Introduction
Exercise is pivotal for the maintenance of physical
and mental wellbeing. Although essential, exercise often results in
stress, particularly in animals. The degree of stress is dependent
on the conditions of the exercise. During exercise, a racehorses
may experience different types of stress, including oxidative
(1), heat (2), hypoxic (3), hormonal (4) and glucose stress (5). As a result of these aforementioned
stress responses, levels of hormones become altered, including
cortisol (6), adrenaline (7) and noradrenaline (8). Among these hormones, the steroidal
hormone cortisol is the primary stress hormone produced by the
adrenal gland. During the onset of stress, the pituitary-adrenal
axis secretes corticotrophin-releasing hormone, which stimulates
the secretion of adrenocorticotropic hormone (ACTH). ACTH then in
turn stimulates the adrenal gland to secrete cortisol (9). Cortisol increases blood glucose levels
and suppresses the digestive system (10,11). In
addition, it affects the brain regions that control fear,
motivation and mood (12). Prolonged
secretion of cortisol may lead to physical and psychological
effects (13). Indeed, serious
mental issues associated with cortisol secretion include
exaggerated negative cognitions, increased feelings of anxiety and
helplessness in response to stress (14).
Prolonged secretion of cortisol occurs with
prolonged stress, and is associated with disease, including heart
disease, weight gain and depression (15). To reduce stress, the level of
cortisol should be controlled or optimized externally, such as
through the use of pharmacological agents that control cortisol
levels. The administration of natural compounds to optimize
cortisol levels in thoroughbred racehorses during stressful
conditions is a convenient approach, as natural compounds may have
fewer off-target effects than other pharmacological agents
(16). Among these natural
compounds, methylsulfonylmethane (MSM) is one such natural organic
sulphur-containing compound that is present in fruit, vegetables
and some beverages (17). MSM has
properties that make it a suitable drug candidate to alleviate
stress, as it has been previously shown to exhibit anti-cancer
(18), anti-inflammatory and
antioxidant activity (19). It has
also been demonstrated to inhibit ketosis in vitro by
regulating the STAT5B signalling cascade (20).
Several genes undergo changes in expression when a
racehorse encounters stress. The most stable reference genes for
the assessment of exercise-induced stress are succinate
dehydrogenase (SDH) complex subunit A (SDHA) and
hypoxanthine phosphoribosyl transferase 1 (HPRT1) (21). SDHA is a flavo-protein located in the
mitochondria, which is involved in the citric acid cycle and the
respiratory chain. It is closely associated with other subunits in
the SDH complex (SDHB, SDHAC, and SDHD). SDHA contains a flavin
adenine dinucleotide (FAD) cofactor-binding site and catalyzes the
transelimination of two hydrogen molecules from succinate to form
fumarate. In this reaction FAD accepts the two hydrogen molecules
and FAD is converted to FADH2 (22). In a previous study that implemented
knockdown of SDH complexes, SDHB knockdown resulted in increased
cytosolic oxidative stress, whereas SDHA knockdown did not,
indicating that SDHA is a stable reference gene under stress
conditions (23).
The HPRT1 enzyme converts hypoxanthine to inosine
monophosphate and guanine to guanosine monophosphate. It serves a
vital role in the production of purine nucleotides via the purine
salvage pathway. HPRT deficiency can lead to replication stress,
which may result in pathological consequences as a result of genome
instability due to inappropriate repair of chromosomal DNA
double-strand breaks (24) and
diseases such as Lesch-Nyhan disease in humans (25,26).
Although these two genes are conventionally considered as
housekeeping or reference genes for stress, the potential
regulation of their expression in racehorse skeletal muscle cells
remains unknown and has not been studied in the presence of
cortisol.
p53 is a tumour suppressor in multicellular
organisms (27). p53 expression is
upregulated in response to oxidative stress, as a result of an
increase in its half-life and stability (28). Oxidative stress activates p53 to
regulate the transcription of genes associated with stress
(29). Although p53 has been found
to transcriptionally regulate the expression of SDHA and
HPRT1 in mice (30,31), the relationship between p53 and the
expression of SDHA and HPRT1 in racehorse skeletal
muscle cells remains unknown.
In the present study, it was hypothesized that MSM
may inhibit cortisol-induced stress in the skeletal muscle cells of
thoroughbred racehorses by regulating the expression levels of
SDHA and HPRT1, by acting as a transcription factor
for these two genes.
Materials and methods
Antibodies and cell culture
reagents
Medium 199 was purchased from Gibco; Thermo Fisher
Scientific, Inc. Penicillin-streptomycin solution and FBS were
purchased from HyClone; GE Healthcare Life Sciences. Trypsin-EDTA
(0.05%) was obtained from Gibco; Thermo Fisher Scientific, Inc.
Antibodies specific for β-actin (cat. no. sc-47778) and horseradish
peroxidase-conjugated secondary antibodies [goat anti-mouse (cat.
no. sc-2005) and anti-rabbit (cat. no. sc-2004)] were obtained from
Santa Cruz Biotechnology, Inc. Anti-SDHA antibody (cat. no.
ab66484) was purchased from Abcam. The primary antibody against
HPRT1 (cat. no. LS-C81245) was purchased from LifeSpan BioSciences,
Inc. and the anti-p53 primary antibody (cat. no. ARP37163_T100) was
obtained from Aviva Systems Biology Corporation. Pifithrin-α
(PFT-α; P4359) was purchased from Sigma-Aldrich; Merck KGaA
(32).
Isolation of racehorse skeletal muscle
cells
A skeletal muscle tissue biopsy was performed on the
leg of a male neonatal thoroughbred racehorse to cultivate primary
horse skeletal muscle cells. The obtained muscle tissue was first
chopped into 1×1 mm sections, washed twice using PBS and
subsequently transferred to 15 ml tubes containing 2 ml
trypsin/EDTA for 18 h at 4°C. The trypsin was then discarded and
the tissue pieces were further incubated with residual trypsin at
37°C for 30 min, followed by the addition of 5 ml media containing
10% FBS. The resulting suspension was centrifuged for 3 min at 670
× g at room temperature to collect the cell pellet. After
centrifugation, a filter was used (200 µM; Cell Strainers; cat. no.
08-771-1; Falcon™; Thermo Fisher Scientific, Inc.) to completely
disperse any remaining tissues and to collect single cells. The
resulting supernatant was centrifuged further (3 min; 670 × g at
room temperature) before the cell pellet was collected and cultured
in media. This cell isolation protocol was as previously described
(33). The Pusan National
University-Institutional Animal Care and Use Committee approved the
study design (approval no. PNU-2015-0864).
Cell culture and treatment
Racehorse skeletal muscle cells were cultured in
Medium 199 supplemented with 10% FBS and 1% antibiotic-antimycotic
(ABAM; Invitrogen; Thermo Fisher Scientific, Inc.). Horse skeletal
muscle cell cultures were incubated in a humidified atmosphere with
5% CO2 at 37°C. For each experiment, at between 70 and
80% confluence, cells were gently washed twice with PBS and then
treated by adding MSM with fresh media. Unless specified otherwise,
cells were treated with 50 mM MSM for 24 h at 37°C.
Cell viability assay
Cell viability was assessed using an MTT assay
(Sigma Aldrich; Merck KGaA). Briefly, cells were resuspended in
Medium 199 and seeded into 24-well culture plates at a density of
1×104 cells/well, 1 day prior to drug treatment. The
next day, culture medium was replaced with fresh Medium 199
(vehicle control) or different concentrations of MSM (5–400 mM),
and the cells were incubated for a further 24 h at 37°C. MTT (5
mg/ml) was subsequently added into each well, and the culture
dishes were incubated at 37°C for 4 h. Formazan crystals in each
well were then dissolved using DMSO (Sigma Aldrich; Merck KGaA),
and the absorbance at 550 nm was measured using an Ultra
multifunctional microplate reader (Tecan Group, Ltd.). Cell
viability was determined from these readings using the calculation
% Viability=(fluorescence value of MSM/fluorescence value of
non-treated control) ×100. All measurements were performed in
triplicate, and experiments were repeated at least three times.
Western blotting
Whole cell lysates were prepared from untreated or
MSM-treated racehorse skeletal muscle cells by incubation with
radioimmunoprecipitation lysis buffer (EMD Millipore) containing
phosphatase and protease inhibitors on ice. Cells were disrupted by
aspiration through a 23-gauge needle with the resultant lysate
centrifuged at 18,300 × g for 10 min at 4°C to remove cellular
debris. Protein concentrations were measured using the Bradford
assay (Thermo Fisher Scientific, Inc.). Equal amounts of protein
(100 µg/lane) were separated on a 10% SDS-PAGE gel, followed by
transferal onto nitrocellulose membranes. The blots were then
blocked for 1 h at room temperature with 5% skim milk dissolved in
TBS buffer supplemented with 0.1X Tween-20 (TBS-T). The membranes
were then probed overnight at 4°C with the relevant primary
antibodies [anti-SDHA (1:500), anti-HPRT1 (1:500), anti-p53 (1:500)
and anti-β-actin (1:1,000)] diluted in 5% bovine serum albumin
(BSA; EMD Millipore) or skim milk (Difco™ skim milk; BD
Biosciences). Membranes were then washed with TBS-T and incubated
for 1 h at room temperature with HRP-conjugated secondary
antibodies (1:2,000). Detection was performed using an Enhanced
Chemiluminescence Plus detection kit (Amersham; GE Healthcare) and
imaged on an ImageQuant™ LAS 4000 imaging device (Fujifilm
Corporation). Blots were stripped using Restore™ Western Blot
Stripping Buffer (Thermo Fisher Scientific, Inc.). Densitometry
values were determined using FUJI FILM Multi Gauge version 3.1
(Fuji Photo Film Co., Ltd.).
Reverse transcription-semiquantitative
polymerase chain reaction (RT-sqPCR)
Total RNA was extracted with the RNeasy Mini kit
(Qiagen GmbH) according to the manufacturer's protocol. RNA was
quantified spectrophotometrically at 260 nm. Subsequently, RT-sqPCR
analyses were performed to detect SDHA, HPRT1 and
GAPDH RNA expression. Briefly, cDNA was synthesized from
total RNA at 42°C for 1 h, and at 95°C for 5 min using first-strand
cDNA synthesis kit (AccuPower® RT PreMix; cat. no.
K-2041; Bioneer Corporation) and oligo d(T) primers. The RT-PCR
Premix kit (AccuPower® PCR PreMix; cat. no. K-2016;
Bioneer Corporation) was used to amplify SDHA, HPRT1 and
GAPDH with primers synthesized by Bioneer Corporation. To
generate a 200-bp SDHA fragment, the following primers were
used: SDHA forward, 5′-CTACAAGGGGCAGGTTCTGA-3′ and reverse,
5′-TCTGCAATACTCAGGGCACA-3′. To generate a 290-bp HPRT1
fragment, the following primers were used: HPRT1 forward,
5′-TCTTTGCTGACCTGCTGGAT-3′ and reverse, 5′-GGGTCCTTTTCACCAGCAAG-3′.
To generate a 211-bp p53 fragment, the following primer pair
was used: p53 forward, 5′-AGGTTGGCTCTGACTGTACC-3′ and
reverse, 5′-TCCTCCTTCTTGCGGAAGTT-3′. Finally, a 320-bp GAPDH
mRNA fragment was generated using the following primer pair:
GAPDH forward, 5′-AAGGCCATCACCATCTTCCA-3′ and reverse,
5′-ACGATGCCAAAGTGGTCATG-3′ and an 18S mRNA fragment was
generated using the following primer pair: 18S forward,
5′-AGCCTTCGGCTGACTGGCTGG-3′ and reverse,
5′-CTGCCCATCATCATGACCTGG-3′. The thermocycling conditions were as
follows: Initial denaturation at 95°C for 10 min, followed by 31
cycles at 95°C for 45 sec, 58°C for 60 sec and 72°C for 60 sec,
followed by final extension at 72°C for 10 min. The PCR products
were resolved by electrophoresis on a 2% agarose gel, and were
visualized using ethidium bromide (cat. no. E7637; Sigma-Aldrich;
Merck KGaA) staining. Quantification was performed using FUJI FILM
Multi Gauge version 3.1 (Fuji Photo Film Co., Ltd.).
p53 binding motif analysis
The p53 binding motif was identified using Geneious
Prime software (Geneious; version R6.1; http://www.geneious.com). The sequences of SDHA
and HPRT1 were screened for the p53 binding motif (AGACAT).
The results obtained showed 4 binding motifs for p53 in the
SDHA sequence and 6 binding motifs for p53 in the
HPRT1 sequence. Primers were designed on the basis of these
sequences.
Chromatin immunoprecipitation (ChIP)
assay
The ChIP assay was performed using the
Imprint® chromatin immunoprecipitation kit
(Sigma-Aldrich; Merck KGaA) according to the manufacturer's
protocol. Briefly, racehorse skeletal muscle cells were fixed using
1% formaldehyde at room temperature for 10 min and quenched using
1.25 M glycine at room temperature. Samples were then mixed and
washed with ice-cold PBS by centrifugation at room temperature for
5 min at 180 × g. After washing, the cells were suspended in nuclei
preparation buffer and shearing buffer prior to their sonication
(30% amplitude for 30 sec followed by 30 Sec rest for 20 cycles) on
ice. The sheared DNA was subsequently centrifuged at 4°C for 5 min
at 180 × g and the cleared supernatant was used for protein/DNA
immunoprecipitation. The clarified supernatant was diluted with
buffer at a 1:1 ratio and 5-µl aliquots of the diluted samples were
used as internal controls. The diluted supernatant was then
incubated in 96 well plates pre-coated with 4 µg/µl anti-p53
antibody for 90 min at room temperature. The negative and positive
controls were incubated with 1 µg normal goat IgG and 1 µg anti-RNA
polymerase II (Sigma-Aldrich; Merck KGaA), respectively. The
unbound DNA was washed using immunoprecipitation wash buffer, and
the bound DNA was collected by applying the crosslink reversal
method, using DNA release buffer containing proteinase K. The
released DNA and DNA from the internal control were subsequently
purified using a GenElute™ Binding Column G (Sigma-Aldrich; Merck
KGaA), following which they were quantified using conventional PCR.
The thermocycling conditions for PCR were as follows: Initial
denaturation at 95°C for 10 min, followed by 35 cycles at 95°C for
40 sec, 58°C for 50 sec and 72°C for 50 sec, followed by final
extension at 72°C for 10 min. The PCR products were resolved by
electrophoresis on a 1.5% agarose gel, and were visualized using
ethidium bromide (cat. no. E7637; Sigma-Aldrich; Merck KGaA)
staining. Quantification was performed using FUJI FILM Multi Gauge
version 3.1 (Fuji Photo Film Co., Ltd.).
Statistical analyses
All experiments were performed at least three times.
Data are presented as the mean ± SEM. Statistical analyses were
conducted with one-way ANOVA and Student's t-test. They were
performed with Duncan's multiple range test as a post hoc test.
Analyses were performed using the SAS 9.3 program (SAS Institute,
Inc.). P<0.05 was considered to indicate a statistically
significant difference.
Results
Effect of cortisol and MSM on horse
skeletal muscle cell viability
A candidate drug for the treatment of stress should
be designed in such a way that it causes little or no toxicity in
racehorse skeletal muscle cells. Therefore, the effect of
increasing concentrations of cortisol and MSM on thoroughbred
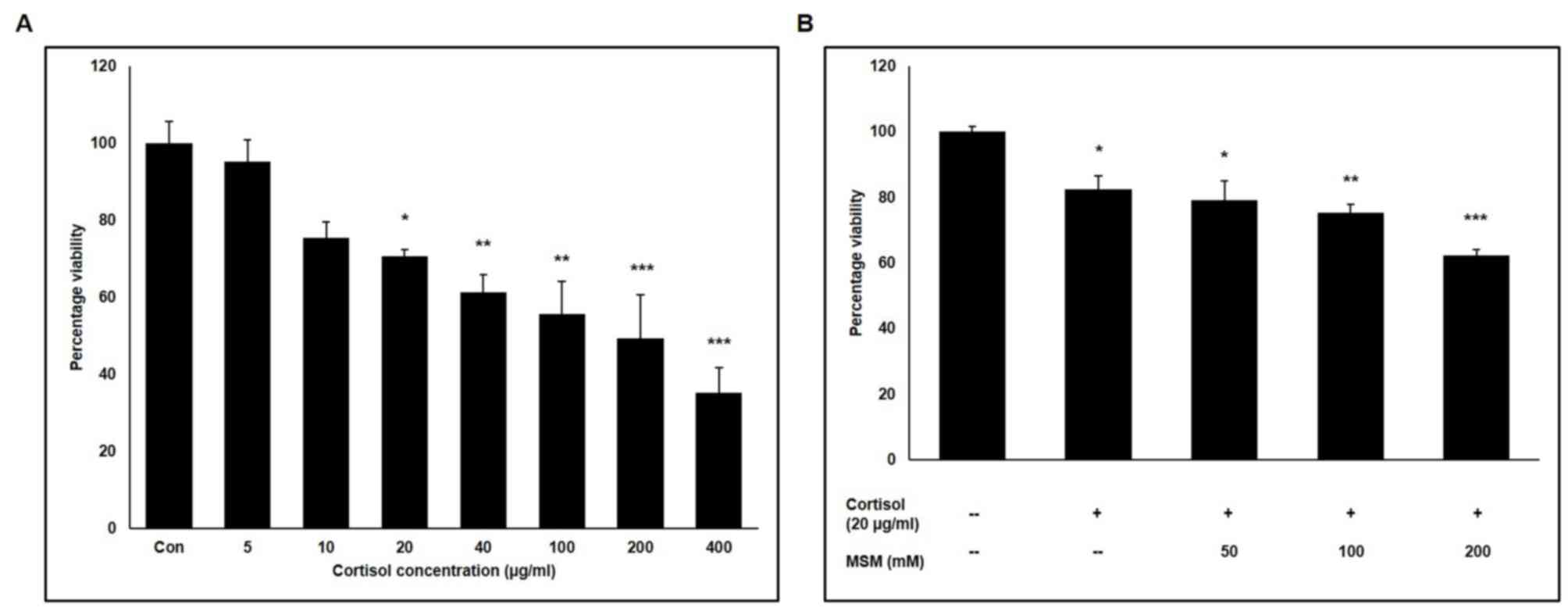
racehorse skeletal muscle cells was analysed using an MTT assay. It
was found that ~70% of the cells remained viable following
treatment with 20 µg/ml cortisol (Fig.
1A). Subsequently, the effect of three ascending concentrations
of MSM on horse skeletal muscle cell viability was checked in the
presence of 20 µg/ml cortisol. Combined with 20 µg/ml cortisol,
little difference was observed in cell viability between 50 mM MSM
treatment and no MSM treatment (Fig.
1B). Therefore, doses of 20 µg/ml cortisol and 50 mM MSM were
selected for subsequent experiments.
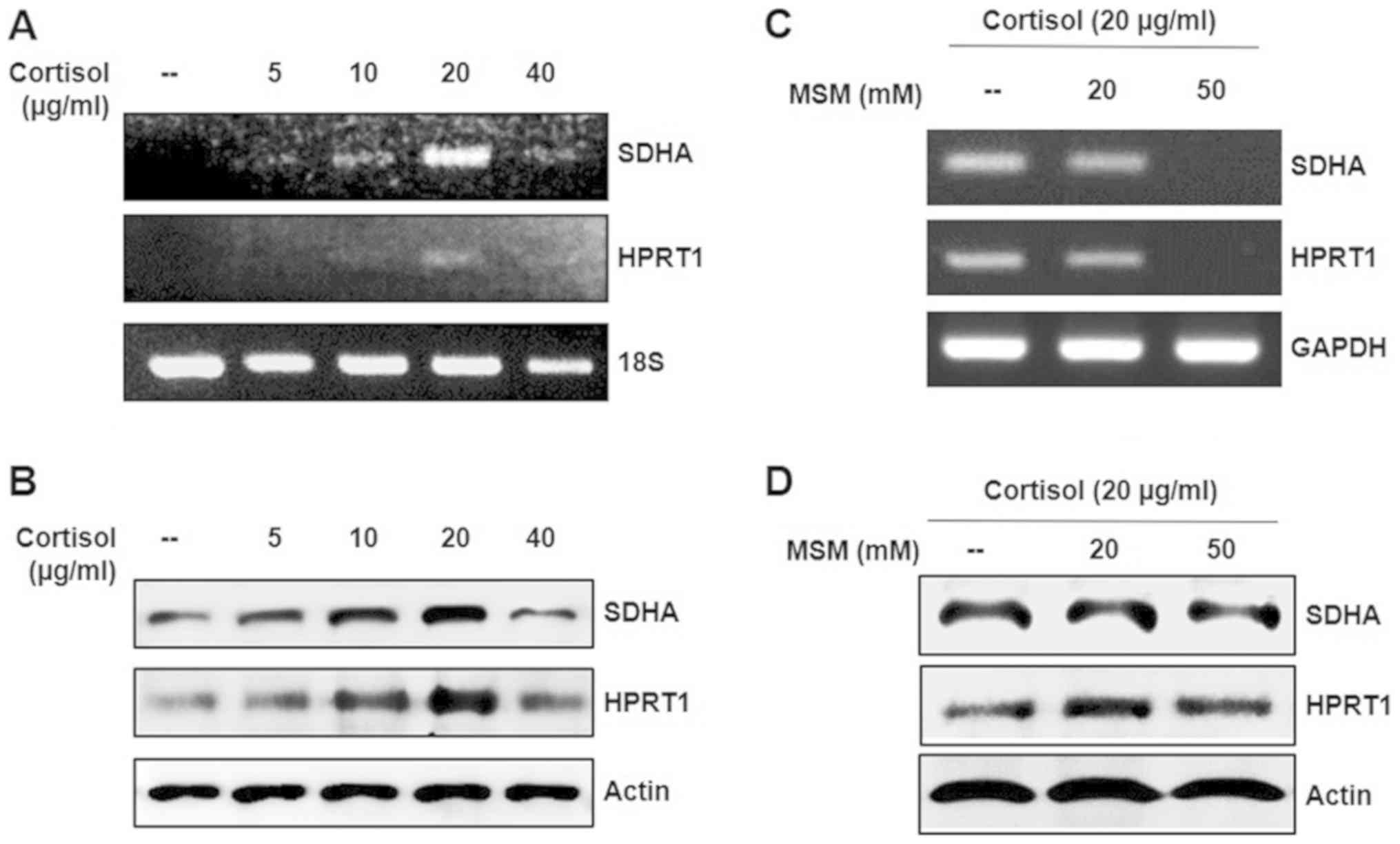
Cortisol-induced expression of SDHA
and HPRT1 is inhibited by MSM
It was hypothesized that the expression pattern of
SDHA and HPRT1 is a significant factor in
cortisol-induced stress. Therefore, in the present study, the
expression of SDHA and HPRT1 following treatment with
increasing concentrations of cortisol was analysed. The expression
levels of SDHA and HPRT1 were elevated in response to 20 µg/ml
cortisol for 24 h, at the mRNA and protein levels (Figs. 2A, B and S1). From this, it was hypothesized that
cortisol treatment induced stress in horse skeletal muscle cells.
To examine whether the impact of cortisol-induced stress, could be
minimized using MSM, increasing concentrations of MSM were applied
in conjunction with cortisol. Treatment of cells with 50 mM MSM or
higher reduced the expression of SDHA and HPRT1
(Figs. 2C, D and S1). MSM at 100 and 200 mM reduced the
expression of GAPDH, which may have been due to the
increased cytotoxicity caused by the synergistic effect of cortisol
and MSM (data not shown). These results suggested that MSM
inhibited cortisol-induced stress.
MSM inhibits cortisol-induced stress
by regulating SDHA/HPRT1 and p53 expression
RT-PCR was performed to assess the expression
patterns of SDHA, HPRT1 and p53 in cortisol-treated
and untreated (control) horse skeletal muscle cells (Fig. 3). MSM treatment appeared to reverse
cortisol-induced increases in SDHA, HPRT1 and p53
expression significantly (Fig. 3A and
B), an observation that was replicated at the protein level
(Fig. 3C and D). These results
suggested that p53 may serve an important role in MSM treatment of
cortisol-induced stress.
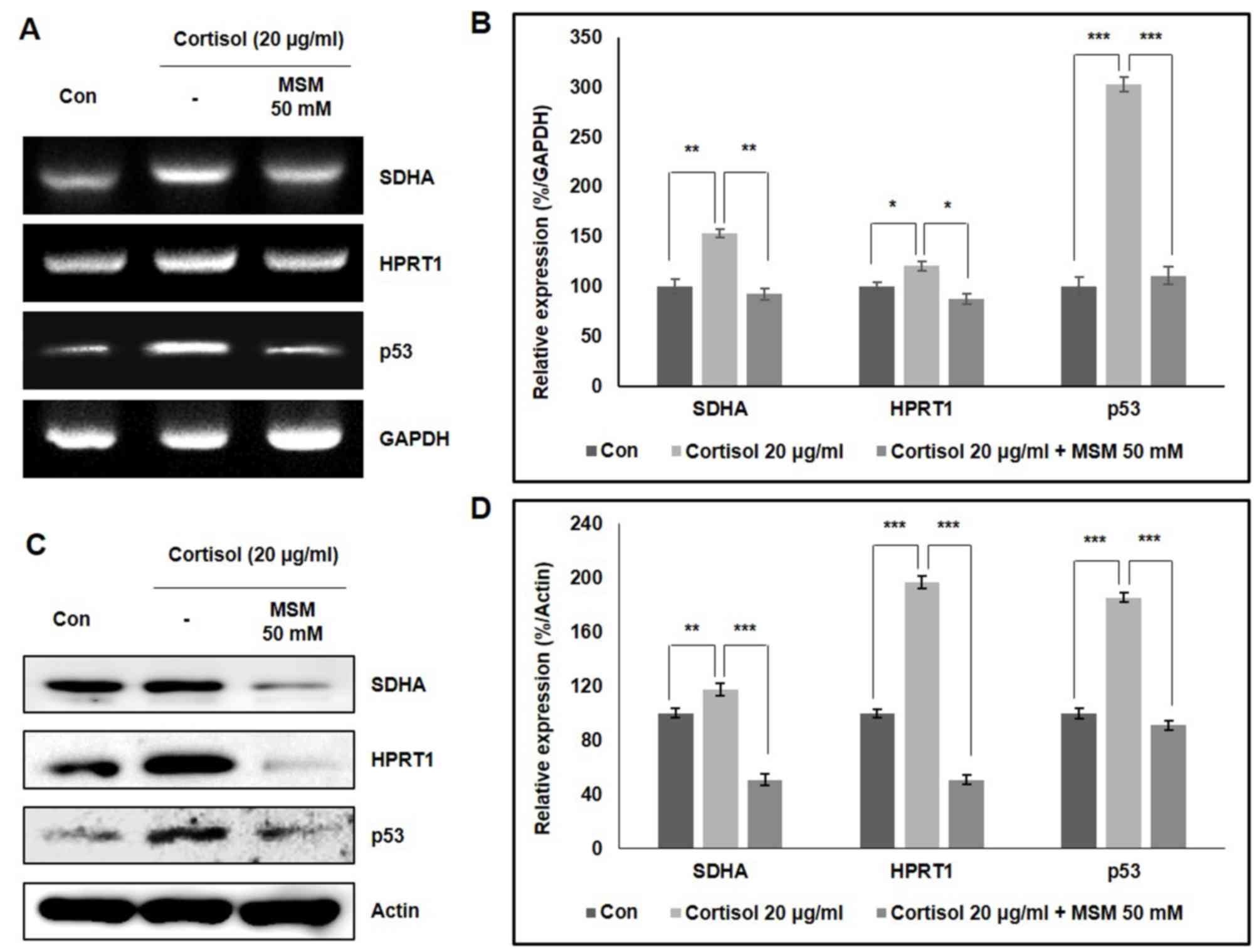 | Figure 3.MSM reverses cortisol-induced
increases in SDHA, HPRT1 and p53 expression levels. (A) RT-PCR
analysis of SDHA, HPRT1 and p53 gene expression in
horse skeletal muscle cells following 24 h treatment with 20 µg/ml
cortisol and 50 mM MSM. (B) Quantified densitometry data, with
comparisons performed using Student's t-test. (C) Western blot
analysis of horse skeletal muscle cells showing the expression
levels of SDHA, HPRT1 and p53 proteins after 24 h treatment with 20
µg/ml cortisol and 50 mM MSM, with respect to β-actin expression.
(D) Quantified densitometry data, with comparisons was performed
using Student's t-test. *P.0.05, **P<0.01 and ***P<0.001 vs.
respective control. *P.0.05, **P<0.01 and ***P<0.001 between
cortisol group and cortisol + MSM group. SDHA, succinate
dehydrogenase complex subunit A; HPRT1, hypoxanthine phosphoribosyl
transferase 1; RT-PCR, reverse transcription-PCR; MSM,
methylsulfonylmethane. |
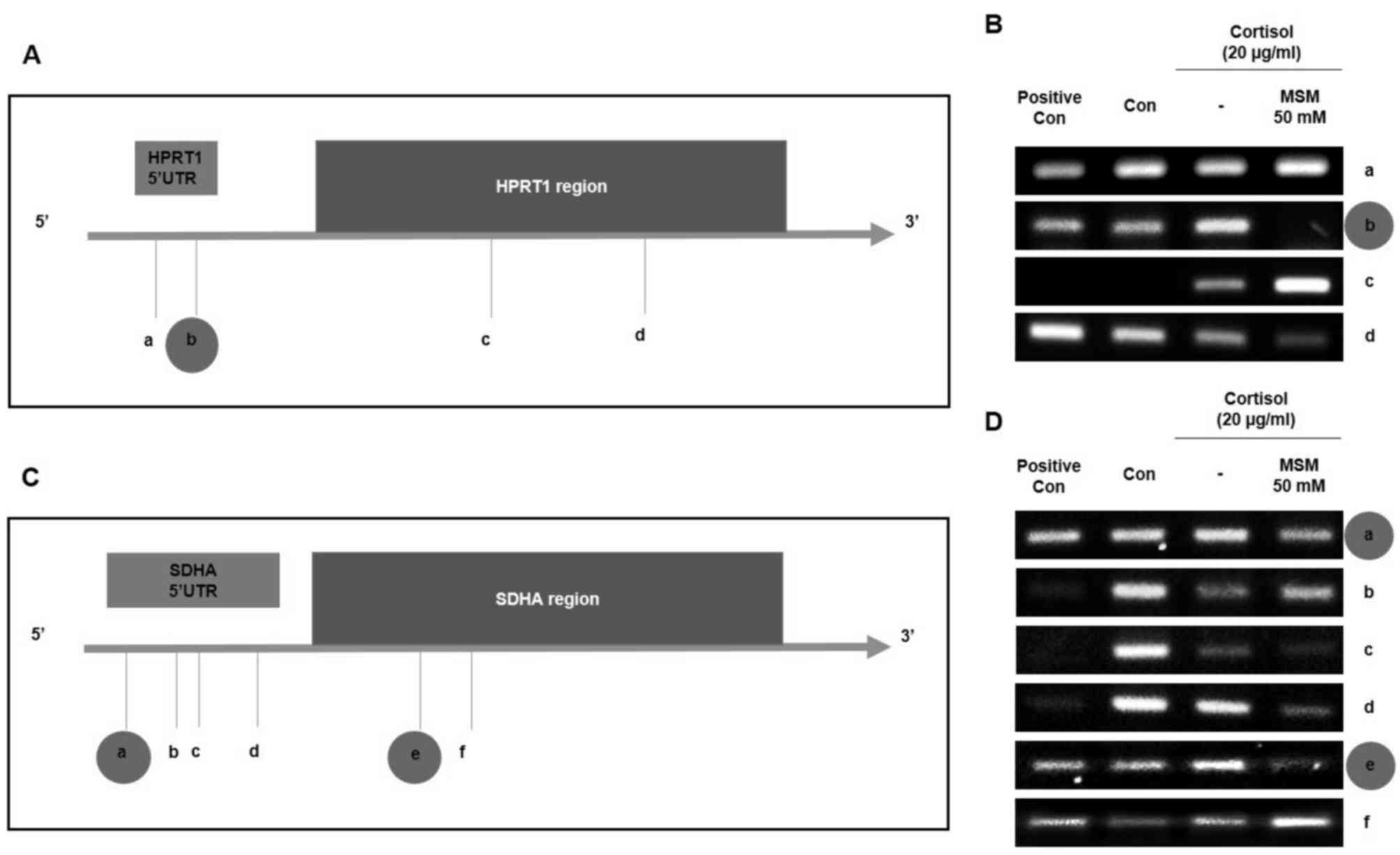
MSM inhibits the binding of p53 to the
SDHA and HPRT1 gene promoter regions
It was hypothesized that p53 serves an important
role in cortisol-induced stress through its interactions with SDHA
and HPRT1 at a post-translational level. Therefore, the ability of
p53 and MSM to transcriptionally regulate SDHA and
HPRT1 genes in horse skeletal muscle cells was investigated.
p53 binding motifs were discovered at four sites (labelled a-d) in
the HPRT1 gene promoter (Fig.
4A) and at six sites (labelled a-f) in the SDHA gene
promoter region using Geneious prime software (Geneious; Version
R6.1) (Fig. 4A and C). These binding
sites were confirmed using a ChIP assay. Positive p53 binding was
found at sites a, b and d in the HPRT1 sequence, whereas no
binding was observed at site c (Fig.
4B). MSM treatment acted on site b, where it inhibited
p53-induced HPRT1 expression (Fig. 4B). Strong positive p53 binding was
also found in the SDHA promoter sequence at sites a, b, c, d
and e, but a negative result was observed at site f (Fig. 4D). Here, it was also found that MSM
reversed cortisol-induced p53 binding to sites a and e in the
SDHA promoter. These results suggest that p53 serves a vital
role in the cortisol-induced expression of SDHA and
HPRT1, which is inhibited by MSM driven regulation of p53
expression.
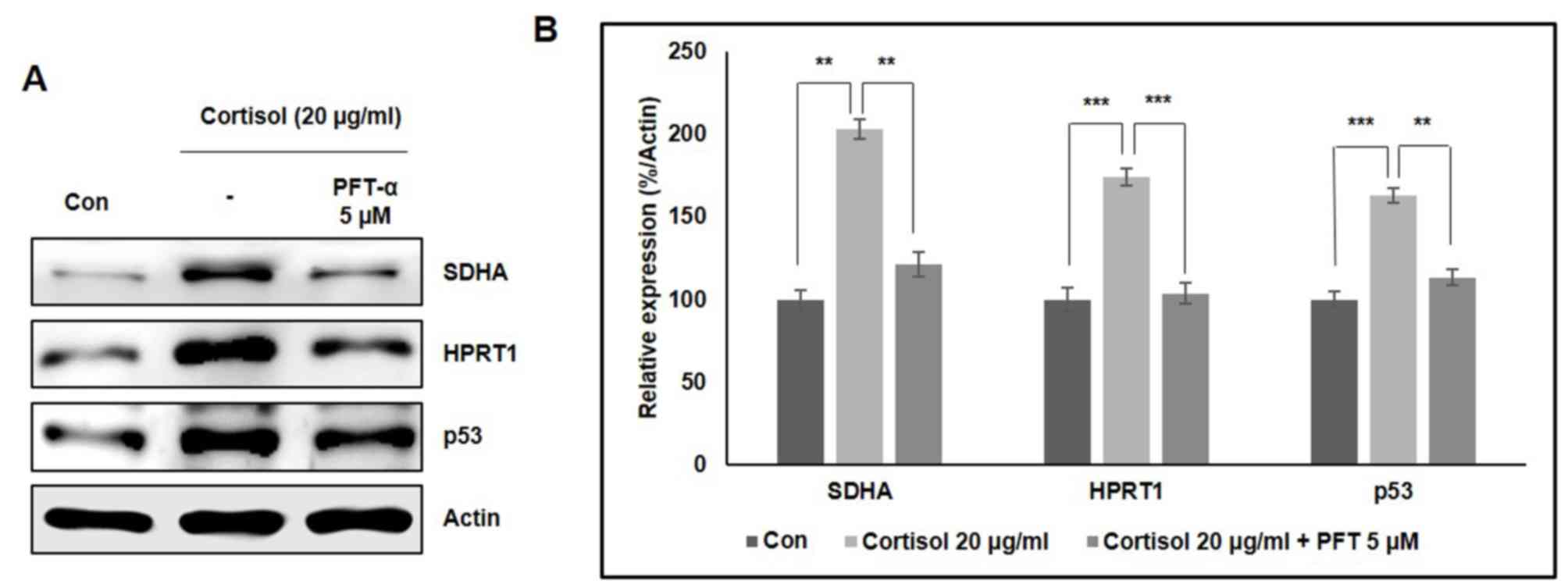
Role of p53 in the cortisol-induced
expression of SDHA and HPRT1
To confirm the relationship between p53 and
SDHA/HPRT1 expression in the presence of cortisol, western blotting
analysis was performed using, PFT-α, a p53 inhibitor. Significant
increases in SDHA and HPRT1 expression were observed in racehorse
skeletal muscle cells in response to 20 µg/ml cortisol, which was
significantly reversed with the concomitant addition of PFT-α
(Fig. 5A). Significantly elevated
p53 expression was also observed in cortisol-treated cells, which
was also reversed by PFT-α treatment (Fig. 5B). These results further support the
role of p53 in cortisol-induced increases in SDHA and HPRT1
expression.
Discussion
When a thoroughbred racehorse participates in a
race, cortisol levels are elevated, as a result of a variety of
factors. To control stress, cortisol levels should be optimized.
Treating the racehorses with drugs may affect performance, since
these drugs typically produce side effects. Therefore, treating
racehorses with natural remedies may provide a better alternative
for overcoming cortisol-induced stress in racehorses. In
particular, MSM is a natural organic sulphur-containing compound,
which has been previously reported to increase growth hormone
receptor expression (34) and bone
growth by regulating bone morphogenetic protein-2 expression
(35). In the present study, it was
demonstrated that 20 µg/ml cortisol induced ~30% cell death
according to the MTT assay, whilst enhancing the expression of SDHA
and HPRT in racehorse muscle cells. By contrast, 40 µg/ml cortisol
reduced the expression of 18S due to ~40% cell death. Therefore, 20
µg/ml cortisol was used for further studies. In the presence of 20
µg/ml cortisol, 50 mM MSM did not aggravate cell death, suggesting
that 50 mM MSM may be non-toxic to racehorse muscle cells. Based on
the findings in the present study, MSM may make a promising
candidate drug for controlling cortisol-induced stress.
The present study was, to the best of our knowledge,
the first to analyse stress in racehorses by culturing racehorse
skeletal muscle cells and inducing stress in vitro using
cortisol. To confirm stress induction, the expression levels of the
two most stable reference genes, SDHA and HPRT, were
analysed (21). These two genes
exhibited altered expression following cortisol treatment. Indeed,
SDHA is a key factor in oxidative stress (36), but, to the best of our knowledge, no
evidence exists that identifies a role for SDHA in cortisol-induced
stress conditions. Additional studies have reported that HPRT is
involved in cellular stress responses (37,38),
HPRT expression in response to stress in the presence of cortisol
remains unclear. Recently, Morgan et al (39) used SDHA and HPRT as
housekeeping genes when studying cortisol metabolism. In the
present study it was hypothesized that if treatment with cortisol
was able to alter the expression of SDHA and HPRT,
then it could be concluded that the cells were in a ‘stressed
state’, since these are often considered to be the most stable
reference genes (25). Results from
the present study showed that the expression levels of both SDHA
and HPRT1 were elevated in 20 µg/ml cortisol-treated cells. These
findings suggested that cortisol treatment induced stress in
thoroughbred racehorse skeletal muscle cells. It was also
hypothesized in the present study that MSM may reduce stress in the
same cell type, and the data showed that 50 mM MSM could reverse
the cortisol-induced elevation of SDHA and HPRT1 expression.
Many studies have provided evidence that p53 serves
an integral part in stress. As a result of cellular stress such as
oxidative stress, p53 becomes activated, which then takes part in
cell cycle arrest and apoptosis induction (28). The amount of stress is directly
proportional to the degree of p53 activation, which becomes
saturated in response to prolonged stress (40). Data from the present study showed
that the addition of cortisol induced the expression of p53, which
was inhibited or normalized to levels comparable to those of
control cells following additive MSM treatment. In addition, p53
expression was found to be directly proportional to the expression
of SDHA and HPRT1. Previous evidence has demonstrated that p53 acts
as a transcription factor for SDHA and HPRT in mice
(30). Therefore, to determine the
relationship between p53 and SDHA/HPRT in the horse
genome, the sequences of horse SDHA and HPRT1 genes
were analysed. A number of novel binding sites for p53 were found
in the promoter regions of the SDHA and HPRT1 genes.
Since these binding sites were new in the horse genome, to the best
of our knowledge, their function was validated by performing ChIP
assays to detect p53/SDHA and p53/HPRT1 binding complexes. Once p53
binds to the promoter region of these genes, it may promote the
transcription of the respective genes.
MSM can induce p53 independent apoptosis in cancer
(41). In the present study, it was
observed that MSM could inhibit or reverse the cortisol-induced
formation of p53/SDHA and p53/HPRT1 complexes. These results
suggested that MSM could be a candidate drug for treating
cortisol-induced stress in thoroughbred racehorse skeletal muscle
cells. In tumour cells, MSM induces p53 independent apoptosis, and
therefore controls tumour growth (41). However, in normal cells, p53 is
activated in response to cellular stress (42). Therefore, the potential anti-stress
activity of MSM was analysed by comparing cellular responses to MSM
treatment with that of a p53 inhibitor, PFT-α. PFT-α inhibited the
cortisol-induced regulation of SDHA and HPRT1
expression in a pattern similar to that observed with MSM,
suggesting that MSM acted as a p53 inhibitor in stressful
conditions. Taken together, these findings suggested that MSM may
serve as a candidate anti-stress drug for treating cortisol-induced
stress conditions.
In conclusion, the present study demonstrated that
MSM inhibited cortisol-induced stress in thoroughbred horse
skeletal muscle cells by regulating p53-mediated SDHA/HPRT1
expression. Novel binding sites for p53 in the SDHA and
HPRT1 gene promoter regions were also found in this cell
type. Therefore, MSM may be a candidate anti-stress drug for
treating stress in racing horses.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was carried out with the support of
‘Cooperative Research Program for Agriculture Science and
Technology Development (grant no. PJ01325702)’ Rural Development
Administration, Republic of Korea.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
NS and DYK conceived and designed the experiments,
performed the experiments and wrote the paper. YMY and KJJ
contributed in designing the experiments and data analysis. DHK,
HGL, YMP, IHK, HKL and BWC analyzed experiments and data along with
KJJ and YMY. All authors contributed to revising the manuscript and
approved the final version for publication.
Ethics approval and consent to
participate
The Pusan National University-Institutional Animal
Care and Use Committee approved the study design (approval no.
PNU-2015-0864).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Williams CA: The effect of oxidative
stress during exercise in the horse. J Anim Sci. 94:4067–4075.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Geor RJ and McCutcheon LJ: Hydration
effects on physiological strain of horses during exercise-heat
stress. J Appl Physiol (1985). 84:2042–2051. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Caillaud C, Connes P, Bouix D and Mercier
J: Does haemorheology explain the paradox of hypoxemia during
exercise in elite athletes or thoroughbred horses? Clin Hemorheol
Microcirc. 26:175–181. 2002.PubMed/NCBI
|
|
4
|
González O, González E, Sánchez C, Pinto
J, González I, Enríquez O, Martínez R, Filgueira G and White A:
Effect of exercise on erythrocyte beta-adrenergic receptors and
plasma concentrations of catecholamines and thyroid hormones in
thoroughbred horses. Equine Vet J. 30:72–78. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tadros EM, Frank N, De Witte FG and Boston
RC: Effects of intravenous lipopolysaccharide infusion on glucose
and insulin dynamics in horses with equine metabolic syndrome. Am J
Vet Res. 74:1020–1029. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Morris MC and Rao U: Cortisol response to
psychosocial stress during a depressive episode and remission.
Stress. 17:51–58. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Álvarez-Diduk R and Galano A: Adrenaline
and noradrenaline: Protectors against oxidative stress or molecular
targets? J Phys Chem B. 119:3479–3491. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Flint MS, Baum A, Episcopo B, Knickelbein
KZ, Liegey Dougall AJ, Chambers WH and Jenkins FJ: Chronic exposure
to stress hormones promotes transformation and tumorigenicity of
3T3 mouse fibroblasts. Stress. 16:114–121. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ranabir S and Reetu K: Stress and
hormones. Indian J Endocrinol Metab. 15:18–22. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Konturek PC, Brzozowski T and Konturek SJ:
Stress and the gut: Pathophysiology, clinical consequences,
diagnostic approach and treatment options. J Physiol Pharmacol.
62:591–599. 2011.PubMed/NCBI
|
|
11
|
Mosavat M, Ooi FK and Mohamed M: Stress
hormone and reproductive system in response to honey
supplementation combined with different jumping exercise
intensities in female rats. Biomed Res Int. 2014:1236402014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Steimer T: The biology of fear- and
anxiety-related behaviors. Dialogues Clin Neurosci. 4:231–249.
2002.PubMed/NCBI
|
|
13
|
McEwen BS: Central effects of stress
hormones in health and disease: Understanding the protective and
damaging effects of stress and stress mediators. Eur J Pharmacol.
583:174–185. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hannibal KE and Bishop MD: Chronic stress,
cortisol dysfunction, and pain: A psychoneuroendocrine rationale
for stress management in pain rehabilitation. Phys Ther.
94:1816–1825. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fioranelli M, Bottaccioli AG, Bottaccioli
F, Bianchi M, Rovesti M and Roccia MG: Stress and inflammation in
coronary artery disease: A review
Psychoneuroendocrineimmunology-Based. Front Immunol. 9:20312018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Butawan M, Benjamin RL and Bloomer RJ:
Methylsulfonylmethane: Applications and safety of a novel dietary
supplement. Nutrients. 9(pii): E2902017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
S P N, Darvin P, Yoo YB, Joung YH, Kang
DY, Kim DN, Hwang TS, Kim SY, Kim WS, Lee HK, et al: The
combination of methylsulfonylmethane and tamoxifen inhibits the
Jak2/STAT5b pathway and synergistically inhibits tumor growth and
metastasis in ER-positive breast cancer xenografts. BMC Cancer.
15:4742015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lim EJ, Hong DY, Park JH, Joung YH, Darvin
P, Kim SY, Na YM, Hwang TS, Ye SK, Moon ES, et al:
Methylsulfonylmethane suppresses breast cancer growth by
down-regulating STAT3 and STAT5b pathways. PLoS One. 7:e333612012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Joung YH, Darvin P, Kang DY, Sp N, Byun
HJ, Lee CH, Lee HK and Yang YM: Methylsulfonylmethane inhibits
RANKL-induced osteoclastogenesis in BMMs by suppressing NF-κB and
STAT3 activities. PLoS One. 11:e01598912016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Preetha NS, Kang DY, Darvin P, Kim DN,
Joung YH, Kim SY, Cho KY, Do CH, Park KD, Lee JH, et al: Induction
of in vitro ketosis condition and suppression using
methylsulfonylmethane by altering ANGPTL3 expression through STAT5b
signaling mechanism. Anim Cells Syst. 19:30–38. 2015. View Article : Google Scholar
|
|
21
|
Cappelli K, Felicetti M, Capomaccio S,
Spinsanti G, Silvestrelli M and Supplizi AV: Exercise induced
stress in horses: Selection of the most stable reference genes for
quantitative RT-PCR normalization. BMC Mol Biol. 9:492008.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cheng VW, Piragasam RS, Rothery RA,
Maklashina E, Cecchini G and Weiner JH: Redox state of flavin
adenine dinucleotide drives substrate binding and product release
in Escherichia coli succinate dehydrogenase. Biochemistry.
54:1043–1052. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guzy RD, Sharma B, Bell E, Chandel NS and
Schumacker PT: Loss of the SdhB, but Not the SdhA, subunit of
complex II triggers reactive oxygen species-dependent
hypoxia-inducible factor activation and tumorigenesis. Mol Cell
Biol. 28:718–731. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gravells P, Ahrabi S, Vangala RK, Tomita
K, Brash JT, Brustle LA, Chung C, Hong JM, Kaloudi A, Humphrey TC
and Porter AC: Use of the HPRT gene to study nuclease-induced DNA
double-strand break repair. Hum Mol Genet. 24:7097–7110.
2015.PubMed/NCBI
|
|
25
|
Ceballos-Picot I, Mockel L, Potier MC,
Dauphinot L, Shirley TL, Torero-Ibad R, Fuchs J and Jinnah HA:
Hypoxanthine-guanine phosphoribosyl transferase regulates early
developmental programming of dopamine neurons: Implications for
Lesch-Nyhan disease pathogenesis. Hum Mol Genet. 18:2317–2327.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kang TH, Park Y, Bader JS and Friedmann T:
The housekeeping gene hypoxanthine guanine
phosphoribosyltransferase (HPRT) regulates multiple developmental
and metabolic pathways of murine embryonic stem cell neuronal
differentiation. PLoS One. 8:e749672013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Surget S, Khoury MP and Bourdon JC:
Uncovering the role of p53 splice variants in human malignancy: A
clinical perspective. Onco Targets Ther. 7:57–68. 2013.PubMed/NCBI
|
|
28
|
Pflaum J, Schlosser S and Müller M: p53
family and cellular stress responses in cancer. Front Oncol.
4:2852014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Han ES, Muller FL, Perez VI, Qi W, Liang
H, Xi L, Fu C, Doyle E, Hickey M, Cornell J, et al: The in vivo
gene expression signature of oxidative stress. Physiol Genomics.
34:112–126. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Alston CL, van der Westhuizen FH, He L,
Wassmer E, Davison JE, Falkous G, McFarland R and Taylor RW: P53
Novel SDHA and SDHB mutations as a cause of isolated mitochondrial
complex II deficiency. Neuromuscular Disord. 22 (Suppl 1):S212012.
View Article : Google Scholar
|
|
31
|
Suzuki T, Kusunoki Y, Tsuyama N, Ohnishi
H, Seyama T and Kyoizumi S: Elevated in vivo frequencies of mutant
T cells with altered functional expression of the T-cell receptor
or hypoxanthine phosphoribosyltransferase genes in p53-deficient
mice. Mutat Res. 483:13–17. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang H, Chi Y, Gao K, Zhang X and Yao J:
p53 protein-mediated up-regulation of MAP kinase phosphatase 3
(MKP-3) contributes to the establishment of the cellular senescent
phenotype through dephosphorylation of extracellular
signal-regulated kinase 1/2 (ERK1/2). J Biol Chem. 290:1129–1140.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Maghsoudlou P, Ditchfield D, Klepacka DH,
Shangaris P, Urbani L, Loukogeorgakis SP, Eaton S and De Coppi P:
Isolation of esophageal stem cells with potential for therapy.
Pediatr Surg Int. 30:1249–1256. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Joung YH, Lim EJ, Darvin P, Chung SC, Jang
JW, Do Park K, Lee HK, Kim HS, Park T and Yang YM: MSM enhances GH
signaling via the Jak2/STAT5b pathway in osteoblast-like cells and
osteoblast differentiation through the activation of STAT5b in
MSCs. PLoS One. 7:e474772012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kim DN, Joung YH, Darvin P, Kang DY, Sp N,
Byun HJ, Cho KH, Park KD, Lee HK and Yang YM: Methylsulfonylmethane
enhances BMP2induced osteoblast differentiation in mesenchymal stem
cells. Mol Med Rep. 14:460–466. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Birch-Machin MA and Bowman A: Oxidative
stress and ageing. Br J Dermatol. 175 (Suppl 2):S26–S29. 2016.
View Article : Google Scholar
|
|
37
|
Walker DM, Patrick O'Neill J, Tyson FL and
Walker VE: The stress response resolution assay. I. Quantitative
assessment of environmental agent/condition effects on cellular
stress resolution outcomes in epithelium. Environ Mol Mutagen.
54:268–280. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Walker DM, Nicklas JA and Walker VE: The
stress response resolution assay. II. Quantitative assessment of
environmental agent/condition effects on cellular stress resolution
outcomes in epithelium. Environ Mol Mutagen. 54:281–293. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Morgan RA, Keen JA, Homer N, Nixon M,
McKinnon-Garvin AM, Moses-Williams JA, Davis SR, Hadoke PWF and
Walker BR: Dysregulation of cortisol metabolism in equine pituitary
pars intermedia dysfunction. Endocrinology. 159:3791–3800. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Devi GR, Alam MJ and Singh RK:
Synchronization in stress p53 network. Math Med Biol. 32:437–456.
2015.PubMed/NCBI
|
|
41
|
Karabay AZ, Koc A, Ozkan T, Hekmatshoar Y,
Sunguroglu A, Aktan F and Buyukbingol Z: Methylsulfonylmethane
induces p53 independent apoptosis in HCT-116 colon cancer cells.
Int J Mol Sci. 17(pii): E11232016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
James A, Wang Y, Raje H, Rosby R and
DiMario P: Nucleolar stress with and without p53. Nucleus.
5:402–426. 2014. View Article : Google Scholar : PubMed/NCBI
|