Introduction
Lung cancer is the leading cause of
cancer-associated death worldwide, of which non-small-cell lung
cancer (NSCLC) accounts for >80% of lung cancer cases (1,2).
Although there have been significant advances in treatment methods,
including surgery combined with radiotherapy and/or chemotherapy,
as a result of a high rate of local recurrence and metastasis, the
5-year survival rate remains poor (3,4).
Therefore, a better understanding of the underlying molecular
mechanisms involved in NSCLC progression is required.
Long non-coding RNAs (lncRNAs) are a type of
non-coding RNA >200 nucleotides in length, with limited or no
coding potential (5,6). There are an increasing number of
studies which have demonstrated that lncRNAs are involved in
multiple cellular processes, including in cancer (7,8). For
example, Zhang et al (9)
demonstrated that that upregulation of lncRNA metastasis associated
lung adenocarcinoma transcript 1 was associated with tumor
progression and poor prognosis in clear cell renal cell carcinoma.
Gao et al (10) demonstrated
that ZNFX1 antisense RNA 1 exhibited an oncogenic role in glioma
progression by regulating epithelial-mesenchymal transition (EMT)
and the Notch signaling pathway. Chen et al (11) showed that lncRNA colon cancer
associated transcript 1 promotes the progression of multiple
myeloma by acting as a molecular sponge of microRNA (miR)-181a-5p,
thus modulating the expression of homeobox A1.
Protein disulfide isomerase family A member 3
pseudogene 1 (PDIA3P1) is a 2,099-nucleotide lncRNA that is mapped
to human chromosome 1q21.1. Sun et al (12) reported that lncRNA PDIA3P was
upregulated and interacted with miR-185-5p to promote the
proliferation of oral squamous cell carcinoma cells by targeting
cyclin D2. Yang et al (13)
showed that lncRNA PDIA3P interacted with c-myc to regulate cell
proliferation by activating the pentose phosphate pathway in
multiple myeloma. The aim of the present study was to reveal the
functions of lncRNA PDIA3P in the progression of NSCLC.
Materials and methods
Tumor specimens
A total of 73 pairs of NSCLC tissues and the
adjacent normal tissues were obtained from patients who received
surgery at The Affiliated Hospital of Hebei University of
Engineering (Handan, China) between January 2013 and December 2016.
All of the specimens were immediately frozen in liquid nitrogen and
stored at −80°C until they were used for RNA extraction. None of
the patients received previous local or systemic treatment prior to
the operation. Clinical information was obtained from the medical
records of the patients. All the patients in the present study
provided written informed consent. The present study was approved
by The Ethics Committee of Hebei University of Engineering
(approval no. HBU-2018-01127).
Cell culture and transfection
The human NSCLC cell lines A549, H1299, H1703, H520
and SK-MES-1, and the normal human bronchial epithelium cell line
BEAS-2B were obtained from The Cell Bank of Type Culture Collection
of the Chinese Academy of Sciences. All cells were cultured in
RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (HyClone; GE Healthcare
Life Sciences) at 37°C with 5% CO2. The concentrations
of siRNA and plasmids were 50 µM and 2 µg/ml, respectively. Small
interfering (si)-PDIA3P and the negative control (NC) were
purchased from Shanghai GenePharma Co., Ltd. The pcDNA3.1-PDIA3P
plasmid was prepared in our previous study using RNA interference
sequences (12) and an empty
pcDNA3.1 vector (Shanghai GenePharma Co., Ltd.) was used as a
negative control. The Wnt pathway inhibitor IWR-1-endo was obtained
from Cayman Chemical Company. Aliquots of 2 mM in DMSO were stored
at −20°C and working concentrations (5 µM) were prepared prior to
use. Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) was used for transfection, according to the
manufacturer's protocol (14). The
sequences of si-PDIA3P were as follows: si-RNA-1,
5′-AACCACTGGGGAGGACTAGG-3′; si-RNA-2, 5′-TGGTAGCAGAGAATTTGAT-3′;
and si-NC, 5′-AATTCTCCGAACGTGTCACGT-3′. Further experiments were
completed 24 h following transfection.
Reverse transcription-quantitative
(RT-q)PCR
Total RNAs were extracted from NSCLC tissues and
cell lines using TRIzol® (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. Total
RNA was reverse transcribed into cDNA using the PrimeScript RT
reagent kit (Takara Bio, Inc.) according to the manufacturer's
protocol. qPCR analysis was performed using a SYBR Green qPCR
Master Mix kit (Promega Corporation), according to the
manufacturer's instructions. The thermocycling conditions were as
follows: Initial denaturation at 95°C for 5 min followed by 40
cycles at 95°C for 10 sec and 60°C for 1 min. Relative expression
was normalized to GAPDH and calculated using the 2−ΔΔCq
method (15). The sequences of the
PCR primers were as follows: PDIA3P forward,
5′-AACCACTGGGGAGGACTAGG-3′ and reverse,
5′-CAGTGCAGCTAAGAAATGGCT-3′; and GAPDH forward,
5′-ATGGGGAAGGTGAAGGTCG-3′ and reverse,
5′-GGGTCATTGATGGCAACAATATC-3′.
Cell proliferation assay
A Cell Counting Kit-8 (Dojindo Molecular
Technologies, Inc.) assay was performed to determine the
proliferative rate of lung cancer cells. Briefly, cells were seeded
into 96-well plates and cultured for the indicated times (24, 48
and 72 h). Subsequently, 10 µl CCK-8 solution was added and
incubated for another 2 h at 37°C. The absorbance at 450 nm was
determined using a microplate reader (Bio-Rad Laboratories,
Inc.).
Colony formation assay
A total of 2 ml complete medium containing
2×103 transfected cells were added to 6-well plates and
incubated for 2 weeks. Subsequently, the supernatant was discarded
and the cells were washed with PBS, fixed using 500 µl methanol at
room temperature for 20 min and stained with 0.1% crystal violet
(Nanjing KeyGen Biotech Co., Ltd.) for 20 min at room temperature.
The number of colonies >10 cells were counted using an optical
light microscope at ×50 magnification (Olympus Corporation).
Cell invasion assay
The invasive capability of the cells was examined
using a Transwell chamber assay with an 8-µM pore (EMD Millipore).
Briefly, 1×104 cells were seeded in the upper chamber
coated with Matrigel (Sigma-Aldrich; Merck KGaA) and incubated for
48 h. Cells which had not invaded were removed using a swab and the
cells which had invaded through to the lower surface were fixed
with methanol for 35 min at room temperature, stained with crystal
violet for 50 min at room temperature, washed with PBS and counted
under a light microscope (magnification, ×50; Nikon
Corporation).
Animal experiments
The transfected A549 cells were subcutaneously
injected into 4 week-old female BALB/c nude mice (Beijing
Experimental Animal Research Center, Beijing, China). Each group
included 4 mice, and they were housed under specific pathogen free
conditions at 20–26°C, 40–70% humidity and a 12/12 h light/dark
cycle. The mice had free access to food and water. At the end of
the 7-week observation period, the mice were sacrificed by cervical
dislocation, and the tumor tissues were removed for subsequent
experiments (16). All experimental
procedures were approved by the committee on animal experimentation
of Hebei University of Engineering.
Western blotting
Proteins were lysed using RIPA buffer (Beyotime
Institute of Biotechnology) and the concentrations were measured
using a bicinchoninic acid protein assay (Beyotime Institute of
Biotechnology). Proteins (20 µg/per lane) were separated using a
10% SDS-PAGE gel and transferred to a PVDF membrane (EMD
Millipore). After incubation with antibodies against β-catenin
(1:1,000; cat. no. ab32572), c-myc (1:1,000; cat. no. ab32072),
glycogen synthase kinase (GSK)-3β (1:1,000; cat. no. ab32391) and
GAPDH (1:5,000; cat. no. ab181602) (all from Abcam) overnight at
4°C, the membranes were incubated with a goat anti-rabbit horse
radish peroxidase-conjugated secondary antibody (1:5,000; cat. no.
ab97051; Abcam) at room temperature for 1 h. The signals were
visualized using an enhanced chemiluminescence reagent (EMD
Millipore). Protein bands were visualized using ImageJ 1.48
software (National Institutes of Health).
Clinical databases
The online database Gene Expression Profiling
Interactive Analysis (GEPIA; http://gepia.cancerpku.cn/index.html.) was used to
analyze the RNA sequencing expression data relevant to the present
study based on The Cancer Genome Atlas and the Genotype-Tissue
Expression databases (17). GEPIA
performs survival analyses based on gene expression levels and uses
a log-rank test for hypothesis evaluation.
Immunohistochemistry
The expression of Ki-67 in nude mice injected with
A549 cells was measured using immunohistochemistry. The
immunohistochemistry assay was conducted according to previous
report (18). The sections were
treated with rabbit polyclonal anti-Ki-67 antibody (1:100; cat. no.
ab833; Abcam) at 4°C overnight. After successfully completing the
previous steps, the sections were incubated with a secondary goat
anti-rabbit antibody (1:1,000; cat. no. ab6721; Abcam) for 20 min
at 37°C. Image acquisition was performed by light microscope
(magnification, ×100; Nikon Corporation) and Image-Pro Plus version
6.0 (Media Cybernetics, Inc.) was used to analyze the integrated
optical density values of the brown area.
Statistical analysis
All data were analyzed using SPSS 18.0 software
(SPSS, Inc.) and are expressed as the mean ± SD from at least three
independent experiments. The differences between the two groups
were evaluated using a Student's t-test or one-way ANOVA followed
by a Bonferroni post hoc test. P<0.05 was considered to indicate
a statistically significant difference.
Results
lncRNA PDIA3P is upregulated in
NSCLC
In the present study, PDIA3P expression in NSCLC
tissues was examined. RT-qPCR analysis showed that PDIA3P
expression was significantly higher in NSCLC tissues (Fig. 1A; P<0.05) compared with the
control. High PDIA3P expression levels were inversely associated
with TNM stages III and IV, and the presence of lymph node
metastasis (Fig. 1B and C; Table I; P<0.05). Furthermore, data from
the GEPIA database showed that PDIA3P expression was increased in
NSCLC (lung adenocarcinoma and lung squamous cell carcinoma)
tissues compared with normal tissues (Fig. 1D; P<0.05). In addition, high
PDIA3P expression was associated with advanced tumor stage and poor
disease-free survival of patients with NSCLC (Fig. 1E and F; P<0.05).
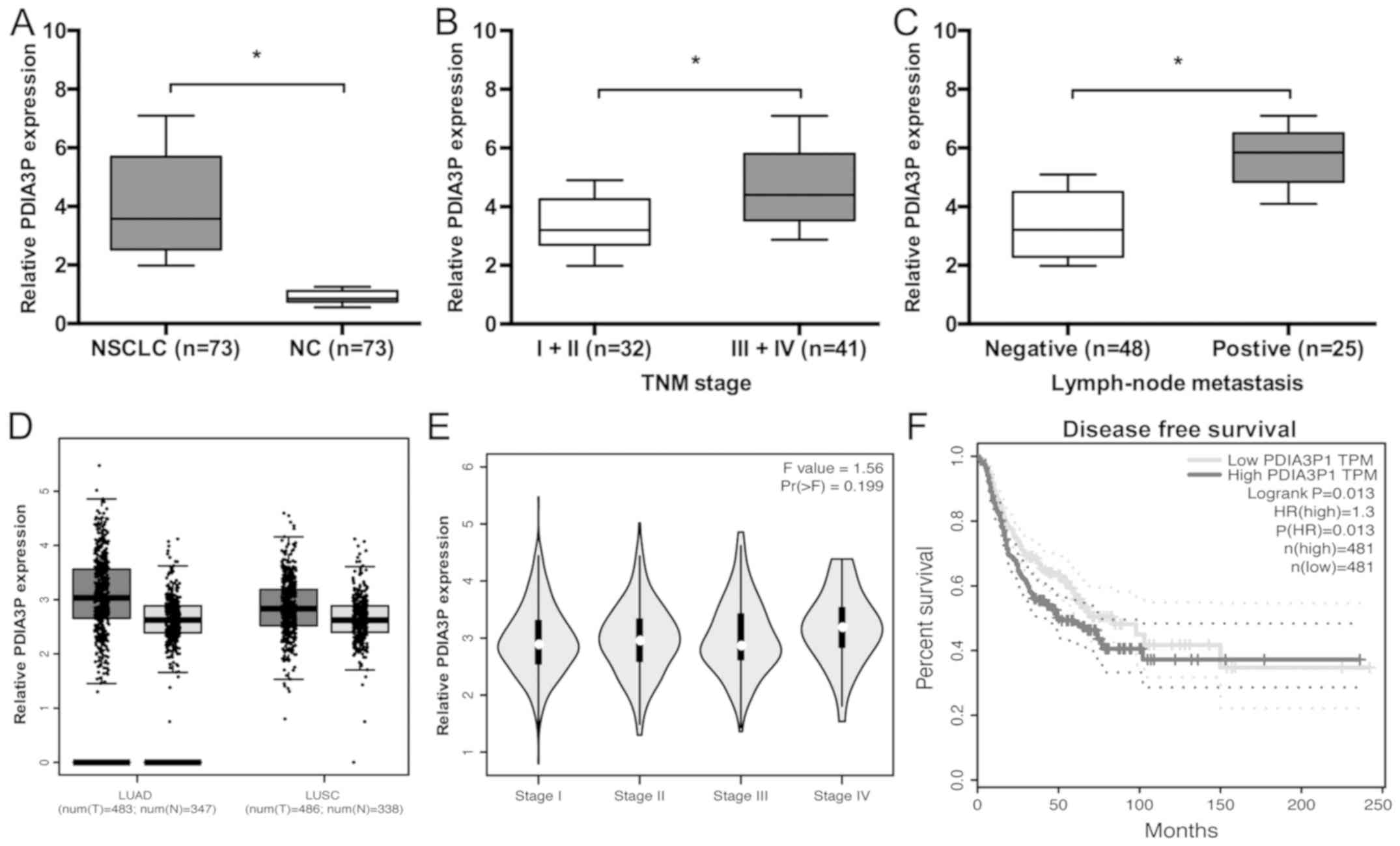 | Figure 1.Expression of lncRNA PDIA3P is
upregulated in NSCLC. (A) PDIA3P expression in 73 paired NSCLC
tissues was analyzed by reverse transcription-quantitative PCR.
*P<0.05. (B) PDIA3P expression was associated with advanced TNM
stage in patients with NSCLC. *P<0.05. (C) PDIA3P expression was
associated with advanced lymph node metastasis in patients with
NSCLC. *P<0.05. (D) PDIA3P expression in NSCLC tissues and
normal tissues was analyzed using GEPIA. (E) The GEPIA database
indicated that high PDIA3P expression was associated with tumor
stage. (F) The GEPIA database showed that high PDIA3P expression
was associated with poor disease-free survival of patients with
NSCLC. P=0.013. lncRNA, long noncoding RNA; LUAD, lung
adenocarcinoma; LUSC, lung squamous cell carcinoma; NSCLC,
non-small cell lung cancer; NC, negative control; HR, hazard ratio;
TNM, Tumor-Node-Metastasis; TPM, transcripts per million; GEPIA,
Gene Expression Profiling Interactive Analysis; PDIA3P, protein
disulfide isomerase family A member 3 pseudogene 1. |
 | Table I.Associations between PDIA3P expression
and clinical features of patients with NSCLC. |
Table I.
Associations between PDIA3P expression
and clinical features of patients with NSCLC.
|
| PDIA3P
expression |
|
|---|
|
|
|
|
|---|
| Characteristics | High, n=37 | Low, n=36 | P-value |
|---|
| Age, years |
|
| 0.415 |
|
>60 | 21 | 17 |
|
| ≤60 | 16 | 19 |
|
| Sex |
|
| 0.736 |
| Male | 22 | 20 |
|
|
Female | 15 | 16 |
|
| TNM stage |
|
| 0.003a |
| I/II | 10 | 22 |
|
|
III/IV | 27 | 14 |
|
| Tumor size, cm |
|
| 0.295 |
|
>3 | 23 | 18 |
|
| ≤3 | 14 | 18 |
|
| Lymph node
metastasis |
|
| 0.009a |
|
Negative | 19 | 29 |
|
|
Positive | 18 | 7 |
|
lncRNA PDIA3P-silencing suppresses the
proliferation and invasion of NSCLC cells
To determine the biological functions of PDIA3P in
NSCLC progression, the expression levels of PDIA3P were first
determined in NSCLC cell lines (A549, H1299, H1703, H520 and
SK-MES-1) and the normal human bronchial epithelium cell line
BEAS-2B, and A549 and H520 cell lines were selected for further
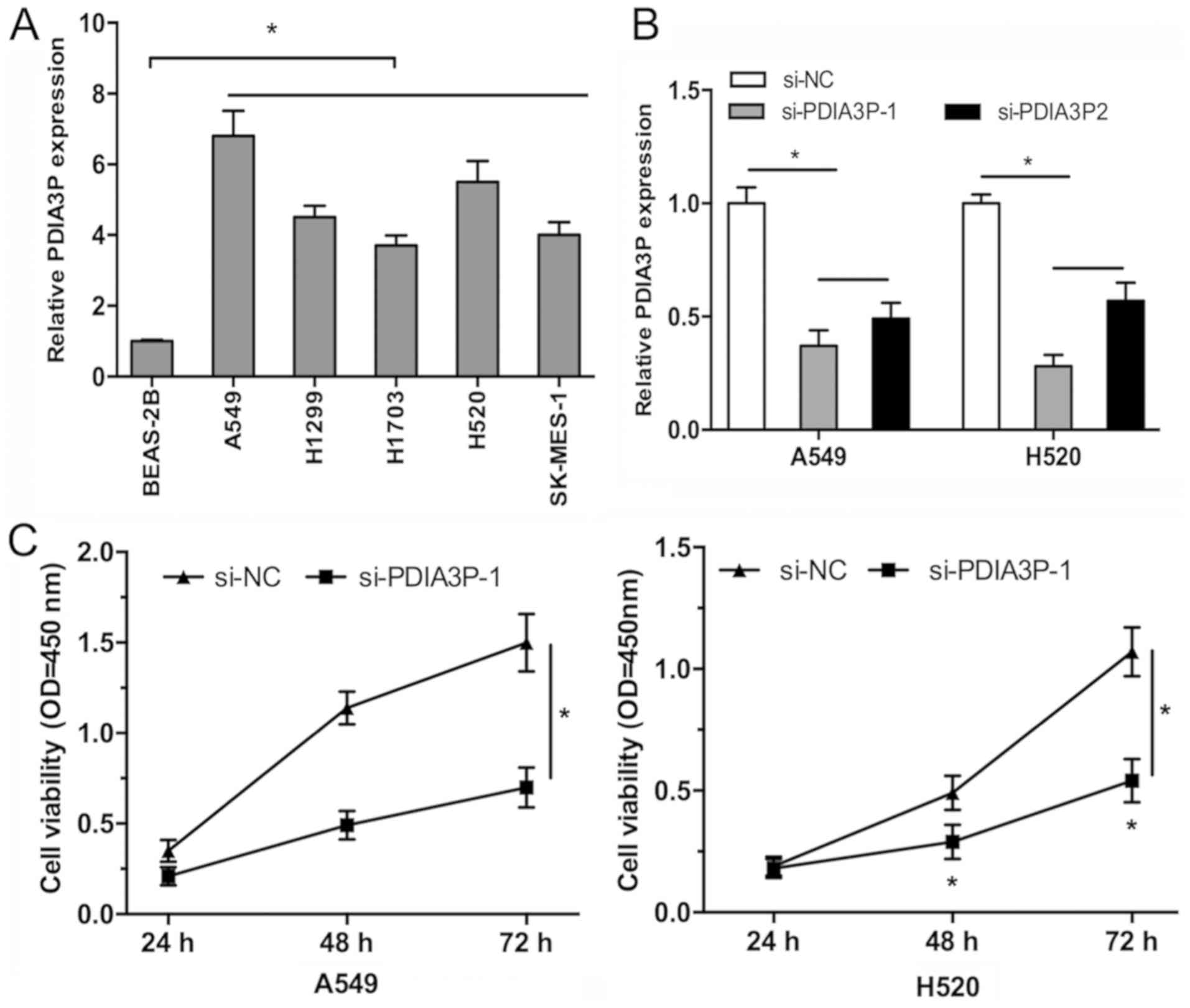
study as they exhibited high PDIA3P expression. RT-qPCR analysis
showed that PDIA3P expression was significantly increased in all
the NSCLC cell lines compared with BEAS-2B cells (Fig. 2A; P<0.05). Therefore, PDIA3P was
silenced in both A549 and H520 cells by si-PDIA3P (Fig. 2B; P<0.05). A CCK-8 assay showed
that PDIA3P knockdown decreased the proliferative rate of A549 and
H520 cells compared with the negative control (Fig. 2C; P<0.05), and the cell colony
formation was significantly decreased following PDIA3P silencing in
A549 and H520 cells compared with the negative control (Fig. 2D; P<0.05). Furthermore, the
migratory and invasive ability of A549 and H520 cells was reduced
following silencing of PDIA3P compared with the control (Fig. 2E and F; P<0.05).
lncRNA PDIA3P silencing reduces tumor
growth in vivo
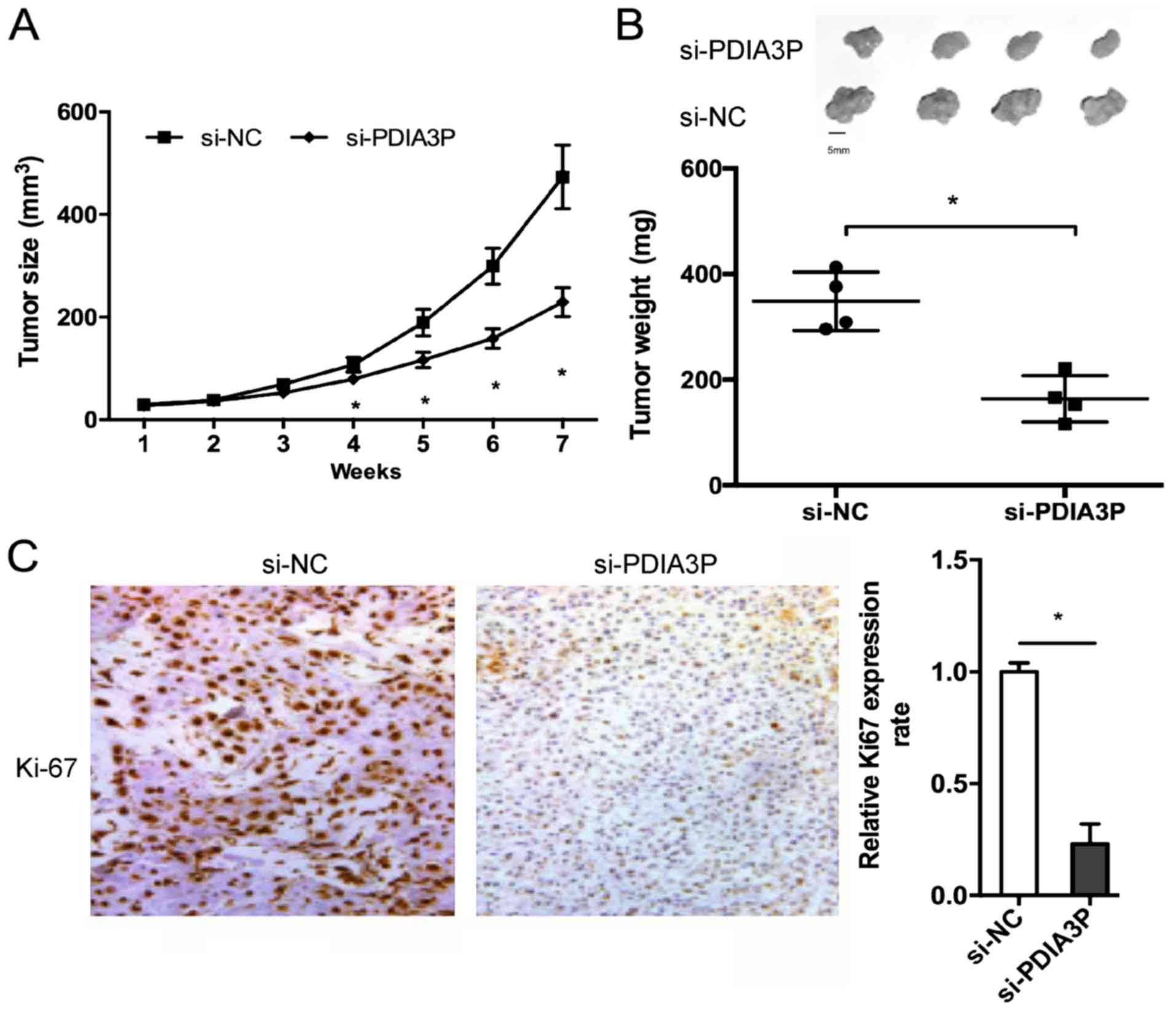
The effects of PDIA3P on NSCLC growth in vivo
were determined. The results showed that PDIA3P inhibition
significantly reduced the tumor volume compared with the negative
control (Fig. 3A; P<0.05). At 7
weeks after the injection, the mice were sacrificed, and tumor
weight was determined. The results showed that PDIA3P silencing
reduced the tumor weight (Fig. 3B;
P<0.05). Furthermore, immunohistochemical staining showed that
the Ki67 expression was decreased in PDIA3P-silenced xenograft
tumor tissues (Fig. 3C;
P<0.05).
lncRNA PDIA3P activates the
Wnt/β-catenin signaling pathway
To determine the underlying mechanism by which
PDIA3P affects NSCLC progression, RT-qPCR and western blotting were
performed to examine the effects of PDIA3P on the Wnt/β-catenin
pathway, which is frequently aberrantly activated in human cancer
(19). The results showed that
PDIA3P inhibition decreased both the mRNA and protein expression
levels of β-catenin and c-myc, and increased the expression levels
of GSK-3β in A549 cells (Fig. 4A and
B; P<0.05). A549 cell line was used as it exhibited the
highest expression. PDIA3P overexpression upregulated β-catenin and
c-myc expression, and decreased GSK-3β expression in A549 cells,
both at the mRNA and protein level (Fig.
4C-E; P<0.05). Furthermore, the Wnt pathway inhibitor
IWR-1-endo partly reversed the effects of PDIA3P expression on
proliferation (Fig. 4F; P<0.05)
and invasion (Fig. 4G; P<0.05) in
A549 cells. Therefore, PDIA3P may promote the progression of NSCLC,
at least partly by regulating the Wnt/β-catenin pathway.
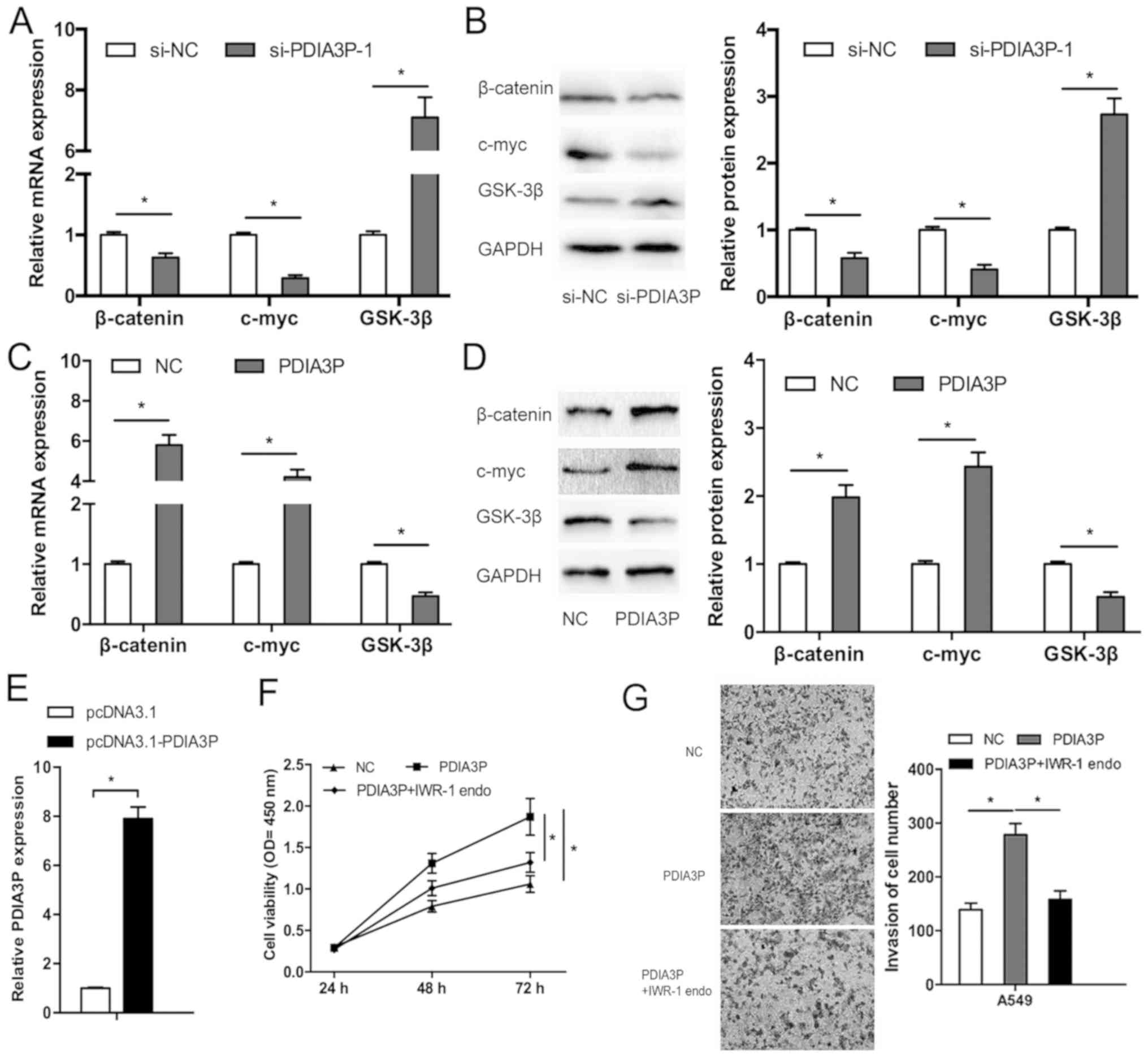 | Figure 4.Long noncoding RNA PDIA3P promotes the
Wnt/β-catenin pathway in NSCLC cells. (A) mRNA expression levels of
β-catenin, c-myc and GSK-3β in A549 cells transfected with
si-PDIA3P. *P<0.05. (B) Western blot analysis of the expression
levels of β-catenin, c-myc and GSK-3β in NSCLC cells transfected
with si-PDIA3P. *P<0.05. (C) mRNA expression levels of
β-catenin, c-myc and GSK-3β in A549 cells transfected with PDIA3P.
*P<0.05. (D) Western blot analysis of the expression levels of
β-catenin, c-myc and GSK-3β in NSCLC cells transfected with PDIA3P.
*P<0.05. (E) PDIA3P expression in NSCLC cells transfected with
plasmid pcDNA3.1-PDIA3P. The Wnt pathway inhibitor IWR-1-endo
partly rescued the effects of PDIA3P overexpression on the (F)
proliferation and (G) invasion ability of NSCLC cells.
Magnification, ×50. *P<0.05. NSCLC, non-small cell lung cancer;
si, small interfering; NC, negative control; OD, optical density;
PDIA3P, protein disulfide isomerase family A member 3 pseudogene 1;
GSK-3β, glycogen synthase kinase-3β. |
Discussion
There has been an increase in the number of studies
demonstrating the potential roles of various lncRNAs as key
regulators of cancer progression over the past decade (20–22), and
genetic and epigenetic alterations of lncRNAs may serve key roles
in tumorigenesis (23). Furthermore,
abnormal expression of lncRNAs may serve important roles in cancer
progression, including in NSCLC. For example, Xie et al
(24) showed that the lncRNA gastric
cancer associated transcript 2 is downregulated and associated with
poor prognosis in patients with NSCLC. Cui et al (25) found that upregulation of the lncRNA
small nucleolar RNA host gene 1 contributes to NSCLC progression
via inhibition of miR-101-3p and activation of the Wnt/β-catenin
signaling pathway. Gao et al (26) found that lncRNA FLVCR1 divergent
transcript contributes to proliferation and invasion by sponging
miR-573 to upregulate E2F transcription factor 3 expression in lung
cancer.
In the present study, the function and underlying
mechanisms of PDIA3P expression in NSCLC progression were
determined. The results showed that PDIA3P expression was
significantly increased in NSCLC tissues compared with adjacent
non-tumor tissues. High PDIA3P expression was associated with
advanced TNM stage cancer, lymph-node metastasis of cancer and poor
disease-free survival of patients with NSCLC. PDIA3P functions in
lung cancer were examined both in vitro and in vivo.
The results showed that PDIA3P knockdown significantly inhibited
the growth of NSCLC cells both in vitro and in vivo.
Similarly, wound healing assays and Transwell assays showed that
PDIA3P inhibition decreased the migration and invasion of lung
cancer cells in vitro. In a further study the effects of
PDIA3P on proliferation markers, such as proliferating cell nuclear
antigen, and migration markers, such as matrix metalloproteinases,
will be determined. Taken together, the results of the present
study showed that PDIA3P may act as an oncogenic lncRNA in the
progression of NSCLC.
The Wnt/β-catenin pathway is frequently activated in
a wide range of different types of cancer, and is known to promote
tumor invasion and metastasis through upregulation of factors
regulating EMT (27,28). Recent studies demonstrated that
certain lncRNAs affect the Wnt/β-catenin signaling pathway and thus
regulate cancer progression. For example, Ma et al (29) showed that the lncRNA CCAL regulates
the progression of colorectal cancer by activating the
Wnt/β-catenin pathway through suppression of activator protein 2α.
Zhao et al (30) demonstrated
that upregulation of the lncRNA HNF1A antisense RNA 1 promotes cell
proliferation and metastasis in osteosarcoma through activation of
the Wnt/β-catenin pathway. However, the association between PDIA3P
expression and the Wnt/β-catenin pathway in NSCLC remains unclear.
In the present study, it was demonstrated that PDIA3P inhibition
significantly reduced β-catenin and c-myc expression, and increased
GSK-3β expression in NSCLC cells, while ectopic PDIA3P expression
resulted in the opposite effects. In addition, in vitro
functional assays showed that IWR-1 endo (Wnt pathway inhibitor)
(31) attenuated the effects of
PDIA3P on the proliferation and invasive ability of NSCLC cells,
suggesting that PDIA3P promotes NSCLC progression at least partly
through the Wnt/β-catenin pathway.
There are certain limitations to the present study.
The effects of IWR-1 alone on b-catenin, or on cell viability and
invasion ability, were not assessed.
In conclusion, PDIA3P is significantly increased in
NSCLC, and promotes the proliferation and invasion of NSCLC by
regulating the Wnt/β-catenin pathway. Future experiments should
examine the effects of PDIA3P on EMT. The findings of the present
study indicated that PDIA3P may serve as a potential therapeutic
target for the treatment of patients with NSCLC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
BY designed the current study. XY and BY performed
the experiments and analyzed the data. BY drafted the manuscript.
XY and BY wrote the manuscript. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
All the patients in the present study provided
written informed consent. The present study was approved by The
Ethics Committee of Hebei University of Engineering (approval no.
HBU-2018-01127). All experimental procedures involving animals were
approved by the committee on animal experimentation of Hebei
University of Engineering.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dela Cruz CS, Tanoue LT and Matthay RA:
Lung cancer: Epidemiology, etiology, and prevention. Clin Chest
Med. 32:605–644. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Didkowska J, Wojciechowska U, Mańczuk M
and Łobaszewski J: Lung cancer epidemiology: Contemporary and
future challenges worldwide. Ann Transl Med. 4:1502016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mattick JS and Makunin IV: Non-coding RNA.
Hum Mol Genet. 15:R17–R29. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research. Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Qi P and Du X: The long non-coding RNAs, a
new cancer diagnostic and therapeutic gold mine. Mod Pathol.
26:155–165. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang HM, Yang FQ, Chen SJ, Che J and
Zheng JH: Upregulation of long non-coding RNA MALAT1 correlates
with tumor progression and poor prognosis in clear cell renal cell
carcinoma. Tumor Biol. 36:2947–2955. 2015. View Article : Google Scholar
|
|
10
|
Gao K, Ji Z, She K, Yang Q and Shao L:
Long non-coding RNA ZFAS1 is an unfavourable prognostic factor and
promotes glioma cell progression by activation of the Notch
signaling pathway. Biomed Pharmacother. 87:555–560. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen L, Hu N, Wang C, Zhao H and Gu Y:
Long non-coding RNA CCAT1 promotes multiple myeloma progression by
acting as a molecular sponge of miR-181a-5p to modulate HOXA1
expression. Cell Cycle. 17:319–329. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sun CC, Zhang L, Li G, Li SJ, Chen ZL, Fu
YF, Gong FY, Bai T, Zhang DY, Wu QM and Li DJ: The lncRNA PDIA3P
interacts with miR-185-5p to modulate oral squamous cell carcinoma
progression by targeting cyclin D2. Mol Ther-Nucleic Acids.
9:100–110. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang X, Ye H, He M, Zhou X, Sun N, Guo W,
Lin X, Huang H, Lin Y, Yao R and Wang H: LncRNA PDIA3P interacts
with c-Myc to regulate cell proliferation via induction of pentose
phosphate pathway in multiple myeloma. Biochem Biophys Res Commun.
498:207–213. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang FQ, Zhang HM, Chen SJ, Yan Y and
Zheng JH: MiR-506 is down-regulated in clear cell renal cell
carcinoma and inhibits cell growth and metastasis via targeting
FLOT1. PLoS One. 10:e01202582015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ou L, Wang D, Zhang H, Yu Q and Hua F:
Decreased expression of MiR-138-5p by LncRNA H19 in cervical cancer
promotes tumor proliferation. Oncol Res. 26:401–410. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gu Y, Xiao X and Yang S: LncRNA MALAT1
acts as an oncogene in multiple myeloma through sponging miR-509-5p
to modulate FOXP1 expression. Oncotarget. 8:101984–101993. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tu J, Yu Y, Liu W and Chen S: Significance
of human epidermal growth factor receptor 2 expression in
colorectal cancer. Exp Ther Med. 9:17–24. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pridgeon MG, Grohar PJ, Steensma MR and
Williams BO: Wnt signaling in ewing sarcoma, osteosarcoma, and
malignant peripheral nerve sheath tumors. Curr Osteoporosis Rep.
15:239–246. 2017. View Article : Google Scholar
|
|
20
|
Yang G, Lu X and Yuan L: LncRNA: A link
between RNA and cancer. Biochim Biophys Acta. 1839:1097–1109. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Peng WX, Koirala P and Mo Y:
LncRNA-mediated regulation of cell signaling in cancer. Oncogene.
36:5661–5667. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tang Q, Ni Z, Cheng Z, Xu J, Yu H and Yin
P: Three circulating long non-coding RNAs act as biomarkers for
predicting NSCLC. Cell Physiol Biochem. 37:1002–1009. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xie X, Liu HT, Mei J, Ding FB, Xiao HB, Hu
FQ, Hu R and Wang MS: LncRNA HMlincRNA717 is down-regulated in
non-small cell lung cancer and associated with poor prognosis. Int
J Clin Exp Pathol. 7:8881–8886. 2014.PubMed/NCBI
|
|
25
|
Cui Y, Zhang F, Zhu C, Geng L, Tian T and
Liu H: Upregulated lncRNA SNHG1 contributes to progression of
non-small cell lung cancer through inhibition of miR-101-3p and
activation of Wnt/β-catenin signaling pathway. Oncotarget.
8:17785–17794. 2017.PubMed/NCBI
|
|
26
|
Gao X, Zhao S, Yang X, Zang S and Yuan X:
Long non-coding RNA FLVCR1-AS1 contributes to the proliferation and
invasion of lung cancer by sponging miR-573 to upregulate the
expression of E2F transcription factor 3. Biochem Biophys Res
Commun. 505:931–938. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
MacDonald BT, Tamai K and He X:
Wnt/Beta-catenin signaling: Components, mechanisms, and diseases.
Dev Cell. 17:9–26. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Clevers H and Nusse R: Wnt/β-catenin
signaling and disease. Cell. 149:1192–1205. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ma Y, Yang Y, Wang F, Moyer MP, Wei Q,
Zhang P, Yang Z, Liu W, Zhang H, Chen N, et al: Long non-coding RNA
CCAL regulates colorectal cancer progression by activating
Wnt/β-catenin signalling pathway via suppression of activator
protein 2α. Gut. 65:1494–1504. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhao H, Hou W, Tao J, Zhao Y, Wan G, Ma C
and Xu H: Upregulation of lncRNA HNF1A-AS1 promotes cell
proliferation and metastasis in osteosarcoma through activation of
the Wnt/β-catenin signaling pathway. Am J Transl Res. 8:3503–3512.
2016.PubMed/NCBI
|
|
31
|
Huang SM, Mishina YM, Liu S, Cheung A,
Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner
S, et al: Tankyrase inhibition stabilizes axin and antagonizes Wnt
signalling. Nature. 461:614–620. 2009. View Article : Google Scholar : PubMed/NCBI
|