Introduction
Glioma is a primary brain tumor in the central
nervous system that is highly aggressive. Although the morbidity of
glioma is relatively low (accounting for ~2% in all adult
malignancies), the number of newly diagnosed patients is increasing
(1). Since gliomas have tendencies
to invade surrounding tissues, current treatment strategies are
ineffective and consequently the prognosis for patients remains
poor (2). Therefore, there is
currently an urgent need to explore the pathological mechanism of
this disease to devise new therapeutic interventions.
With the advancing knowledge and understanding of
microRNAs (miRNAs), miRNAs are now known as an important class of
post-transcriptional regulators of gene expression by translational
repression and mRNA breakdown (3–5). It has
been shown to participate in a variety of biological processes,
including cell proliferation, differentiation, migration,
apoptosis, cell-cycle regulation and invasion, development and
metabolism by targeting the 3′-untranslated regions (3′UTR) of
target genes (6). Accumulating
evidence have also indicated aberrant expression of miRNAs were
found in a number of diseases, particularly in cancers (7). Indeed, a wide range of miRNAs have been
reported to function as biomarkers for diagnosis and prognosis of
human glioma (8). Decreased miR-374a
expression was associated with glioma progression (9), whereas increased expressions of
miRNA-21, miR-128 or miR-342-3p were positively correlated with
histopathological grades of glioma (10,11). In
fact, >97 miRNAs were found to be aberrantly expressed through
miRNA microarrays and involved in the pathogenesis of glioma
through major signaling pathways (12). However, the role of miR-454-3p and
underlying mechanism in glioma remains poorly understood.
Early growth response 3 (EGR3) is a member of the
EGR family of C2H2-type zinc-finger proteins (13). It regulates gene transcription and
has been found to be involved in multiple biological processes,
including the development of muscle, lymphocyte and neurons, in
addition to the growth, migration, invasion and metastasis of
cancer cells (14–16). Therefore, associations between
abnormal EGR3 expression and schizoaffective disorder,
schizophrenia, systemic autoimmunity, and a variety of cancers has
been previously demonstrated (14–16). In
hepatocellular carcinoma, downregulation of EGR3 expression of
promoted cell proliferation by upregulating Fas ligand expression
(17). In contrast, reduction in
EGR3 expression by KH-type splicing regulatory protein suppressed
cell invasion and metastasis in non-small cell lung cancer (NSCLC)
(14). These previous findings
suggest that the function of EGR3 may differ depending on the type
of cancer in question.
In the present study, the aim is to elucidate the
potential role of miR-454-3p in glioma and explore the potential
mechanism. The results can deepen the understanding on the
pathogenesis of glioma which may provide novel molecular targets
for treating this disease.
Materials and methods
Ethics statement
The present study was approved by the Research
Ethics Committee of The First People's Hospital of Taizhou. All
patients provided written informed consent prior to enrollment in
the study.
Tissue specimens
Clinical samples were obtained from 40 patients
(sex, 20 males and 20 females; age range, 28–71 years; mean age,
58.5±14.1 years) who were diagnosed with glioma according to World
Health Organization (WHO) classification without preoperative
radiation or chemotherapy, by surgery at the The First People's
Hospital of Taizhou from January 2015 to December 2016 (Taizhou,
China). Patients who had received neoadjuvant or adjuvant therapy
were excluded from the present study. Tumors and adjacent non-tumor
intracranial tissue samples were collected and immediately frozen
in liquid nitrogen.
Cell line and culture
Human glioma cell lines U138MG (cat. no.
ATCC® HTB-16™) and U251 (cat. no. ATCC®
HTB-17), together with 293T cells (cat. no. ATCC®
CRL-11268™) were purchased from the American Type Culture
Collection. And normal human astrocytes (NHA; cat. no. BNCC341796),
A172 (cat. no. BNCC341782) cells were purchased from BeNa culture
collection. U138MG, NHA, A172 and 293T cells were cultured in DMEM
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal
bovine serum (Sigma-Aldrich; Merck KGaA) and penicillin (100
IU/ml)/streptomycin (100 µg/ml), whilst U251 cells were cultured in
RPMI-1640 media (Gibco; Thermo Fisher Scientific, Inc.),
supplemented with 10% fetal bovine serum (Sigma-Aldrich; Merck
KGaA) and penicillin (100 IU/ml)/streptomycin (100 µg/ml) in a
humidified incubator containing 5% CO2 at 37°C.
Cell transfection with mimics,
inhibitor, siRNA and plasmids
miR-454-3p mimic and its negative control (NC),
inhibitor of miR-454-3p and its negative control (NC-in), siRNA for
EGR3 (siEGR3) and its negative control (siNC) were synthesized by
Shanghai GenePharma Co., Ltd., with their respective sequences
listed in Table II. For the
overexpression of EGR3, the coding sequence for EGR3 was amplified
from NHA cells using KOD-Plus-Neo Polymerase (cat. no. KOD-401;
Toyobo Life Science) using the forward primer containing a
BamHI restriction site,
5′-GGGGATCCATGACCGGCAAACTCGCCGAGAAGC-3′ and the reverse primer,
containing a EcoRI restriction site,
5′-GGGAATTCTCAGGCGCAGGTGGTGACCACGG-3′ and then subcloned into the
pcDNA3.1 plasmid (Invitrogen; Thermo Fisher Scientific, Inc.). The
mimics (50 nM) and inhibitor (100 nM) of miR-454-3p, siEGR3 (100
nM), plasmids (1 ng/µl) and their negative controls were
transfected into cells using Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to
manufacturer's protocol.
 | Table II.Sequences. |
Table II.
Sequences.
| Targets | Sequences
(5′-3′) |
|---|
| Duplex of mimics of
miR-454-3p |
UAGUGCAAUAUUGCUUAUAGGGU |
|
|
CCUAUAAGCAAUAUUGCACUAUU |
| Duplex of NC |
UUCUCCGAACGUGUCACGUTT |
|
|
ACGUGACACGUUCGGAGAATT |
| Inhibitor of
miR-454-3p |
ACCCUAUAAGCAAUAUUGCACUA |
| NC-in |
UUGUACUACACAAAAGUACUG |
| siEGR3 |
AGAUCCACCUCAAGCAAAAUU |
| siNC |
AAUUCUCCGAACGUGUCACGU |
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from the cells or tissues
using mirVana™ miRNA Isolation Kit (Ambion; Thermo Fisher
Scientific, Inc.) according to manufacturer's protocol. To measure
the levels of miR-454-3p expression, TaqMan™ MicroRNA Reverse
Transcription Kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.) were used to perform reverse transcription according to
manufacturer's protocol. To determine the mRNA levels of EGR3,
total RNA was reverse transcribed into cDNA using PrimeScript™ RT
reagent Kit with gDNA Eraser (Takara Biotechnology Co., Ltd.). qPCR
was performed using SYBR® PrimeScript™ RT-PCR kit
(Takara Biotechnology Co., Ltd.) according to manufacturers'
protocols using Applied Biosystems® 7500 Real-Time PCR
Systems (Applied Biosystems; Thermo Fisher Scientific, Inc.) under
the following thermocycling conditions: Initial denaturation at
95°C for 10 min, followed by 40 cycles of 95°C for 5 sec and 60°C
for 30 sec. U6 snRNA and GAPDH were used as reference genes for
miR-454-3p and EGR3 respectively. Relative expression levels were
calculated using the 2−ΔΔCq method (18). All PCRs were performed in triplicate
for analysis and the sequences of primers used were listed in
Table III.
 | Table III.Primer sequences. |
Table III.
Primer sequences.
| Primers | Sequences
(5′-3′) |
|---|
| miRNA reverse
transcription primer |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACCCTA |
| miRNA-454-3p-F |
ACCCTATCAATATTGTCTCTGC |
| miRNA-454-3p-R |
GCGAGCACAGAATTAATACGAC |
| U6-F |
CTCGCTTCGGCAGCACA |
| U6-R |
AACGCTTCACGAATTTGCGT |
| EGR3-F |
TTCGCTTTCGACTCTCC |
| EGR3-R |
CTCCGAGTAGAGATCGC |
| GAPDH-F |
CCACCCATGGCAAATTCC |
| GAPDH-R |
CAGGAGGCATTGCTGATGAT |
Cell viability assay
Cell growth was measured using MTT (Sigma-Aldrich;
Merck KGaA) and colony formation assays. For MTT assays,
approximately 3,000 cells/well were seeded into 96-well plates,
following which 20 µl MTT dissolved in PBS (5 mg/ml) was added to
each well at indicated times and incubated for a further 4 h. The
supernatant was then carefully removed and 150 µl DMSO was added to
each well and plates were subsequently shaken for 10 min to
dissolve the formazan crystals. Optical density at 490 nm was then
measured per well using a Thermo Scientific Multiskan (Thermo
Fisher Scientific, Inc.).
For colony formation assays, 1,000 cells were plated
into each well of a 6-well plate and cultured for 14 days. Colonies
were subsequently fixed using 10% formaldehyde for 5 min at room
temperature (RT) and stained using 0.1% crystal violet for 30 sec
at RT. The numbers of colonies were counted for each condition
using light microscopy. Each experiment was repeated three times
independently.
Prediction targets of miR-454-3p
The targets of miR-454-3p were predicted using the
TargetScan website (Version 7.1; http://www.targetscan.org/vert_71/) (19).
Dual-luciferase reporter assay
For dual-luciferase reporter assays, 4,000 293T
cells were seeded into 96-well plates and pMirGLO plasmids (1
ng/µl) (Promega Corporation) encoding wild-type or mutant 3′UTR of
EGR3 were co-transfected into cells with 20 nM either miR-454-3p
mimics or NC using Lipofectamine 3000. Firefly and Renilla
luciferase activities were assayed 24 h after transfection using
Dual-Luciferase® Reporter Assay System (Promega
Corporation) according to manufacturer's protocols. Firefly
luciferase activities were normalized to Renilla luciferase
activities. Each experiment was repeated three times
independently.
Western blot analysis
For western blotting, tissues were homogenized in
RIPA buffer (Cell Signaling Technology, Inc.) from which protein
samples were isolated. After quantification using
Pierce® Bicinchoninic Acid Protein Assay Kit (Thermo
Fisher Scientific, Inc.), 40 µg total protein samples per lane were
subjected to 10% SDS-PAGE and were subsequently transferred to a
PVDF membrane. The membranes were then blocked with 5% skimmed milk
dissolved in PBS for 1 h at room temperature followed by incubation
with primary antibodies against EGR3 (1:100; cat. no. sc-390967;
Santa Cruz Biotechnology, Inc.) and GAPDH (1:500; cat. no. D16H11;
Cell Signaling Technology, Inc.) at 4°C overnight. The membranes
were subsequently washed using tris-buffered saline with Tween-20
(0.05%) three times for 5 min each and subsequently incubated in
horseradish peroxidase-conjugated secondary antibody (1:2,000; cat.
no. 7076 for EGR3 detection and cat. no. 7074 for GAPDH detection;
Cell Signaling Technology, Inc.) for 1 h at room temperature.
Protein bands were visualized using an enhanced ECL-kit (Thermo
Fisher Scientific, Inc.) and captured using the ChemiDoc™ XRS+
System (Version: 5.2.1; Bio-Rad Laboratories, Inc.). Bands were
quantified using ImageJ (National Institutes of Health) and
normalized to GAPDH.
Statistical analysis
Statistical analyses were performed using the
GraphPad Prism 6 software (GraphPad Software, Inc.) and data were
presented as mean ± SD. Differences between two groups were
analyzed using Student's t-test, whereas Tukey's multiple
comparisons test after ANOVA was applied for >3 groups. And the
survival curve was constructed using the Kaplan-Meier method and
compared using the log-rank test, high- and low-expressing groups
were divided according to the median expression of miR-454-3p.
P<0.05 was considered indicate a statistically significant
difference.
Results
miR-454-3p is upregulated in human
glioma tissues
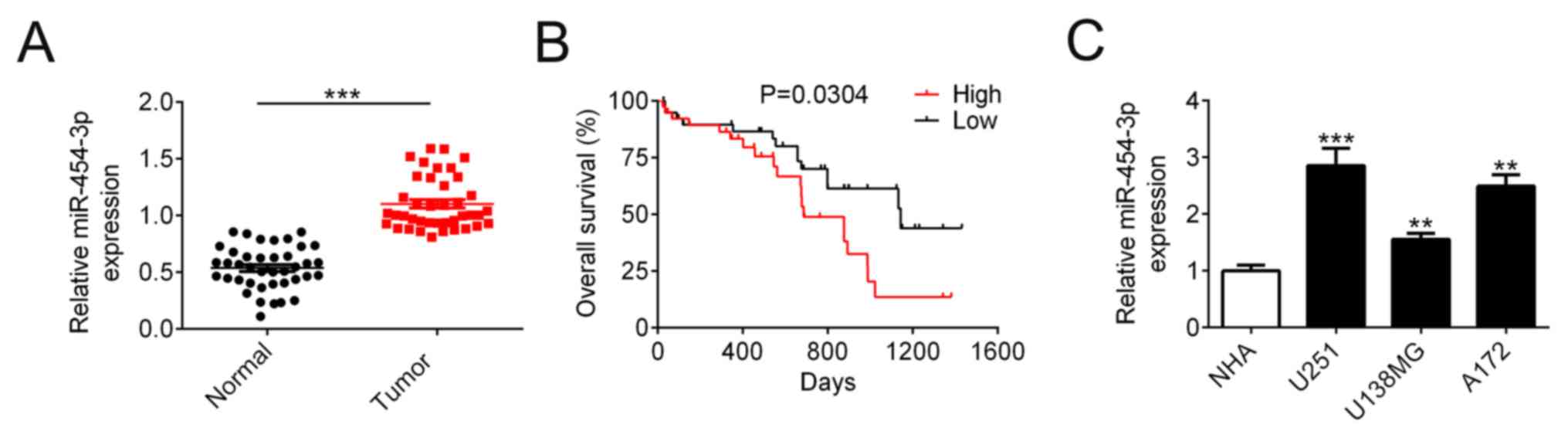
In order to investigate the role of miR-454-3p in
human glioma pathogenesis, expression levels of miR-454-3p were
compared between glioma and adjacent non-tumor samples in
intracranial tissues from 40 patients using RT-qPCR. The baseline
patient and tumor characteristics were summarized in Table I. The expression levels of miR-454-3p
were associated with the tumor size (P=0.024) and WHO stage
(P=0.011). As shown in Fig. 1A,
miR-454-3p expression in glioma tissues (0.54±0.03, n=40) was
significantly higher compared with that in adjacent normal tissues
(1.10±0.04, n=40). Overall survival analysis revealed that patients
with higher expression levels of miR-454-3p expression were
associated with significantly shorter overall survival (median
survival: 687 vs. 1,143 days, n=40, Fig.
1B). The results indicated that miR-453-3p may be involved in
glioma tumorigenesis. To determine the function of miR-454-3p
further in glioma cells, the expression levels of miR-454-3p were
measured in U251, U138MG and A172 glioma cell lines and NHA cells.
When compared with NHA cells, miR-454-3p expression was found to be
significantly increased in glioma cells, with the magnitude the
greatest in U251 cells (Fig.
1C).
 | Table I.Association between
clinicopathological features and expression of miR-454-3p. |
Table I.
Association between
clinicopathological features and expression of miR-454-3p.
|
|
| Relative miR-454-3p
expression |
|
|---|
|
|
|
|
|
|---|
| Characteristic | n | Low | High | P-value |
|---|
| Age |
|
|
| 0.301 |
|
<45 | 16 | 6 | 10 |
|
| ≥45 | 24 | 13 | 11 |
|
| Sex |
|
|
| 0.113 |
|
Female | 20 | 12 | 8 |
|
| Male | 20 | 7 | 13 |
|
| Tumor size |
|
|
| 0.024 |
| <5
cm | 22 | 14 | 8 |
|
| ≥5
cm | 18 | 5 | 13 |
|
| Peritumoral brain
edema |
|
|
| 0.342 |
| <1
cm | 20 | 8 | 12 |
|
| ≥1
cm | 20 | 11 | 9 |
|
| WHO stage |
|
|
| 0.011 |
| I | 9 | 8 | 1 |
|
| II | 8 | 5 | 3 |
|
|
III | 8 | 2 | 6 |
|
| IV | 15 | 4 | 11 |
|
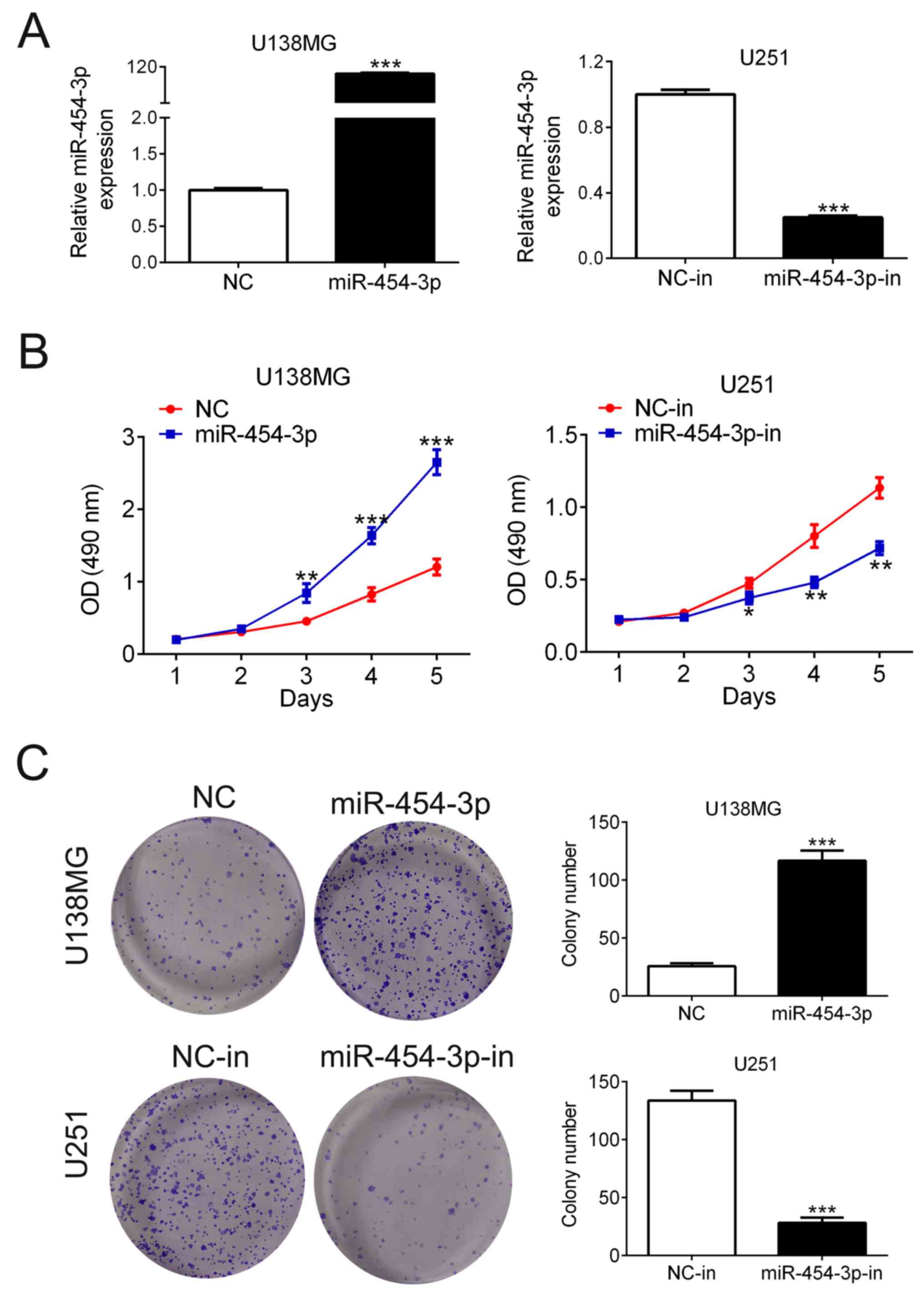
miR-454-3p promotes human glioma cell
proliferation
MTT and colony formation assay were performed to
determine the effects of miR-454-3p on cell proliferation.
Considering the different expression levels of miR-454-3p in U138MG
and U251 cells, U138MG and U251 cells were first transfected with
miR-454-3p mimics or inhibitor, respectively. The expression levels
of miR-454-3p were significantly increased in U138MG cells
following transfection with miR-454-3p mimics compared with its
corresponding negative control (NC); in contrast, miR-454-3p
expression was significantly reduced in U251 cells following
transfection with the miR-454-3p inhibitor compared with its
corresponding NC-in (n=3; Fig. 2A).
MTT assay showed that miR-454-3p overexpression significantly
increased U138MG cell viability, whereas miR-454-3p knockdown
significantly reduced U251 cell viability (n=5; Fig. 2B). Colony formation assay showed
consistent results (Fig. 2C) U138MG
cells transfected with miR-454-3p mimics exhibited significantly
more colonies compared with those transfected with NC (26±3 vs.
117±9, n=3), whilst U251 cells transfected with miR-454-3p
inhibitor resulted in significantly decreased colony numbers (114±9
vs. 28±8, n=3). These results suggested that the overexpression of
miR-454-3p enhanced human glioma cell growth in vitro.
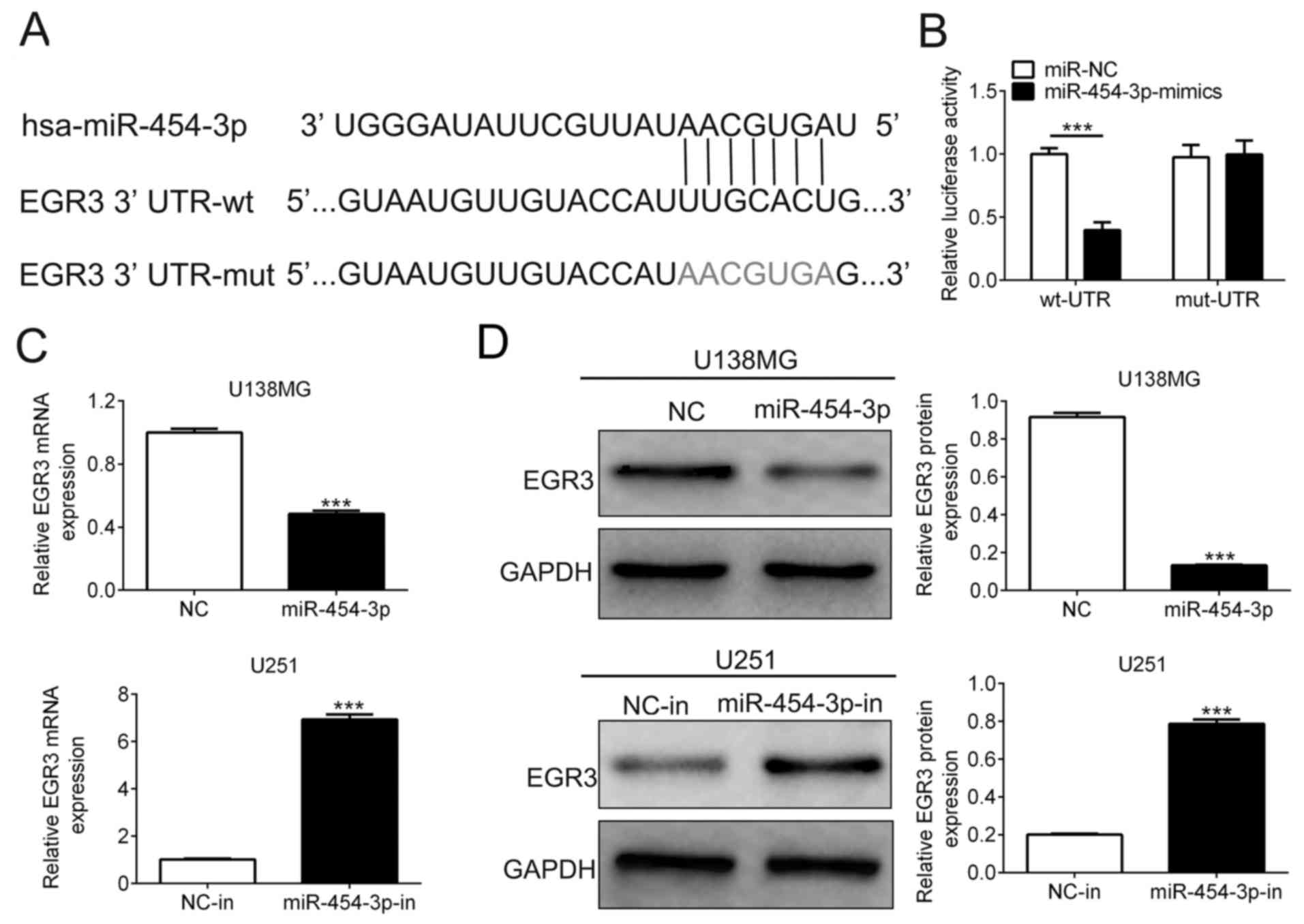
miR-454-3p directly targets the 3′-UTR
of the EGR3 gene
Potential target genes of miR-454-3p were next
predicted using the TargetScan Database. As shown in Fig. 3A, a potential binding site on the
3′UTR of EGR3 was detected for miR-454-3p. To verify this, a
dual-luciferase reporter assay we performed using plasmids encoding
the wild-type or mutant sequence of the EGR3 3′UTR. The relative
luciferase activity of cells co-transfected with wild-type EGR3
3′UTR and miR-454-3p mimics was significantly lower compared with
that co-transfected with NC and wild-type EGR3 3′UTR (0.40±0.04 vs.
1.00±0.03, n=3). However, the relative luciferase activity of cells
co-transfected with miR-454-3p mimics and the mutant sequence of
EGR3 3′UTR showed no marked difference compared with that
co-transfected with NC and mutant sequence of the EGR3 3′UTR
(0.98±0.06 vs. 1.0±0.06; n=3; Fig.
3A). According to RT-qPCR analysis, the expression levels of
EGR3 mRNA in U138MG cells transfected with miR-454-3p mimics was
significantly decreased compared with cells transfected with NC. On
the contrary, transfection with the miR-454-3p inhibitor
significantly reduced EGR3 expression in U251 cells compared with
cells transfected with NC-in (Fig.
3B). Transfection with miR-454-3p mimics significantly reduced
the expression level of EGR3 proteins compared with those
transfected with NC in U138MG cells (0.13±0.01 vs. 0.92±0.04, n=3),
whilst transfection with the miR-454-3p inhibitor significantly
increased EGR3 protein expression in U251 cells (0.79±0.02 vs.
0.20±0.01, n=3) compared with those transfected with NC-in
(Fig. 3C). These results suggest
EGR3 to be a target gene of miR-454-3p.
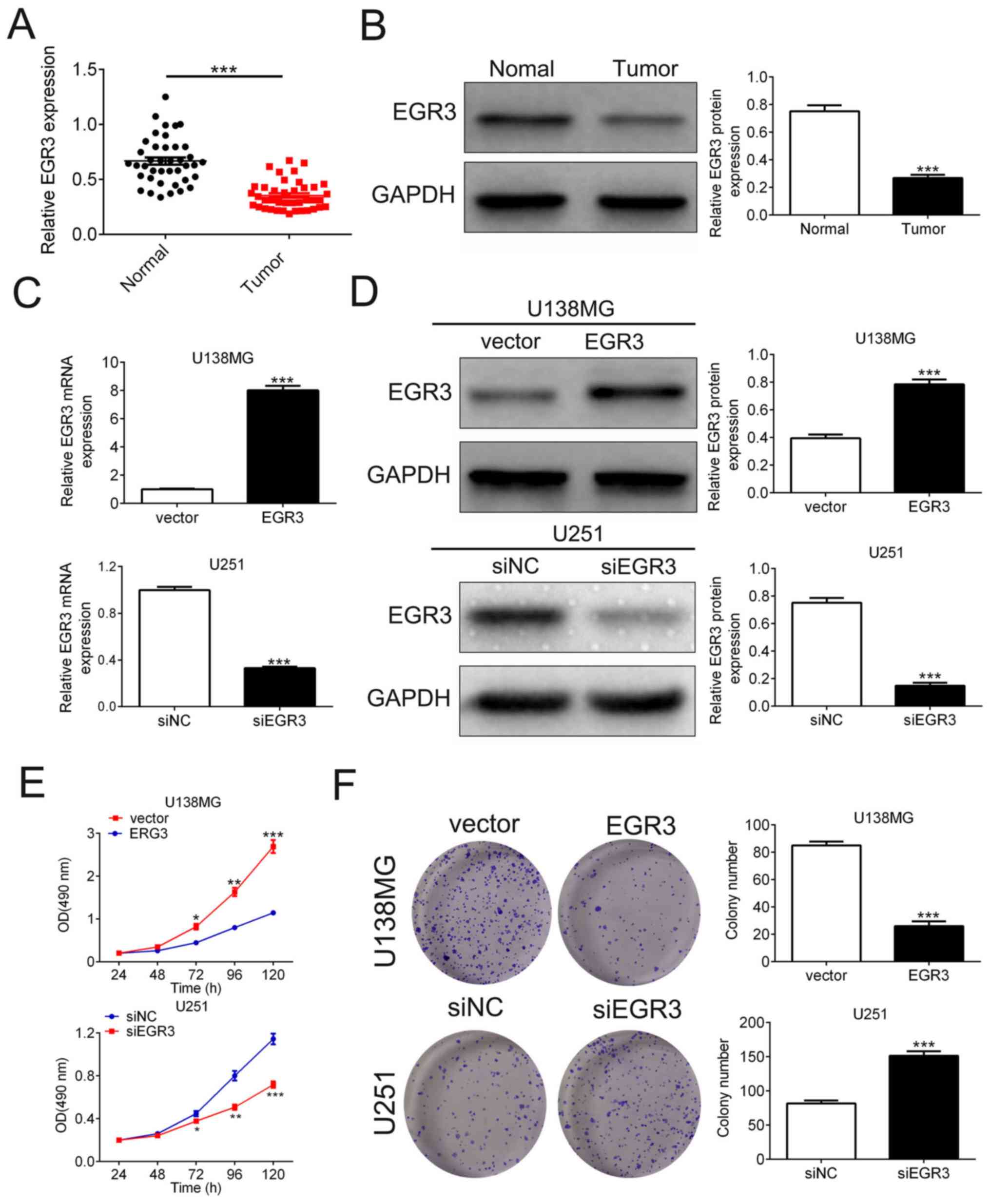
EGR3 is downregulated in glioma
tissues and its overexpression inhibits glioma cell
proliferation
To reveal the function and significance of EGR3 in
glioma tumors, the expression levels of EGR3 in glioma and adjacent
normal tissues were tested using RT-qPCR and western blot analysis.
The expression of EGR3 mRNA (0.67±0.03 vs. 0.35±0.02, n=40) and
protein (0.75±0.04 vs. 0.27±0.02, n=3) in glioma tissues were
significantly decreased compared with adjacent normal tissues
(Fig. 4A and B). The effects of EGR3
on glioma cell proliferation were investigated further using MTT
and colony formation assays. EGR3 was first overexpressed in U138MG
cells by transfection with plasmids expressing EGR3 and knocked
down in U251 cells by siRNA transfection. Transfection efficiency
was confirmed using RT-qPCR and western blot analyses (n=3,
Fig. 4C and D). EGR3 overexpression
significantly reduced the viability of U138MG cells (n=5, Fig. 4E), in addition to significantly
reducing the colony number of U138MG cells compared with cells
transfected with negative control vector (85±3 vs. 36±3; n=3;
Fig. 4F). In contrast, following
transfection with siEGR3, U251 cell viability was significantly
increased (n=5) and colony number was also significantly higher
compared with those transfected with siNC (81±4 vs. 151±7, n=3;
Fig. 4E and F).
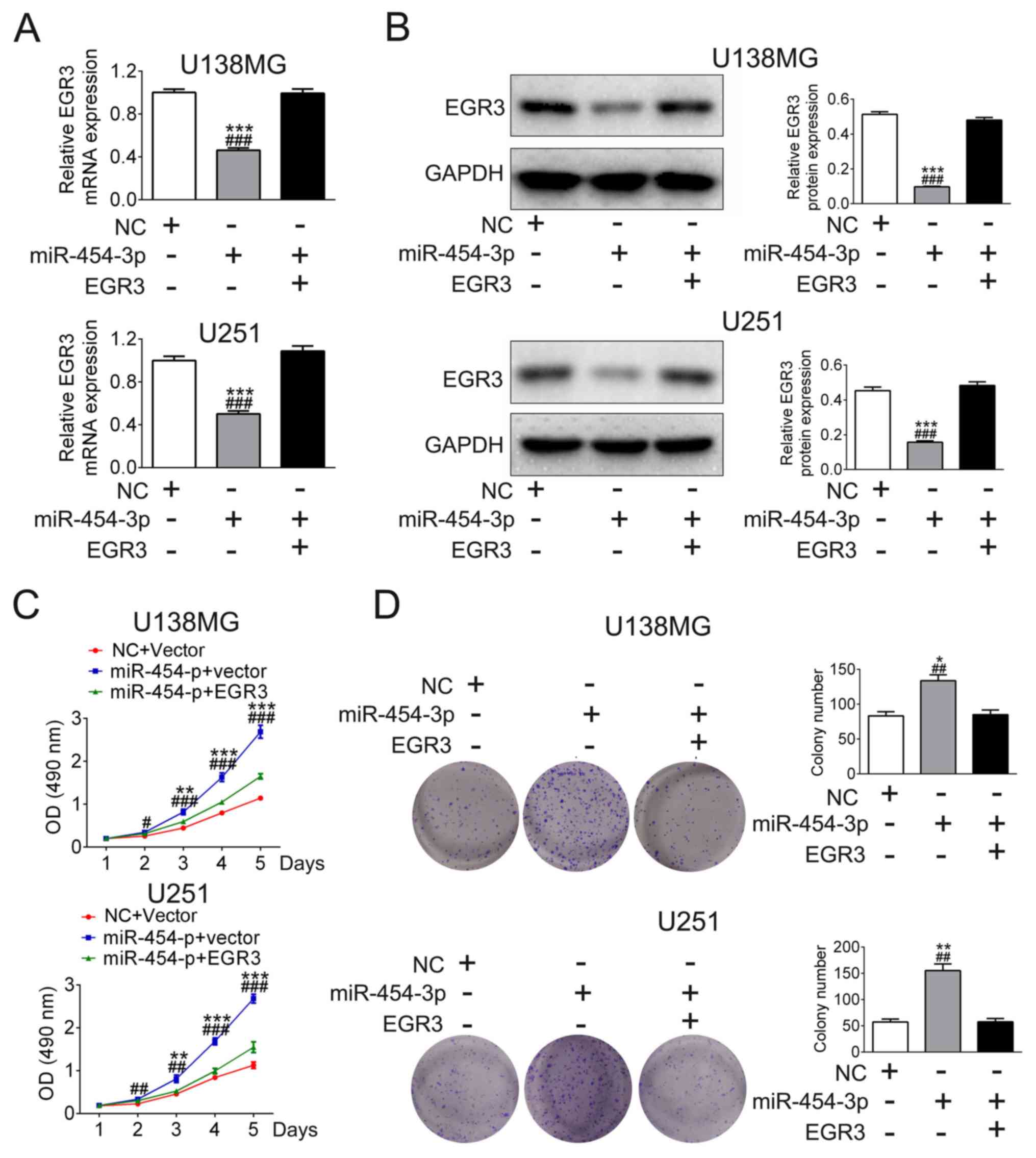
EGR3 is involved in miR-454-3p-induced
elevation of glioma cell proliferation
Based on the findings already obtained from the
present study, it was hypothesized that miR-454-3p may promote
glioma cell proliferation of by targeting and regulating EGR3
expression. U138MG and U251 cells were either transfected with
miR-454-3p mimics only or co-transfected with miR-454-3p mimics and
EGR3 plasmids. Transfection with miR-454-3p mimics significantly
reduced EGR3 expression, which was reversed by co-transfection with
EGR3 plasmids (n=3, Fig. 5A and B).
Besides, the results of MTT and colony formation assays showed that
transfection with miR-454-3p mimics increased cell viability (n=5)
and colony formation (n=3), but was reversed by co-transfection
with EGR3 plasmids in U138MG and U251 cells (Fig. 5C and D). These results indicated that
miR-454-3p promote glioma cell proliferation in by negatively
regulating EGR3 expression.
Discussion
Glioma is a common malignancy in the central nervous
system with the overall prognosis of ~1 year for most patients from
the time of diagnosis (20).
Combined therapies currently available for glioma have limited
benefits due to the high rates of invasion (21). It is therefore imperative to explore
the pathological mechanism and new therapeutic strategies for the
diagnosis and treatment of this disease.
A previous study have demonstrated that plasma
levels of miR-454-3p were upregulated in patients with glioma
compared with healthy controls, where the prognosis of glioma with
high miR-454-3p expression was significantly worse compared with
that of glioma with low miR-454-3p expression (22). Indeed, there is accumulating evidence
that aberrant expression of miRNA is involved in the pathogenesis
of many diseases including cancers (23). A number of studies have previously
found that miR-454-3p was downregulated in gastric cancer and
osteosarcomas (23,24), whereas it was upregulated in prostate
cancer, hepatocellular carcinoma and non-small cell lung cancer
(25–27). These results suggest that expression
profiles of miR-454-3p are distinct in different types of cancers.
In the present study, it was found that miR-454-3p expression was
upregulated in glioma tissues compared with adjacent normal
tissues, which was concordant with previous results found in the
plasma (22). In addition,
association between increased miR-454-3p expression with poor
prognosis were also previously found in hepatocellular carcinoma
(26) and triple-negative breast
cancer (28). Taken together, these
results suggest that miR-454-3p may serve as a potential prognostic
biomarker for glioma, though the role of miR-4543-3p in glioma
remains unknown.
Since cell proliferation is one of the most
important hallmarks of cancer pathophysiology (11), the present study evaluated the effect
of miR-454-3p on glioma cell growth and found that miR-454-3p
overexpression could significantly promote cell growth. EGR3 was
subsequently identified as a direct target of miR-454-3p, where the
overexpression of miR-454-3p inhibited EGR3 mRNA and protein
expression. Overexpression of EGR3 was also found to reverse
miR-454-3p mimic-induced cell proliferation in vitro.
Previous studies reported that EGR3 was ubiquitously expressed in
the cerebral cortex, substrate ganglion and neuromuscular spindle
(13). Increased EGR3 expression
induced expression of IL-6 and IL-8 has been previously
demonstrated in prostate cancer (29), suggesting that EGR3 also serves an
important role in tumorigenesis. Of note, EGR3 were also shown to
be downregulated in glioma tissues in comparison with adjacent
normal tissues, and EGR3 knockdown in vitro promoted glioma
cell proliferation.
It was previously found that overexpression of
miR-454-3p in hepatic stellate cells by transfection with
miR-454-3p mimics could deactivate the hepatic stellate cells by
targeting SMAD4, which is involved in transforming growth factor-β
signaling (30). Overexpression of
miR-454-3p could also promote prostate cancer cell growth by
targeting N-myc downstream-regulated gene 2 (25), whereas the upregulation miR-454-3p
expression promoted the progression of NSCLCs by directly targeting
PTEN (27). These results suggest
that miR-454-3p is involved in the regulation of a number of
signaling pathways by binding to multiple targets. Therefore, the
role of miR-454-3p in glioma may also be through other signaling
pathway, which require further study.
To conclude, the present study was the first to
demonstrate that miR-454-3p is in involved in the cell
proliferation of glioma, by negatively regulating EGR3 expression.
These findings provide valuable insights into the underlying
biological features of this disease and may provide an avenue for
the development of possible novel therapeutic targets.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
GS and JW designed all the experiments and revised
the paper. ZM, DC, LL and XL performed the experiments and wrote
the paper.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of The First People's Hospital of Taizhou. All
patients provided written informed consent prior to enrollment in
the study.
Patient consent for publication
All patients provided written informed consent prior
to enrollment in the study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Pan Y, Yuan F, Li Y, Wang G, Lin Z and
Chen L: Bromodomain PHD-finger transcription factor promotes glioma
progression and indicates poor prognosis. Oncol Rep. 41:246–256.
2019.PubMed/NCBI
|
|
2
|
Yan J, Xu C, Li Y, Tang B, Xie S, Hong T
and Zeng E: Long non-coding RNA LINC00526 represses glioma
progression via forming a double negative feedback loop with AXL. J
Cell Mol Med. 23:5518–5531. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Guo Y, Hong W, Wang X, Zhang P, Körner H,
Tu J and Wei W: MicroRNAs in microglia: How do micrornas affect
activation, inflammation, polarization of microglia and mediate the
interaction between microglia and glioma? Front Mol Neurosci.
12:1252019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Petrescu GED, Sabo AA, Torsin LI, Calin GA
and Dragomir MP: MicroRNA based theranostics for brain cancer:
Basic principles. J Exp Clin Cancer Res. 38:2312019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Si W, Shen J, Zheng H and Fan W: The role
and mechanisms of action of microRNAs in cancer drug resistance.
Clin Epigenetics. 11:252019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mohr AM and Mott JL: Overview of microRNA
biology. Semin Liver Dis. 35:3–11. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Omar HA, El-Serafi AT, Hersi F, Arafa ESA,
Zaher DM, Madkour M, Arab HH and Tolba MF: Immunomodulatory
MicroRNAs in cancer: Targeting immune checkpoints and the tumor
microenvironment. FEBS J. Jul 15–2019.(Epub ahead of print) doi:
10.1111/febs.15000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ye X, Wei W, Zhang Z, He C, Yang R, Zhang
J, Wu Z, Huang Q and Jiang Q: Identification of microRNAs
associated with glioma diagnosis and prognosis. Oncotarget.
8:26394–26403. 2017.PubMed/NCBI
|
|
9
|
Zhang Y, Zhang R, Sui R, Chen Y, Liang H,
Shi J and Piao H: MicroRNA-374a governs aggressive cell behaviors
of glioma by targeting prokineticin 2. Technol Cancer Res Treat.
18:15330338188214012019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Barciszewska AM: MicroRNAs as efficient
biomarkers in high-grade gliomas. Folia Neuropathol. 54:369–374.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liang HL, Hu AP, Li SL, Xie JP, Ma QZ and
Liu JY: MiR-454 prompts cell proliferation of human colorectal
cancer cells by repressing CYLD expression. Asian Pac J Cancer
Prev. 16:2397–2402. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Piwecka M, Rolle K, Belter A, Barciszewska
AM, Żywicki M, Michalak M, Nowak S, Naskręt-Barciszewska MZ and
Barciszewski J: Comprehensive analysis of microRNA expression
profile in malignant glioma tissues. Mol Oncol. 9:1324–1340. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Meyers KT, Marballi KK, Brunwasser SJ,
Renda B, Charbel M, Marrone DF and Gallitano AL: The immediate
early gene Egr3 is required for hippocampal induction of Bdnf by
electroconvulsive stimulation. Front Behav Neurosci. 12:922018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chien MH, Lee WJ, Yang YC, Li YL, Chen BR,
Cheng TY, Yang PW, Wang MY, Jan YH, Lin YK, et al: KSRP suppresses
cell invasion and metastasis through miR-23a-mediated EGR3 mRNA
degradation in non-small cell lung cancer. Biochim Biophys Acta
Gene Regul Mech. 1860:1013–1024. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Maple A, Lackie RE, Elizalde DI, Grella
SL, Damphousse CC, Xa C, Gallitano AL and Marrone DF: Attenuated
late-phase Arc transcription in the dentate gyrus of mice lacking
Egr3. Neural Plast. 2017:60630482017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Oliveira Fernandes M and Tourtellotte WG:
Egr3-dependent muscle spindle stretch receptor intrafusal muscle
fiber differentiation and fusimotor innervation homeostasis. J
Neurosci. 35:5566–5578. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang S, Xia C, Xu C, Liu J, Zhu H, Yang
Y, Xu F, Zhao J, Chang Y and Zhao Q: Early growth response 3
inhibits growth of hepatocellular carcinoma cells via upregulation
of Fas ligand. Int J Oncol. 50:805–814. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar
|
|
20
|
Zhang Y, Chen J, Xue Q, Wang J, Zhao L,
Han K, Zhang D and Hou L: Prognostic significance of MicroRNAs in
glioma: A systematic review and meta-analysis. Biomed Res Int.
2019:40159692019.PubMed/NCBI
|
|
21
|
Beadle C, Assanah MC, Monzo P, Vallee R,
Rosenfeld SS and Canoll P: The role of myosin II in glioma invasion
of the brain. Mol Biol Cell. 19:3357–3368. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shao N, Wang L, Xue L, Wang R and Lan Q:
Plasma miR-454-3p as a potential prognostic indicator in human
glioma. Neurol Sci. 36:309–313. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kabekkodu SP, Shukla V, Varghese VK, Adiga
D, Vethil Jishnu P, Chakrabarty S and Satyamoorthy K: Cluster
miRNAs and cancer: Diagnostic, prognostic and therapeutic
opportunities. Wiley Interdiscip Rev RNA. e15632019.PubMed/NCBI
|
|
24
|
Wang X, Liu B, Wen F and Song Y:
MicroRNA-454 inhibits the malignant biological behaviours of
gastric cancer cells by directly targeting mitogen-activated
protein kinase 1. Oncol Rep. 39:1494–1504. 2018.PubMed/NCBI
|
|
25
|
Fu Q, Gao Y, Yang F, Mao T, Sun Z, Wang H,
Song B and Li X: Suppression of microRNA-454 impedes the
proliferation and invasion of prostate cancer cells by promoting
N-myc downstream-regulated gene 2 and inhibiting WNT/β-catenin
signaling. Biomed Pharmacother. 97:120–127. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou L, Qu YM, Zhao XM and Yue ZD:
Involvement of miR-454 overexpression in the poor prognosis of
hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 20:825–829.
2016.PubMed/NCBI
|
|
27
|
Zhu DY, Li XN, Qi Y, Liu DL, Yang Y, Zhao
J, Zhang CY, Wu K and Zhao S: MiR-454 promotes the progression of
human non-small cell lung cancer and directly targets PTEN. Biomed
Pharmacother. 81:79–85. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cao ZG, Li JJ, Yao L, Huang YN, Liu YR, Hu
X, Song CG and Shao ZM: High expression of microRNA-454 is
associated with poor prognosis in triple-negative breast cancer.
Oncotarget. 7:64900–64909. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang P, Yang X, Wang L, Zhang D, Luo Q
and Wang B: Overexpressing miR-335 inhibits DU145 cell
proliferation by targeting early growth response 3 in prostate
cancer. Int J Oncol. 54:1981–1994. 2019.PubMed/NCBI
|
|
30
|
Zhu D, He X, Duan Y, Chen J, Wang J, Sun
X, Qian H, Feng J, Sun W, Xu F and Zhang L: Expression of
microRNA-454 in TGF-β1-stimulated hepatic stellate cells and in
mouse livers infected with Schistosoma japonicum. Parasit Vectors.
7:1482014. View Article : Google Scholar : PubMed/NCBI
|