Introduction
Bone morphogenetic proteins (BMP) are reported to be
associated with proliferation and differentiation of numerous types
of cells (1,2). In a previous in vitro study,
bone morphogenetic protein-7 (BMP-7) was shown to promote
odontogenic differentiation of stem cells derived from dental pulp
(3). BMP-7 at concentrations of 50
and 100 ng/ml was shown to produce dental pulp stem cells that
tended toward odontogenic differentiation through the Smad5
signaling pathway and did not significantly interrupt cell
proliferation in vitro (3). A
previous report showed that BMP-7 made the fibroblasts derived from
a human dermis differentiate into osteoblasts and promoted
osteogenesis (4). Similarly, BMP-7
enhanced the human-induced pluripotent stem cells' differentiation
potential (5).
Mesenchymal stromal cells from intraoral region drew
great attention in tissue regeneration because they can be achieved
with a minimally invasive maneuver (6). For several years, gingiva-derived stem
cells have been used for various purposes (7–9).
Mesenchymal gingiva-derived stem cells repaired calvarial and
mandibular defects, and gingival tissue can serve as a source for
stem cell therapy (10).
Gingiva-derived stem cells can aggregate into tight
three-dimensional spheroids, which can produce their own matrix
(8). Gingiva-derived stem cells have
demonstrated to have immunoregulatory effects by promoting Treg
cell polarization, inducing T-cell apoptosis and suppressing the
in vitro polarization of Th17 cell (9). Gingiva-derived stem cells encapsulated
in an alginate/hyaluronic acid three-dimensional scaffold have been
applied (7). Gingiva-derived stem
cells have been used for bone and nerve regeneration (11). This study was aimed at examining the
time-dependent impact of BMP-7 on changes of gene expression in the
stem cells. mRNA sequencing and data analysis were performed along
with gene ontology and pathway analysis. Quantitative analysis of
mRNA using real-time polymerase chain reactions of ColI, Sp7 and
IBSP and western blot analysis of collagen I, osterix and bone
sialoprotein as well as β-actin were performed. The purpose of this
study was to demonstrate the effects of BMP-7 on the
gingiva-derived stem cells.
Materials and methods
Stem cells collected from human
gingivae
Human gingivae were collected from healthy
participants following the method used in a previous study
(12). Approval of this study was
obtained from the Institutional Review Board at Seoul St Mary's
Hospital (KC18SESI0199). The participants signed the written
consent, and all of the procedures completed in this study followed
the relevant regulations and guidelines.
Concisely, de-epithelialization of the obtained
tissues was performed, and the tissues were dissected into
1–2-mm2 fragments. The tissues were dissolved using
collagenase type IV (Sigma-Aldrich Co., St. Louis, MO, USA) at 2
mg/ml and dispase (Sigma-Aldrich Co.) at 1 mg/ml. The resultant
products was filtered with a 70-µm cell strainer (Falcon, BD
Biosciences, Franklin Lakes, NJ, USA), and sterile
phosphate-buffered saline (PBS, Welgene, Daegu, Korea) was applied
to remove the non-adherent cells after incubation for 24 h.
Treatment of the stem cells with
BMP-7
The gingivae-derived stem cells were then treated
with BMP-7 (CYT-333; ProSpec Co., Nagoya, Japan) at a final
concentration of 100 ng/ml. The control group was loaded with the
same concentration of the dissolving media of acetic acid. The
cells were obtained at 3 or 24 h.
Evaluation of the secretion of human
vascular endothelial growth factor for paracrine effect
Secretion of human vascular endothelial growth
factor was determined at 3 and 24 h using a kit
(Quantikine® ELISA, cat. no. DVE00; R&D Systems,
Inc., Minneapolis, MN, USA). Preparation of the samples and
reagents were performed following the manufacturer's
recommendations. The resulting products were diluted ten times. The
differences of absorbance levels at 450 and 570 nm were used for
the evaluation of paracrine effects.
RNA isolation
Isolation of total RNA was done using Trizol reagent
(Invitrogen Corp., Carlsbad, CA, USA). Quality of RNA was evaluated
by bioanalyzer (Agilent 2100; Agilent Technologies, Amstelveen, The
Netherlands) using the nanochip (RNA 6000, Agilent Technologies),
and quantification of RNA was carried out using a spectrophotometer
(ND-2000, Thermo Fisher Scientific Inc., Waltham, MA, USA).
Library preparation and
sequencing
Library of control and test RNAs were built using
SMARTer Stranded RNA-Seq Kit (Clontech Laboratories, Inc., Palo
Alto, CA, USA) following the manufacturer's instructions.
Concisely, each 2 µg total RNA was prepared and reacted with
magnetic beads coated with oligo-dT. Washing solution was used to
remove other RNAs except mRNA. Initiation of library production was
performed by the random hybridization of starter/stopper
heterodimers to the poly(A) RNA still attached to the magnetic
beads. Illumina-compatible linker sequences were part of these
starter/stopper heterodimers. The starter was extended to the next
hybridized heterodimer by a single-tube reverse transcription and
ligation reaction. The stopper ligated the newly-synthesized cDNA
insert. The library was released by second strand synthesis from
the beads and the library was amplified with introduction of
barcodes. Paired-end 100 sequencing performed high-throughput
sequencing using HiSeq 2500 (Illumina, Inc., San Diego, CA,
USA).
Data analysis
TopHat software tool was used to map mRNA-Seq reads
tool in order to gain the alignment file (13). Counts from unique and multiple
alignments were used to determine differentially expressed gene
from coverage in Bedtools (14).
Quantile-quantile normalization method was applied to process the
read count data were processed from EdgeR within Rusing
Bioconductor (15). Cufflinks were
used for assembling transcripts, estimating their abundances and
revealing differential expression of genes or isoforms with the
alignment files. The expression level of the gene regions was
determined by applying method of fragments per kilobase of exon per
million fragments. Gene classification was based on searches
performed by DAVID (http://david.abcc.ncifcrf.gov/) (16). Pathway analysis was performed on
differentially expressed genes basted on the Kyoto encyclopedia of
genes and genome pathway databases (17).
Quantification by real-time polymerase
chain reaction
Total RNA was isolated from cultured stem cells
using Trizol (Invitrogen Corp.) and was reverse transcribed. Primer
sequences were as follows: Collagen I Forward
5′:CCAGAAGAACTGGTACATCAGCAA-3′, Reverse
5′:CGCCATACTCGAACTGGAATC-3′, Sp7 Forward
5′:TTGCCAATCACTCTCTTTACC-3′, Reverse 5′: GAGATACCCAAGCCCAGAAT-3′,
IBSP Forward 5′:AGGACTGCCAGAGGAAGCAA-3′, Reverse 5′:
CACAGGCCATTCCCAAAATG-3′ and β-actin Forward 5′:
TGGCACCCAGCACAATGAA-3′ and Reverse 5′:
CTAAGTCATAGTCCGCCTAGAAGCA-3′. The housekeeping gene was β-actin in
this study, which was used for normalization. SYBR Green Real-Time
PCR Master Mixes (Applied Biosystems, Carlsbad, CA, USA) was used
to detect mRNA expression through a real-time polymerase chain
reaction following the manufacturer's manual.
Western blot analysis
Samples were rinsed with ice-cold PBS twice and
treated with a lysis buffer at 3 and 24 h for 30 min. The lysates
were centrifuged at 13,500 rpm for 15 min at 4°C . Separation of
the samples were performed by gel electrophoresis
(Mini-PROTEAN® TGX™ Precast Gels; Bio-Rad, Hercules, CA,
USA), transblotted to the membranes (Immun-Blot®;
Bio-Rad) and immunoblotted with the corresponding antibodies and
the detection kits. Primary antibodies against collagen I (ab6308;
Abcam, MA, USA), RUNX2 (ab76956; Abcam), OCN (ab13418; Abcam), Sp7
transcription factor (ab22552; Abcam), bone sialoprotein (ab52128,
Abcam), β-actin (SC-516102; Santa Cruz Biotechnology, Inc., Dallas,
TX, USA) and secondary antibodies were purchased from Santa Cruz
Biotechnology. The protein expressions of Collagen I, Sp7, bone
sialoprotein and β-actin was quantitatively analyzed using with
image processing program (ImageJ, National Institutes of Health,
Bethesda, MD, USA).
Alkaline phosphatase activity
In the test groups, the stem cells were treated with
BMP-7 for 3 or 24 h. Then media were changed to osteogenic media
composed of α-MEM (Gibco, Grand Island, NY, USA), containing 200 mM
of L-glutamine (Sigma-Aldrich Co.), 10 mM of ascorbic acid
2-phosphate (Sigma-Aldrich Co.), 100 µg/ml of streptomycin
(Sigma-Aldrich Co.), 15% fetal bovine serum (Gibco), 100 U/ml of
penicillin, 2 mg/ml of glycerophosphate disodium salt hydrate, and
38 µg/ml of dexamethasone. A commercially available kit (K412-500;
BioVision, Inc., Milpitas, CA, USA) were used for the analysis of
alkaline phosphatase activity on Day 7.
Statistical analysis
The data were presented as mean ± standard
deviations of the results. A test of normality was conducted with
Shapiro-Wilk, and differences among the groups were analyzed by
one-way analysis of variance with Tukey's post hoc test using a
statistical package (SPSS 12 for Windows; SPSS Inc., Chicago, IL,
USA).P<0.05 was considered to indicate a statistically
significant difference.
Results
Evaluation of gene ontology and
pathway analysis
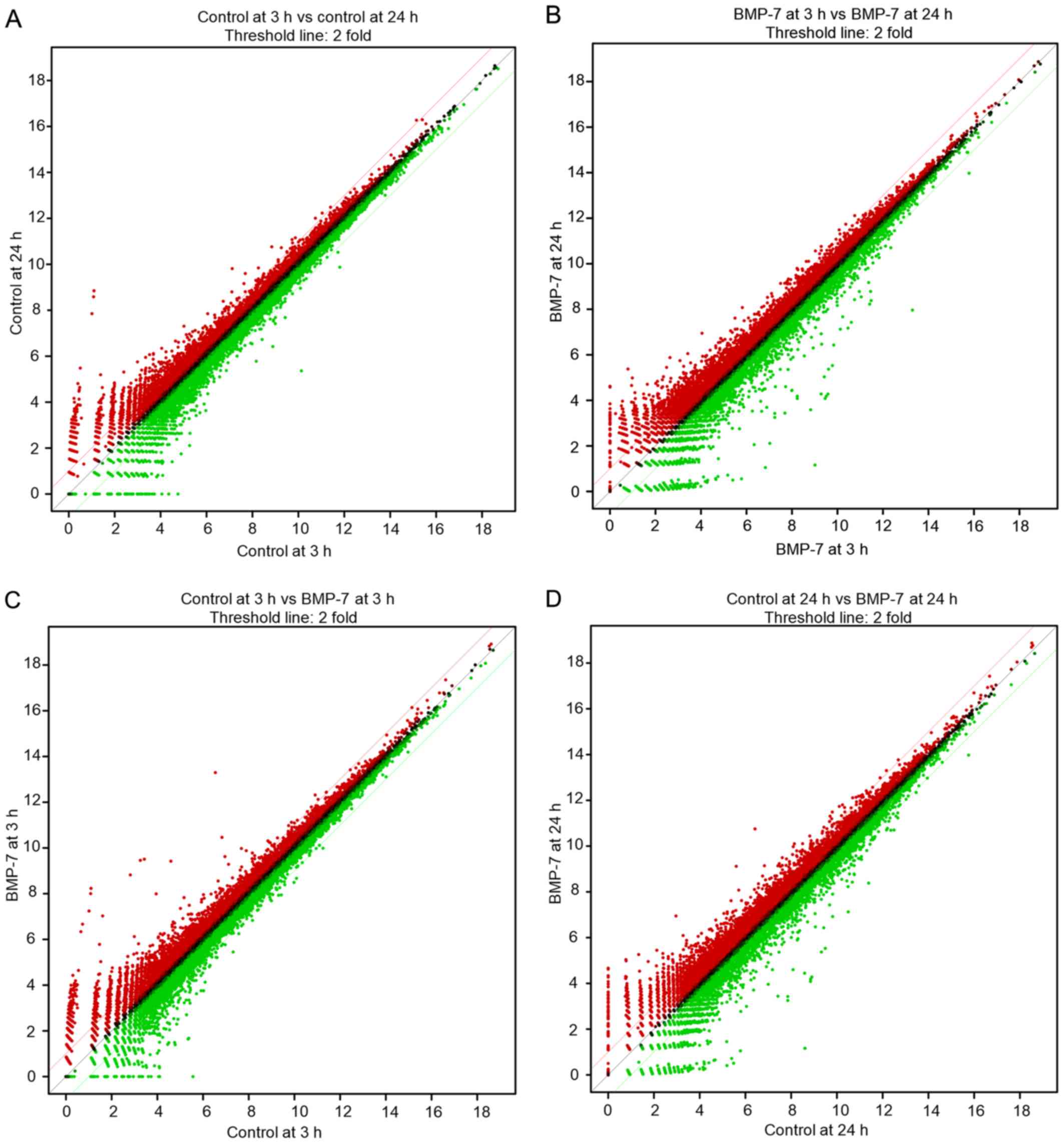
Differentially expressed mRNAs in this study is a
total of 25,737. The scatter plots displaying differentially
expressed mRNAs, are illustrated in Fig.
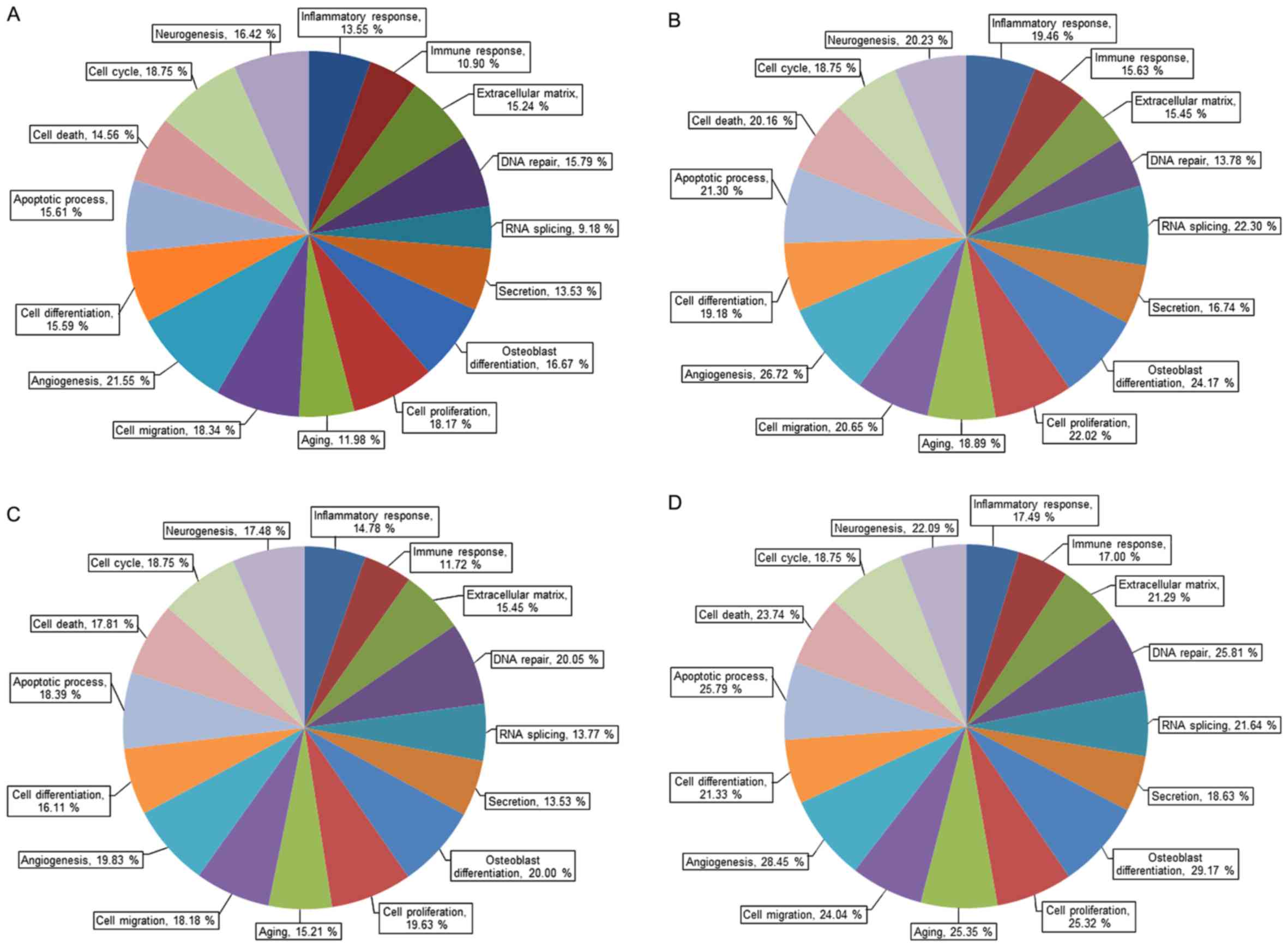
1. Gene ontology analysis of mRNA expression is categorized by
its importance and the selection criteria were fold change ≥1.3 and
log2 normalized read counts ≥4 (Fig.
2). Fig. 3 shows gene ontology
analysis of mRNA expression by upregulation and downregulation
(fold change of 1.3 or greater, log2 normalized read counts of 4 or
greater were selected).
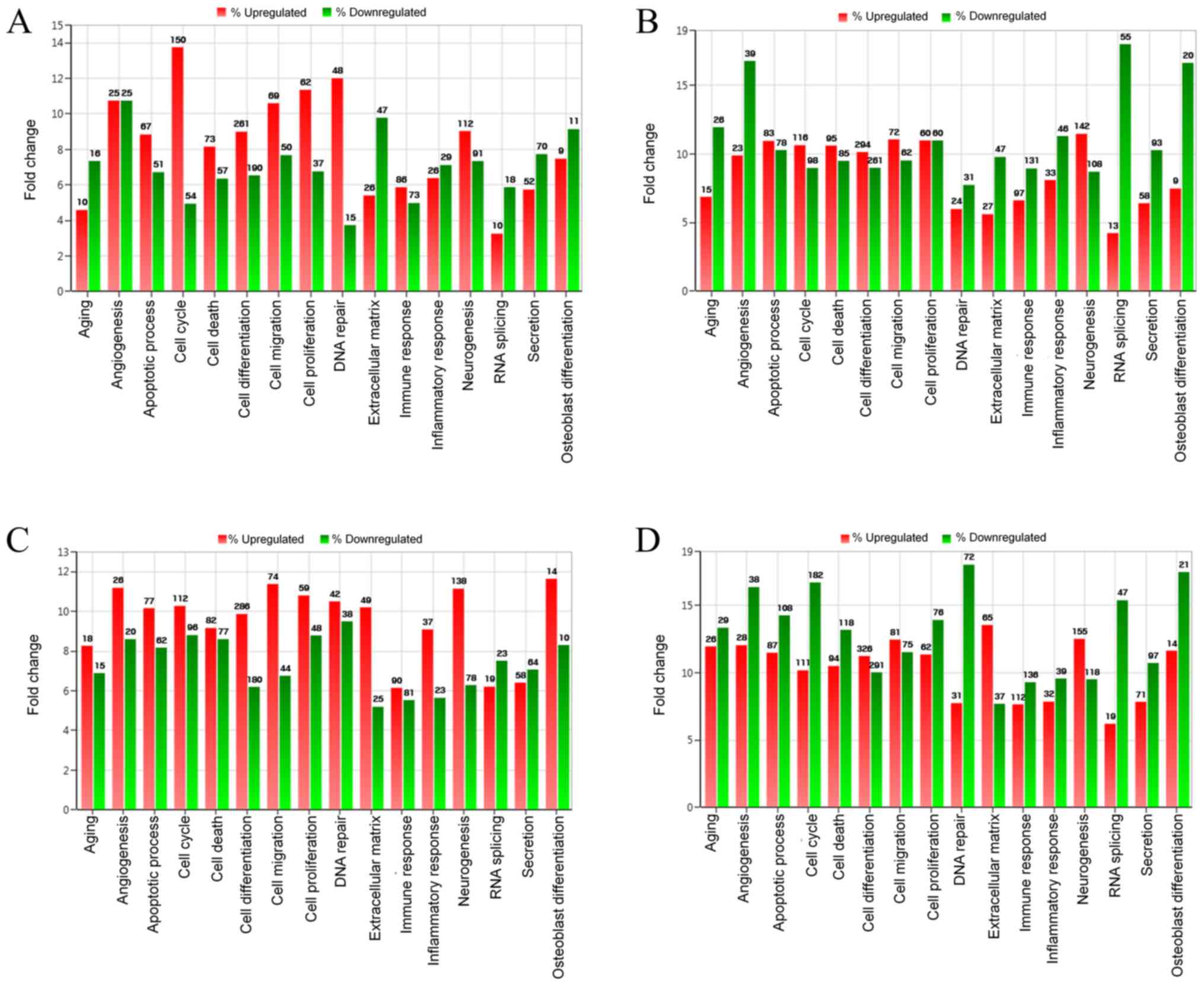
The results of differentially expressed mRNA related
to osteoblast differentiation, are displayed in Table I. To analyze the effects of
incubation time on the cultured cells, the comparisons were
performed between the control at 24 and at 3 h. The investigation
demonstrated that upregulation was seen in 9 mRNAs and
downregulation was noted in 11 mRNAs. Comparisons between BMP-7 at
24 h and BMP-7 at 3 h showed that 9 mRNAs were upregulated and 20
were downregulated. Comparisons between BMP-7 at 3 h and the
control at 3 h exhibited upregulation of 14 mRNAs and
downregulation of 10 mRNAs. When the results of BMP-7 at 24 h were
compared with the control at 24 h, upregulation of 14 mRNAs and
downregulation of 21 mRNAs were seen.
 | Table I.Differentially expressed mRNA related
to osteoblast differentiation (fold change of 1.3 or greater, log2
normalized read counts ≥4 were selected). |
Table I.
Differentially expressed mRNA related
to osteoblast differentiation (fold change of 1.3 or greater, log2
normalized read counts ≥4 were selected).
| Gene symbol | Control-24
h/Control-3 h | Gene symbol | BMP7-24 h/BMP7-3
h | Gene symbol | BMP7-3 h/Control-3
h | Gene symbol | BMP7-24
h/Control-24 h |
|---|
| AMELX | 2.167 | BMP4 | 2.073 | LRRC17 | 3.748 | WNT11 | 3.428 |
| SOX8 | 1.932 | ALPL | 2.069 | WNT11 | 2.907 | ALPL | 2.946 |
| HDAC4 | 1.743 | WNT11 | 1.900 | SOX8 | 2.304 | BMP4 | 2.805 |
| CHRD | 1.523 | SHOX2 | 1.800 | BMP3 | 2.150 | SHOX2 | 2.684 |
| NF1 | 1.478 | RUNX2 | 1.734 | HDAC4 | 1.808 | RUNX2 | 1.829 |
| DNAJC13 | 1.415 | TMSB4Y | 1.592 | AMELX | 1.741 | TWIST1 | 1.723 |
| BMP2 | 1.407 | WNT3 | 1.476 | PTH1R | 1.568 | NOG | 1.709 |
| GLI2 | 1.365 | CREB3L1 | 1.418 | DNAJC13 | 1.447 | SMO | 1.518 |
| SMAD3 | 1.301 | SNAI2 | 1.309 | BMP4 | 1.445 | PTHLH | 1.514 |
| PENK | 0.761 | SMAD3 | 0.752 | NF1 | 1.440 | IFT80 | 1.513 |
| SNAI2 | 0.691 | HSPE1 | 0.731 | VCAN | 1.434 | ITGA11 | 1.390 |
| SEMA7A | 0.655 | IGFBP5 | 0.718 | SHOX2 | 1.400 | WNT3 | 1.389 |
| HSPE1 | 0.654 | FASN | 0.706 | BMP2 | 1.390 | SNAI2 | 1.386 |
| SNAI1 | 0.638 | RSL1D1 | 0.674 | IFT80 | 1.321 | PENK | 1.347 |
| EPHA2 | 0.634 | IGFBP3 | 0.661 | SNAI2 | 0.732 | PHB | 0.749 |
| PTHLH | 0.592 | FHL2 | 0.639 | TMSB4Y | 0.718 | LIMD1 | 0.749 |
| JUNB | 0.585 | FZD1 | 0.636 | JUNB | 0.693 | RSL1D1 | 0.724 |
| IGFBP3 | 0.585 | SYNCRIP | 0.628 | EPHA2 | 0.662 | GLI2 | 0.714 |
| NOG | 0.371 | ALYREF | 0.626 | FBL | 0.659 | SMAD3 | 0.682 |
| ALPL | 0.312 | MYBBP1A | 0.596 | MEF2D | 0.643 | FBL | 0.680 |
|
|
| GTPBP4 | 0.579 | NOG | 0.581 | SNAI1 | 0.666 |
|
|
| AMELX | 0.577 | SEMA7A | 0.516 | SYNCRIP | 0.661 |
|
|
| SEMA7A | 0.519 | ALPL | 0.444 | FASN | 0.656 |
|
|
| BMP3 | 0.511 | SNAI1 | 0.439 | FHL2 | 0.638 |
|
|
| DDX21 | 0.507 |
|
| MYBBP1A | 0.637 |
|
|
| MSX2 | 0.440 |
|
| CHRD | 0.629 |
|
|
| CBFB | 0.427 |
|
| FZD1 | 0.623 |
|
|
| LRRC17 | 0.268 |
|
| ALYREF | 0.606 |
|
|
| SOX8 | 0.057 |
|
| GTPBP4 | 0.562 |
|
|
|
|
|
|
| DDX21 | 0.526 |
|
|
|
|
|
|
| CBFB | 0.519 |
|
|
|
|
|
|
| MSX2 | 0.512 |
|
|
|
|
|
|
| AMELX | 0.464 |
|
|
|
|
|
|
| SEMA7A | 0.409 |
|
|
|
|
|
|
| SOX8 | 0.068 |
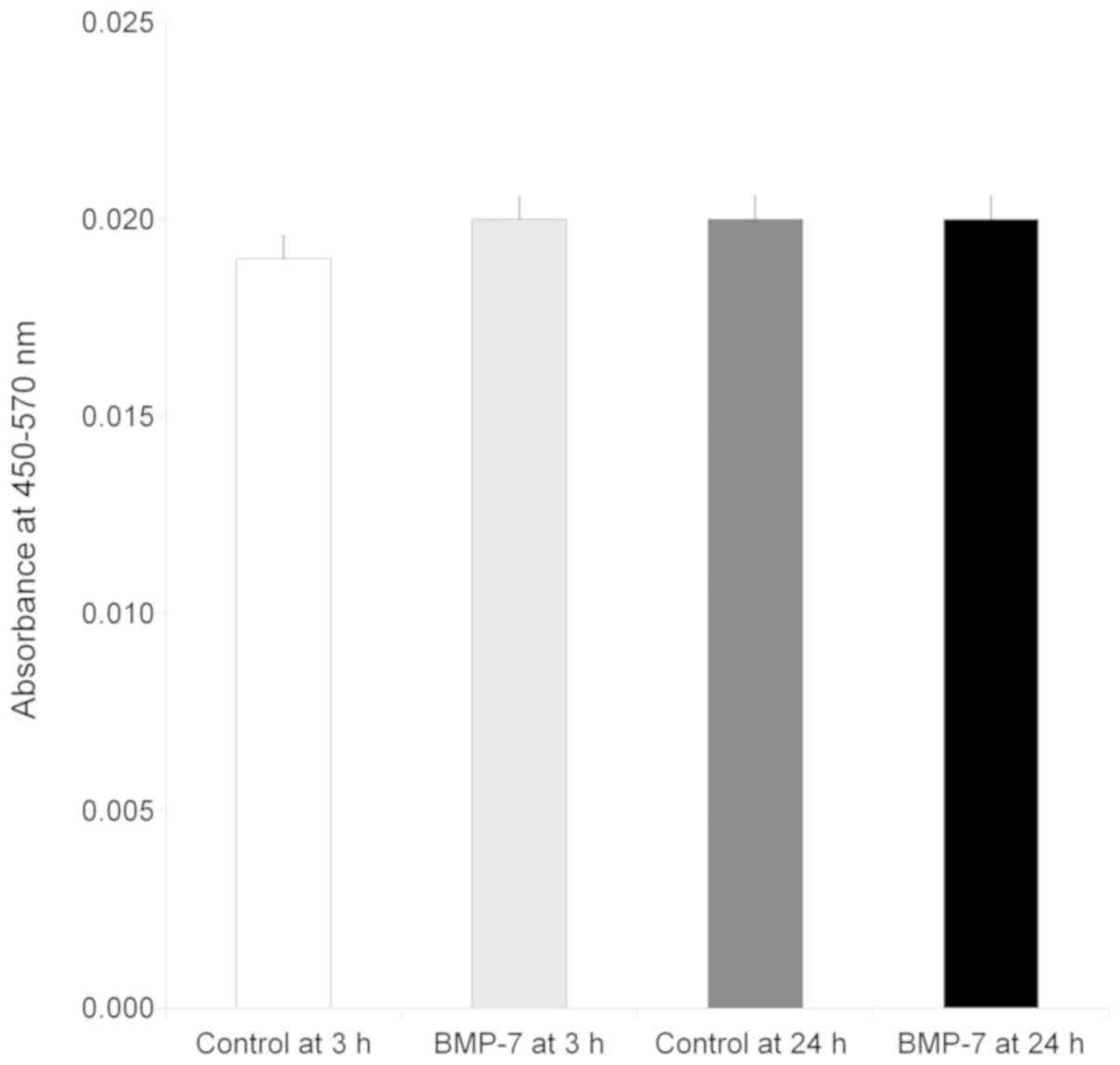
Secretion of human vascular
endothelial growth factor
The results clearly demonstrated that the vascular
endothelial growth factor was secreted at 3 and 24 h irrespective
of culture period (Fig. 4). No
significant change in secretion of the vascular endothelial growth
factor was seen at 3 h with the addition of bone morphogenetic
protein (P>0.05). Similarly, no statistically significant
changes were noted with the loading of bone morphogenetic protein
at 24 h (P>0.05).
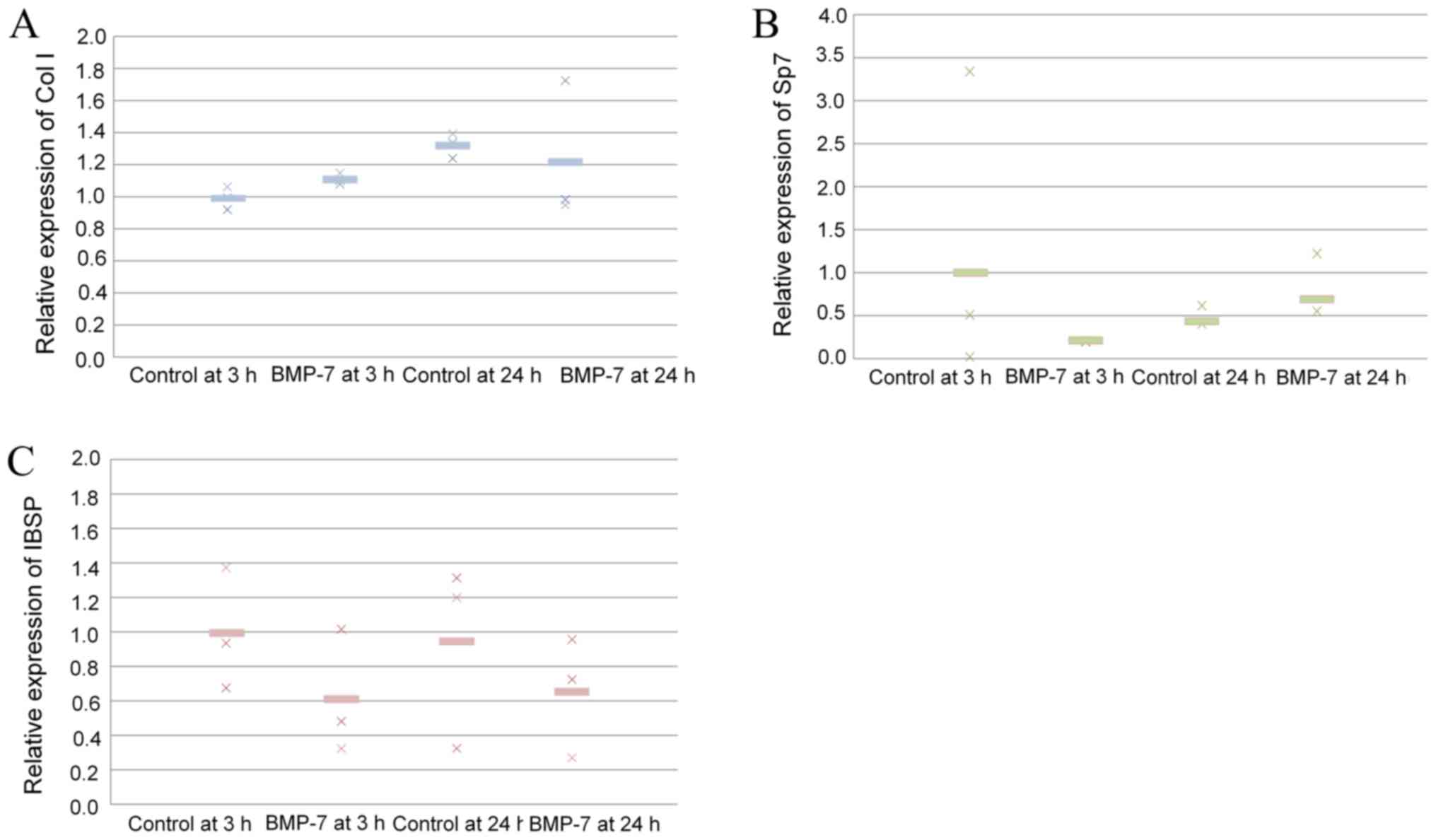
Validation of mRNA expression
Quantitative real-time PCR revealed that mRNA levels
of collagen I were higher in the 24-h control group than in 3-h
control group (P>0.05; Fig.
5A). Application of BMP-7 increased the expression of collagen
I at 3 and 24 h. The results showed that application of BMP-7 at 24
h produced a decrease of Sp7 (P>0.05; Fig. 5B). The results showed that
application of BMP-7 at 24 h showed a decrease of IBSP, but no
significant differences were noted (P>0.05; Fig. 5C).
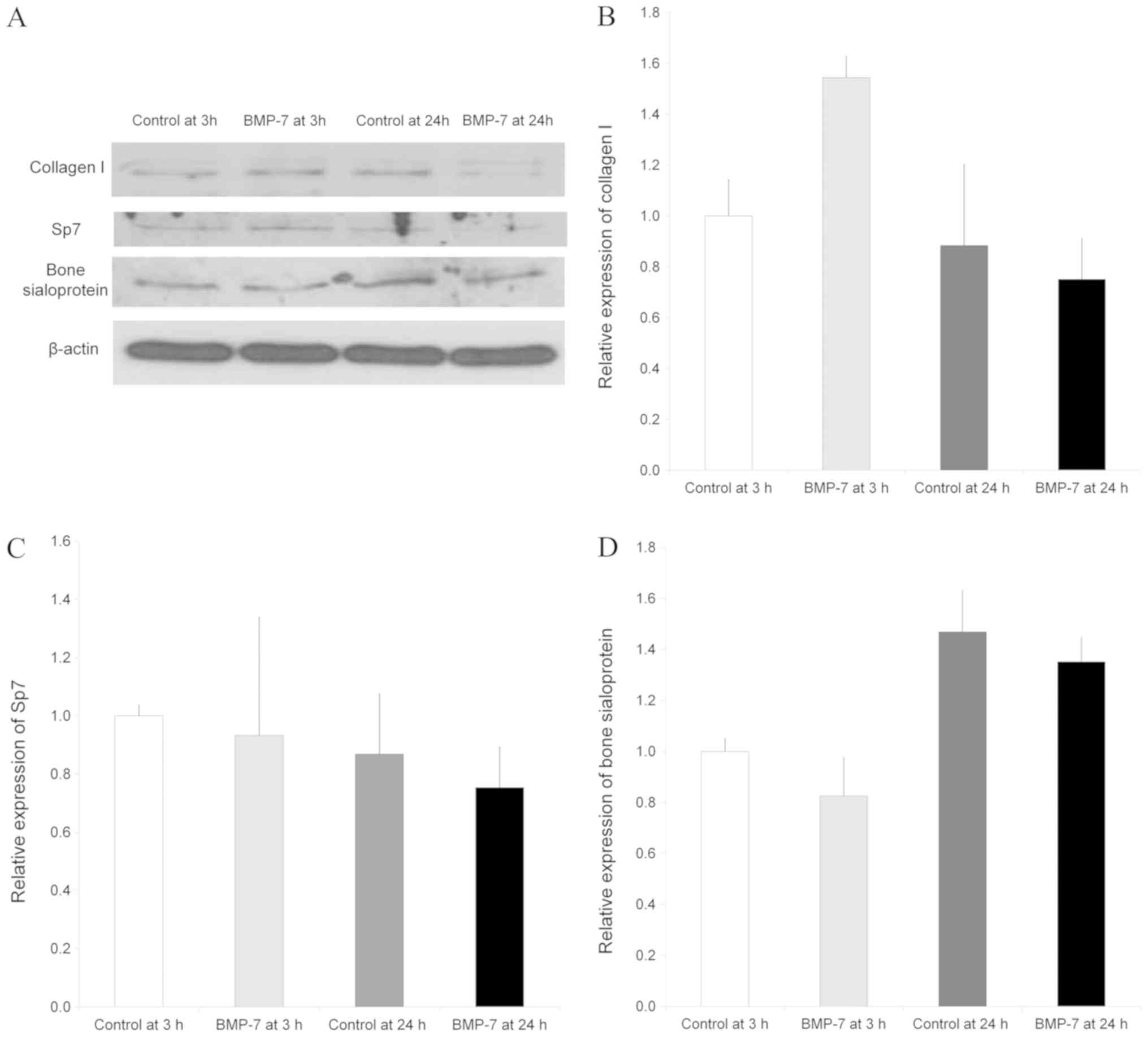
Western blot analysis
Western blot analysis was done to analyze protein
expression of collagen I, Sp7 and bone sialoprotein following the
application with BMP-7 at 3 and 24 h compared with the untreated
control group at 3 and 24 h (Fig.
6A). Normalization of the protein expressions showed that the
control at 24 h showed 88.4±31.8% expression of collagen I, and the
group treated with BMP-7 yielded 154.5±8.2% and 75.1±16.0% of
expression of collagen I at 3 and 24 h, respectively, compared to
control values at 3 h as a baseline of 100% (100.0±14.3%)
(P>0.05, Fig. 6B).
The expression of Sp7 in the control group at 24 h
did not show significant change compared with the control at 3 h.
Normalization of the protein expressions revealed that the control
at 24 h showed 86.8±20.7% expression of Sp7, and the group treated
with BMP-7 yielded 93.2±40.6% and 75.4±13.9% of expression of Sp7
at 3 and 24 h, respectively, compared to control values at 3 h as a
baseline of 100% (100.0±3.6%) (P>0.05, Fig. 6C).
The relative expression of bone sialoprotein is
shown in Fig. 6C. Normalization of
the protein expressions demonstrated that the control at 24 h
showed 146.8±16.4% expression of bone sialoprotein, and the group
treated with BMP-7 yielded 82.5±15.1% and 135.1±9.6% of expression
of bone sialoprotein at 3 and 24 h, respectively, when the control
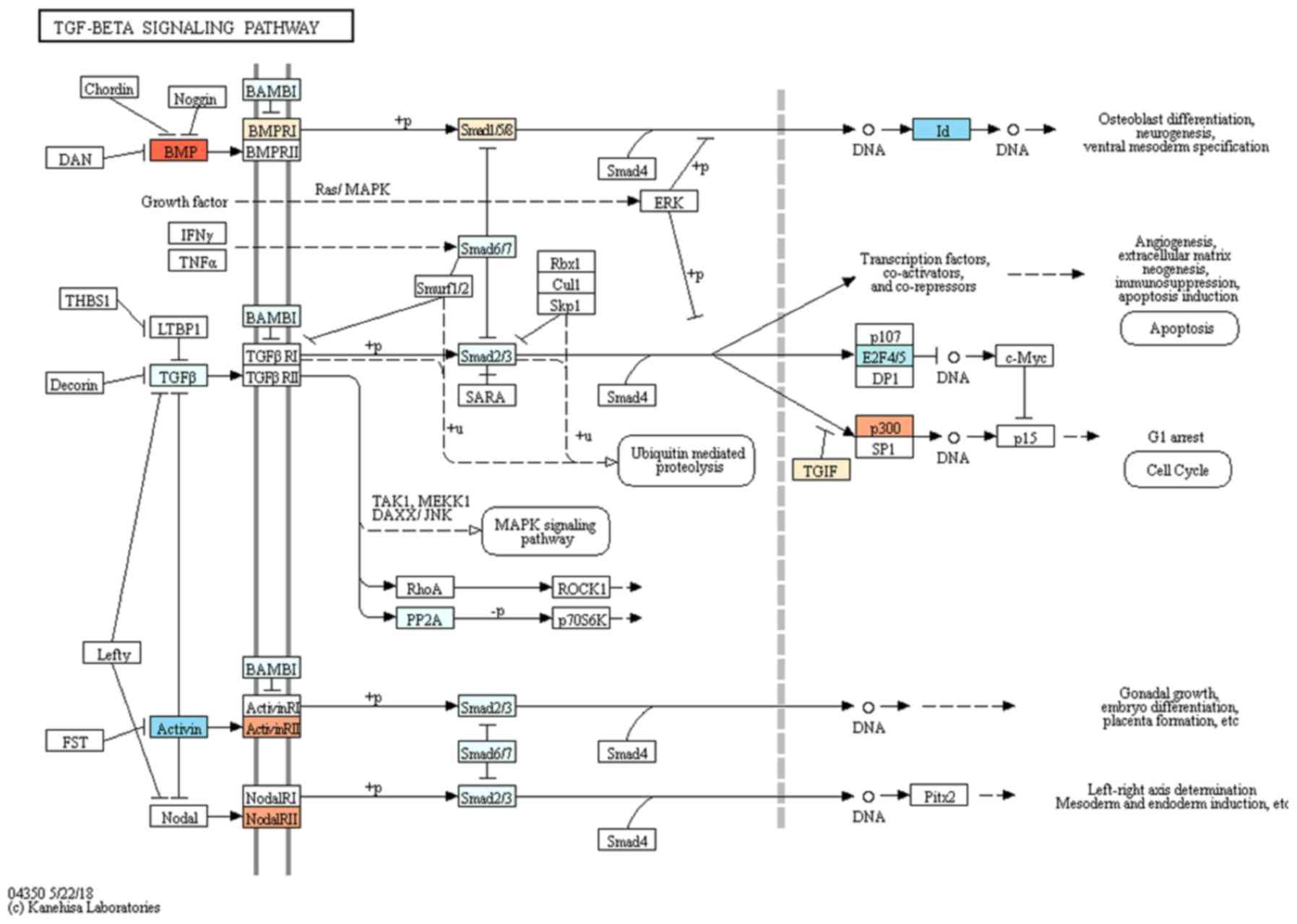
value at 3 h were considered 100% (100.0±5.0%). TGF-β signaling
pathway was associated with the target genes selected for
osteoblast differentiation (Fig.
7).
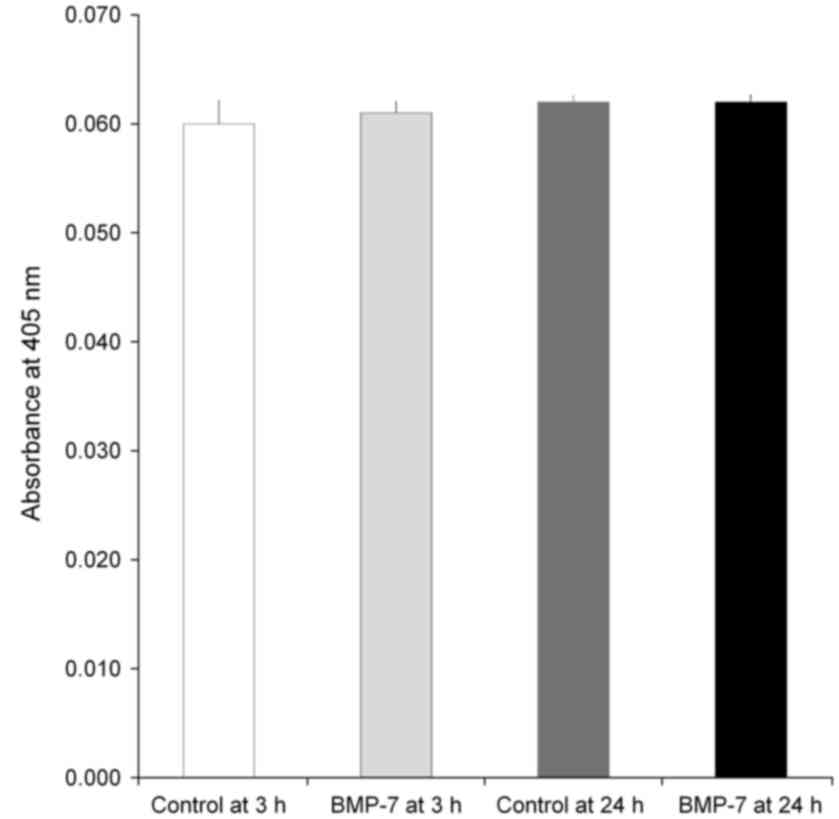
Alkaline phosphatase activity
The data of the alkaline phosphatase activity assays
at Day 7 are demonstrated in Fig. 8.
The absorbance values at 405 nm at Day 7 for control at 3 h, BMP-7
at 3 h, control at 24 h and BMP-7 at 24 h were 0.060±0.002,
0.061±0.001, 0.062±0.001, and 0.062±0.001, respectively
(P>0.05).
Discussion
In this report, we evaluated the effects of BMP-7 on
stem cells under predetermined concentrations at 3 and 24 h. mRNA
sequencing and validation of the expression was done with
qualitative real-time PCR and Western blot analysis. It was seen
that the application of BMP-7 produced increased expression of
collagen I of human gingiva-derived mesenchymal stem cells.
Application with gingiva-derived stem cell inhibited
macrophage foam cell formation and reduction of inflammatory
macrophage activation (18).
Previous reports showed that long incubation of gingiva-derived
stem cells lead to neural precursor cells in vitro (19). Three-dimensional bioprinted
constructs made with gingiva-derived stem cells promoted facial
nerve regeneration (8).
BMP-7 expressing stem-cell sheets were applied in
the previous studies (20–22). In a previous report, BMP-7
overexpressing adenovirus vector was used to transfect mesenchymal
stem cells (22). In a non-union
fracture model, BMP-7 expressing mesenchymal stem cells enhanced
bone regeneration (20). Similarly,
BMP-7 expressing stem cells produced enhanced bone healing in
critical-sized defects when compared with the control stem cell
sheets (23). Moreover, application
of BMP-7 overexpressing stem cells derived from bone marrow
decelerated the development of disc degeneration (21).
It is widely accepted that differentiation of
mesenchymal stem cells is associated with gene transcription and
courses of molecular signaling (24). Researchers previously used RNA
sequencing to analyze transcriptional profiling of osteoblast
differentiation (25). Moreover, RNA
sequencing and quantitative reverse transcription polymerase chain
reactions were applied for characterization of three-dimensional
organoids (26). RNA Sequencing has
also been applied to compare the differences between mesenchymal
stem cells derived from bone marrow from healthy control and
diseased participants (27).
Researchers previously tested the functionality of mesenchymal stem
cells, the precursors of osteoblasts, using RNA sequencing to
measure the epigenetic changes with aging (28). This study tested gingiva-derived stem
cells' responsiveness to BMP-7 showed differentially expressed mRNA
related to osteoblast differentiation in each group.
Researchers previously tested the expression of
osteoblast-associated genes and proteins, including COL1A1, Runx2,
IBSP, SPARC, SPP1 and BGALP (29).
In this study, several gene expressions were tested in mesenchymal
stem cells, including Runx2, Sp7 and collagen I (29–31).
Researchers previously demonstrated that romidepsin promoted
osteogenic differentiation of human mesenchymal stem cells and that
it showed increased SP7 (Osterix) and alkaline phosphatase
expression (32). They also showed
that transfection of Runx2/SP7 enhanced the osteogenic
differentiation of mesenchymal stem cells (33). This study showed that BMP-7
upregulates genes related to osteoblast differentiation, including
collagen I, in gingiva-derived stem cells. Alkaline phosphatase
activity is considered an early-stage marker for osteoblast
differentiation, the lack of difference may be dues to various
reasons including the treatment time of BMP-7 and the stage of
tested cells (34,35).
Conclusively, the effects of BMP-7 on stem cells
were assessed using mRNA sequencing, and the validation of the
expression was done with quantitative real-time polymerase chain
reactions and Western blot analysis. The short-term application of
BMP-7 produced an increased expression of collagen I, which was
related to target genes chosen for osteoblast differentiation. This
study may provide new insights into the role of BMP-7 using mRNA
sequencing.
Acknowledgements
Not applicable.
Funding
Thε present study was supported by Basic Science
Research Program through the National Research Foundation of Korea
(NRF) funded by the Ministry of Science, Information and
Communication Technology & Future Planning (Grant no.
NRF-2017R1A1A1A05001307).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
HL, SM, YS, YP, and JP contributed to study
conception and design. HL, SM, YS, YP, and JP performed the
experiments; HL, SM, YS, YP, and JP analyzed the data and HL, SM,
YS, YP, and JP wrote the manuscript. All authors reviewed the
manuscript.
Ethical approval and consent to
participate
The protocol of the study was reviewed and the
Institutional Review Board of the Catholic University of Korea,
College of Medicine approved the design of this study
(KC18SESI0199). Written informed consent was gathered from the
participants according to the Act on Legal Codes for Biomedical
Ethics and Safety and the Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Park JB: Use of bone morphogenetic
proteins in sinus augmentation procedure. J Craniofac Surg.
20:1501–1503. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Park JB: Effects of the combination of
fibroblast growth factor-2 and bone morphogenetic protein-2 on the
proliferation and differentiation of osteoprecursor cells. Adv Clin
Exp Med. 23:463–467. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhu L, Ma J, Mu R, Zhu R, Chen F, Wei X,
Shi X, Zang S and Jin L: Bone morphogenetic protein 7 promotes
odontogenic differentiation of dental pulp stem cells in vitro.
Life Sci. 202:175–181. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen F, Bi D, Cao G, Cheng C, Ma S, Liu Y
and Cheng K: Bone morphogenetic protein 7-transduced human
dermal-derived fibroblast cells differentiate into osteoblasts and
form bone in vivo. Connect Tissue Res. 59:223–232. 2018.PubMed/NCBI
|
|
5
|
Jovanovic VM, Salti A, Tilleman H, Zega K,
Jukic MM, Zou H, Friedel RH, Prakash N, Blaess S, Edenhofer F and
Brodski C: BMP/SMAD pathway promotes neurogenesis of midbrain
dopaminergic neurons in vivo and in human induced pluripotent and
neural stem cells. J Neurosci. 38:1662–1676. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Soundara Rajan T, Giacoppo S, Scionti D,
Diomede F, Grassi G, Pollastro F, Piattelli A, Bramanti P, Mazzon E
and Trubiani O: Cannabidiol activates neuronal precursor genes in
human gingival mesenchymal stromal cells. J Cell Biochem.
118:1531–1546. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ansari S, Diniz IM, Chen C, Sarrion P,
Tamayol A, Wu BM and Moshaverinia A: Human periodontal ligament-
and gingiva-derived mesenchymal stem cells promote nerve
regeneration when encapsulated in alginate/hyaluronic acid 3D
scaffold. Adv Healthc Mater. 6:2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang Q, Nguyen PD, Shi S, Burrell JC,
Cullen DK and Le AD: 3D bio-printed scaffold-free nerve constructs
with human gingiva-derived mesenchymal stem cells promote rat
facial nerve regeneration. Sci Rep. 8:66342018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang R, Yu T, Liu D, Shi S and Zhou Y:
Hydrogen sulfide promotes immunomodulation of gingiva-derived
mesenchymal stem cells via the Fas/FasL coupling pathway. Stem Cell
Res Ther. 9:622018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang F, Yu M, Yan X, Wen Y, Zeng Q, Yue W,
Yang P and Pei X: Gingiva-derived mesenchymal stem cell-mediated
therapeutic approach for bone tissue regeneration. Stem Cells Dev.
20:2093–2102. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee SI, Ko Y and Park JB: Evaluation of
the shape, viability, stemness and osteogenic differentiation of
cell spheroids formed from human gingiva-derived stem cells and
osteoprecursor cells. Exp Ther Med. 13:3467–3473. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jin SH, Lee JE, Yun JH, Kim I, Ko Y and
Park JB: Isolation and characterization of human mesenchymal stem
cells from gingival connective tissue. J Periodontal Res.
50:461–467. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Trapnell C, Pachter L and Salzberg SL:
TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics.
25:1105–1111. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Quinlan AR and Hall IM: BEDTools: A
flexible suite of utilities for comparing genomic features.
Bioinformatics. 26:841–842. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gentleman RC, Carey VJ, Bates DM, Bolstad
B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al:
Bioconductor: Open software development for computational biology
and bioinformatics. Genome Biol. 5:R802004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang DW, Sherman BT, Tan Q, Collins JR,
Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID gene functional classification tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8:R1832007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: new perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45:D353–D361. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang X, Huang F, Li W, Dang JL, Yuan J,
Wang J, Zeng DL, Sun CX, Liu YY, Ao Q, et al: Human gingiva-derived
mesenchymal stem cells modulate monocytes/macrophages and alleviate
atherosclerosis. Front Immunol. 9:8782018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rajan TS, Scionti D, Diomede F, Piattelli
A, Bramanti P, Mazzon E and Trubiani O: Prolonged expansion induces
spontaneous neural progenitor differentiation from human
gingiva-derived mesenchymal stem cells. Cell Reprogram. 19:389–401.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Song J, Kim Y, Kweon OK and Kang BJ: Use
of stem-cell sheets expressing bone morphogenetic protein-7 in the
management of a nonunion radial fracture in a Toy Poodle. J Vet
Sci. 18:555–558. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liao JC: Cell therapy using bone
marrow-derived stem cell overexpressing BMP-7 for degenerative
discs in a rat tail disc model. Int J Mol Sci. 17:E1472016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yan X, Zhou Z, Guo L, Zeng Z, Guo Z, Shao
Q and Xu W: BMP7-overexpressing bone marrow-derived mesenchymal
stem cells (BMSCs) are more effective than wild-type BMSCs in
healing fractures. Exp Ther Med. 16:1381–1388. 2018.PubMed/NCBI
|
|
23
|
Kim Y, Kang BJ, Kim WH, Yun HS and Kweon
OK: Evaluation of mesenchymal stem cell sheets overexpressing BMP-7
in canine critical-sized bone defects. Int J Mol Sci. 19:E20732018.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fakhry M, Hamade E, Badran B, Buchet R and
Magne D: Molecular mechanisms of mesenchymal stem cell
differentiation towards osteoblasts. World J Stem Cells. 5:136–148.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Khayal LA, Grünhagen J, Provazník I,
Mundlos S, Kornak U, Robinson PN and Ott CE: Transcriptional
profiling of murine osteoblast differentiation based on RNA-seq
expression analyses. Bone. 113:29–40. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wiener DJ, Basak O, Asra P, Boonekamp KE,
Kretzschmar K, Papaspyropoulos A and Clevers H: Establishment and
characterization of a canine keratinocyte organoid culture system.
Vet Dermatol. 29:375–e126. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sargent A, Shano G, Karl M, Garrison E,
Miller C and Miller RH: Transcriptional profiling of mesenchymal
stem cells identifies distinct neuroimmune pathways altered by CNS
disease. Int J Stem Cells. 11:48–60. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Del Real A, Pérez-Campo FM, Fernández AF,
Sañudo C, Ibarbia CG, Pérez-Núñez MI, Criekinge WV, Braspenning M,
Alonso MA, Fraga MF and Riancho JA: Differential analysis of
genome-wide methylation and gene expression in mesenchymal stem
cells of patients with fractures and osteoarthritis. Epigenetics.
12:113–122. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Baird A, Lindsay T, Everett A, Iyemere V,
Paterson YZ, McClellan A, Henson FMD and Guest DJ: Osteoblast
differentiation of equine induced pluripotent stem cells. Biol
Open. 7:bio0335142018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Barone A, Toti P, Funel N, Campani D and
Covani U: Expression of SP7, RUNX1, DLX5, and CTNNB1 in human
mesenchymal stem cells cultured on xenogeneic bone substitute as
compared with machined titanium. Implant Dent. 23:407–415.
2014.PubMed/NCBI
|
|
31
|
Hajizadeh N, Madani ZS, Zabihi E, Golpour
M, Zahedpasha A and Mohammadnia M: Effect of MTA and CEM on
mineralization-associated gene expression in stem cells derived
from apical papilla. Iran Endod J. 13:94–101. 2018.PubMed/NCBI
|
|
32
|
Ali D, Chalisserry EP, Manikandan M, Hamam
R, Alfayez M, Kassem M, Aldahmash A and Alajez NM: Romidepsin
promotes osteogenic and adipocytic differentiation of human
mesenchymal stem cells through inhibition of histondeacetylase
activity. Stem Cells Int. 2018:23795462018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim HJ, Park JS, Yi SW, Oh HJ, Kim JH and
Park KH: Sequential transfection of RUNX2/SP7 and ATF4 coated onto
dexamethasone-loaded nanospheresenhances osteogenesis. Sci Rep.
8:14472018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Prins HJ, Braat AK, Gawlitta D, Dhert WJ,
Egan DA, Tijssen-Slump E, Yuan H, Coffer PJ, Rozemuller H and
Martens AC: In vitro induction of alkaline phosphatase levels
predicts in vivo bone forming capacity of human bone marrow stromal
cells. Stem Cell Res. 12:428–440. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lu C, Xing Z, Yu YY, Colnot C, Miclau T
and Marcucio RS: Recombinant human bone morphogenetic protein-7
enhances fracture healing in an ischemic environment. J Orthop Res.
28:687–696. 2010.PubMed/NCBI
|