
study indicated that probiotics may provide a means of improving
the gut microbiota of HFD populations naturally, and that this
effect may be enhanced by combining probiotics with dietary
intervention. To a certain extent, the gut microbiota may be a
suitable target of therapeutic intervention to prevent those
individuals on an HFD from developing the above-mentioned
diseases.
First, the present results indicated that the
diversity of gut microbiota of individuals receiving an HFD may be
altered after probiotic supplementation with three live bacterial
strains and dietary intervention. Probiotic supplementation in two
groups led to a more distinct microbial clustering that was closer
in the PCoA plot, irrespective of dietary patterns. This combined
effect on adults on an HFD has been rarely reported. However, the
topic of changes in the diversity of the gut microbiota remains
controversial (40). It was found
that daily consumption of yogurt including lactobacilli and
bifidobacteria by healthy medical students increased the
α-diversity of the intestinal microbiome (41). However, other studies were unable to
identify any significant changes in gut microbial diversity, either
on the basis of terminal restriction fragments or next-generation
sequencing analysis, when comparing day 0 and day 42 of yogurt
consumption (42). Therefore, the
overall composition of the gut microbial community and its
diversity may be governed and affected by the employed detection
method of the bacteria. On the other hand, a systematic review of
randomized controlled trials were observed no effects of probiotics
on the fecal microbiota composition in terms of α-diversity in any
of the included studies when compared with a placebo, whereas only
one study identified that probiotics were able to significantly
modify the fecal bacterial community in terms of β-diversity
(43). The consumption of
Lactobacillus casei Zhang et al (44) markedly altered the composition of the
intestinal microbiota and the gut microbiota diversity. Park et
al (45), also demonstrated that
mice in a probiotic treatment group had a lower gut microbiota
diversity. Although low gut microbiota diversity is usually a
hallmark of intestinal dysbiosis, factors including ecological
stability, idealized composition or favorable functional profile
have recently been suggested as hallmarks of a healthy gut
microbiota (46). Certain probiotic
strains may diminish the diversity of the gut microbiota either by
production of anti-microbial peptides, or alternatively, by
increasing competition for nutrients, processes that are able to
reduce microbial growth. Hanifi et al (47) demonstrated that supplementation with
Bacillus subtilis R0179 did not appear to reverse the
overall microbiota diversity, as it simultaneously inhibited the
growth of certain opportunistic pathogens. In the present study, it
was observed that simply replacing an HFD with an LFD did not alter
the α-diversity of the fecal microbiota. A murine (RELMβ) knockout
model study revealed that alterations in gut microbiome composition
induced by dietary fat were independent of obesity (48). In the present study, it was revealed
that ~50% of all detected microbial species were shared by 44% of
the volunteers, regardless of diet or treatment. However, divergent
results have been identified in other studies. Intestinal
microbiota diversity was reduced in HFD- and high sugar diet-fed
mice, whereas a control diet was able to prevent these changes.
Heinsen et al (49)
demonstrated that a very low-calorie diet beneficially altered the
gut microbiome diversity in obese human subjects, but that these
changes were not sustained during weight maintenance.
At the phylum level, the ratio of
Bacteroidetes to Firmicutes was indicated to be
decreased in diet-induced obesity, which could be reversibly
increased by diet adaptation (50,51). By
contrast, various studies have suggested that the ratio of
Bacteroidetes to Firmicutes is not a contributing factor in
human obesity, and it appears not to be associated with diet
(52,53). In line with this, in the present
study, no significant differences were observed in the relative
abundance of Bacteroidetes and Firmicutes among the
four treatment groups, which may be due to differences in genetic
background, age and sex. Furthermore, dietary intervention and
probiotics supplementation were unable to change the bacterial
composition at the phylum level, since Bacteroidetes and
Firmicutes are the phyla predominantly present in the gut
microbiota of humans (53–56).
At the species level, significant differences among
the treatment groups were identified in the present study, which
was consistent with the results of the animal study by Park et
al (45). The relative abundance
of 30 species was altered by dietary intervention and/or probiotic
supplementation, 25 of which were changed in subjects who received
dietary intervention and probiotic treatment. In addition,
increased unclassified species mainly belonged to two
butyrate-producing families, Ruminococcaceae and
Lachnospiraceae (57), for
which three taxa have been identified at the genus level, including
Oscillibacter, Butyricicoccus and Coprococcus, which
may be beneficial for colonic health (58–63). One
study has indicated that two Lactobacillus strains,
Lactobacillus plantarum HAC01 and L. rhamnosus GG,
exert a beneficial effect on Lachnospiraceae and
Ruminococcaceae at the bacterial family level, rather than
Firmicutes and Bacteroidetes at the phylum level
(64). The relative abundance of the
Lachnospiraceae (phylum Firmicutes) was significantly
higher in the Lactobacillus-treated groups compared with
that in the PBS-treated control group. Amelioration of
obesity-associated dysbiosis by alteration of the gut microbiota
appears to be associated with ‘indicator’ bacterial taxa, including
the family Lachnospiraceae. Among those eight species whose
populations were decreased following probiotic supplementation and
dietary intervention compared with the natural HFD group, one
species identified was B. ovatus, which was considered to
belong to a group of Bacteroides species sharing similar
phenotypic characteristics to those of B. fragilis and B.
vulgatus that were frequently identified in patients with IBDs,
including Crohn's disease and ulcerative colitis (65,66). To
a certain extent, probiotic supplementation and dietary
intervention may protect an HFD population from suffering from
gastrointestinal inflammation disorders, and help to remold a
healthy gut bacterial symbiosis through inhibition of
IBD-associated specific bacterial taxa. Various bacterial families
associated with unclassified species reduced by probiotic and
dietary intervention have been identified as Clostridium
XVIII and Anaerostipes. Certain evidence suggests that
Clostridium XVIII may serve as the next ‘smart’ probiotics
(67), and the genus
Anaerostipes is associated with potential butyrate-producing
bacteria (58). Why these beneficial
bacteria are also reduced in number following probiotic
intervention remains to be elucidated. A plausible hypothesis
explaining this is that the potential of certain probiotic strains
to ‘rebalance’ butyrate concentrations may protect the host under
those physiological conditions associated with altered butyrate
concentrations (58). One unknown
species belonging to the genus Flavonifractor, which is
capable of cleaving the flavonoid C-ring (68), was determined to decline after
combined probiotic and dietary intervention. Depletion of a species
belonging to Flavonifractor has been demonstrated in obese
individuals (69) and another study
suggested that treatment with Bifidobacterium catenulatum
LI10 was able to attenuate this depletion (70), which was not in agreement with the
results of the present study. Another study published by Toscano
et al (71) indicated that,
after one month of probiotic intake, a reduction in the population
of Flavonifractor was observed, which was consistent with
the result of the present study. In addition, other unknown species
from the genera Veillonella and Fusobacterium, which
may act as pathogens and carcinoma-associated taxa (71–77),
were significantly reduced by probiotic and dietary intervention,
suggesting an improvement of the gut bacterial community induced by
an HFD. Of note, a higher abundance of the supplemented probiotic
strains in fecal samples was not observed following supplementation
of these probiotics, probably since they did not appear to colonize
the intestine themselves within such a short study duration.
In conclusion, the present study revealed that the
diversity of gut microbiota was promoted in HFD populations
receiving probiotic treatment and dietary intervention, along with
an increase in the populations of numerous beneficial species, and
a reduction in the number of certain detrimental species. Taken
together, the present results suggest that Lactobacillus
acidophilus, Bifidobacterium longum and Enterococcus
faecalis supplementation and dietary intervention may modulate
the gut microbiota, and may provide a natural alternative to treat
HFD-associated disorders.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by grants from the National
Natural Science Foundation of China (grant. nos. 81472262 and
81230057) and the Emerging Cutting-Edge Technology Joint Research
Projects of Shanghai (grant. no. SHDC12012106).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
LMQ, RYG and HLQ designed the study; data collection
and analysis was performed by LMQ, RYG and JMH; the manuscript was
written by LMQ; and revisions of the manuscript were made by RYG
and HLQ; All of the authors provided intellectual input for the
study and approved the final version of the manuscript.
Ethics approval and consent to
participate
All procedures performed involving human
participants were in accordance with the ethical standards of the
Institutional and National Research Committee and with the 1964
Helsinki Declaration and its later amendments or comparable ethical
standards. The present study was approved by the Ethics Committee
of the Institutional Review Boards of Shanghai Tenth People's
Hospital (Shanghai, China) and Jiangyin People's Hospital
(Jiangyin, China). Written informed consent was obtained from the
patients or their guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zoetendal EG, Rajilic-Stojanovic M and de
Vos WM: High-throughput diversity and functionality analysis of the
gastrointestinal tract microbiota. Gut. 57:1605–1615. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zoetendal EG, Raes J, van den Bogert B,
Arumugam M, Booijink CC, Troost FJ, Bork P, Wels M, de Vos WM and
Kleerebezem M: The human small intestinal microbiota is driven by
rapid uptake and conversion of simple carbohydrates. ISME J.
6:1415–1426. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hooper LV, Midtvedt T and Gordon JI: How
host-microbial interactions shape the nutrient environment of the
mammalian intestine. Annu Rev Nutr. 22:283–307. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Backhed F, Ley RE, Sonnenburg JL, Peterson
DA and Gordon JI: Host-bacterial mutualism in the human intestine.
Science. 307:1915–1920. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Qin J, Li R, Raes J, Arumugam M, Burgdorf
KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al: A
human gut microbial gene catalogue established by metagenomic
sequencing. Nature. 464:59–65. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lin CS, Chang CJ, Lu CC, Martel J, Ojcius
DM, Ko YF, Young JD and Lai HC: Impact of the gut microbiota,
prebiotics, and probiotics on human health and disease. Biomed J.
37:259–268. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Brown K, DeCoffe D, Molcan E and Gibson
DL: Diet-induced dysbiosis of the intestinal microbiota and the
effects on immunity and disease. Nutrients. 4:1095–1119. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Abu-Shanab A and Quigley EM: The role of
the gut microbiota in nonalcoholic fatty liver disease. Nat Rev
Gastroenterol Hepatol. 7:691–701. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chiu CC, Ching YH, Li YP, Liu JY, Huang
YT, Huang YW, Yang SS, Huang WC and Chuang HL: Nonalcoholic fatty
liver disease is exacerbated in High-Fat Diet-Fed gnotobiotic mice
by colonization with the Gut Microbiota from patients with
nonalcoholic steatohepatitis. Nutrients. 9:E12202017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Federico A, Dallio M, Caprio GG, Ormando
VM and Loguercio C: Gut microbiota and the liver. Minerva
Gastroenterol Dietol. 63:385–398. 2017.PubMed/NCBI
|
|
11
|
Budden KF, Gellatly SL, Wood DL, Cooper
MA, Morrison M, Hugenholtz P and Hansbro PM: Emerging pathogenic
links between microbiota and the gut-lung axis. Nat Rev Microbiol.
15:55–63. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bercik P: The microbiota-gut-brain axis:
Learning from intestinal bacteria? Gut. 60:288–289. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Collins SM and Bercik P: Gut microbiota:
Intestinal bacteria influence brain activity in healthy humans. Nat
Rev Gastroenterol Hepatol. 10:326–327. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bienenstock J, Kunze W and Forsythe P:
Microbiota and the gut-brain axis. Nutr Rev. 73 (Suppl 1):S28–S31.
2015. View Article : Google Scholar
|
|
15
|
Dinan TG and Cryan JF: Gut-brain axis in
2016: Brain-gut-microbiota axis-mood, metabolism and behaviour. Nat
Rev Gastroenterol Hepatol. 14:69–70. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Arumugam M, Raes J, Pelletier E, Le
Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto
JM, et al: Enterotypes of the human gut microbiome. Nature.
473:174–180. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu GD, Chen J, Hoffmann C, Bittinger K,
Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R,
et al: Linking long-term dietary patterns with gut microbial
enterotypes. Science. 334:105–108. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bushman FD, Lewis JD and Wu GD: Diet, gut
enterotypes and health: Is there a link? Nestle Nutr Inst Workshop
Ser. 77:65–73. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim KA, Gu W, Lee IA, Joh EH and Kim DH:
High fat diet-induced gut microbiota exacerbates inflammation and
obesity in mice via the TLR4 signaling pathway. PLoS One.
7:e477132012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
David LA, Maurice CF, Carmody RN,
Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y,
Fischbach MA, et al: Diet rapidly and reproducibly alters the human
gut microbiome. Nature. 505:559–563. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schulz MD, Atay C, Heringer J, Romrig FK,
Schwitalla S, Aydin B, Ziegler PK, Varga J, Reindl W, Pommerenke C,
et al: High-fat-diet-mediated dysbiosis promotes intestinal
carcinogenesis independently of obesity. Nature. 514:508–512. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ou JH, Ridlon JM, DeLany JP, Vipperla K,
Newton K and O'Keefe SJ: Obesity and colon cancer risk: Is it the
Fat? Gastroenterology. 142:S3132012. View Article : Google Scholar
|
|
23
|
Gerritsen J, Smidt H, Rijkers GT and de
Vos WM: Intestinal microbiota in human health and disease: The
impact of probiotics. Genes Nutr. 6:209–240. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hu X, Wang T, Li W, Jin F and Wang L:
Effects of NS Lactobacillus strains on lipid metabolism of rats fed
a high-cholesterol diet. Lipids Health Dis. 12:672013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liboredo JC, Anastacio LR, Peluzio Mdo C,
Valente FX, Penido LC, Nicoli JR and Correia MI: Effect of
probiotics on the development of dimethylhydrazine-induced
preneoplastic lesions in the mice colon. Acta Cir Bras. 28:367–372.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bertkova I, Hijova E, Chmelarova A,
Mojzisova G, Petrasova D, Strojny L, Bomba A and Zitnan R: The
effect of probiotic microorganisms and bioactive compounds on
chemically induced carcinogenesis in rats. Neoplasma. 57:422–428.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang W, Shi LP, Shi L and Xu L: Efficacy
of probiotics on the treatment of non-alcoholic fatty liver
disease. Zhonghua Nei Ke Za Zhi. 57:101–106. 2018.(In Chinese).
PubMed/NCBI
|
|
28
|
Fan YJ, Chen SJ, Yu YC, Si JM and Liu B: A
probiotic treatment containing Lactobacillus, bifidobacterium and
enterococcus improves IBS symptoms in an open label trial. J
Zhejiang Univ Sci B. 7:987–991. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang Y, Xia Y, Chen H, Hong L, Feng J,
Yang J, Yang Z, Shi C, Wu W, Gao R, et al: The effect of
perioperative probiotics treatment for colorectal cancer:
Short-term outcomes of a randomized controlled trial. Oncotarget.
7:8432–8440. 2016.PubMed/NCBI
|
|
30
|
Liang S, Xu L, Zhang D and Wu Z: Effect of
probiotics on small intestinal bacterial overgrowth in patients
with gastric and colorectal cancer. Turk J Gastroenterol.
27:227–232. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Knights D, Ward TL, McKinlay CE, Miller H,
Gonzalez A, McDonald D and Knight R: Rethinking ‘enterotypes’. Cell
Host Microbe. 16:433–437. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xia F, Chen J, Fung WK and Li H: A
logistic normal multinomial regression model for microbiome
compositional data analysis. Biometrics. 69:1053–1063. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Qian L, Gao R, Hong L, Pan C, Li H, Huang
J and Qin H: Association analysis of dietary habits with gut
microbiota of a native Chinese community. Exp Ther Med. 16:856–866.
2018.PubMed/NCBI
|
|
34
|
Wan Y, Wang F, Yuan J, Li J, Jiang D,
Zhang J, Huang T, Zheng J, Mann J and Li D: Effects of
macronutrient distribution on weight and related cardiometabolic
profile in healthy non-obese Chinese: A 6-month, randomized
controlled-feeding trial. Ebiomedicine. 22:200–207. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li W and Godzik A: Cd-hit: A fast program
for clustering and comparing large sets of protein or nucleotide
sequences. Bioinformatics. 22:1658–1659. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Deng W, Wang Y, Liu Z, Cheng H and Xue Y:
HemI: A toolkit for illustrating heatmaps. PLoS One. 9:e1119882014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Weichselbaum E: Potential benefits of
probiotics-main findings of an in-depth review. Br J Community
Nurs. 15(110): 112–114. 2010.
|
|
38
|
Iannitti T and Palmieri B: Therapeutical
use of probiotic formulations in clinical practice. Clin Nutr.
29:701–725. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gareau MG, Sherman PM and Walker WA:
Probiotics and the gut microbiota in intestinal health and disease.
Nat Rev Gastroenterol Hepatol. 7:503–514. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chan YK, Brar MS, Kirjavainen PV, Chen Y,
Peng J, Li D, Leung FC and El-Nezami H: High fat diet induced
atherosclerosis is accompanied with low colonic bacterial diversity
and altered abundances that correlates with plaque size, plasma
A-FABP and cholesterol: A pilot study of high fat diet and its
intervention with Lactobacillus rhamnosus GG (LGG) or telmisartan
in ApoE(−/-) mice. BMC Microbiol. 16:2642016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kato-Kataoka A, Nishida K, Takada M, Kawai
M, Kikuchi-Hayakawa H, Suda K, Ishikawa H, Gondo Y, Shimizu K,
Matsuki T, et al: Fermented milk containing Lactobacillus casei
strain shirota preserves the diversity of the gut microbiota and
relieves abdominal dysfunction in healthy medical students exposed
to academic stress. Appl Environ Microbiol. 82:3649–3658. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lisko DJ, Johnston GP and Johnston CG:
Effects of dietary yogurt on the healthy human gastrointestinal
(GI) microbiome. Microorganisms. 5:E62017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kristensen NB, Bryrup T, Allin KH, Nielsen
T, Hansen TH and Pedersen O: Alterations in fecal microbiota
composition by probiotic supplementation in healthy adults: A
systematic review of randomized controlled trials. Genome Med.
8:522016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang J, Wang L, Guo Z, Sun Z, Gesudu Q,
Kwok L, Menghebilige and Zhang H: 454 pyrosequencing reveals
changes in the faecal microbiota of adults consuming Lactobacillus
casei Zhang. FEMS Microbiol Ecol. 88:612–622. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Park DY, Ahn YT, Park SH, Huh CS, Yoo SR,
Yu R, Sung MK, McGregor RA and Choi MS: Supplementation of
Lactobacillus curvatus HY7601 and Lactobacillus plantarum KY1032 in
diet-induced obese mice is associated with gut microbial changes
and reduction in obesity. PLoS One. 8:e594702013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Backhed F, Fraser CM, Ringel Y, Sanders
ME, Sartor RB, Sherman PM, Versalovic J, Young V and Finlay BB:
Defining a healthy human gut microbiome: Current concepts, future
directions, and clinical applications. Cell Host Microbe.
12:611–622. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hanifi A, Culpepper T, Mai V, Anand A,
Ford AL, Ukhanova M, Christman M, Tompkins TA and Dahl WJ:
Evaluation of bacillus subtilis R0179 on gastrointestinal viability
and general wellness: A randomised, double-blind,
placebo-controlled trial in healthy adults. Benef Microbes.
6:19–27. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hildebrandt MA, Hoffmann C, Sherrill-Mix
SA, Keilbaugh SA, Hamady M, Chen YY, Knight R, Ahima RS, Bushman F
and Wu GD: High-fat diet determines the composition of the murine
gut microbiome independently of obesity. Gastroenterology.
137:1716–1724.e1-2. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Heinsen FA, Fangmann D, Muller N, Schulte
DM, Ruehlemann MC, Turk K, Settgast U, Lieb W, Baines JF, Schreiber
S, et al: Beneficial effects of a dietary weight loss intervention
on human gut microbiome diversity and metabolism are not sustained
during weight maintenance. Obes Facts. 9:379–391. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Bell DS: Changes seen in gut bacteria
content and distribution with obesity: Causation or association?
Postgrad Med. 127:863–868. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Shang Y, Khafipour E, Derakhshani H, Sarna
LK, Woo CW, Siow YL and O K: Short term high fat diet induces
Obesity-Enhancing changes in mouse gut microbiota that are
partially reversed by cessation of the high fat diet. Lipids.
52:499–511. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Schwiertz A, Taras D, Schafer K, Beijer S,
Bos NA, Donus C and Hardt PD: Microbiota and SCFA in lean and
overweight healthy subjects. Obesity (Silver Spring). 18:190–195.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Duncan SH, Lobley GE, Holtrop G, Ince J,
Johnstone AM, Louis P and Flint HJ: Human colonic microbiota
associated with diet, obesity and weight loss. Int J Obes (Lond).
32:1720–1724. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Human Microbiome Project Consortium:
Structure, function and diversity of the healthy human microbiome.
Nature. 486:207–214. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhang J, Guo Z, Xue Z, Sun Z, Zhang M,
Wang L, Wang G, Wang F, Xu J, Cao H, et al: A phylo-functional core
of gut microbiota in healthy young Chinese cohorts across
lifestyles, geography and ethnicities. ISME J. 9:1979–1990. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Mariat D, Firmesse O, Levenez F, Guimaraes
V, Sokol H, Dore J, Corthier G and Furet JP: The
Firmicutes/Bacteroidetes ratio of the human microbiota changes with
age. BMC Microbiol. 9:1232009. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Kang C, Wang B, Kaliannan K, Wang X, Lang
H, Hui S, Huang L, Zhang Y, Zhou M, Chen M and Mi M: Gut microbiota
mediates the protective effects of dietary capsaicin against
chronic low-grade inflammation and associated obesity induced by
high-fat diet. MBio. 8:e00470–e00417. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ferrario C, Taverniti V, Milani C, Fiore
W, Laureati M, De Noni I, Stuknyte M, Chouaia B, Riso P and
Guglielmetti S: Modulation of fecal clostridiales bacteria and
butyrate by probiotic intervention with Lactobacillus paracasei DG
varies among healthy adults. J Nutr. 144:1787–1796. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Song JJ, Tian WJ, Kwok LY, Wang YL, Shang
YN, Menghe B and Wang JG: Effects of microencapsulated
Lactobacillus plantarum LIP-1 on the gut microbiota of
hyperlipidaemic rats. Brit J Nutr. 118:481–492. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Eeckhaut V, Machiels K, Perrier C, Romero
C, Maes S, Flahou B, Steppe M, Haesebrouck F, Sas B, Ducatelle R,
et al: Butyricicoccus pullicaecorum in inflammatory bowel disease.
Gut. 62:1745–1752. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Devriese S, Eeckhaut V, Geirnaert A, Van
den Bossche L, Hindryckx P, Van de Wiele T, Van Immerseel F,
Ducatelle R, De Vos M and Laukens D: Reduced Mucosa-associated
butyricicoccus activity in patients with ulcerative colitis
correlates with aberrant claudin-1 expression. J Crohns Colitis.
11:229–236. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Walker AW, Ince J, Duncan SH, Webster LM,
Holtrop G, Ze X, Brown D, Stares MD, Scott P, Bergerat A, et al:
Dominant and diet-responsive groups of bacteria within the human
colonic microbiota. ISME J. 5:220–230. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
An C, Kuda T, Yazaki T, Takahashi H and
Kimura B: Caecal fermentation, putrefaction and microbiotas in rats
fed milk casein, soy protein or fish meal. Appl Microbiol
Biotechnol. 98:2779–2787. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Park S, Ji Y, Jung HY, Park H, Kang J,
Choi SH, Shin H, Hyun CK, Kim KT and Holzapfel WH: Lactobacillus
plantarum HAC01 regulates gut microbiota and adipose tissue
accumulation in a diet-induced obesity murine model. Appl Microbiol
Biotechnol. 101:1605–1614. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Saitoh S, Noda S, Aiba Y, Takagi A,
Sakamoto M, Benno Y and Koga Y: Bacteroides ovatus as the
predominant commensal intestinal microbe causing a systemic
antibody response in inflammatory bowel disease. Clin Diagn Lab
Immunol. 9:54–59. 2002.PubMed/NCBI
|
|
66
|
Dicksved J, Halfvarson J, Rosenquist M,
Jarnerot G, Tysk C, Apajalahti J, Engstrand L and Jansson JK:
Molecular analysis of the gut microbiota of identical twins with
Crohn's disease. ISME J. 2:716–727. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
El Hage R, Hernandez-Sanabria E and Van de
Wiele T: Emerging trends in ‘Smart Probiotics’: Functional
consideration for the development of novel health and industrial
applications. Front Microbiol. 8:18892017. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Winter J, Moore LH, Dowell VR Jr and
Bokkenheuser VD: C-ring cleavage of flavonoids by human intestinal
bacteria. Appl Environ Microbiol. 55:1203–1208. 1989.PubMed/NCBI
|
|
69
|
Kasai C, Sugimoto K, Moritani I, Tanaka J,
Oya Y, Inoue H, Tameda M, Shiraki K, Ito M, Takei Y and Takase K:
Comparison of the gut microbiota composition between obese and
non-obese individuals in a Japanese population, as analyzed by
terminal restriction fragment length polymorphism and
next-generation sequencing. BMC Gastroenterol. 15:1002015.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Fang D, Shi D, Lv L, Gu S, Wu W, Chen Y,
Chen Y, Guo J, Li A, Xinjun Hu, et al: Bifidobacterium
pseudocatenulatum LI09 and Bifidobacterium catenulatum LI10
attenuate D-galactosamine-induced liver injury by modifying the gut
microbiota. Sci Rep. 7:87702017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Toscano M, De Grandi R, Miniello VL,
Mattina R and Drago L: Ability of Lactobacillus kefiri LKF01
(DSM32079) to colonize the intestinal environment and modify the
gut microbiota composition of healthy individuals. Dig Liver Dis.
49:261–267. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Jimenez MB: Treatment of irritable bowel
syndrome with probiotics. An etiopathogenic approach at last? Rev
Esp Enferm Dig. 101:553–564. 2009.PubMed/NCBI
|
|
73
|
van den Bogert B, Meijerink M, Zoetendal
EG, Wells JM and Kleerebezem M: Immunomodulatory properties of
Streptococcus and Veillonella isolates from the human small
intestine microbiota. PLoS One. 9:e1142772014. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Garrido D, Suau A, Pochart P, Cruchet S
and Gotteland M: Modulation of the fecal microbiota by the intake
of a Lactobacillus johnsonii La1-containing product in human
volunteers. FEMS Microbiology Letters. 248:249–256. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Zhu Q, Jin Z, Wu W, Gao R, Guo B, Gao Z,
Yang Y and Qin H: Analysis of the intestinal lumen microbiota in an
animal model of colorectal cancer. PLoS One. 9:e908492014.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Hibberd AA, Lyra A, Ouwehand AC, Rolny P,
Lindegren H, Cedgard L and Wettergren Y: Intestinal microbiota is
altered in patients with colon cancer and modified by probiotic
intervention. BMJ Open Gastroenterol. 4:e0001452017. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Wong SH, Kwong TNY, Chow TC, Luk AKC, Dai
RZW, Nakatsu G, Lam TYT, Zhang L, Wu JCY, Chan FKL, et al:
Quantitation of faecal Fusobacterium improves faecal immunochemical
test in detecting advanced colorectal neoplasia. Gut. 66:1441–1418.
2017. View Article : Google Scholar : PubMed/NCBI
|