Introduction
Lung cancer is one of the most common and aggressive
cancer types and is associated with a high mortality rate. It was
reported to be a leading cause of cancer-associated death in the US
in 2017 (1), and in China, it was
the most frequent cause of cancer-associated mortality in males
aged ≥75 years in 2015 (2). While
numerous advancements have been made in the treatment of lung
cancer, therapy failure and low survival rates remain serious
issues (3,4). Thus, it is important to develop novel
treatments to eliminate lung cancer cells, which are resistant to
conventional therapies.
Lung cancer-initiating cells (also called lung
cancer stem cells) are considered to be the seeds of lung cancer;
they are self-renewing cancer cells with the ability to undergo
multipotential differentiation (5).
Treatment resistance and cancer metastasis frequently occur during
cancer therapy due to the presence of lung cancer-initiating cells.
Therefore, development of innovative treatments that target and
eliminate lung cancer-initiating cells is crucial for improving
outcomes for patients with lung cancer. CD133 is a marker for
identifying lung cancer-initiating cells (5,6).
Previous studies confirmed that CD133+ lung cancer cells
are able to generate tumorigenic spheres, which were able to
initiate tumor growth in mice (5,6). In
addition, CD133+ lung cancer cells have been indicated
to highly express a variety of stemness genes (5,6). A
previous study by our group also confirmed that CD133+
lung cancer cells expressed high levels of stemness genes and had
increased tumorigenicity in mice compared with CD133-lung cancer
cells, suggesting that CD133+ lung cancer cells
exhibited lung cancer-initiating cell properties (7). However, tumors may consist of multiple
genetically or phenotypically distinct types of cancer-initiating
cells. For instance, breast cancer and ovarian cancer contain
distinct populations of cancer-initiating cells that regenerate the
phenotype and heterogeneity of the initial tumor (8,9). In
addition to CD133, CD44 is also a specific marker for lung
cancer-initiating cells, and CD44+ lung cancer cells
also have certain properties of stem cells (10). Considering that tumors may consist of
heterogeneous populations of cancer-initiating cells, it was
hypothesized that it is imperative to target multiple subsets of
cancer-initiating cells to increase the cancer-therapeutic
efficacy.
Nanomedicines are characterized by controlled and
targeted delivery of drugs and may markedly increase the
therapeutic index of common chemotherapy drugs (11). Nanomicelles, a type of nanomedicine
prepared by self-assembly of various types of block copolymers,
possess several unique features, including simplicity and high
efficiency of drug loading (12,13).
Polyethylene glycol 2000-distearoyl phosphatidylethanolamine
(DSPE-PEG2000) nanomicelles are promising due to their particularly
small size (~20 nm), good biocompatibility and superior penetration
into solid tumors (14–16). In a previous study by our group,
CD133 aptamer-conjugated gefitinib DSPE-PEG2000 nanomicelles
(CD133-NM-Gef) were constructed to enhance the delivery of
gefitinib into lung tumors (7). In
the present study, to achieve simultaneous targeting of
nanomicelles to CD44+ and CD133+ cancer
cells, CD44 aptamers were further conjugated to CD133-NM-Gef to
develop CD133 and CD44 aptamer-conjugated nanomicelles loaded with
gefitinib (CD133/CD44-NM-Gef). After characterization of
CD133/CD44-NM-Gef, the in vitro targeting properties,
treatment efficacy and mechanism of action of CD133/CD44-NM-Gef
were investigated.
Materials and methods
Culture and passage of lung cancer
cells
Two human lung cancer cell lines, namely the H446
small cell lung cancer cell line and the A549 non-small cell lung
cancer cell line, were purchased from the American Type Culture
Collection. Cells were cultured in RPMI-1640 medium (Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum (Thermo
Fisher Scientific, Inc.) and antibiotics (100 µg/ml streptomycin
and 100 U/ml penicillin) at 37°C in 5% CO2/95% air. The
cell culture medium was replaced three times per week and cell
maintenance was performed by serial passage after
trypsinization.
Lipids, aptamers, antibodies,
cytokines and kits
The following lipids were purchased from Avanti
Polar Lipids:
1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-maleimide-
PEG-2000 (DSPE-PEG2000-Mal) for sulfhydryl conjugation,
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-carboxyfluorescein
(PECF) to label nanomicelles and 1,2-distearoyl
-sn-glycero-3-phosphoethanolamine-N- (methoxy PEG2000)
(DSPE-PEG2000). Thiolated CD133 aptamers with a sulfhydrylgroup at
the 5′-end (5′-SH-CCCUCCUACAUAGGG-3′) and thiolated CD44 aptamers
with a sulfhydryl group at the 5′-end
(5′-SH-GGGAUGGAUCCAAGCUUACUGGCAUCUGGAUUUGCGCGUGCCAGAAUAAAGAGUAUAACGUGUGAAUGGGAAGCUUCGAUAGGAAUUCGG-3′)
were synthesized and purchased from Ruibo Co., Ltd.
Phycoerythrin-labeled CD133 antibodies and Alexa Fluor®
488-labeled CD44 antibodies were purchased from R&D Systems,
Inc. The CD133 MicroBead Kits (cat. no. 130-100-857) and CD44
MicroBead Kits (cat. no. 130-095-194) used to isolate
CD133+ and CD44+ lung cancer cells were
purchased from Miltenyi Biotec. Dalian Meilun Biotech provided
gefitinib. Thermo Fisher Scientific, Inc. provided SuperScript III
reverse transcriptase and reagents for culturing lung
cancer-initiating cells, including human epidermal growth factor
[EGF; freeze-dried powder re-suspended in bovine serum albumin
(Thermo Fisher Scientific, Inc.)-containing buffer], human basic
fibroblast growth factor (bFGF freeze-dried powder, resuspended in
bovine serum albumin-containing buffer), B27 and
insulin-transferrin-selenium (ITS). Rat plasma was purchased from
Innovative Research, Inc.
Flow cytometry-based analysis of CD133
and CD44 expression and magnetic sorting-based separation
After lung cancer cells were cultured overnight, the
cells were trypsinized, washed and suspended in PBS. The cells were
then incubated with fluorescent antibody (phycoerythrin-labeled
CD133 antibodies; cat. no. FAB11331P-025; and Alexa
Fluor® 488-labeled CD44 antibodies; cat. no. FAB6127G;
R&D Systems, Inc.) at a final concentration of 1 µg/ml on ice
in a refrigerator. After 1 h, the cells were washed with PBS to
eliminate any unbound fluorescent antibody. Finally, the washed
cells were suspended in PBS for immediate analysis by
fluorescence-activated cell sorting (FACS) using a FACSCalibur (BD
Biosciences). CD133+ or CD44+ cells were
separated using a magnetic column included in the MicroBead kit
according to the manufacturer's protocol [CD133 MicroBeadkit (cat.
no. 130-100-857) and CD44 MicroBeadkits (cat. no. 130-095-194);
both Miltenyi Biotec). The cells were centrifuged and the
supernatant was removed. Beads were added and incubated with the
cells. Prior to sorting, the column was placed in a magnetic field
and rinsed, and the cells were then loaded onto the column. The
column was then added to another tube and marker-negative cells
were collected. Finally, the proportion of positively-stained cells
was analyzed as described above. The rat IgG2B Alexa
Fluor® 488-conjugated (cat. no. MAB0061; R&D
Systems, Inc.) or phycoerythrin-labeled isotype (cat. no. IC013P;
R&D Systems, Inc.) control antibodies with a dilution of 1:500
were used as the negative controls.
In vivo tumorf ormation analysis
The tumor formation assay was performed by
inoculating mice with increasing numbers of lung cancer cells.
BALB/c nude mice (total number, 240) were purchased from the
Shanghai Experimental Animal Center of Chinese Academy of Sciences.
All of the mice were 4–5 week-old males weighing ~20 g and housed
in a specific pathogen-free environment. All procedures were
performed in line with permission from, and within the guidelines
of the Animal Administrative Committee of the Naval Medical
University (Shanghai, China). The tumor formation assay was
performed as follows: Lung cancer cells were washed and
re-suspended in PBS. Aliquots of cells (2.5×103,
4×103, 1×104, 2×104,
2.5×105 and 2.5×106 cells, all suspended in
0.1 ml PBS) were completely mixed with BD Matrigel™ (0.1 ml). Then
the cell suspension (0.2 ml) was implanted subcutaneously into the
right flank of the mice. After implantation, tumor formation in the
animals was observed for 15 weeks. Animal health and behavior were
monitored once every 3 days during the 15-week period. The mice
were euthanized by carbon dioxide inhalation with a flow rate of 2
l/min after 15 weeks or when the tumor volume exceeded 1,500
mm3. The flow rate of carbon dioxide used for euthanasia
of rodents did not displace >30% of the chamber volume/min.
Heartbeat and respiration were checked to verify the death of the
mice.
Analysis of tumorsphere formation by
lung cancer cells
A tumorsphere is a spherical and solid structure,
which is derived from a single cancer stem cell. The tumorsphere
formation assay was performed in specific serum-free cell culture
medium to assess the self-renewal capability of cancer-initiating
cells. In brief, lung cancer cells were cultured overnight and
washed to eliminate any residues of medium. The cells were
trypsinized, washed and cultured in serum-free cell medium composed
of Dulbecco's modified Eagle's medium/F12 (Thermo Fisher
Scientific, Inc.), 1X B27, 1X ITS, 0.4% bovine serum albumin, 20
ng/mlEGF and 20 ng/ml bFGF. The cells were then cultured in
Corning® ultra-low adherent 6-well dishes (Corning,
Inc.) at a density of 5,000 cells/well. The cells were cultured for
one week and the number of tumorspheres was calculated by direct
observation under a conventional microscope. To obtain
second-passage tumorspheres, first-passage tumorspheres were washed
and dissociated into single cells using the cell dissociation
reagent StemPro®Accutase® (Thermo Fisher
Scientific, Inc.). Subsequently, the cells were propagated to form
second-passage tumorspheres.
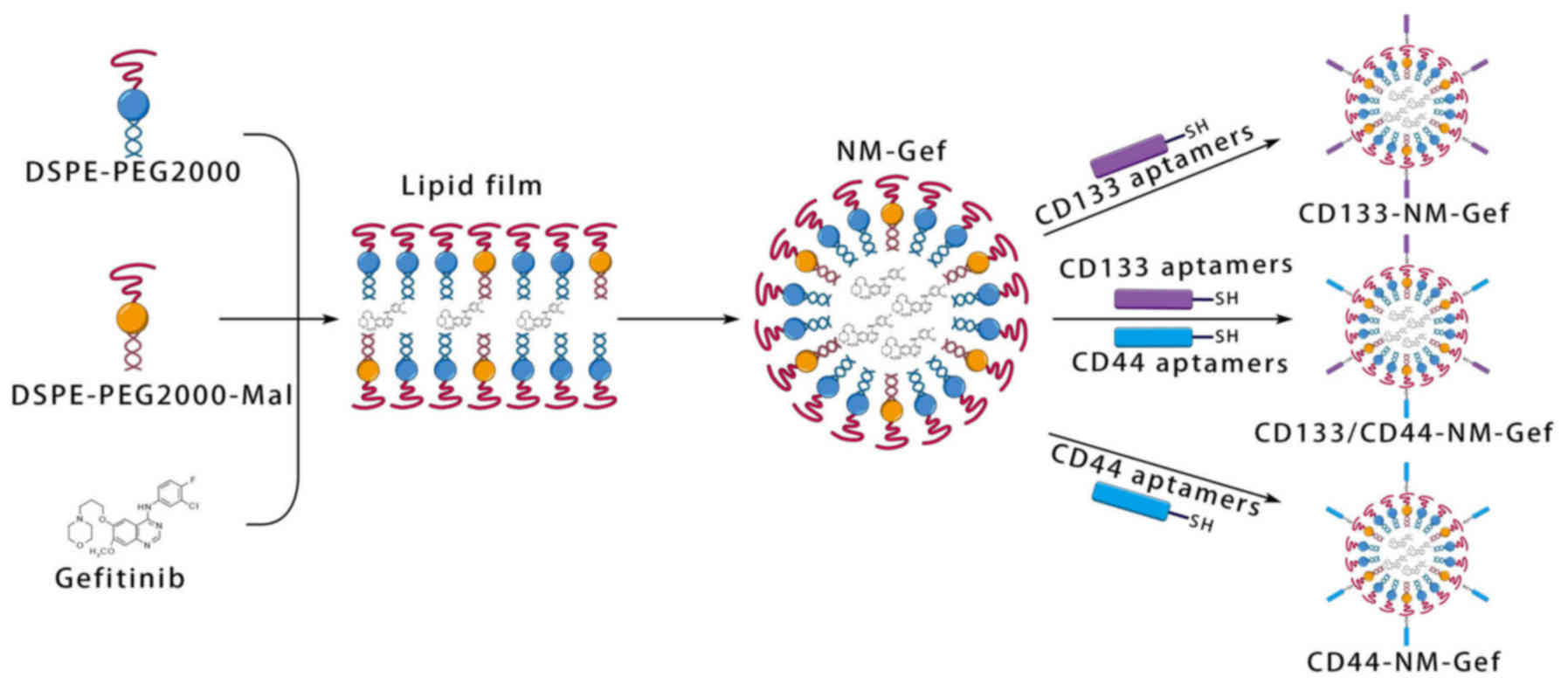
Construction of nanomicelles loaded
withgefitinib (NM-Gef)
Nanomicelles loaded with gefitinib were constructed
using a lipid film-based approach as follows: A total of 2 mg
gefitinib and 10 mg of a lipid mixture composed of 8 mg
DSPE-PEG2000 and 2 mg DSPE-PEG2000-Mal were dissolved in
chloroform. To generate fluorescent nanomicelles, 1% PECF (mass
ratio) was added to the lipid mixture. Subsequently, the lipid
solution was aspirated and added to a flask, which was then placed
on a vacuum rotary evaporator. The lipid mixture was dried
completely to obtain a lipid film. The lipid film was then
rehydrated by addition of 4 ml PBS. Subsequently, a hand-held
extruder (Avanti Polar Lipids) with polycarbonate membranes (100
and 50 nm) was used to obtain small and homogeneous nanomicelles.
After preparation of nanomicelles, the following aptamers were
incubated with the nanomicelles: CD133 aptamer (0.1 mg), CD44
aptamer (0.1 mg) or CD133 aptamer (0.1 mg) combined with CD44
aptamer (0.1 mg). The mixture was stirred for 4 h. After the
reaction, unconjugated aptamers were removed by ultrafiltration
using centrifugal filter units (Amicon® Ultra-4, 50 kDa
nominal molecular weight limit; EMD Millipore). The following
nanomicelles were produced: NM-Gef, CD133-NM-Gef, CD44-NM-Gef,
CD133/CD44-NM-Gef and CD133/CD44-NM.
Evaluation of aptamer conjugation
efficiency of nanomicelles
After preparation of aptamer-conjugated
nanomicelles, the concentration of unconjugated aptamers was
measured at 260 nm using an ultraviolet/visible light
spectrophotometer. The aptamer conjugation efficiency was
calculated using the following formula: Efficiency =
(Qt-Qu)/Qt, where Qt is
the quantity of total added aptamers and QU the quantity
of unconjugated aptamers.
Nanomicelle characteristics
After dilution of 100 µl nanomicelles in distilled
water (1.9 ml), the diluted samples were placed in sample cells in
a Zetasizer Nano ZS90 (Malvern Instruments) to measure the size and
zeta potential of the nanomicelles according to a standard
protocol. AJEM2100F high-resolution transmission electron
microscope (TEM; JEOL Ltd.) was used to visualize the detailed
morphology of the phosphotungstic acid-stained nanomicelles.
Reverse-phase high-performance liquid chromatography
(HPLC; Agilent 1200L; Agilent Technologies Inc.) was used to
determine gefitinib drug loading. In brief, 1 ml of nanomicelle
solution was placed in a vacuum oven and completely dried. The
dried nanomicelles were dissolved in1 ml methanol for HPLC
analysis. A reverse-phase C18 column (Diamonsil®; 250 mm
× 4.6 mm, 5 µm particle size; Dikma Technologies, Inc.) was used.
The mobile phase consisted of methanol and 0.02 M dipotassium
hydrogen orthophosphate 10:90 (v/v) and was maintained at a flow
rate of 1.0 ml/min. The detection wavelength of gefitinib was 246
nm. A gefitinib calibration curve was used to determine the
gefitinib concentration in the nanomicelles. The drug encapsulation
efficacy was determined as the amount of loaded drug divided by the
amount of total added drug. The drug loading was calculated as the
mass of loaded drug divided by the amount of nanomicelles. A PECF
calibration curve was used to determine the PECF concentration in
the nanomicelles with BioTek Synergy 4 multimode reader (excitation
495 nm/emission 525 nm; BioTek; Agilent Technologies, Inc.).
Gefitinib release fromn
anomicelles
Gefitinib release from nanomicelles was measured in
PBS or PBS with 10% rat plasma. The release assay was performed as
follows: After the preparation of 5 ml of nanomicelles solution, it
was added to a molecular weight cut-off 1,000
Spectra/Por® dialysis membrane (Repligen Corp.). The
membrane was sealed and placed in a vessel containing 500 ml
release medium. This vessel was placed in a 37°C water bath with
gentle stirring at 90 × g using a magnetic stir bar. An aliquot of
0.5 ml dialysate was used for analysis and was replaced with 0.5 ml
fresh medium. The amount of gefitinib released was analyzed as
described above. The following formula was used to determine
gefitinib release: Release rate = (Mi/Mt)
×100%, where Mi represents the amount of released
gefitinib and Mt indicates the total amount of
gefitinib.
Flow cytometric analysis of targeting
of fluorescent nanomicelles
The targeting of nanomicelles to lung cancer cells
was evaluated by flow cytometry. In brief, lung cancer cells were
cultured overnight in 10-cm culture plates. The cells were then
washed and dissociated into single cells using trypsin. After
trypsinization and washing, the cells were cultured at a density of
5×105 cells per well on 12-well cell culture plates
overnight. After overnight incubation, the old medium was aspirated
and replaced with fresh medium. Subsequently, 0.01 ml PECF-labeled
nanomicelles (0.5 µg/ml PECF) were prepared and incubated with the
cells (5×105 cells; 1 ml) at 37°C. After 2 h, the lung
cancer cells were trypsinized into single cells and a FACSCalibur
flow cytometer was used to analyze the fluorescence of the lung
cancer cells.
Effect of nanomicelles on the
proliferation of lung cancer cells
Cells in the logarithmic growth phase were seeded in
96-well plates at 3×103 cells/well. The plates were
incubated overnight at 37°C. After incubation, different
concentrations of gefitinib and nanomicelles were added to the
wells. The experimental groups were as follows: Gefitinib, NM-Gef,
CD133-NM-Gef, CD44-NM-Gef, CD133/CD44-NM-Gef and CD133/CD44-NM.
After adding the drug to the wells, the plates were returned to the
incubator and cultured for 72 h. Subsequently, the plates were
removed from the incubator and 10 µl Cell Counting Kit (CCK)-8
solution was added to each of the wells, taking care not to
introduce any bubbles. The plates were returned to the incubator
for another 2 h and the absorbance at 450 nm was then measured
using a microplate reader (Multiskan MK3; Thermo Fisher Scientific,
Inc.). Data were processed with GraphPad Prism 5.0 (GraphPad
Software, Inc.) to calculate IC50 values.
Tumorsphere assay and flow
cytometry-based analysis of the percentages of lung
cancer-initiating cells
Tumorsphere assay and flow cytometry-based analysis
of the percentages of lung cancer-initiating cells was performed as
follows: Lung cancer cell lines containing heterogeneous
populations of cells in the logarithmic growth phase were
inoculated overnight in 12-well cell culture plates. The cell
density was adjusted to 1×105 cells per well and the
cells were treated for 24 h with nanomicelles (5 µg/ml equivalent
gefitinib concentration) or 5 µg/ml gefitinib dissolved in fresh
medium. The groups in this experiment were as follows: NM-Gef,
CD133-NM-Gef, CD44-NM-Gef, CD133/CD44-NM-Gef and CD133/CD44-NM. The
treatment was terminated by aspiration of the old cell culture
medium. Subsequently, the cells were washed with PBS and fresh
medium was added. After 72 h, the cells were enzymatically
dissociated into single cells and tumorsphere formation was
assessed as described above. The cells were cultured in serum-free
cell medium in Corning® ultra-low adherence 6-well
dishes at a density of 5,000 cells/well. The images of tumorspheres
were captured with an EuromexbScope Trinocular Phase Contrast
Microscope (Euromex Microscopen bv). Flow cytometry was used to
analyze the percentage of CD133+ or CD44+
cells as described above.
Reverse transcription-quantitative
(RT-q) PCR
TRIzol reagent (Takara Bio, Inc.) was applied to
extract RNA using a standard approach. SuperScript III reverse
transcriptase (Thermo Fisher Scientific, Inc.) was used to
synthesize first-strand complementary DNA according to the
manufacturer's protocol. ASYBR Green PCR kit (cat. no. RR420L;
Takara Bio, Inc.) was used to quantify specific transcripts using
LightCycler® 1.5 (Roche Applied Science). The following
thermocycling conditions were used: Initial denaturation at 95°C
for 10 sec; 40 cycles of 95°C for 5 sec and 60°C for 20 sec. The
mRNA expression levels, which were normalized against β-actin, were
calculated using the 2−ΔΔCq method, as described
previously (17). The sequences of
the PCR primers were as follows: β-actin forward,
5′-CGTGGACATCCGTAAAGACC-3′ and reverse, 5′-ACATCTGCTGGAAGGTGGAC-3′;
CD133 forward, 5′-TCAATTTTGGATTCATATGCCTT-3′ and reverse,
5′-ACTCCCATAAAGCTGGACCC-3′; CD44 forward,
5′-TTACAGCCTCAGCAGAGCAC-3′ and CD44 reverse,
5′-AAGGACACACCCAAGCAAGG-3′.
Statistical analysis
GraphPad Prism 5.0 (GraphPad Software, Inc., USA)
was used for data analysis. Values are expressed as the mean ±
standard deviation. Differences between two groups were determined
using anunpaired Student's t-test. Differences among three or more
groups were determined using one-way analysis of variance with the
Newman-Keuls post hoc test. P<0.05 was considered to indicate
statistical significance.
Results
Construction, characterization and
drug release properties of nanomicelles
Gefitinib-loaded nanomicelles were constructed by
lipid film formation, hydration and aptamer conjugation (Fig. 1). Nanomicelles were formed after
hydration of the lipid films. To conjugate aptamers to the
nanomicelles, the maleimide groups in the nanomicelles were reacted
with the sulfhydryl groups in the aptamers. The properties of the
nanomicelles are summarized in Fig.
2. TEM indicated that all nanomicelles were spherical in shape
and uniformly distributed in size (Fig.
2A). As indicated in Fig. 2B,
the nanomicelles in each groupwere~20 nm in size with a
polydispersity index (PDI) of <0.2. The small size and PDI of
the nanomicelles indicated that the nanomicelles prepared were
homogeneously distributed. The drug loading of the nanomicelles in
each group was 7–9% and the encapsulation efficiency was ~80%,
suggesting the nanomicelles efficiently encapsulated gefitinib.
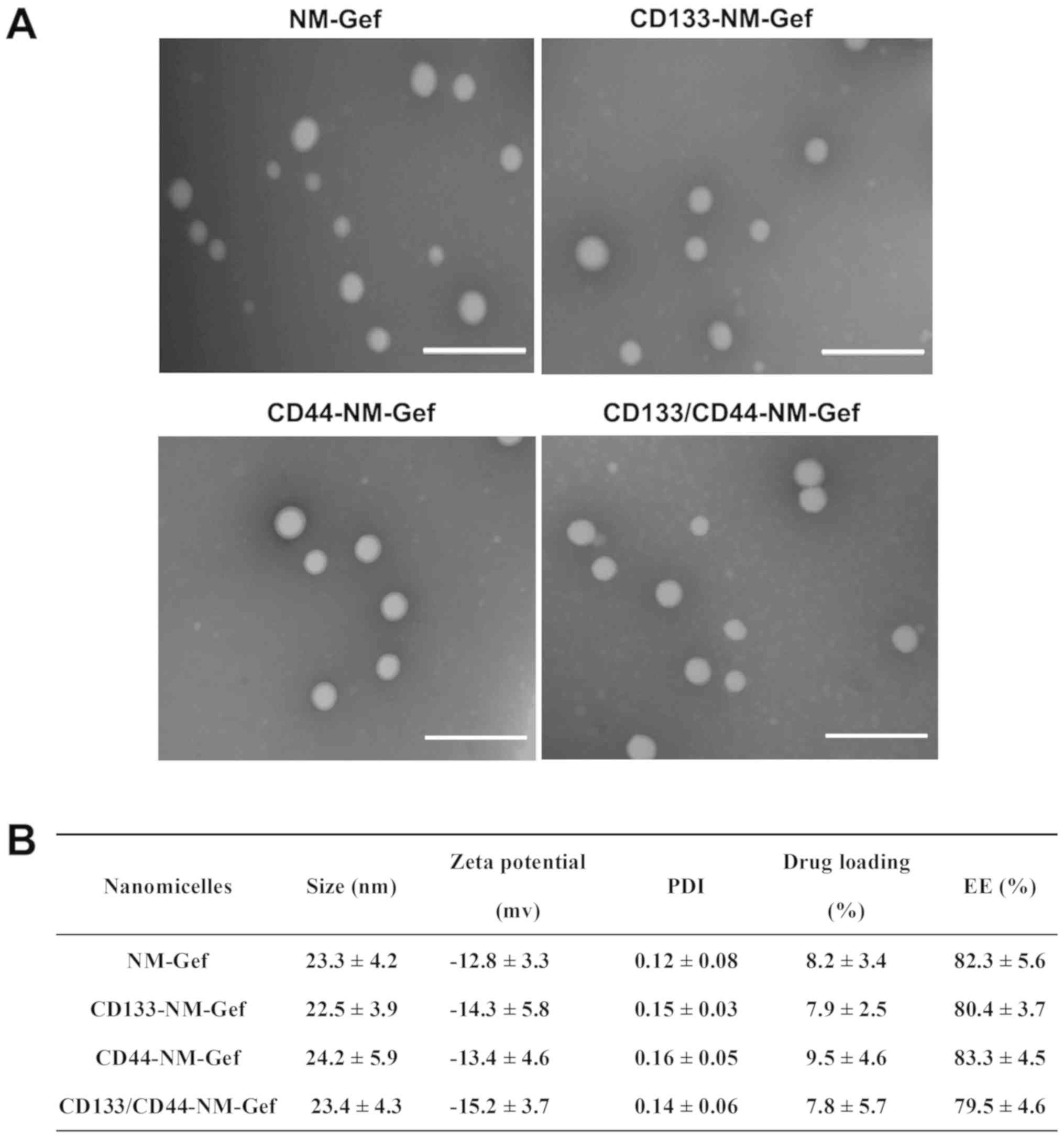 | Figure 2.Characteristics of various
nanomicelles. (A) TEM images of nanomicelles that were negatively
stained by PTA. After staining with PTA, the nanomicelles were
visualized using TEM at room temperature (scale bars, 100 nm). (B)
Size, zeta potential, PDI, drug loading and EE of nanomicelles.
Values are expressed as the mean ± standard deviation (n=3). PTA,
phosphotungstic acid; TEM, transmission electron microscopy; EE,
encapsulation efficiency; PDI, polydispersity index; CD133-NM-Gef,
CD133 aptamer-conjugated gefitinib
1,2-distearoyl-sn-glycero-3-phosphoethanolamine-polyethylene
glycol-2000 nanomicelles. |
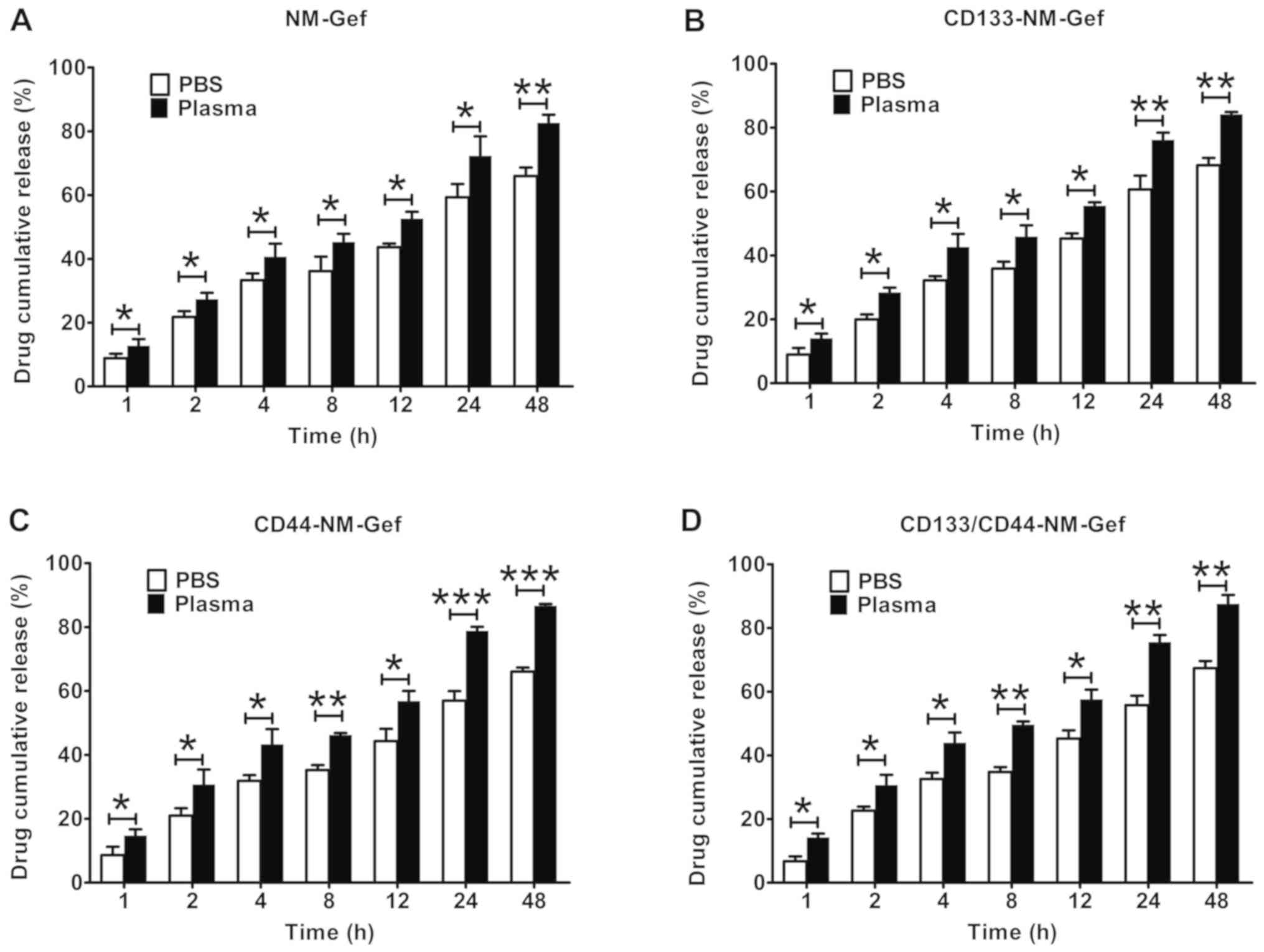
All of the nanomicelles exhibited sustained drug
release over a period of 48 h (Fig.
3). Of note, in comparison to gefitinib release in PBS,
gefitinib release from the nanomicelles was markedly higher in PBS
with plasma (P<0.05). These results may indicate that plasma had
the ability to destabilize the structure of the nanomicelles,
facilitating the release of gefitinib.
Identification of lung
cancer-initiating cells by tumorsphere formation and tumorigenicity
assay in mice
After isolation of lung cancer-initiating cells with
the MicroBead Kit, the purity of CD133+ or
CD44+ cells was determined to be >95%. As indicated
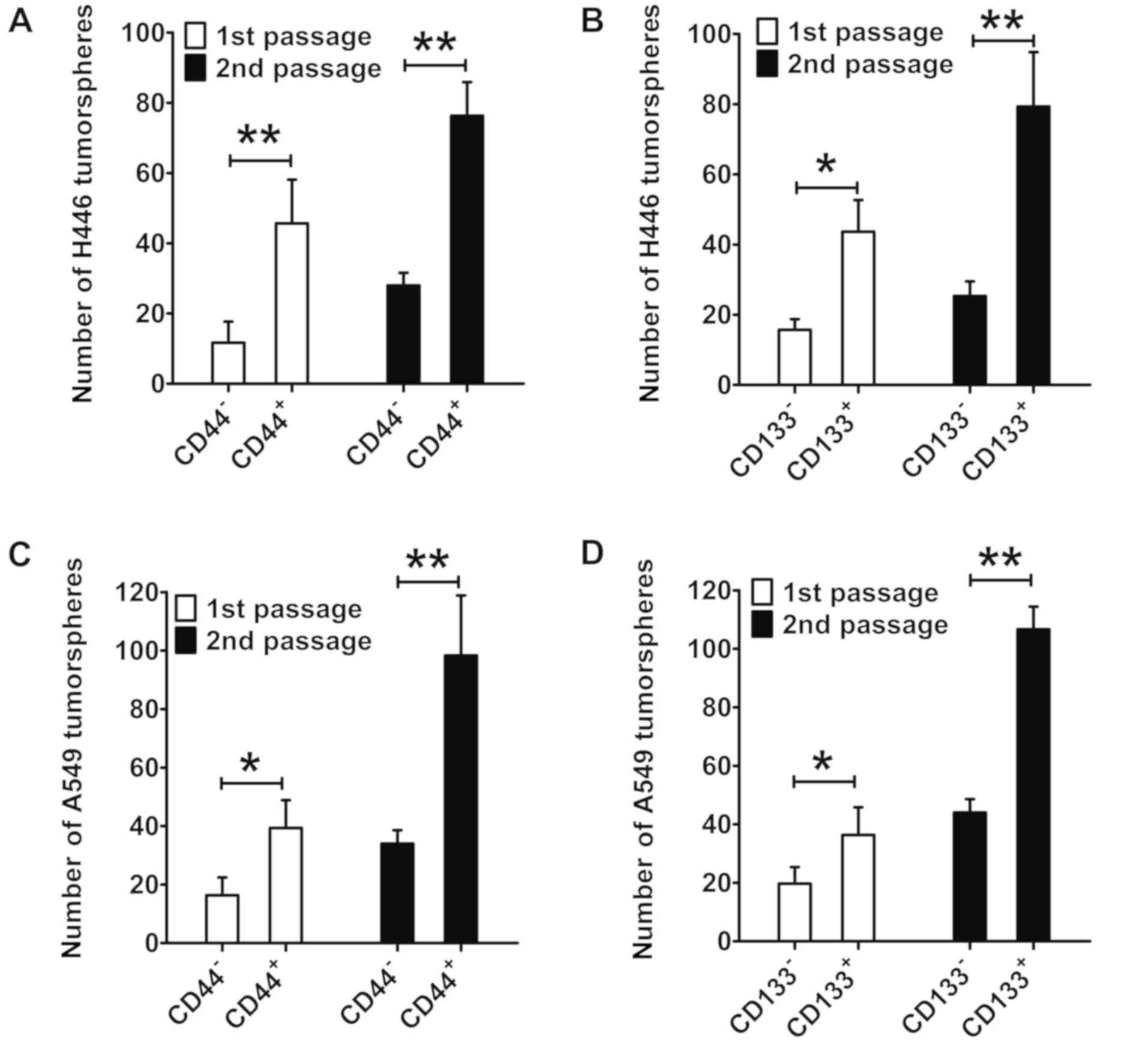
in Fig. 4A, the number of
first-passage and second-passage tumorspheres of CD44+
H446 cells was markedly greater than that of CD44− H446
cells (P<0.01). In addition, the number of first-passage and
second-passage tumorspheres of CD133+ H446 cells was
markedly greater than that of CD133− H446 cells
(P<0.05; Fig. 4B). For A549
cells, the number of tumorspheres of CD44+ cells was
also markedly greater than that of CD44- cells (first-passage
tumorspheres: P<0.05; second-passage tumorspheres: P<0.01;
Fig. 4C). In summary, the capacity
of tumorsphere formation of CD133+ and CD44+
lung cancer cells was markedly greater than that of
CD133− and CD44− lung cancer cells.
As presented in (Table
I), in the in vivo tumor formation experiment,
inoculation of ≥2×104 CD133+ H446 cells had a
tumor formation rate of 100%. By contrast, for CD133−
H446 cells, inoculation of 2×104 cells hada tumor
formation rate of onl y10%. Furthermore, CD133− H446
cells induced tumor formation at a rate of 60% when
2.5×106 cells were inoculated. Similar results were
obtained for CD133+ and CD133− A549 cells:
When the number of cells inoculated was ≥2×104,
CD133+ A549 cells had a tumor formation rate of 100%. By
contrast, for CD133− A549 cells, 2×104 cells
induced tumor formation in 30% of animals. Even at
2.5×106 cells, CD133− H446 cells were only
able to achieve a tumor formation rate of 60%. Similarly, the
tumorigenicity of CD44+ lung cancer cells was greater
than that of CD44− lung cancer cells. In the present
study, each animal had only one tumor and did not present with
multiple tumors.
 | Table I.The in vivo tumorigenic assay
of isolated lung cancer cells based on different markers. |
Table I.
The in vivo tumorigenic assay
of isolated lung cancer cells based on different markers.
|
| Implanted cell
number |
|---|
|
|
|
|---|
| Type |
2.5×106 |
2.5×105 |
2×104 |
1×104 |
4×103 |
2.5×103 |
|---|
| CD133−
H466 | 6/10 | 2/10 | 1/10 | 0/10 | 0/10 | 0/10 |
| CD133+
H466 | 10/10 | 10/10 | 10/10 | 8/10 | 6/10 | 5/10 |
| CD133−
A549 | 6/10 | 3/10 | 3/10 | 1/10 | 0/10 | 0/10 |
| CD133+
A549 | 10/10 | 10/10 | 10/10 | 9/10 | 7/10 | 3/10 |
| CD44−
H466 | 5/10 | 3/10 | 2/10 | 1/10 | 0/10 | 0/10 |
| CD44+
H466 | 10/10 | 9/10 | 9/10 | 7/10 | 5/10 | 4/10 |
| CD44−
A549 | 7/10 | 3/10 | 3/10 | 2/10 | 1/10 | 0/10 |
| CD44+
A549 | 10/10 | 10/10 | 8/10 | 7/10 | 6/10 | 3/10 |
The tumorsphere assays and tumorigenicity
experiments in mice confirmed that CD133+ and
CD44+ cells had characteristics of lung
cancer-initiating cells and exhibited a greater tumorigenic ability
than CD133− and CD44− cells.
Flow cytometry-based analysis of
targeting of fluorescent nanomicelles in vitro
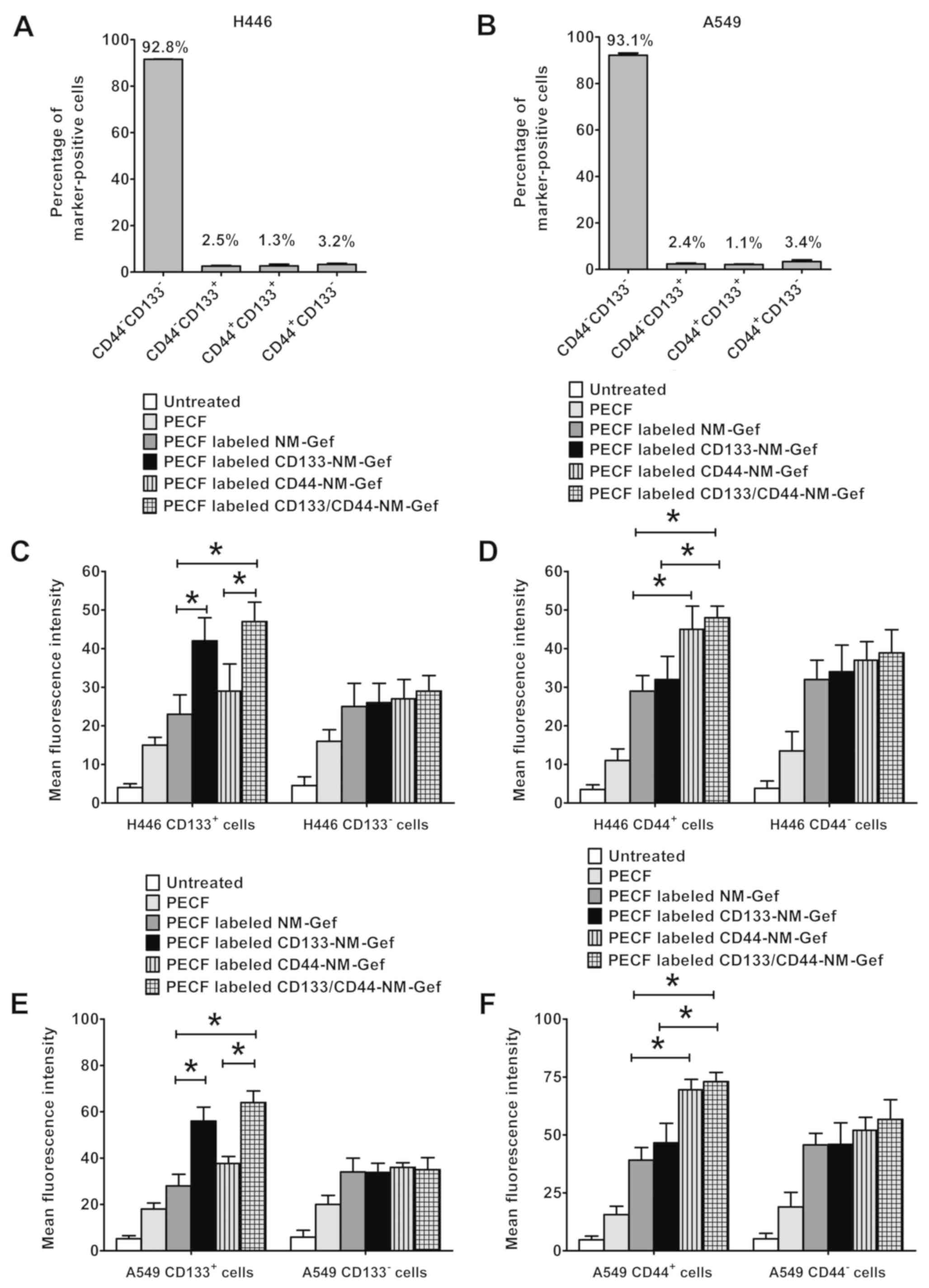
Flow cytometry was used to assess CD133 and CD44
expression in H446 and A549 cells (Fig.
5A and B). In H446 cells, CD44-CD133− cells
accounted for >90% of all cells, whereas the percentages of
CD44-CD133+, CD44+CD133+ and
CD44+CD133− cells were <4%. Similar
results were obtained for A549 cells, with percentages of
CD44−CD133−, CD44-CD133+,
CD44+CD133+ and
CD44+CD133− cells of 93.1, 2.4, 1.1 and 3.4%,
respectively.
Targeting of nanomicelles to lung cancer cells was
then evaluated and the results were presented in Fig. 5C-F. PECF-labeled CD133-NM-Gef
demonstrated increased targeting to H446 CD133+ cells as
compared with PECF-labeled NM-Gef (P<0.05; Fig. 5C). Furthermore, PECF-labeled
CD133/CD44-NM-Gef exhibited increased targeting to H446
CD133+ cells as compared with PECF-labeled NM-Gef and
CD44-NM-Gef (P<0.05). However, the targeting efficiency was not
markedly different between PECF-labeled NM-Gef, CD133-NM-Gef,
CD44-NM-Gef and CD133/CD44-NM-Gef in H446 CD133− cells.
In H446 CD44+ (Fig. 5D),
PECF-labeled CD44-NM-Gef demonstrated increased targeting to H446
CD44+ cells as compared with PECF-labeled NM-Gef
(P<0.05). Furthermore, PECF-labeled CD133/CD44-NM-Gef exhibited
increased targeting to H446 CD44+ cells as compared with
PECF-labeled NM-Gef and CD133-NM-Gef (P<0.05). However, the
targeting efficiency was not markedly different between
PECF-labeled NM-Gef, CD133-NM-Gef, CD44-NM-Gef and
CD133/CD44-NM-Gef in H446 CD44- cells.
In A549 cells, targeting with nanomicelles was
similar to that in H446 cells (Fig. 5E
and F). PECF-labeled CD133-NM-Gef exhibited greater targeting
to CD133+A549 cells than PECF-labeled NM-Gef
(P<0.05), and PECF-labeled CD133/CD44-NM-Gef had a greater
targeting efficiency toward CD133+A549 cells than
PECF-labeled NM-Gef and CD44-NM-Gef (P<0.05; Fig. 5E). However, in A549 CD133−
cells, the targeting efficiency did not differ significantly among
PECF-labeled NM-Gef, CD133-NM-Gef, CD44-NM-Gef and
CD133/CD44-NM-Gef. In CD44+ A549 cells, PECF-labeled
CD44-NM-Gef exhibited a greater targeting efficiency than
PECF-labeled NM-Gef (P<0.05), and PECF-labeled CD133/CD44-NM-Gef
had a greater targeting efficiency than PECF-labeled NM-Gef and
CD133-NM-Gef (P<0.05). However, in CD44-A549 cells, the
targeting efficiency did not differ significantly between
PECF-labeled NM-Gef, CD133-NM-Gef, CD44-NM-Gef and
CD133/CD44-NM-Gef (Fig. 5F).
Effect of nanomicelles on lung cancer
cell proliferation
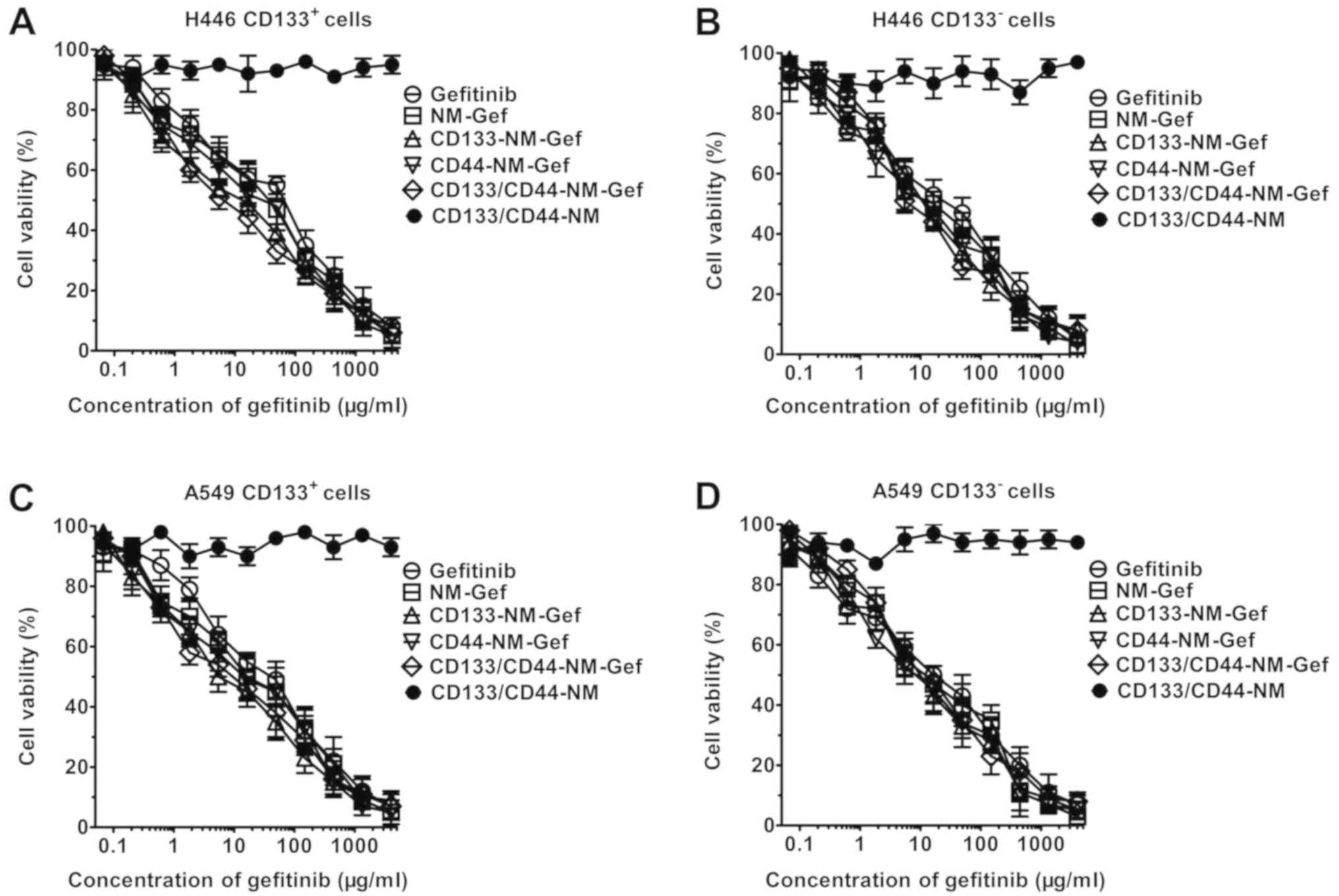
As indicated in Fig.
6, CD133/CD44-NM did not exhibit any toxic effect on lung
cancer cells, as evidenced by a zero-slope survival curve. By
contrast, gefitinib and gefitinib-loaded nanomicelles produced
dose-dependent cytotoxicity toward lung cancer cells. The
IC50 values for gefitinib and the nanomicelles are
listed in Tables II and III. In H446 CD133+ cells,
NM-Gef exhibited greater cytotoxicity than gefitinib (33.2±6.4 vs.
73.2±22.5 µg/ml). In addition, CD133/CD44-NM-Gef (6.3±4.3 µg/ml)
and CD133-NM-Gef (7.9±4.4 µg/ml) were similarly cytotoxic and were
significantly more cytotoxic than NM-Gef (33.2±6.4 µg/ml) and
CD44-NM-Gef (31.3±8.5 µg/ml; P<0.01). By contrast, the
IC50 values of NM-Gef, CD133-NM-Gef, CD44-NM-Gef, and
CD133/CD44-NM-Gef did not differ markedly in H446 CD133−
cells, whereas their IC50 was lower than gefitinib. In
A549 CD133+ cells, CD133/CD44-NM-Gef (5.3±2.8 µg/ml)
exerted a greater cytotoxic effect than gefitinib (32.8±8.1 µg/ml),
NM-Gef (16.3±7.5 µg/ml) and CD44-NM-Gef (15.4±4.3µg/ml; P<0.01).
By contrast, in A549 CD133− cells, the cytotoxic effect
of CD133/CD44-NM-Gef (14.3±6.4 µg/ml) was similar to that of
gefitinib (15.4±4.3/ml), NM-Gef (12.6±6.4 µg/ml) and CD44-NM-Gef
(16.6 µg/ml). Similar results were obtained in CD44+ and
CD44- lung cancer cells (Table II).
In summary, CD133/CD44-NM-Gef exerts the most potent cytotoxic
effects towards CD133+ and CD44+ lung cancer
cells, indicating that CD133/CD44-NM-Gef exhibited dual targeting
to CD133+ and CD44+ lung cancer cells. In the
future, the cytotoxic effect of gefitinib and nanomicelles on
double-positive cells will be investigated.
 | Table II.IC50 values (µg/ml) of
gefitinib and nanomicelles in lung cancer cells by CD133 expression
status at 72 h. |
Table II.
IC50 values (µg/ml) of
gefitinib and nanomicelles in lung cancer cells by CD133 expression
status at 72 h.
|
| H446 | A549 |
|---|
|
|
|
|
|---|
| Treatment |
CD133+ |
CD133− |
CD133+ |
CD133− |
|---|
| Gefitinib | 73.2±22.5 | 38.2±6.5 | 32.8±8.1 | 15.4±4.3 |
| NM-Gef |
33.2±6.4a | 18.5±8.4 |
16.3±7.5a | 12.6±6.4 |
| CD133-NM-Gef |
7.9±4.4c | 16.5±5.8 |
3.6±2.8c | 12.1±4.7 |
| CD44-NM-Gef | 31.3±8.5 | 19.5±8.7 | 15.4±8.4 | 16.6±7.5 |
|
CD133/CD44-NM-Gef |
6.3±4.3b,c | 19.3±6.6 |
5.3±2.8b,c | 14.3±6.4 |
| CD133/CD44-NM | >4,000 | >4,000 | >4,000 | >4,000 |
 | Table III.IC50 values (µg/ml) of
gefitinib and nanomicelles in lung cancer cells by CD44 expression
status at 72 h. |
Table III.
IC50 values (µg/ml) of
gefitinib and nanomicelles in lung cancer cells by CD44 expression
status at 72 h.
|
| H446 | A549 |
|---|
|
|
|
|
|---|
| Treatment |
CD44+ |
CD44− |
CD44+ |
CD44−− |
|---|
| Gefitinib | 87.3±15.4 | 33.3±8.4 | 44.5±9.5 | 15.2±7.7 |
| NM-Gef |
43.2±8.1a | 19.4±9.5 |
21.5±8.5a | 13.3±8.3 |
| CD133-NM-Gef |
38.5±11.4c | 22.4±6.6 |
24.4±9.2c | 19.1±4.3 |
| CD44-NM-Gef | 6.3±4.3 | 23.5±8.4 | 7.6±4.4 | 18.8±6.5 |
|
CD133/CD44-NM-Gef |
5.5±3.5b | 26.4±5.7 |
4.1±2.9b | 21.7±9.5 |
| CD133/CD44-NM | >4,000 | >4,000 | >4,000 | >4,000 |
Percentage of lung cancer-initiating
cells as determined by tumorsphere assay and flow cytometry
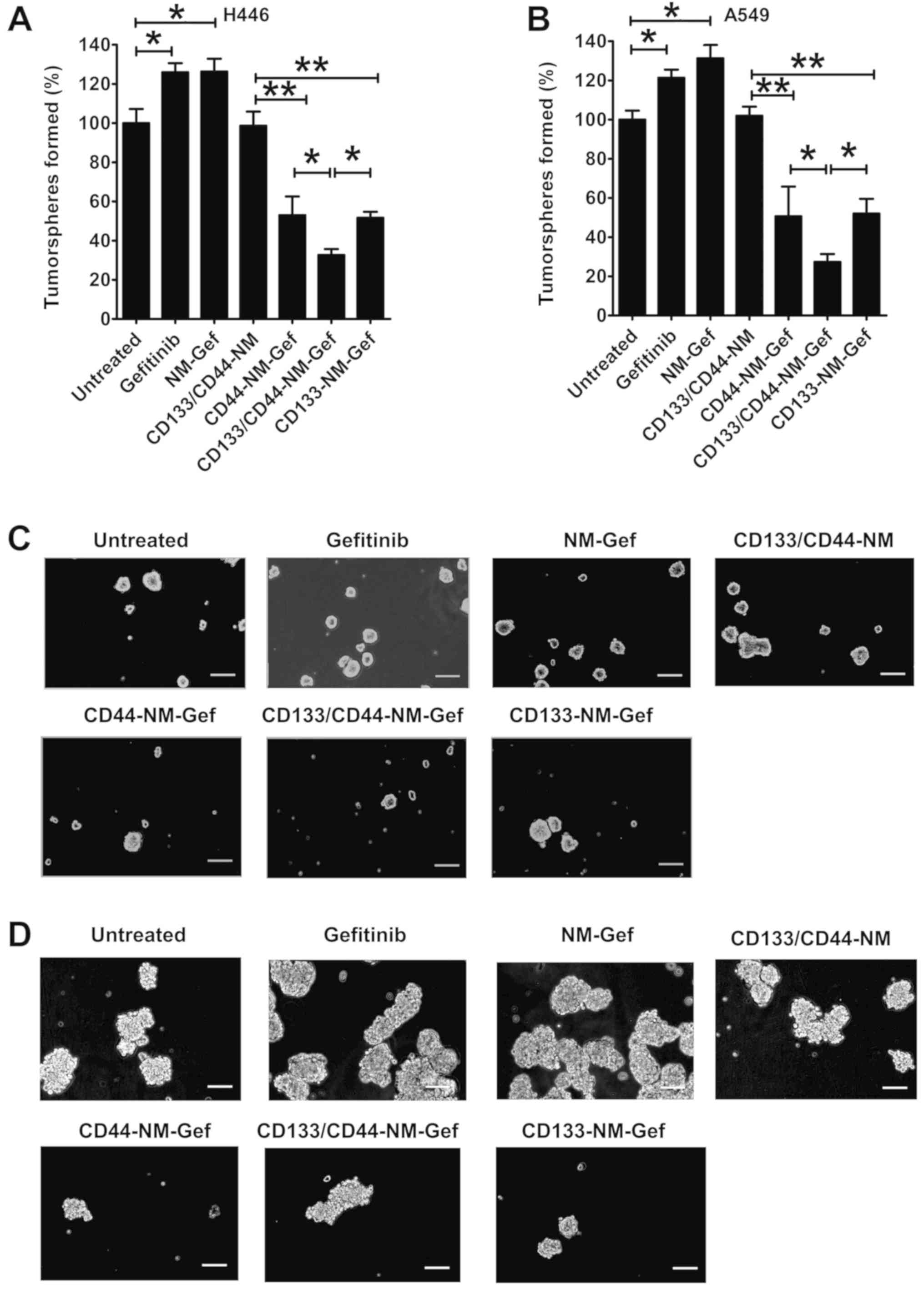
Treatment with gefitinib and NM-Gef markedly
increased the number of H446 tumorspheres (P<0.05; Fig. 7), indicating that gefitinib and
NM-Gef exhibited preferential cytotoxic effects toward lung cancer
cells but not lung cancer-initiating cells, resulting in an
increased percentage of lung cancer-initiating cells. As expected,
due to a lack of drug loading, the blank nanomicelles CD133/CD44-NM
did not affect the number of tumorspheres generated by H446 cells
(Fig. 7A). The inhibitory ability of
CD44-NM-Gef and CD133-NM-Gef was greater than that of CD133/CD44-NM
(P<0.01). Of note, CD133/CD44-NM-Gef treatment exerted greater
inhibitory effects than CD44-NM-Gef and CD133-NM-Gef (P<0.05).
Similarly, in A549 cells, CD133/CD44-NM-Gef treatment inhibited
tumorsphere formation to the greatest extent (Fig. 7B). Representative images of
tumorspheres are provided in Fig. 7C and
D.
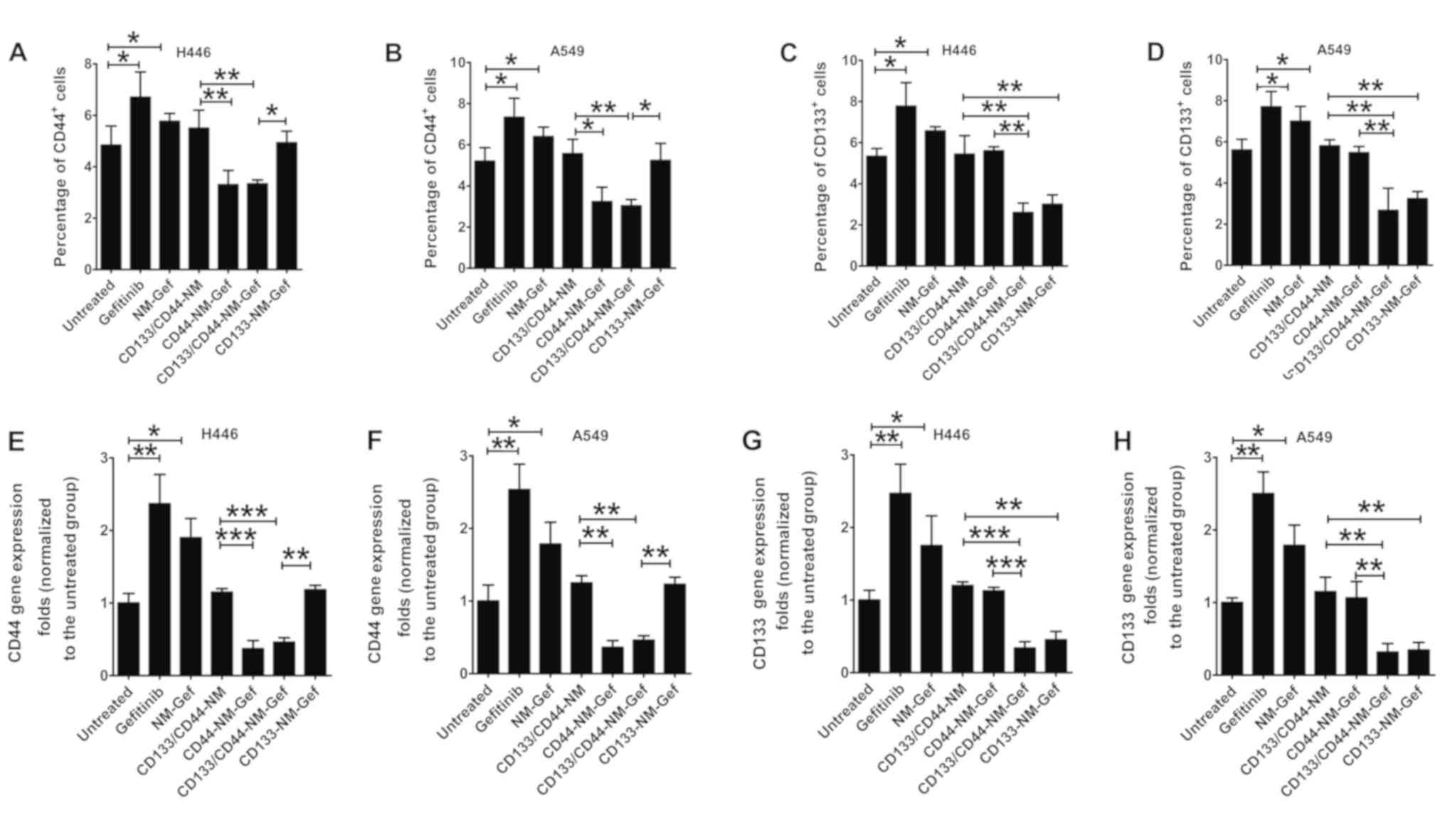
Consistent results were obtained by flow cytometric
analysis of the percentage of CD44+ or CD133+
H446 and A549 cells (Fig. 8A-D). In
H446 cells, the percentage of CD44+ cells was markedly
increased after gefitinib and NM-Gef treatment (P<0.05) and was
markedly decreased after treatment with CD44-NM-Gef or
CD133/CD44-NM-Gef (P<0.05; Fig.
8A). For A549 CD44+ cells, similar results were
obtained (Fig. 8B). The percentage
of CD133+ H446 cells was markedly increasedafter
treatment with gefitinib or NM-Gef (P<0.05; Fig. 8C). CD133-NM-Gef and CD133/CD44-NM-Gef
exerted similar inhibitory effects on the percentage of
CD133+ H446 cells. For CD133+ A549 cells,
similar results were obtained (Fig.
8D). Representative flow cytometric images are provided in
Fig. S1. In general, the proportion
of marker-positive cancer stem cells in cancer cell lines is low.
In line with this, Fig. S1
indicated that the majority of the cells were negative for these
markers. In summary, CD133/CD44-NM-Gef decreased the percentages of
CD133+ and CD44+ lung cancer-initiating
cells, indicating better therapeutic efficacy toward lung
cancer-initiating cells than CD44-NM-Gef and CD133-NM-Gef. The
RT-qPCR analysis results of mRNA expression of CD44 and CD133 in
H446 and A549 cells indicated similar trends to the flow cytometry
results (Fig. 8E-H).
Discussion
Targeting and elimination of lung cancer-initiating
cells, which are considered the seeds of lung cancer, may be able
to effectively eradicate lung cancer. Int he present study,
CD133/CD44-NM-Gef was developed to target CD133+ and
CD44+ lung cancer-initiating cells. CD133/CD44-NM-Gef
was demonstrated to target CD133+ and CD44+
lung cancer-initiating cells and exhibited a better therapeutic
efficacy toward lung cancer-initiating cells in comparison to
single-target and non-targeted nanomicelles, suggesting that
CD133/CD44-NM-Gef represents a promising treatment for lung
cancer-initiating cells.
The safety of nanomedicines is a critical factor for
determining their suitability for clinical application (18). Inorganic nanomedicines, including
silicon or noble metal nanoparticles, in contrast to biodegradable
organic nanomicelles, do not undergo degradation (18). Since biodegradable organic
nanomedicines exhibit superior biocompatibility and safety, they
offer a more promising strategy for clinical use (18–20). In
the present study, the components of CD133/CD44-NM-Gef included
gefitinib, DSPE-PEG2000 and aptamers. Gefitinib, which was
initially heralded as a breakthrough in lung cancer therapy, was
approved by the US Food and Drug Administration (FDA) as the first
tyrosine kinase inhibitor (TKI) for lung cancertherapy (21). DSPE-PEG2000 is an FDA-approved lipid
material and has been widely used for the preparation of various
long-circulation liposomes. Aptamers are oligonucleotide molecules
that bind to a specific target, and pegaptanib, a targeted
anti-vascular endothelial growth factor aptamer, has been
FDA-approved for intraocular injection for the treatment of ocular
vascular diseases (22). The
cytotoxicity assays in the present study demonstrated that the
blank drug delivery system did not induce toxicity in
vitro.
Several chemotherapy drugs, including salinomycin
and metformin, have been demonstrated to eliminate
cancer-initiating cells (23).
Several studies have previously confirmed the selective activity of
salinomycin toward cancer-initiating cells (24–26).
Although several clinical pilot studies initially confirmed the
potent activity of salinomycin toward therapy-resistant cancers, it
is highly toxic and has a narrow therapeutic window (27). In addition, there are no FDA-approved
chemotherapeutic drugs for targeting cancer-initiating cells. Thus,
the present study aimed to construct a novel drug delivery system
to target cancer-initiating cells. Although gefitinib was the first
approved TKI for lung cancer, the present results indicated that it
markedly increased tumorsphere formation and the percentage of
CD44+ and CD133+ cells, indicating that lung
cancer-initiating cells are resistant to gefitinib. However,
CD133/CD44-NM-Gef targeted CD133+ and CD44+
lung cancer-initiating cells, suggesting that CD133/CD44-NM-Gef may
overcome the resistance of lung cancer-initiating cells to
gefitinib and demonstrating the therapeutic potential of these
nanomicelles. As indicated by Gao et al (23), nanomedicines may be able to eradicate
cancer-initiating cells by utilizing cancer-initiating cell
phenotype-specific ligands to deliver drugs to these cells.
Ligand-conjugated nanomedicines are promising agents
for cancer therapy, as they have the ability to markedly increase
therapeutic effects (20,28). Of note, two ligand-conjugated
nanomedicines (doxorubicin- or docetaxel-loaded nanomedicines) have
undergone successful clinical trials for safety and efficacy
(29,30). In the present study, the CD133 and
CD44 aptamers were demonstrated to be crucial for specific
targeting of CD133/CD44-NM-Gef to CD133+ and
CD44+ lung cancer-initiating cells. In CD133+
and CD44+ lung cancer-initiating cells,
CD133/CD44-NM-Gef demonstrated markedly enhanced targeting compared
with single-target and non-targeted nanomicelles, resulting in
enhanced therapeutic effects. It was therefore proven that CD44 and
CD133 aptamers promoted targeting of nanomicelles to lung
cancer-initiating cells. To the best of our knowledge, the present
study was the first to investigate the promotion of drug delivery
via nanomedicines to multiple populations of cancer-initiating
cells using aptamers. Considering that tumors consist of multiple
phenotypically distinct types of cancer-initiating cell, the
present nanomicelle-based multiple targeting strategy is promising
for targeting multiple subsets of cancer-initiating cells within a
tumor to increase the therapeutic efficacy.
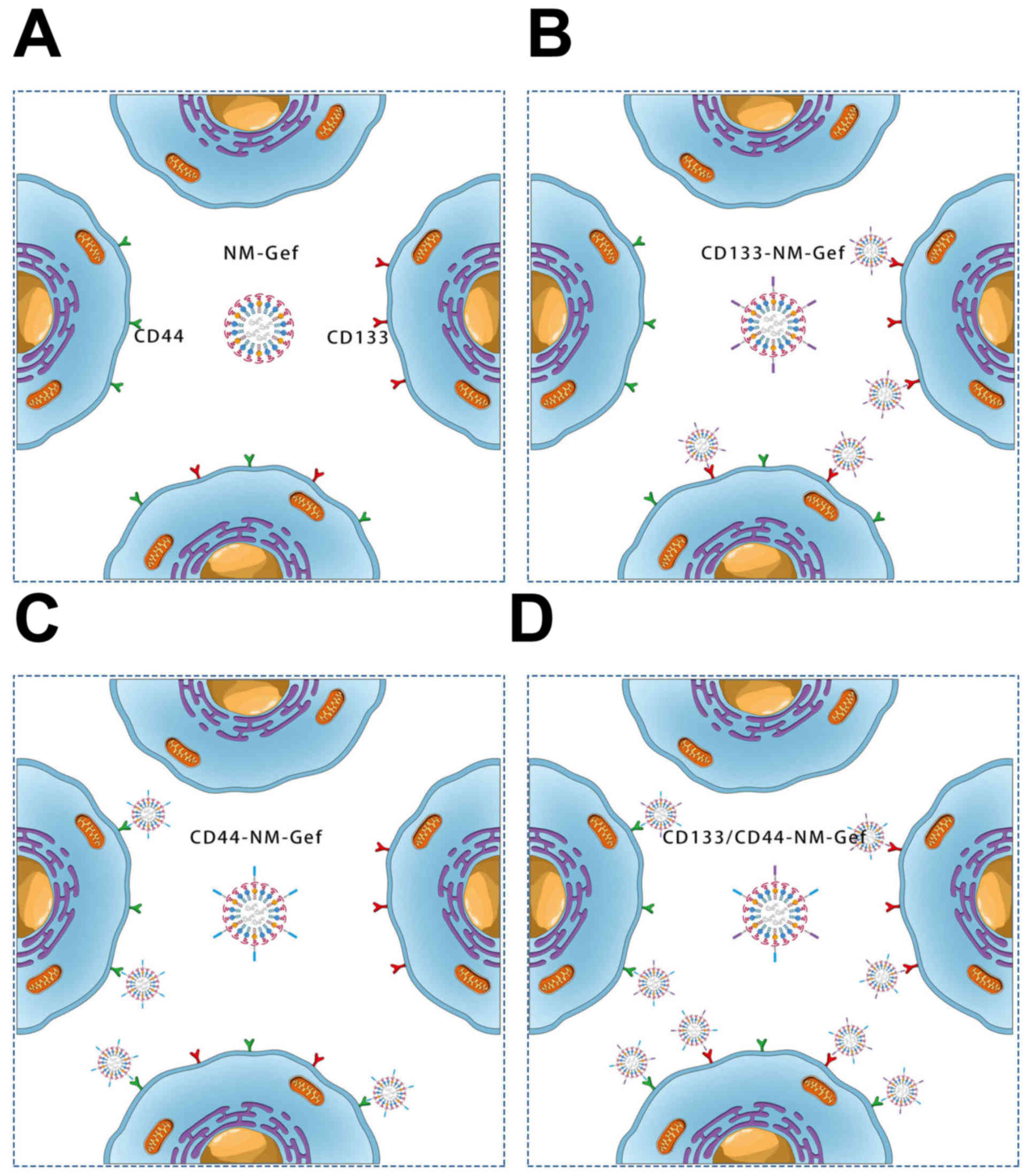
The present study also elucidated the mechanisms
underlying the superior anticancer efficacy of CD133/CD44-NM-Gef
toward lung cancer-initiating cells (Fig. 9). NM-Gef did not specifically target
any cell population due to the absence of CD44 or CD133 aptamers.
By contrast, CD133-NM-Gef exhibited enhanced targeting to
CD44+/CD133+ and
CD44−/CD133+ cells, whereas CD44-NM-Gef
exhibited enhanced targeting to CD44+/CD133+
and CD44+/CD133− cells, suggesting that
CD133-NM-Gef and CD44-NM-Gef could show enhanced targeting of
CD44+/CD133+ cells or single positive cells
(CD44 or CD133) than NM-Gef. Of note, targeting of
CD133/CD44-NM-Gef to CD44+/CD133+,
CD44+/CD133− and
CD44−/CD133+ cells in the whole cell
population was enhanced due to conjugation of the two aptamers.
Although combination of CD133-NM-Gef + CD44-NM-Gef may achieve the
same effect as CD133/CD44-NM-Gef, administration of one drug may be
feasible than the combination of two nanoparticles (26). The present study demonstrated that
CD133/CD44-NM-Gef exerted a significantly greater therapeutic
efficacy toward CD44+ and CD133+ lung
cancer-initiating cells than nanomicelles that did not contain any
aptamers.
One limitation of the present study was that no
clone formation assay was performed in the present study to further
confirm the effect of the drugs on lung cancer-initiating cells
(25). The tumorsphere assay in our
study was used to evaluate the effects of drugs on cell renewal
ability. In future experiments, the clone formation assay will be
performed to substantiate the results obtained in the present
study. Another limitation is that the cytotoxic effects of drugs on
double-positive cells, single-positive/single-negative cells have
not been evaluated, and experiments to evaluate the cytotoxic
effects of drugs on double-positive cells should be performed in
the future.
In conclusion, the present study was the first to
demonstrate targeted drug delivery via nanomicelles to three
populations of cancer-initiating cells using aptamers, to the best
of our knowledge. CD133/CD44-NM-Gef represents a promising agent
for the treatment of lung cancer by targeting cancer-initiating
cells. Considering that eradication of lung cancer-initiating
cellsis crucial for improving the therapeutic efficacy of lung
cancer treatments, patients with lung cancer may substantially
benefit from this dual-targeted therapeutic approach to treatthree
populations of lung cancer-initiating cells.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by the Wuhan Municipal
Health Planning Commission Young Project (project no. WX18Q47) and
the National Science Foundation of China (project no.
81300008).
Availability of data and materials
All data generated or analyzed during this study
are included in this published article.
Authors' contributions
XH, JW and DL contributed to the design of the
study and wrote the manuscript. DL and YZ performed the
experiments. SY analyzed the data. All authors have read and
approved the manuscript.
Ethics approval and consent to
participate
The animal experimental protocols were approved by
the Animal Administrative Committee of the Naval Medical University
(Shanghai, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Eramo A, Lotti F, Sette G, Pilozzi E,
Biffoni M, Di Virgilio A, Conticello C, Ruco L, Peschle C and De
Maria R: Identification and expansion of the tumorigenic lung
cancer stem cell population. Cell Death Differ. 15:504–514. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim JJ and Tannock IF: Repopulation of
cancer cells during therapy: An important cause of treatment
failure. Nat Rev Cancer. 5:516–525. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zakaria N, Satar NA, Abu Halim NH, Ngalim
SH, Yusoff NM, Lin J and Yahaya BH: Targeting lung cancer stem
cells: Research and clinical impacts. Front Oncol. 7:802017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bertolini G, Roz L, Perego P, Tortoreto M,
Fontanella E, Gatti L, Pratesi G, Fabbri A, Andriani F, Tinelli S,
et al: Highly tumorigenic lung cancer CD133+ cells
display stem-like features and are spared by cisplatin treatment.
Proc Natl Acad Sci USA. 106:16281–16286. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang X, Huang J, Leng D, Yang S, Yao Q,
Sun J and Hu J: Gefitinib-loaded DSPE-PEG2000 nanomicelles with
CD133 aptamers target lung cancer stem cells. World J Surg Oncol.
15:1672017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Meyer MJ, Fleming JM, Lin AF, Hussnain SA,
Ginsburg E and Vonderhaar BK: CD44posCD49fhiCD133/2hi defines
xenograft-initiating cells in estrogen receptor-negative breast
cancer. Cancer Res. 70:4624–4633. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Stewart JM, Shaw PA, Gedye C, Bernardini
MQ, Neel BG and Ailles LE: Phenotypic heterogeneity and instability
of human ovarian tumor-initiating cells. Proc Natl Acad Sci USA.
108:6468–6473. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Leung EL, Fiscus RR, Tung JW, Tin VP,
Cheng LC, Sihoe AD, Fink LM, Ma Y and Wong MP: Non-small cell lung
cancer cells expressing CD44 are enriched for stem cell-like
properties. PLoS One. 5:e140622010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jain RK and Stylianopoulos T: Delivering
nanomedicine to solid tumors. Nat Rev Clin Oncol. 7:653–664. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li W, Li J, Gao J, Li B, Xia Y, Meng Y, Yu
Y, Chen H, Dai J, Wang H, et al: The fine-tuning of thermosensitive
and degradable polymer micelles for enhancing intracellular uptake
and drug release in tumors. Biomaterials. 32:3832–3844. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li W, Zhao H, Qian W, Li H, Zhang L, Ye Z,
Zhang G, Xia M, Li J, Gao J, et al: Chemotherapy for gastric cancer
by finely tailoring anti-Her2 anchored dual targeting
immunomicelles. Biomaterials. 33:5349–5362. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ashok B, Arleth L, Hjelm RP, Rubinstein I
and Onyüksel H: In vitro characterization of PEGylated phospholipid
micelles for improved drug solubilization: Effects of PEG chain
length and PC incorporation. J Pharm Sci. 93:2476–2487. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao BJ, Ke XY, Huang Y, Chen XM, Zhao X,
Zhao BX, Lu WL, Lou JN, Zhang X and Zhang Q: The antiangiogenic
efficacy of NGR-modified PEG-DSPE micelles containing paclitaxel
(NGR-M-PTX) for the treatment of glioma in rats. J Drug Target.
19:382–390. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mao X, Liu J, Gong Z, Zhang H, Lu Y, Zou
H, Yu Y, Chen Y, Sun Z, Li W, et al: iRGD-conjugated DSPE-PEG2000
nanomicelles for targeted delivery of salinomycin for treatment of
both liver cancer cells and cancer stem cells. Nanomedicine (Lond).
10:2677–2695. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2 (-Delta Delta C (T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cushing BL, Kolesnichenko VL and O'Connor
CJ: Recent advances in the liquid-phase syntheses of inorganic
nanoparticles. Chem Rev. 104:3893–3946. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gao J, Ochyl LJ, Yang E and Moon JJ:
Cationic liposomes promote antigen cross-presentation in dendritic
cells by alkalizing the lysosomal pH and limiting degradation of
antigens. J Nanomedicine. 12:1251–1264. 2017.
|
|
20
|
Gao J, Chen H, Song H, Su X, Niu F, Li W,
Li B, Dai J, Wang H and Guo Y: Antibody-targeted immunoliposomes
for cancer treatment. Mini Rev Med Chem. 13:2026–2035. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Passiglia F, Listì A, Castiglia M, Perez
A, Rizzo S, Bazan V and Russo A: EGFR inhibition in NSCLC: New
findings and opened questions? Crit Rev Oncol Hematol. 112:126–135.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ng EW, Shima DT, Calias P, Cunningham ET
Jr, Guyer DR and Adamis AP: Pegaptanib, a targeted anti-VEGF
aptamer for ocular vascular disease. Nat Rev Drug Discov.
5:123–132. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gao J, Li W, Guo Y and Feng SS:
Nanomedicine strategies for sustained, controlled and targeted
treatment of cancer stem cells. Nanomedicine (Lond). 11:3261–3282.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gong Z, Chen D, Xie F, Liu J, Zhang H, Zou
H, Yu Y, Chen Y, Sun Z, Wang X, et al: Codelivery of salinomycin
and doxorubicin using nanoliposomes for targeting both liver cancer
cells and cancer stem cells. Nanomedicine (Lond). 11:2565–2579.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xie F, Zhang S, Liu J, Gong Z, Yang K,
Zhang H, Lu Y, Zou H, Yu Y, Chen Y, et al: Codelivery of
salinomycin and chloroquine by liposomes enables synergistic
antitumor activity in vitro. Nanomedicine (Lond). 11:1831–1846.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang M, Xie F, Wen X, Chen H, Zhang H, Liu
J, Zhang H, Zou H, Yu Y, Chen Y, et al: Therapeutic PEG-ceramide
nanomicelles synergize with salinomycin to target both liver cancer
cells and cancer stem cells. Nanomedicine (Lond). 12:1025–1042.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Naujokat C and Steinhart R: Salinomycin as
a drug for targeting human cancer stem cells. J Biomed Biotechnol.
2012:9506582012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gao J, Feng SS and Guo Y: Antibody
engineering promotes nanomedicine for cancer treatment.
Nanomedicine (Lond). 5:1141–1145. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mamot C, Ritschard R, Wicki A, Stehle G,
Dieterle T, Bubendorf L, Hilker C, Deuster S, Herrmann R and
Rochlitz C: Tolerability, safety, pharmacokinetics, and efficacy of
doxorubicin-loaded anti-EGFR immunoliposomes in advanced solid
tumours: A phase 1 dose-escalation study. Lancet Oncol.
13:1234–1241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Miller K, Cortes J, Hurvitz SA, Krop IE,
Tripathy D, Verma S, Riahi K, Reynolds JG, Wickham TJ, Molnar I, et
al: HERMIONE: A randomized Phase 2 trial of MM-302 plus trastuzumab
versus chemotherapy of physician's choice plus trastuzumab in
patients with previously treated, anthracycline-naïve,
HER2-positive, locally advanced/metastatic breast cancer. BMC
Cancer. 16:3522016. View Article : Google Scholar : PubMed/NCBI
|