Introduction
Breast cancer is the most common malignant tumor of
females and is the leading cause of cancer-associated death in
women, worldwide (1). Despite great
improvements in surgical resection combined with advances in
chemotherapy and radiotherapy, the overall survival time for
patients with advanced stage breast cancer remains very poor,
primarily due to the rapid proliferation and metastasis of cancer
cells (1–3). Understanding the molecular mechanisms
underlying breast cancer growth and metastasis is important, as it
may contribute to the discovery of novel molecular targets for
breast cancer treatment (4,5).
Long non-coding RNAs (lncRNAs) are a class of
non-coding RNAs >200 nucleotides in length that have been
demonstrated to be involved in various physiological and
pathological processes by regulating cell proliferation, apoptosis,
cell cycle progression, migration and invasion (3,6,7). Recently, increasing numbers of lncRNAs
have been identified to be deregulated in various malignant tumors,
including breast cancer (8,9). Furthermore, lncRNAs have been reported
to serve important roles in cancer by affecting the malignant
phenotypes of breast cancer cells (3,9–11). For instance, the lncRNA nuclear
enriched abundant transcript 1 (NEAT1) is significantly upregulated
in breast cancer, whilst knockdown of NEAT1 significantly inhibits
the growth of breast cancer cells (10). The lncRNA small nucleolar RNA host
gene 20 promotes the proliferation, invasion and migration of
breast cancer cells by inhibiting the expression of microRNA
(miR)-495 (9). The downregulation of
lncRNA growth arrest-specific 5 induces trastuzumab resistance in
breast cancer (11).
Prostate cancer-associated non-coding RNA 1 (PRNCR1)
is localized to chromosome 8q24 and was originally reported to be
associated with prostate and colorectal cancer susceptibility
(12,13). PRNCR1 directly binds to androgen
receptors (ARs) to enhance ligand-dependent and -independent AR
gene expression as well as the proliferation of prostate cancer
cells (14). Yang et al
(15) reported that PRNCR1 is
upregulated in colorectal cancer and promotes cancer cell
proliferation and cell cycle progression. Cheng et al
(16) identified that PRNCR1
upregulated hes related family bHLH transcription factor with YRPW
motif 2 to promote non-small-cell lung carcinoma (NSCLC)
progression by competitively binding miR-448 (16). In addition to being associated with
cancer, PRNCR1 also affects osteogenic differentiation and
contributes to osteolysis following hip replacement (17). Additionally, PRNCR1 promotes the
progression of eclampsia by regulating the mitogen activated
kinase-like signal pathway (18).
However, the detailed role of PRNCR1 in breast
cancer remains unknown. Therefore, the present study aimed to
examine the clinical significance of PRNCR1 expression in breast
cancer and to explore the role of PRNCR1 in breast cancer cell
proliferation, apoptosis, migration and invasion. The findings may
provide a potential therapeutic target for patients with breast
cancer.
Materials and methods
Clinical tissue samples
A total of 52 paired breast cancer tissues and
adjacent normal tissues, which were cut ≥3 cm from cancer tissues,
were obtained from 52 female patients (mean age, 54.4±13.5 years;
age range, 33–76 years) with primary breast cancer at the
Department of Breast Neoplasm Surgery, Inner Mongolia People's
Hospital between June 2012 and September 2013 (Table I). The present study was approved by
the Ethics Committee of Inner Mongolia People's Hospital and
written consent was obtained from all participants. The inclusion
criteria were that these patients were primary patients with breast
cancer. Patients with preoperative chemotherapy and/or radiotherapy
were excluded from the current study. Luminal cancers were defined
based on PAM50 (19). These tissues
were immediately snap-frozen in liquid nitrogen following surgery
and stored at −80°C until required. The patients were followed up
once every 2–3 months post-surgery for 5 years.
 | Table I.Association between PRNCR1 expression
and clinicopathological characteristics in patients with breast
cancer. |
Table I.
Association between PRNCR1 expression
and clinicopathological characteristics in patients with breast
cancer.
| Variables | Number (n=52) | Low expression
(n=29) | High expression
(n=23) | P-value |
|---|
| Age (years) |
|
|
| 0.680 |
| ≤50 | 22 | 13 | 9 |
|
|
>50 | 30 | 16 | 14 |
|
| Subtype |
|
|
| 0.591 |
| Lunimal A
type | 28 | 18 | 10 |
|
| Lunimal B
type | 5 | 2 | 3 |
|
| HER2
positive | 8 | 4 | 4 |
|
| TNBC | 11 | 5 | 6 |
|
| Differentiation |
|
|
| 0.077 |
| Well and
moderately | 36 | 23 | 13 |
|
| Poor | 16 | 6 | 10 |
|
| Lymph node
metastasis |
|
|
| 0.017a |
|
Present | 38 | 25 | 13 |
|
|
Absent | 14 | 4 | 10 |
|
| Distant
metastasis |
|
|
| 0.004a |
|
Present | 6 | 0 | 6 |
|
|
Absent | 46 | 29 | 17 |
|
| TNM stage |
|
|
| 0.008a |
|
I–II | 35 | 24 | 11 |
|
|
III–IV | 17 | 5 | 12 |
|
Cell culture
A normal human breast cell line, MCF-10A, and 4
human breast cancer cell lines, BT-549, MCF-7, SK-BR-3 and
MDA-MB-231, were obtained from Cell Bank of Type Culture Collection
of Chinese Academy of Sciences. All cell lines were cultured in
DMEM (Thermo Fisher Scientific, Inc.) with 10% FBS (Thermo Fisher
Scientific, Inc.) at 37°C with 5% CO2.
Cell transfection
Cell transfection was performed using
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) in
accordance with the manufacturer's protocol. SK-BR-3 and BT-549
cells were transfected with 100 nM scrambled (NC) small interfering
(si)RNA (cat. no. 12935200, Thermo Fisher Scientific, Inc.), PRNCR1
siRNA1 (cat. no. n541275, Thermo Fisher Scientific, Inc.) and
PRNCR1 siRNA2 (cat. no. n541276, Thermo Fisher Scientific, Inc.).
The following experiments were performed after cell transfection
for 48 h.
Reverse transcription-quantitative PCR
(RT-qPCR) assay
Total RNA was extracted from cells or tissues using
TRIzol® reagent (Thermo Fisher Scientific, Inc.). cDNA
was synthesized from 1 µg RNA using a PrimeScript RT Master Mix kit
(Takara Bio, Inc.) in accordance with the manufacturer's protocols.
qPCR was subsequently performed to determine mRNA expression using
SYBR Green qPCR Master Mix (Thermo Fisher Scientific, Inc.) on an
ABI 7300 Plus Real-Time PCR System (Thermo Fisher Scientific,
Inc.). The thermocycling conditions were as follows: 95°C for 30
sec, followed by 40 cycles of 95°C for 15 sec and 60°C for 30 sec.
The relative mRNA expression was quantified by the
2−ΔΔCq method and normalized to GAPDH (20). The primers for GAPDH were forward,
5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse,
5′-GGCTGTTGTCATACTTCTCATGG-3′. The primers for PRNCR1 were forward,
5′-CCAGATTCCAAGGGCTGATA-3′ and reverse,
5′-GATGTTTGGAGGCATCTGGT-3′.
Western blot analysis
Transfected SK-BR-3 and BT-549 cells were lysed
using radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology). Proteins (50 µg per lane) were separated via
SDS-PAGE on a 10% gel then transferred onto polyvinylidene
difluoride membranes (Thermo Fisher Scientific, Inc.). The
membranes were blocked with 5% dry milk in Tris-buffered saline
(TBS) with 0.2% of Tween-20 at 4°C overnight. The membranes were
subsequently incubated with primary antibodies against E-cadherin
(1:500; ab15148; Abcam), N-cadherin (1:500; ab18203; Abcam),
Vimentin (1:500; ab8978; Abcam), and GAPDH (1:250; ab9485; Abcam)
at room temperature for 3 h. Membranes were then incubated with
horseradish peroxidase-conjugated secondary antibody (1:5,000;
ab6721; Abcam) at room temperature for 1 h. The protein signals
were detected using an Enhanced Chemiluminescence Western Blotting
Substrate kit (Thermo Fisher Scientific, Inc.) and quantified using
ImageJ software v1.46 (National Institutes of Health). GAPDH was
used as an internal control.
CCK-8 assay
Transfected SK-BR-3 and BT-549 cells were
resuspended in DMEM with 10% FBS and seeded into 96-well plates
(10,000 cells per well). Following incubation at 37°C for 0, 24, 48
or 72 h, 10 µl CCK-8 reagent (Thermo Fisher Scientific, Inc.) was
added to cells. Then cells were incubated at 37°C for another 2 h.
The optical density absorbance at 450 nm was measured using a
Varioskan LUX Multimode Microplate Reader (Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocols.
Colony formation assay
Transfected SK-BR-3 and BT-549 cells were
resuspended in DMEM with 10% FBS and seeded into 6-well plates (150
cells/well). Cells were then cultured at 37°C for 14 days. Colonies
were washed with PBS, fixed with 10% formalin at room temperature
for 10 min and stained with 0.1% (w/v) crystal violet (Amresco LLC)
at room temperature for 10 min. The number of cell colonies
(containing >50 cells) were counted using a light microscope
(magnification, ×40).
Cell apoptosis assay
Cell apoptosis was analyzed using an Annexin
V-fluorescein isothiocyanate (FITC) Apoptosis kit (BD Biosciences)
in accordance with the manufacturer's protocol. Transfected SK-BR-3
and BT-549 cells were stained with Annexin V-FITC and propidium
iodide (PI) solutions at 4°C for 30 min. Then, the cell apoptosis
rate was detected by a FACSCalibur flow cytometer (Becton,
Dickinson and Company) and analyzed using BD Accuri C6 software
(version 1.0; Becton, Dickinson and Company).
Cell cycle analysis
Transfected SK-BR-3 and BT-549 cells were cultured
in DMEM with 10% FBS at 37°C for 24 h. Cells were fixed in 70%
ethanol at 4°C overnight and incubated in 1 ml PBS with 0.1%
TritonX-100, 60 µg/ml PI, 0.1 mg/ml DNase free RNase and 0.1%
trisodium citrate at 4°C for 30 min. Cell cycle distribution was
measured using a FACSCalibur flow cytometer and analyzed using BD
Accuri C6 software 1.0 (Becton, Dickinson and Company).
Wound healing assay
After transfection for 48 h, a wound healing assay
was performed to assess cell migration. A wound was introduced to
transfected SK-BR-3 and BT-549 cells with a sterile 100 µl
micropipette tip. Cells were then washed twice with PBS to remove
cellular debris and cultured at 37°C in serum-free DMEM for 24 h.
Cells were photographed using an inverted microscope (IX71; Olympus
Corporation; magnification, ×40) at 0 and 24 h, and the wound width
was analyzed using ImageJ software 1.46 (National Institutes of
Health).
Transwell assay
A transwell assay was performed to assess cell
invasion. Transfected SK-BR-3 and BT-549 cells (10,000 cells/well)
were resuspended in 200 µl serum-free DMEM, which were then added
into the upper chambers (pore size; 8 µm) precoated with Matrigel
(EMD Millipore; Merck KGaA). The lower chambers were filled with
500 µl DMEM with 10% FBS. Cells were then incubated at 37°C for 48
h. A cotton swab was used to carefully remove non-invading cells
from the top chamber. Cells on the lower surface were fixed in 75%
ethanol at room temperature for 30 min and stained with 0.1%
crystal violet at room temperature for 5 min. Invasive cells were
photographed and counted under a phase-contrast inverted microscope
(magnification, ×200).
Statistical analysis
All experiments were performed in triplicate. Data
are presented as the mean ± standard deviation and analyzed using
GraphPad Prism 5 (GraphPad Software, Inc.). Student's t-tests were
used to analyze differences between two groups and one-way analysis
of variance followed by a Tukey's post-hoc test was used for the
comparisons of multiple groups. A χ2 test was used for
analyzing the association between PRNCR1 expression and clinical
characteristics of breast cancer patients. Survival analysis was
conducted using Kaplan-Meier survival curves and a log-rank test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Upregulation of PRNCR1 in breast
cancer predicts poor prognosis
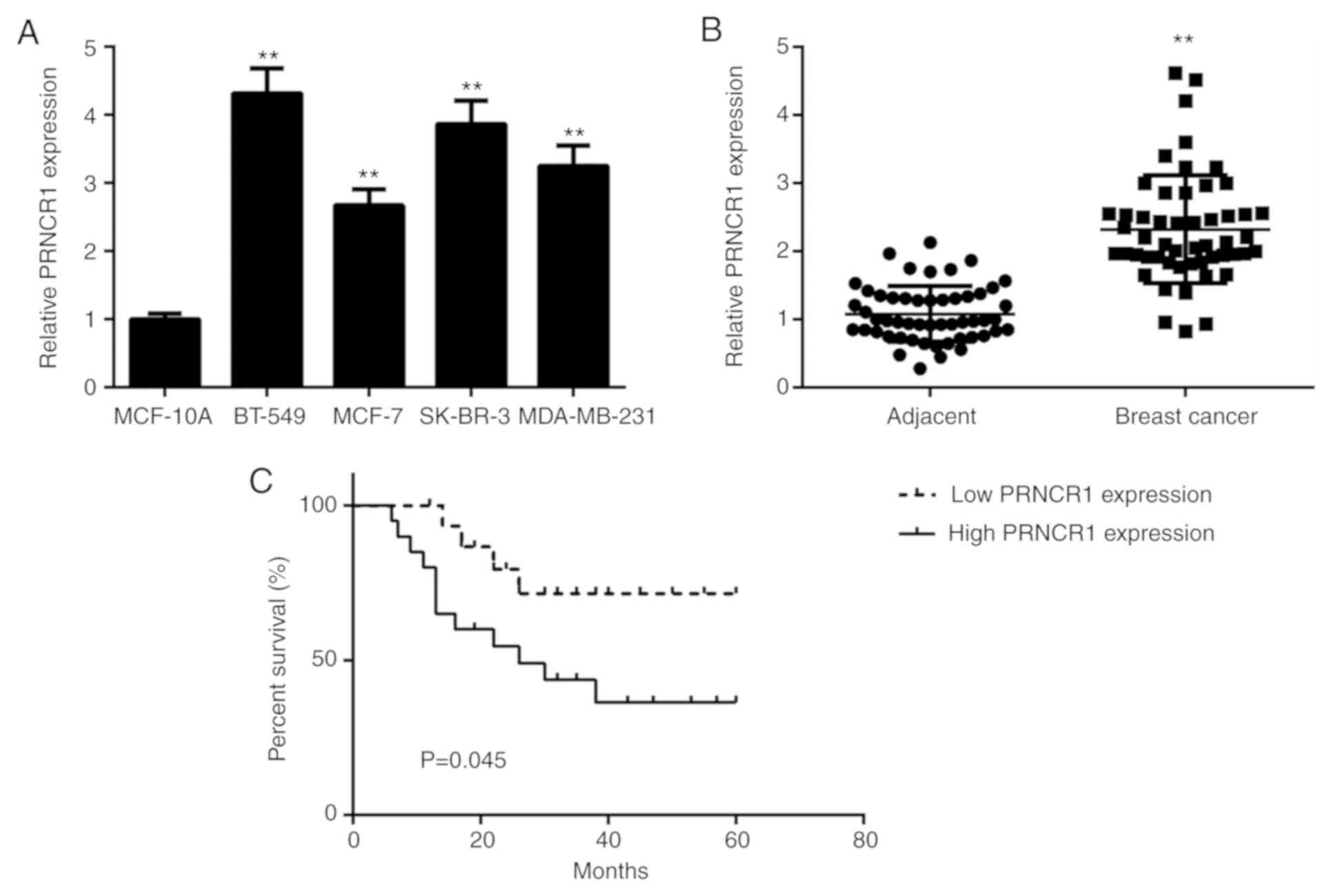
A qPCR assay was performed to determine the
expression of PRNCR1 in breast cancer cell lines and the normal
human breast cell line, MCF-10A. The expression of PRNCR1 was
significantly higher in breast cancer cell lines compared with
MCF-10A cells (Fig. 1A). To confirm
these findings in vivo, PRNCR1 expression in 52 paired
breast cancer tissues and adjacent normal tissues were
investigated. It was demonstrated that PRNCR1 was significantly
upregulated in breast cancer tissues compared with adjacent normal
tissues (Fig. 1B). The 52 patients
with breast cancer were divided into low and high PRNCR1 expression
groups based on the median expression level (median value, 2.38) of
PRNCR1 as a cutoff. Further investigation demonstrated that high
PRNCR1 expression was significantly associated with positive
metastasis and advanced TNM Classification of Malignant Tumors
(TNM) stage (Table I), indicating
that the upregulation of PRNCR1 may serve a role in breast cancer
progression. Survival analysis indicated that the patients with
high PRNCR1 expression exhibited shorter survival times compared
with those with low PRNCR1 expression (Fig. 1C). The results indicated that a high
upregulation of PRNCR1 predicts a poor prognosis in patients with
breast cancer.
PRNCR1 silencing inhibits breast
cancer cell proliferation and colony formation
The role of PRNCR1 in breast cancer in vitro
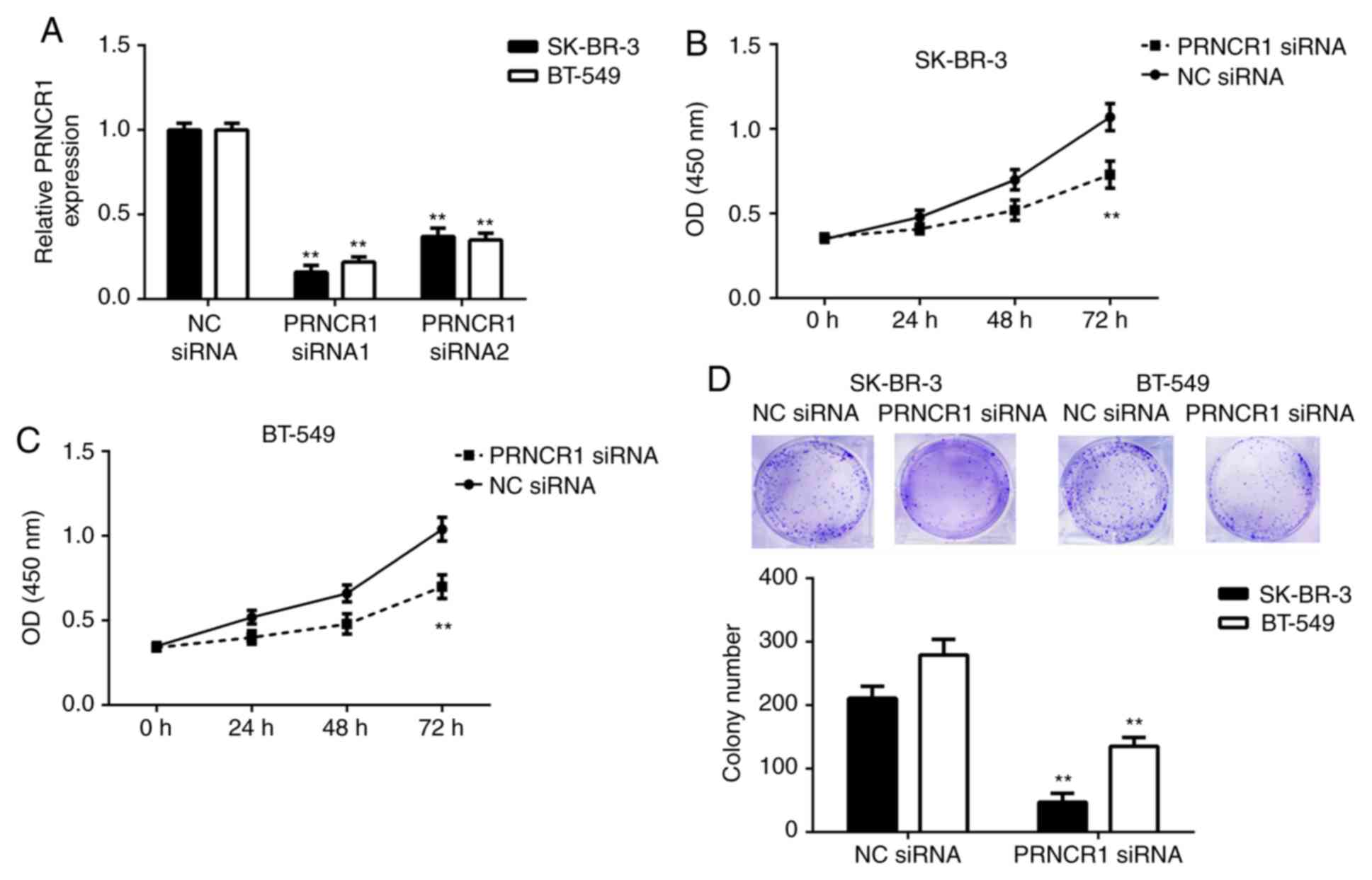
was assessed in the current study. SK-BR-3 and BT-549 cells were
transfected with NC siRNA or two different PRNCR1 siRNAs,
respectively. As presented in Fig.
2A, RT-qPCR data indicated that PRNCR1 levels were
significantly decreased following transfection with PRNCR1 siRNA1
or siRNA2 compared with the NC siRNA group. As PRNCR1 siRNA1
demonstrated the highest inhibitory effect on the expression of
PRNCR1 in SK-BR-3 and BT-549 cells, it was selected for subsequent
experimentation. A CCK-8 assay was then performed to assess the
effects of PRNCR1 downregulation on breast cancer cell
proliferation. The proliferation of SK-BR-3 and BT-549 cells was
significantly reduced in the PRNCR1 siRNA group compared with the
NC siRNA group (Fig. 2B and C). A
colony formation assay was subsequently performed, which determined
that the colony formation capacities of SK-BR-3 and BT-549 cells
were significantly inhibited following the silencing of PRNCR1
expression (Fig. 2D).
PRNCR1 knockdown represses cell cycle
progression and promotes SK-BR-3 and BT-549 cell apoptosis
The present study hypothesized that the role of
PRNCR1 in breast cancer cell proliferation may occur by affecting
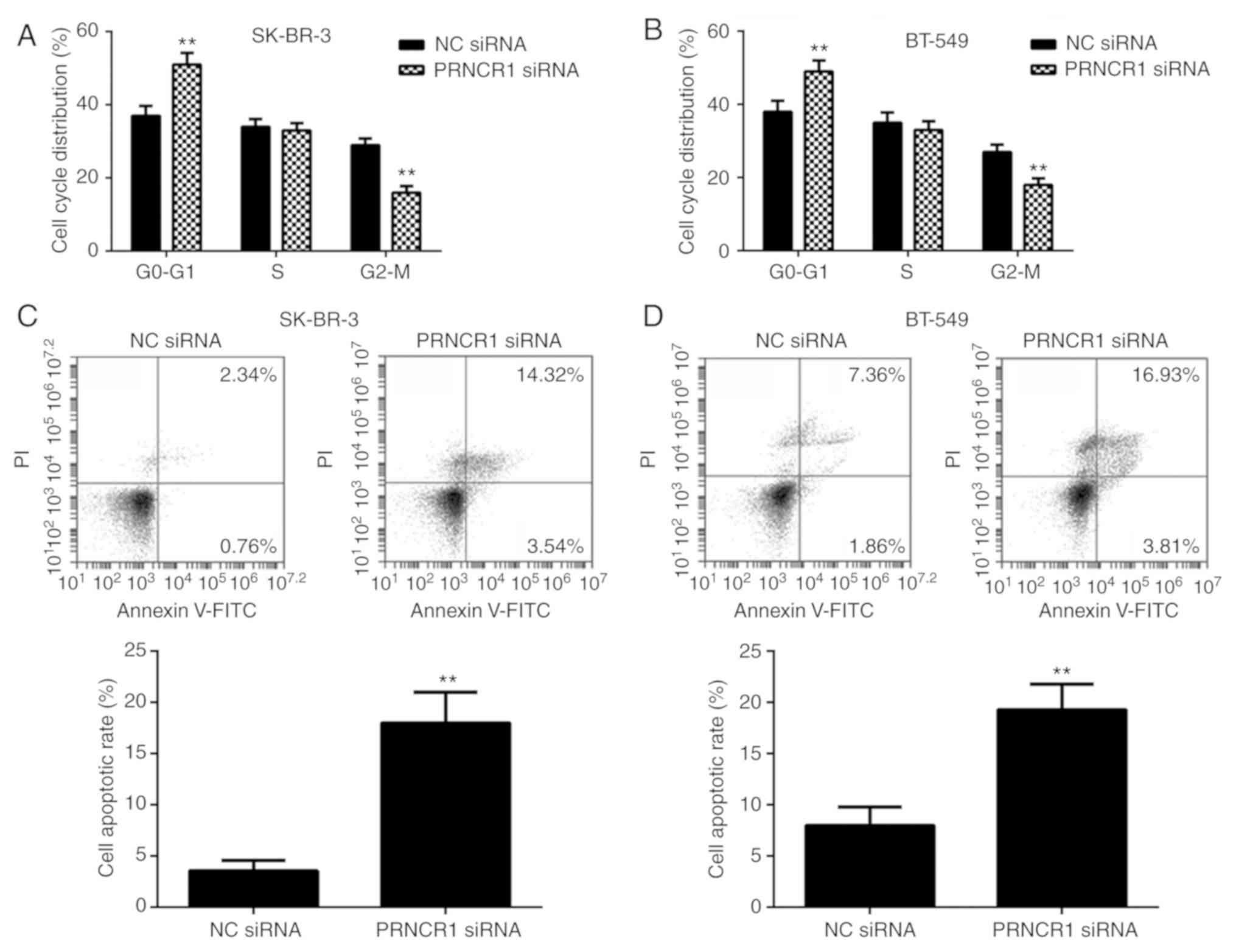
cell cycle progression and/or cell apoptosis. Thus, flow cytometry
was performed to determine the effects of PRNCR1 downregulation on
cell cycle progression and apoptosis in SK-BR-3 and BT-549 cells.
The percentage of SK-BR-3 and BT-549 cells in the G1 stage was
significantly increased, while the percentage of cells in the G2/M
stage was significantly decreased in the PRNCR1 siRNA group
compared with the NC siRNA group (Fig.
3A and B). These results indicated that the silencing of PRNCR1
caused a cycle arrest at the G1 stage in SK-BR-3 and BT-549 cells.
Furthermore, the apoptotic rate of SK-BR-3 and BT-549 cells was
significantly higher in the PRNCR1 siRNA group compared with the NC
siRNA group (Fig. 3C and D).
Therefore, the knockdown of PRNCR1 was demonstrated to inhibit cell
cycle progression and promote the apoptosis of breast cancer
cells.
PRNCR1 knockdown inhibits migration,
invasion and epithelial-mesenchymal transition (EMT) of breast
cancer cells
The function of PRNCR1 in in vitro breast
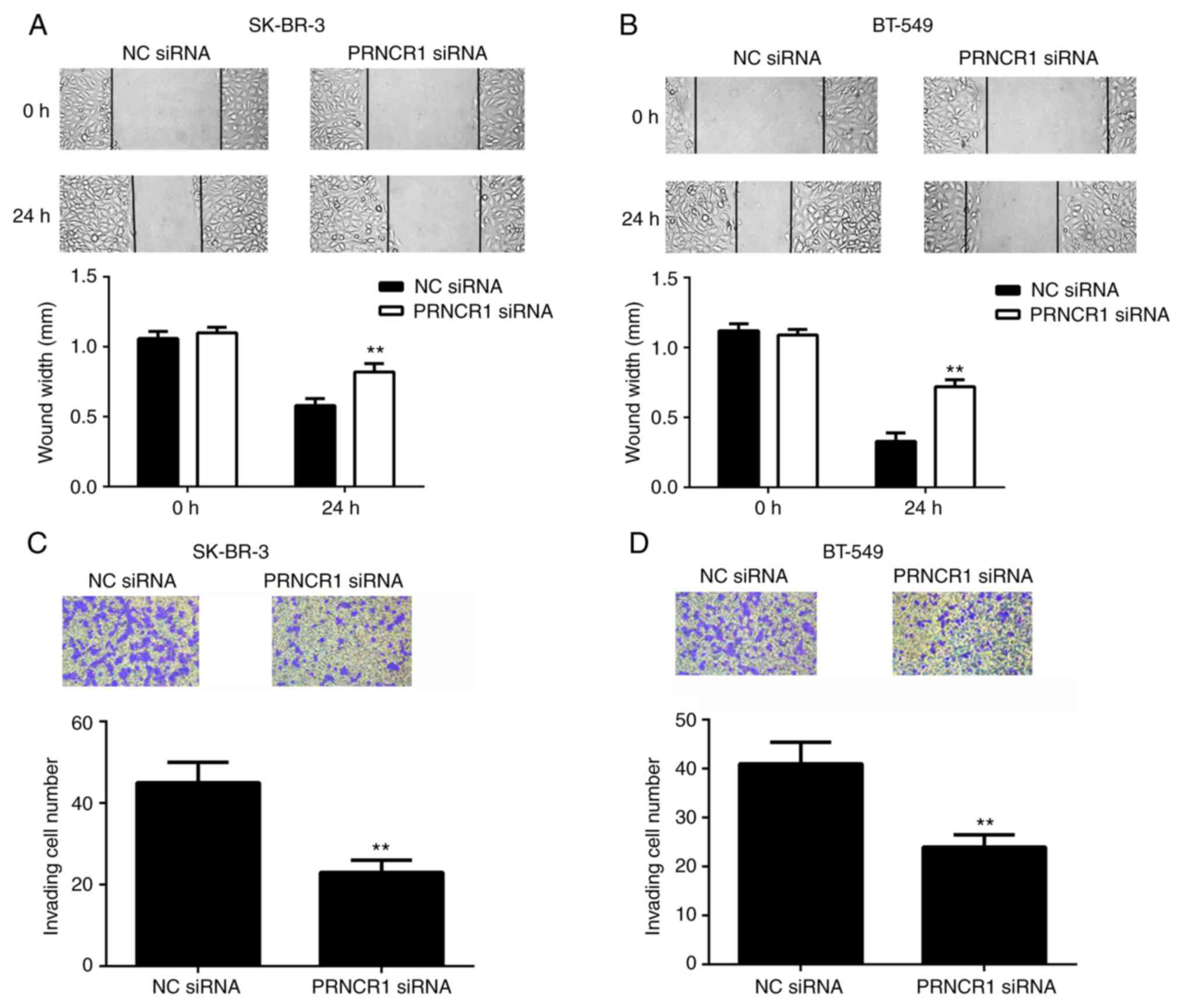
cancer metastasis was assessed. Wound healing and Transwell assays
were performed to examine the effects of PRNCR1 downregulation on
the migration and invasion of breast cancer cells. The results
indicated that the migration of SK-BR-3 and BT-549 cells was
significantly decreased following PRNCR1 silencing compared with
the control group (Fig. 4A and B).
In agreement, the invasive capacity of cells was also significantly
reduced in the PRNCR1 siRNA group compared to the NC siRNA group
(Fig. 4C and D). These results
indicated that PRNCR1 may have a promoting role in breast cancer
metastasis.
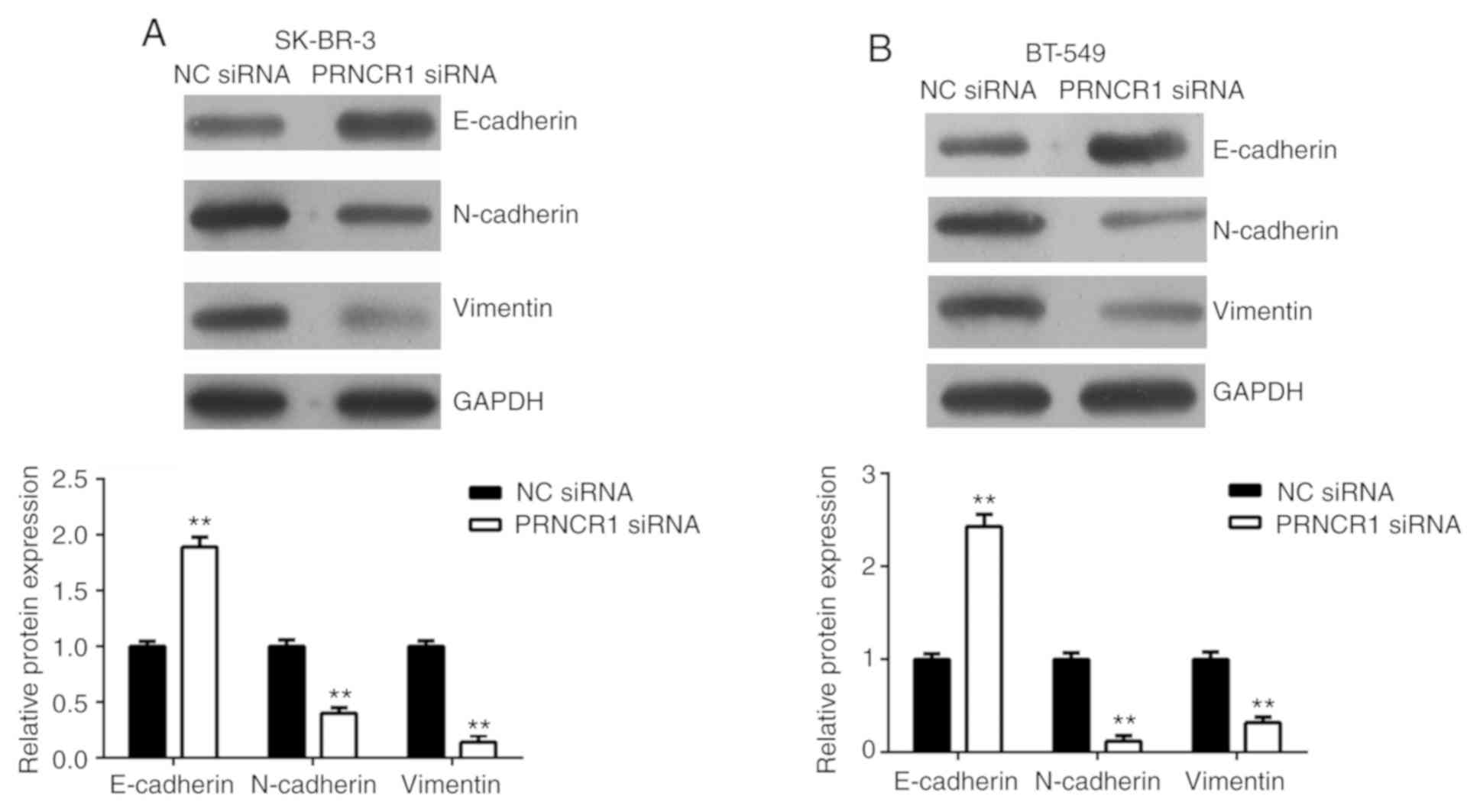
EMT is essential for cancer cell migration and
invasion (21). Therefore, the
effects of PRNCR1 on the EMT of breast cancer cells were
determined. The protein expressions of E-cadherin (an epithelial
marker), N-cadherin (a mesenchymal marker) and vimentin (a
mesenchymal marker) were examined in SK-BR-3 and BT-549 cells.
Silencing of PRNCR1 expression resulted in a significant increase
in E-cadherin expression with significant reductions in N-cadherin
and vimentin compared with the NC siRNA group (Fig. 5). Taken together, these results
indicated that the inhibition of PRNCR1 repressed the migration and
invasion of breast cancer cells by downregulating EMT.
Discussion
To the best of our knowledge, the expression and
function of lncRNA PRNCR1 in breast cancer has not previously been
investigated. In the present study, it was determined that PRNCR1
was significantly upregulated in breast cancer, with high
expression of PRNCR1 significantly associated with breast cancer
progression and poor prognosis. For in vitro experiments,
the knockdown of PRNCR1 inhibited breast cancer cell proliferation,
colony formation, cell cycle progression, migration, invasion, EMT
and promoted breast cancer cell apoptosis.
Recently, accumulating evidence has revealed that
lncRNAs serve key roles in breast cancer progression, with certain
lncRNAs exhibiting potential as biomarkers and therapeutic targets
for this disease (22). For
instance, the lncRNA, DSCAM Antisense RNA 1, regulates the G1/S
cell cycle transition and is an independent prognostic factor for
poor survival in luminal breast cancer patients treated with
endocrine therapy (22). Knockdown
of the lncRNA, HOX transcript antisense RNA, sensitized breast
cancer cells to ionizing radiation by activating miR-218 (23). The lncRNA, long intergenic
non-protein coding RNA 1116, has an oncogenic role in breast cancer
by binding to enhancer of zeste 2 polycomb repressive complex 2
subunit to target p57 and is involved in the mitogen activated
protein kinase and NF-κB signaling pathways (24). However, no previous study has focused
on the expression of PRNCR1 in breast cancer. The present study
observed that PRNCR1 was significantly upregulated in breast cancer
cell lines and clinical tissues compared with normal breast MCF-10A
cells and adjacent normal tissues, respectively. In addition, it
was identified that the increased expression of PRNCR1 was
significantly associated with positive lymph node metastasis and
advanced TNM stage in breast cancer patients, indicating that
upregulation of PRNCR1 promoted breast cancer progression.
Furthermore, patients with high PRNCR1 expression exhibited shorter
survival times compared with those with low PRNCR1 expressions,
indicating that the expression of PRNCR1 may be used as a promising
predicator of prognosis in breast cancer.
The role of PRNCR1 in breast cancer progression
in vitro was investigated via a loss-of-function assay in
SK-BR-3 and BT-549 cells. The results demonstrated that PRNCR1
silencing led to a significant reduction in breast cancer cell
proliferation and colony formation. These results indicated that
PRNCR1 may have a promoting role in breast cancer growth. As the
cell cycle is essential for cell proliferation, the function of
PRNCR1 in cell cycle progression in breast cancer cells was
investigated. The results determined that silencing of PRNCR1
expression led to significant cell cycle arrest in the G1 stage.
This indicated that the suppressive effect of PRNCR1 downregulation
on breast cancer cell proliferation may be due to cell cycle arrest
in the G1 stage. Similarly, Yang et al (15) demonstrated that PRNCR1 upregulation
promotes colorectal cancer cell proliferation and cell cycle
progression. In addition, it was observed that the cell apoptosis
rate was significantly increased following silencing of PRNCR1 in
breast cancer cells, which may also be involved in the
downregulation of cell proliferation induced by PRNCR1
inhibition.
The function of PRNCR1 in breast cancer metastasis
was assessed in the current study, as tumor cell migration and
invasion are crucial for tumor metastasis (21). The results demonstrated that
knockdown of PRNCR1 inhibited breast cancer cell migration and
invasion. These results indicated that PRNCR1 may serve a promoting
role in breast cancer metastasis. Thus, future studies should be
performed to further confirm the function of PRNCR1 in breast
cancer metastasis in vivo via animal experiments. EMT is
characterized by the loss of an epithelial phenotype and
acquisition of mesenchymal properties. It has been widely accepted
that EMT is essential for cell migration and invasion, and thus
serves a crucial role in cancer metastasis (25). Previous studies have identified that
lncRNAs are involved during the EMT process in different types of
human cancer. For instance, the lncRNA, X-inactive specific
transcript, promotes TGF-β-induced EMT by regulating the
miR-367/141-zinc finger E-box binding homeobox 2 axis in NSCLC
(21). The lncRNA, colorectal
neoplasia differentially expressed, enhances EMT in intrahepatic
cholangiocarcinoma (26). However,
to the best of our knowledge, no previous study has reported the
effects of PRNCR1 on EMT. In the present study, it was identified
that the inhibition of PRNCR1 significantly increased E-cadherin
expression while decreasing N-cadherin and vimentin expressions,
indicating that EMT was suppressed. These results suggested that
PRNCR1 may have promoting effects on EMT in breast cancer cells and
that the inhibitory effect of PRNCR1 knockdown in breast cancer
cell migration and invasion may be due to the inhibition of EMT.
The limitations of the present study are the lack of a mechanistic
assessment. Therefore, future studies should involve RNA sequencing
followed by the knockdown of PRNCR1 in breast cancer cells to
analyze the pathways involved in PRNCR1 regulation, as well as EMT.
In addition, the targets of PRNCR1 in breast cancer should also be
investigated.
To the best of our knowledge, the present study was
the first to report that the lncRNA PRNCR1 was significantly
upregulated in breast cancer. The study also determined that PRNCR1
was strongly associated with cancer progression and poor patient
prognosis. In vitro experiments identified that the
knockdown of its expression effectively inhibited malignant
phenotypes of breast cancer cells. Taken together, these results
suggested that PRNCR1 could be used as a potential therapeutic
target for breast cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
BJ designed the study. RZ and WB collected clinical
tissues and performed the statistical analyses. QG, SL, BW, YL and
NC performed the experiments. QG and SL and wrote the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Inner Mongolia People's Hospital. All patients
provided written informed consents.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vishal M, Swetha R, Thejaswini G, Arumugam
B and Selvamurugan N: Role of Runx2 in breast cancer-mediated bone
metastasis. Int J Biol Macromol. 99:608–614. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xu S, Kong D, Chen Q, Ping Y and Pang D:
Oncogenic long noncoding RNA landscape in breast cancer. Mol
Cancer. 16:1292017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Luo T, Yan Y, He Q, Ma X and Wang W:
miR-328-5p inhibits MDA-MB-231 breast cancer cell proliferation by
targeting RAGE. Oncol Rep. 39:2906–2914. 2018.PubMed/NCBI
|
|
5
|
Zhou Y, Meng X, Chen S, Li W, Li D, Singer
R and Gu W: IMP1 regulates UCA1-mediated cell invasion through
facilitating UCA1 decay and decreasing the sponge effect of UCA1
for miR-122-5p. Breast Cancer Res. 20:322018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Peng Z, Liu C and Wu M: New insights into
long noncoding RNAs and their roles in glioma. Mol Cancer.
17:612018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Taylor DH, Chu ET, Spektor R and Soloway
PD: Long non-coding RNA regulation of reproduction and development.
Mol Reprod Dev. 82:932–956. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang O, Yang F, Liu Y, Lv L, Ma R, Chen C,
Wang J, Tan Q, Cheng Y, Xia E, et al: C-MYC-induced upregulation of
lncRNA SNHG12 regulates cell proliferation, apoptosis and migration
in triple-negative breast cancer. Am J Transl Res. 9:533–545.
2017.PubMed/NCBI
|
|
9
|
Guan YX, Zhang ZM, Chen XZ, Zhang Q, Liu
SZ and Zhang YL: Lnc RNA SNHG20 participated in proliferation,
invasion and migration of breast cancer cells via miR-495. J Cell
Biochem. 119:7971–7981. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jiang X, Zhou Y, Sun AJ and Xue JL: NEAT1
contributes to breast cancer progression through modulating miR-448
and ZEB1. J Cell Physiol. 233:8558–8566. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li W, Zhai L, Wang H, Liu C, Zhang J, Chen
W and Wei Q: Downregulation of LncRNA GAS5 causes trastuzumab
resistance in breast cancer. Oncotarget. 7:27778–27786.
2016.PubMed/NCBI
|
|
12
|
Chung S, Nakagawa H, Uemura M, Piao L,
Ashikawa K, Hosono N, Takata R, Akamatsu S, Kawaguchi T, Morizono
T, et al: Association of a novel long non-coding RNA in 8q24 with
prostate cancer susceptibility. Cancer Sci. 102:245–252. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li L, Sun R, Liang Y, Pan X, Li Z, Bai P,
Zeng X, Zhang D, Zhang L and Gao L: Association between
polymorphisms in long non-coding RNA PRNCR1 in 8q24 and risk of
colorectal cancer. J Exp Clin Cancer Res. 32:1042013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pestell RG and Yu Z: Long and noncoding
RNAs (lnc-RNAs) determine androgen receptor dependent gene
expression in prostate cancer growth in vivo. Asian J Androl.
16:268–269. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang L, Qiu M, Xu Y, Wang J, Zheng Y, Li
M, Xu L and Yin R: Upregulation of long non-coding RNA PRNCR1 in
colorectal cancer promotes cell proliferation and cell cycle
progression. Oncol Rep. 35:318–324. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cheng D, Bao C, Zhang X, Lin X, Huang H
and Zhao L: LncRNA PRNCR1 interacts with HEY2 to abolish
miR-448-mediated growth inhibition in non-small cell lung cancer.
Biomed Pharmacother. 107:1540–1547. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gong ZM, Tang ZY and Sun XL: LncRNA PRNCR1
regulates CXCR4 expression to affect osteogenic differentiation and
contribute to osteolysis after hip replacement. Gene. 673:251–261.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiao S, Wang SY and Huang Y: LncRNA PRNCR1
promoted the progression of eclampsia by regulating the MAPK signal
pathway. Eur Rev Med Pharmacol Sci. 22:3635–3642. 2018.PubMed/NCBI
|
|
19
|
Liu MC, Pitcher BN, Mardis ER, Davies SR,
Friedman PN, Snider JE, Vickery TL, Reed JP, DeSchryver K, Singh B,
et al: PAM50 gene signatures and breast cancer prognosis with
adjuvant anthracycline- and taxane-based chemotherapy: Correlative
analysis of C9741 (Alliance). NPJ Breast Cancer. 2:e150232016.
View Article : Google Scholar
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li C, Wan L, Liu Z, Xu G, Wang S, Su Z,
Zhang Y, Zhang C, Liu X, Lei Z and Zhang HT: Long non-coding RNA
XIST promotes TGF-β-induced epithelial-mesenchymal transition by
regulating miR-367/141-ZEB2 axis in non-small-cell lung cancer.
Cancer Lett. 418:185–195. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sun W, Li AQ, Zhou P, Jiang YZ, Jin X, Liu
YR, Guo YJ, Yang WT, Shao ZM and Xu XE: DSCAM-AS1 regulates the
G1/S cell cycle transition and is an independent prognostic factor
of poor survival in luminal breast cancer patients treated with
endocrine therapy. Cancer Med. 7:6137–6146. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hu X, Ding D, Zhang J and Cui J: Knockdown
lncRNA HOTAIR sensitize breast cancer cells to ionizing radiation
through activating miR-218. Biosci Rep. 39:BSR201810382019.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Qiao E, Chen D, Li Q, Feng W, Yu X, Zhang
X, Xia L, Jin J and Yang H: Long noncoding RNA TALNEC2 plays an
oncogenic role in breast cancer by binding to EZH2 to target
p57(KIP2) and involving in p-p38 MAPK and NF-κB pathways. J Cell
Biochem. 120:3978–3988. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Smith BN and Bhowmick NA: Role of EMT in
metastasis and therapy resistance. J Clin Med. 5:E172016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xia XL, Xue D, Xiang TH, Xu HY, Song DK,
Cheng PG and Wang JQ: Overexpression of long non-coding RNA CRNDE
facilitates epithelial-mesenchymal transition and correlates with
poor prognosis in intrahepatic cholangiocarcinoma. Oncol Lett.
15:4105–4112. 2018.PubMed/NCBI
|