Introduction
Asthma, also known as bronchial asthma, refers to
chronic airway inflammation involving a variety of cells and cell
components. This inflammation often causes an increase in airway
response, which can cause repeated attacks of wheezing, shortness
of breath, chest distress, cough and other symptoms (1). Asthma is a very common chronic
respiratory disease at present, with a high morbidity in all ages
(2). According to statistics, the
morbidity of asthma worldwide is approximately 3.8/1,000 (3). Previous studies have confirmed that the
pathogenic factors of asthma are complex, factors like heredity,
inhalation allergen, air pollution and respiratory virus infection
all may cause asthma (4). Asthma can
cause respiratory failure, respiratory arrest, pneumothorax or
mediastinal emphysema, and lung infection, and more serious cases
will directly cause sudden death (5). Ebmeier et al (6) further indicated that the mortality
caused by asthma was on the rise. At present, the diagnosis and
treatment of asthma are complicated and difficult. In clinical
practice, multiple comprehensive tests such as blood routine,
sputum, lung function, blood gas, imaging technology and allergen
are required to achieve the purpose of diagnosis, while treatment
requires multi-functional treatment and extremely long treatment
period, and the complete cure rate is relatively low (7,8).
Therefore, in order to effectively control the clinical harm of
asthma, researchers are constantly working to explore and find new
and effective diagnosis and treatment programs.
With the deepening of research, the application of
long-chain non-coding RNA (lncRNA) has gradually become a hot spot
(9–11). As a tumor suppressor, lncRNA
MEG3 has been proved to play a role in a number of tumor
diseases (12,13), and Devadoss et al (14) explained the importance of MEG3
in chronic obstructive pulmonary disease. However, there is no
study that has yet confirmed the effect of MEG3 on asthma.
Because chronic obstructive pulmonary disease was consistent with
some pathogenic mechanisms of asthma, we speculated that
MEG3 also had special expression in asthma. In order to
verify our conjecture, this experiment provided reference and
guidance for future clinical diagnosis and treatment of asthma by
exploring and analyzing the expression of MEG3 in asthma
patients and the situation of MEG3 in asthma with different
inflammatory phenotypes.
Materials and methods
General information
A total of 119 asthma patients admitted to Wuhan No.
1 Hospital (Wuhan, China) from March 2017 to March 2019 and 125
healthy people undergoing physical examination at the same time
were selected as the research objects for prospective analysis.
This experiment was approved by the Ethics Committee
of Wuhan No. 1 Hospital. Patients who participated in this research
had complete clinical data. The signed informed consents were
obtained from the patients or the guardians.
Inclusion and exclusion criteria
Inclusion criteria were as follows: Those in
compliance with clinical manifestations of asthma; people who were
diagnosed as asthma after a series of examinations in Wuhan No. 1
Hospital; those who had complete case data; those who agreed to
cooperate and participate in the investigation of medical staff in
Wuhan No. 1 Hospital; those who were 10–70 years old.
Exclusion criteria were as follows: Patients with
other respiratory and lung diseases; patients with tumor; patients
with autoimmune deficiency diseases; patients with systemic
infection; patients with organ dysfunction; patients with other
cardiovascular and cerebrovascular diseases; patients with drug
allergy; patients with low treatment compliance for mental
disorders; patients transferred to another hospital and patients
with physical disabilities who could not take care of
themselves.
Methods
Altogether 5 ml of fasting venous blood from the two
groups was drawn respectively into EDTA anticoagulants, left at
room temperature for 30 min and centrifuged at 800 × g for 10 min
to obtain upper serum, which was stored in a refrigerator at −80°C
for testing. PCR method was used to detect the expression of
MEG3 in serum of patients. The collected serum was extracted
with TRIzol kit (kit and corollary reagents were from Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) and the extracted
total RNA was detected by ultraviolet spectrophotometer and agarose
gel electrophoresis for purity, concentration and integrity of
total RNA. TransScript® miRNA RT Enzyme Mix and 2X TS
miRNA Reaction Mix were used to reverse transcription with total
RNA, and the operation steps were strictly in accordance with the
manufacturer's kit. Then PrimeScript RT Master Mix kit (kit and
corollary reagents were from Takara Bio, Inc., Otsu, Japan) was
used for PCR amplication experiment. PCR reaction system was as
follows: cDNA 1 µl, upstream and downstream primers 0.4 µl each
(Table I), 2X TransTaq®
Tip Green qPCR SuperMix 10 µl, Passive Reference Dye (50X) 0.4 µl,
finally ddH2O supplemented to 20 µl. PCR reaction
conditions were as follows: pre-denaturation at 94°C for 30 sec,
denaturation at 94°C for 5 sec, annealing at 60°C for 30 sec, with
a total of 40 cycles. Each sample was provided with 3 repeated
wells, and the experiment was carried out 3 times. GAPDH was used
as internal reference and 2−∆Cq was used for data
analysis (14).
 | Table I.Primer sequence. |
Table I.
Primer sequence.
| Genes | Upstream (5′-3′) | Downstream
(5′-3′) |
|---|
| MEG3 |
TCGCTCTTCTCCATCGAACCG |
GTAGGGCGACGACTTTGAGT |
| GAPDH |
ATGGTGAAGGTCGGTGTGA |
CCATGTAGTTGAGGTCAATGAG |
Observation indicators
Expression of MEG3 in two groups, the
predictive value of MEG3 for asthma, and expression of
MEG3 in asthma with different inflammatory phenotypes were
observed. Patients in the research group were followed up for 3
months to record the recurrence of asthma and analyze the risk
factors affecting it.
Statistical analysis
SPSS24.0 (IBM Corp., Armonk, NY, USA) was used to
process and analyze all data, and GraphPad 8 was used to draw all
graphical results. The counting data were expressed in the form of
(rate), and Chi-square test was used for comparison between groups.
The measurement data were expressed in the form of (mean ± standard
deviation), and the comparison between groups adopted t-test.
One-way analysis of variance and LSD back testing were used for
comparison among groups, and ROC curve was used to analyze the
predicted value. Pearson's correlation coefficient was used for
correlation analysis, and logistic regression analysis was used to
analyze the risk factors. P-value <0.050 was considered
statistically significant.
Results
Comparison of general information
Age, BMI, sex, smoking, living environment and
nationality of the two groups were compared, there was no
significant difference (P>0.050), while there was significant
difference between the two groups in family medical history and
history of respiratory disease (P<0.001) (Table II).
 | Table II.Comparison of general information of
patients in the two groups [n (%)]. |
Table II.
Comparison of general information of
patients in the two groups [n (%)].
| Characteristics | Research group
(n=119) | Control group
(n=125) | t or χ2
value | P-value |
|---|
| Age (years) |
31.5±16.4 | 32.8±15.8 | 0.631 | 0.529 |
| Course of disease
(years) |
2.42±0.78 |
|
|
|
| BMI
(kg/cm2) | 22.81±2.18 | 22.97±2.16 | 0.576 | 0.565 |
| Sex |
|
| 0.186 | 0.666 |
|
Male | 68 (57.14) | 68 (54.40) |
|
|
|
Female | 51 (42.86) | 57 (45.60) |
|
|
| Smoking |
|
| 1.870 | 0.172 |
|
Yes | 75 (63.03) | 68 (54.40) |
|
|
| No | 44 (36.97) | 57 (45.60) |
|
|
| Living
environment |
|
| 0.264 | 0.608 |
| Cities
and towns | 96 (80.67) | 104 (83.20) |
|
|
|
Countryside | 23 (19.33) | 21
(16.80) |
|
|
| Family medical
history |
|
| 22.240 | <0.001 |
|
Yes | 35 (29.41) | 8
(6.40) |
|
|
| No | 84 (70.59) | 117 (93.60) |
|
|
| History of
respiratory disease |
|
| 13.350 | <0.001 |
|
Yes | 24 (20.17) | 6
(4.80) |
|
|
| No | 95 (79.83) | 119 (95.20) |
|
|
| Nationality |
|
| 0.462 | 0.497 |
|
Han | 112 (94.12) | 120 (96.00) |
|
|
| Ethnic
minorities | 7
(5.88) | 5
(4.00) |
|
|
| Working and
learning environment |
|
| 0.422 | 0.516 |
|
Outdoor | 79 (66.39) | 78 (62.40) |
|
|
|
Indoor | 40 (33.61) | 47 (37.60) |
|
|
| Inflammatory
phenotype |
|
Eosinophilic asthma | 49 (41.18) |
|
|
|
|
Neutrophilic asthma | 32 (26.89) |
|
|
|
| Mixed
granulocytic asthma | 26 (21.85) |
|
|
|
|
Paucigranulocytic asthma | 12 (10.08) |
|
|
|
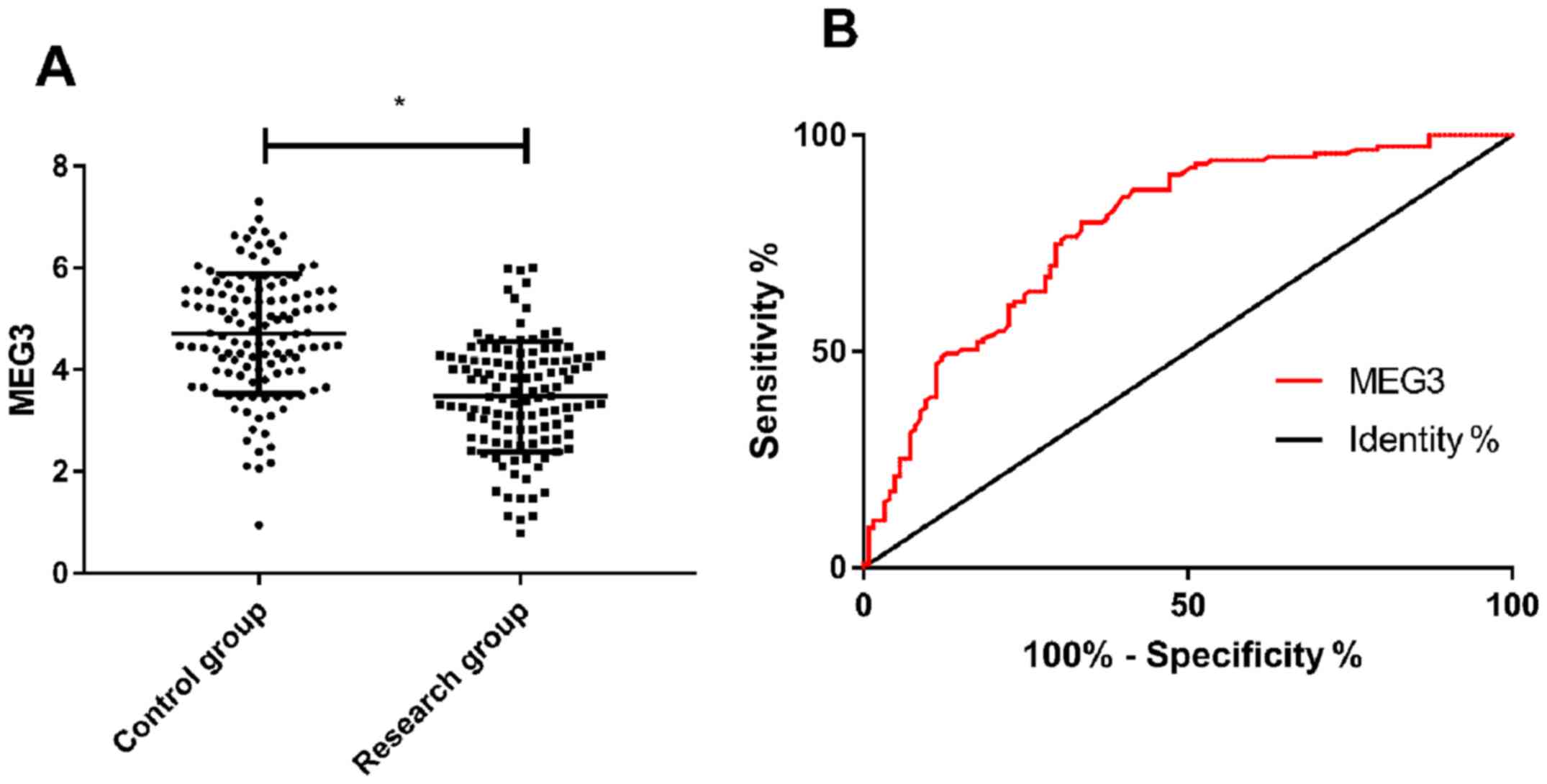
Comparison of the expression level of
MEG3
The expression level of MEG3 in the research
group was significantly lower than that in the control group
(P<0.001), and ROC curve analysis showed that when cut-off value
was 4.295, the predictive sensitivity, specificity, AUC and 95% CI
for detection of MEG3 were 79.83%, 66.40%, 0.783 and
0.725–0.840, respectively (P<0.001) (Fig. 1).
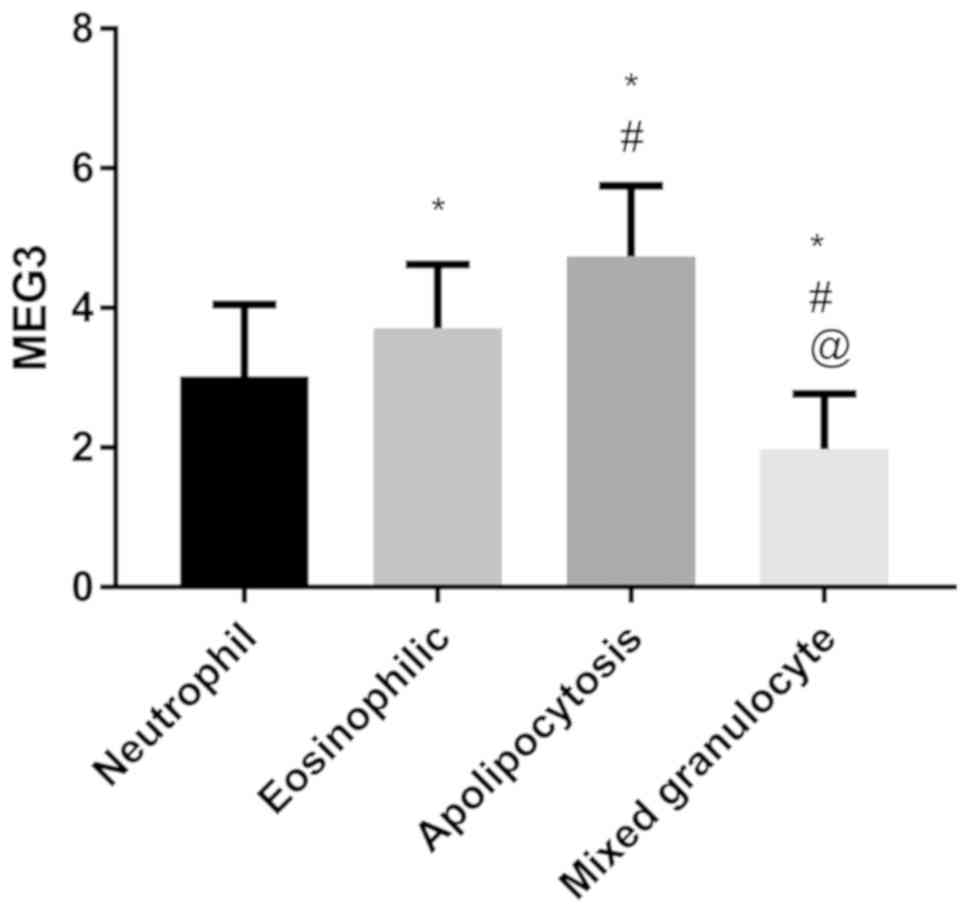
Expression of MEG3 in different
inflammatory phenotypes
The expression level of MEG3 in patients with
mixed granulocytic asthma was the lowest, followed by neutrophilic
asthma and eosinophilic asthma, and the expression level of
MEG3 in paucigranulocytic asthma was the highest
(P<0.050) (Fig. 2).
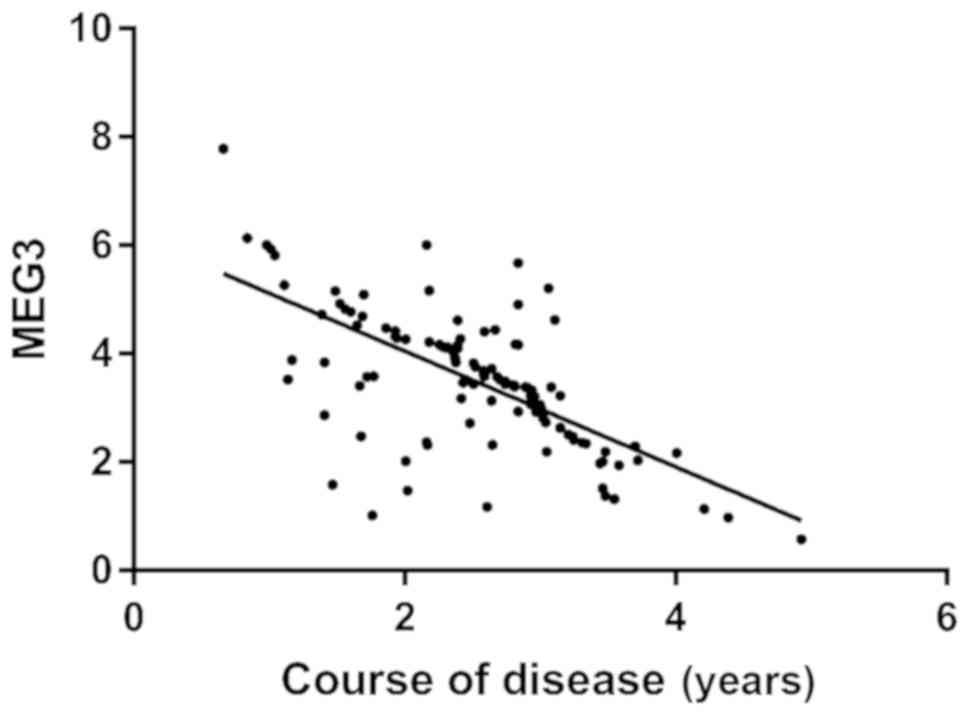
Correlation analysis between MEG3 and
course of disease
Pearson correlation coefficient analysis showed that
the expression level of MEG3 in the research group was
negatively correlated with the course of disease (r=−0.666,
P<0.001) (Fig. 3).
Univariate analysis of factors
affecting recurrence of asthma
A total of 119 patients in the research group were
successfully followed up for 3 months, with a follow-up success
rate of 100.0%. Among them, 42 patients who had recurrence of
asthma were regarded as recurrence group and the other 77 patients
who did not recur were treated as non-recurrence group. Univariate
analysis showed that age, BMI, sex, family medical history and
nationality were not the single factors affecting recurrence of
asthma (P>0.050), while course of disease, smoking, living
environment, history of respiratory, working (learning)
environment, inflammatory phenotype and MEG3 were the single
factors affecting recurrence of asthma (P<0.050) (Table III).
 | Table III.Univariate analysis |
Table III.
Univariate analysis
|
Characteristics | Recurrence group
(n=42) | Non-recurrence
group (n=77) | χ2
value | P-value |
|---|
| Age |
|
|
0.483 | 0.487 |
|
≤31.5 | 24 (57.14) | 49 (63.64) |
|
|
|
>31.5 | 18 (42.86) | 28 (36.36) |
|
|
| Course of disease
(years) |
|
| 11.180 | <0.001 |
|
≤2.42 | 15 (35.71) | 52 (67.53) |
|
|
|
>2.42 | 27 (64.29) | 25 (32.47) |
|
|
| BMI
(kg/cm2) |
|
|
0.150 | 0.698 |
|
≤22.81 | 23 (54.76) | 45 (58.44) |
|
|
|
>22.81 | 19 (45.24) | 32 (41.56) |
|
|
| Sex |
|
|
0.150 | 0.698 |
|
Male | 23 (54.76) | 45 (58.44) |
|
|
|
Female | 19 (45.24) | 32 (41.56) |
|
|
| Smoking |
|
| 14.340 | <0.001 |
|
Yes | 36 (85.71) | 39 (50.65) |
|
|
| No | 6
(14.29) | 38 (49.35) |
|
|
| Living
environment |
|
|
0.184 | 0.668 |
| Cities
and towns | 33 (78.57) | 63 (81.82) |
|
|
|
Countryside | 9
(21.43) | 14 (18.18) |
|
|
| Family medical
history |
|
|
0.074 | 0.785 |
|
Yes | 13 (30.95) | 22 (28.57) |
|
|
| No | 29 (69.05) | 55 (71.43) |
|
|
| History of
respiratory disease |
|
| 30.380 | <0.001 |
|
Yes | 20 (47.62) | 4 (5.19) |
|
|
| No | 22 (52.38) | 73 (94.81) |
|
|
| Nationality |
|
|
0.147 | 0.701 |
|
Han | 40 (95.24) | 72 (93.51) |
|
|
| Ethnic
minorities | 2 (4.76) | 5 (6.49) |
|
|
| Working (learning)
environment |
|
|
6.171 | 0.013 |
|
Outdoor | 34 (80.95) | 45 (58.44) |
|
|
|
Indoor | 8
(19.05) | 32 (41.56) |
|
|
| Inflammatory
phenotype |
|
|
9.698 | 0.021 |
|
Eosinophilic asthma | 16 (38.10) | 33 (42.86) |
|
|
|
Neutrophilic asthma | 10 (23.81) | 22 (28.57) |
|
|
| Mixed
granulocytic asthma | 16 (38.10) | 10 (12.99) |
|
|
|
Paucigranulocytic asthma | 2 (4.76) | 10 (12.99) |
|
|
| MEG3 |
|
| 22.800 | <0.001 |
|
≤3.48 | 35 (83.33) | 29 (37.66) |
|
|
|
>3.48 | 7
(16.67) | 48 (62.34) |
|
|
Multivariate analysis of factors
affecting recurrence of asthma
The indicators with differences in univariate
analysis were included in the assignment (Table IV), and the results were input into
SPSS and then selected to go forward: LR was used for logistic
regression analysis, and the results revealed that the living
environment, history of respiratory disease and working (learning)
environment were not independent factors influencing recurrence of
asthma (P>0.050), while the course of disease and inflammatory
phenotype were independent risk factors influencing recurrence of
asthma (P<0.050), and MEG3 was independent protective
factor influencing recurrence of asthma (P<0.050) (Table V).
 | Table IV.Assignment table. |
Table IV.
Assignment table.
| Factors | Assignment |
|---|
| Course of
disease | ≤2.42=0;
>2.42=1 |
| Smoking | No =0; yes =1 |
| Living
environment | Countryside =0;
cities and towns =1 |
| History of
respiratory disease | No =0; yes =1 |
| Working (learning)
environment | Indoor =0; outdoor
=1 |
| Inflammatory
phenotype | Paucigranulocytic
asthma =0; eosinophilic asthma =1; neutrophilic asthma =2; mixed
granulocytic asthma =3 |
| MEG3 | ≤3.48=0;
>3.48=1 |
 | Table V.Results of multivariate analysis. |
Table V.
Results of multivariate analysis.
| Factors | B | S.E. | Wald | P-value | Exp (B) | 95% CI |
|---|
| Course of
disease | 2.543 | 0.043 | 12.542 | 0.012 | 12.621 |
12.162–13.425 |
| Smoking | 0.007 | 0.785 |
1.242 | 0.214 |
1.241 | −0.624–2.572 |
| Living
environment | 0.004 | 0.852 |
1.026 | 0.128 |
1.007 | −0.715–2.624 |
| History of
respiratory disease | 0.248 | 0.242 |
4.162 | 0.274 |
1.624 |
1.124–2.128 |
| Working (learning)
environment | 1.358 | 0.776 |
5.242 | 0.324 |
3.242 |
1.135–2.165 |
| Inflammatory
phenotype | 3.684 | 0.097 | 14.684 | 0.011 | 35.242 |
34.162–35.985 |
| MEG3 | 1.492 | 0.687 |
4.732 | 0.030 |
7.862 |
1.157–16.527 |
Discussion
Asthma is airway inflammation involving T
lymphocytes, mast cells and other inflammatory mediator cells,
which is mainly manifested by symptoms such as wheezing, cough and
dyspnea (15). Asthma is a
respiratory disease caused by the combination of internal and
external environment of the body (16). The degree of air turbidity and
inhaled particles in the external environment are the keys to
asthma (17), while lncRNA may play
a crucial role in the internal environment of the body. lncRNA is a
kind of RNA molecule with transcripts over 200nt in length and no
protein coding, and it can exert its biological function through a
variety of mechanisms (18).
MEG3 has been proved to normally play an anti-cancer role in
tumor diseases (19), but its role
in asthma is not yet clear. Facing the increasingly serious asthma
diseases, it is of great significance for us to study the situation
of MEG3 in asthma for its clinical diagnosis and treatment
in the future.
The results of this experiment showed that
MEG3 was low expressed in asthma patients, suggesting that
MEG3 might be involved in the occurrence of asthma. Zhou
et al (20) also stated that
MEG3 was also low expressed in bronchial epithelial cells,
which could support our experimental results. Therefore, we
speculated that the role of MEG3 in asthma might be similar
to that of tumor suppressor genes. Some studies claimed that
MEG3 inhibited cell growth, reduced protein synthesis of
cells, and down-regulated apoptosis-related protein Bcl-xl to
accelerate apoptosis through P13K/m-TOR signaling pathway (21). However, Wang et al (22) indicated that FIZZ1 was the key to
cause asthma through airway P13K/Akt signaling, and Zhang et
al (23) studied that P13K and
Notch signaling pathway were the keys to regulate activation and
proliferation of T lymphocytes in asthma patients. Zhang et
al (24) confirmed that the
onset of asthma required activation of the m-TOR signaling pathway.
Based on the above, we suspected that the role of MEG3 in
asthma might also be carried out through P13K/m-TOR signaling
pathway. However, due to the failure to carry out basic
experiments, we still could not verify our hypothesis, and this
would also be a focus of our future research for further analysis.
Furthermore, we analyzed the predictive value of MEG3 for
asthma, and the results showed that when cut-off value was 4.295,
the predictive sensitivity and specificity of MEG3 for
asthma were 79.83 and 66.40%, respectively. It indicated that
MEG3 could be used as an auxiliary diagnostic indicator for
asthma in the future and could be used for early screening
clinically. Compared with the traditional detection of asthma (such
as sputum smear, imaging and analysis of blood gas) (25), the advantage of detecting MEG3
in serum lies in the convenient acquisition of detection samples.
Peripheral blood, as the most convenient and minimally invasive
detection sample in clinical detection, has a longer preservation
period and is conducive to clinical follow-up at any time. However,
results of serum test are more objective than imaging results and
do not need to rely on the subjective judgment of clinicians for
prediction, which also reduces mis-diagnosis and missed diagnosis
caused by human factors to a certain extent. However, by comparing
the expression level of MEG3 in asthma with different
inflammatory phenotypes, it could be seen that the expression level
of MEG3 was the lowest in patients with mixed granulocytic
asthma and the highest in patients with paucigranulocytic asthma,
which was basically consistent with the pathological condition of
eliminating inflammatory phenotypes (26). Mixed granulocytic asthma is the most
serious type of asthma, which is characterized by recurrent attacks
and refractory treatment, while paucigranulocytic asthma is a
well-controlled or intermittent dyspnea. Such patients usually have
a short course of disease and a mild condition, and can basically
recover after treatment (27). The
differences in expression of MEG3 in asthma with different
phenotypes also indicated that MEG3 might be closely related
to the development of the disease, and the further analysis of the
relationship between MEG3 and the course of disease in the
research group also proved the same. This also suggested that
monitoring of MEG3 in asthma patients could be used to judge
the course of illness or rehabilitation of patients in the future.
However, through logistic regression analysis, we found that the
course of disease and inflammatory phenotype were independent risk
factors for recurrence of asthma, and MEG3 was an
independent protective factor for recurrence of asthma. As the
pathological process of asthma, the course of disease and
inflammatory phenotype had been recognized unanimously in clinical
practice, and the effect of MEG3 on recurrence of asthma
indicated that MEG3 might be a potential indicator for
future treatment of asthma and had extremely high clinical
application prospect.
The results of this experiment revealed that
MEG3 was low expressed in asthma patients, while Chen et
al (28) discovered MEG3
was highly expressed in hepatocytes. The reason for the differences
with our results also showed that MEG3 played different
biological effects in different diseases. However, since we have
not been able to determine the exact mechanism of action of
MEG3 on asthma through basic experiments, this is the
limitation to our research. However, due to the short experimental
period, it is impossible to judge the impact of MEG3 on the
long-term prognosis of asthma patients. In the future, we will
expand the research period and sample size to obtain more detailed
and representative experimental results.
In conclusion, MEG3 is low expressed in
asthma and has good predictive value for it, while its expression
is the lowest and course of disease is closely related in mixed
granulocytic asthma, which may be the key to the diagnosis and
treatment of asthma in the future.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YF wrote the manuscript, interpreted and analyzed
the patient data. CY and WY designed the study and performed the
experiment. WY was responsible for the analysis and discussion of
the data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Wuhan No. 1 Hospital (Wuhan, China). Patients who participated in
this research had complete clinical data. The signed informed
consents were obtained from the patients or the guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Pavord ID, Beasley R, Agusti A, Anderson
GP, Bel E, Brusselle G, Cullinan P, Custovic A, Ducharme FM, Fahy
JV, et al: After asthma: Redefining airways diseases. Lancet.
391:350–400. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nunes C, Pereira AM and Morais-Almeida M:
Asthma costs and social impact. Asthma Res Pract. 3:12017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Winer RA, Qin X, Harrington T, Moorman J
and Zahran H: Asthma incidence among children and adults: Findings
from the Behavioral Risk Factor Surveillance system asthma
call-back survey - United States, 2006–2008. J Asthma. 49:16–22.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Loftus PA and Wise SK: Epidemiology of
asthma. Curr Opin Otolaryngol Head Neck Surg. 24:245–249. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
D'Amato G, Vitale C, Molino A, Stanziola
A, Sanduzzi A, Vatrella A, Mormile M, Lanza M, Calabrese G,
Antonicelli L, et al: Asthma-related deaths. Multidiscip Respir
Med. 11:372016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ebmeier S, Thayabaran D, Braithwaite I,
Bénamara C, Weatherall M and Beasley R: Trends in international
asthma mortality: Analysis of data from the WHO Mortality Database
from 46 countries (1993–2012). Lancet. 390:935–945. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Horak F, Doberer D, Eber E, Horak E, Pohl
W, Riedler J, Szépfalusi Z, Wantke F, Zacharasiewicz A and
Studnicka M: Diagnosis and management of asthma - Statement on the
2015 GINA Guidelines. Wien Klin Wochenschr. 128:541–554. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ichinose M, Sugiura H, Nagase H, Yamaguchi
M, Inoue H, Sagara H, Tamaoki J, Tohda Y and Munakata M; Japanese
Society of Allergology, : Japanese guidelines for adult asthma
2017. Allergol Int. 66:163–189. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Paralkar VR, Taborda CC, Huang P, Yao Y,
Kossenkov AV, Prasad R, Luan J, Davies JO, Hughes JR, Hardison RC,
et al: Unlinking an lncRNA from its associated cis element. Mol
Cell. 62:104–110. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nie W, Ge HJ, Yang XQ, Sun X, Huang H, Tao
X, Chen WS and Li B: LncRNA-UCA1 exerts oncogenic functions in
non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett.
371:99–106. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li W, Zhai L, Wang H, Liu C, Zhang J, Chen
W and Wei Q: Downregulation of LncRNA GAS5 causes trastuzumab
resistance in breast cancer. Oncotarget. 7:27778–27786.
2016.PubMed/NCBI
|
|
12
|
Boon RA, Hofmann P, Michalik KM,
Lozano-Vidal N, Berghäuser D, Fischer A, Knau A, Jaé N, Schürmann C
and Dimmeler S: Long noncoding RNA Meg3 controls endothelial cell
aging and function: Implications for regenerative angiogenesis. J
Am Coll Cardiol. 68:2589–2591. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang J, Yao T, Wang Y, Yu J, Liu Y and
Lin Z: Long noncoding RNA MEG3 is downregulated in cervical cancer
and affects cell proliferation and apoptosis by regulating miR-21.
Cancer Biol Ther. 17:104–113. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Devadoss D, Long C, Langley RJ, Manevski
M, Nair M, Campos MA, Borchert G, Rahman I and Chand HS: Long
noncoding transcriptome in chronic obstructive pulmonary disease.
Am J Respir Cell Mol Biol. Sep 5–2019.(Epub ahead of print).
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Borish L: The immunology of asthma: Asthma
phenotypes and their implications for personalized treatment. Ann
Allergy Asthma Immunol. 117:108–114. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang IV, Lozupone CA and Schwartz DA: The
environment, epigenome, and asthma. J Allergy Clin Immunol.
140:14–23. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Burbank AJ, Sood AK, Kesic MJ, Peden DB
and Hernandez ML: Environmental determinants of allergy and asthma
in early life. J Allergy Clin Immunol. 140:1–12. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Quinn JJ, Zhang QC, Georgiev P, Ilik IA,
Akhtar A and Chang HY: Rapid evolutionary turnover underlies
conserved lncRNA-genome interactions. Genes Dev. 30:191–207. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chak WP, Lung RW, Tong JH, Chan SY, Lun
SW, Tsao SW, Lo KW and To KF: Downregulation of long non-coding RNA
MEG3 in nasopharyngeal carcinoma. Mol Carcinog. 56:1041–1054. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou C, Huang C, Wang J, Huang H, Li J,
Xie Q, Liu Y, Zhu J, Li Y, Zhang D, et al: LncRNA MEG3
downregulation mediated by DNMT3b contributes to nickel malignant
transformation of human bronchial epithelial cells via modulating
PHLPP1 transcription and HIF-1α translation. Oncogene.
36:3878–3889. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu DH, Chi GN, Zhao CH and Li DY: Long
noncoding RNA MEG3 inhibits proliferation and migration but induces
autophagy by regulation of Sirt7 and PI3K/AKT/mTOR pathway in
glioma cells. J Cell Biochem. 120:7516–7526. 2018. View Article : Google Scholar
|
|
22
|
Wang J, Li F, Yang M, Wu J, Zhao J, Gong
W, Liu W, Bi W and Dong L: FIZZ1 promotes airway remodeling through
the PI3K/Akt signaling pathway in asthma. Exp Ther Med.
7:1265–1270. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang W, Nie Y, Chong L, Cai X, Zhang H,
Lin B, Liang Y and Li C: PI3K and Notch signal pathways
coordinately regulate the activation and proliferation of T
lymphocytes in asthma. Life Sci. 92:890–895. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang Y, Jing Y, Qiao J, Luan B, Wang X,
Wang L and Song Z: Activation of the mTOR signaling pathway is
required for asthma onset. Sci Rep. 7:45322017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bos LD, Sterk PJ and Fowler SJ:
Breathomics in the setting of asthma and chronic obstructive
pulmonary disease. J Allergy Clin Immunol. 138:970–976. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chung S, Lee TJ, Reader BF, Kim JY, Lee
YG, Park GY, Karpurapu M, Ballinger MN, Qian F, Rusu L, et al:
FoxO1 regulates allergic asthmatic inflammation through regulating
polarization of the macrophage inflammatory phenotype. Oncotarget.
7:17532–17546. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Taylor SL, Leong LEX, Choo JM, Wesselingh
S, Yang IA, Upham JW, Reynolds PN, Hodge S, James AL, Jenkins C, et
al: Inflammatory phenotypes in patients with severe asthma are
associated with distinct airway microbiology. J Allergy Clin
Immunol. 141:94–103.e15. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen DL, Shen DY, Han CK and Tian Y:
LncRNA MEG3 aggravates palmitate-induced insulin resistance by
regulating miR-185-5p/Egr2 axis in hepatic cells. Eur Rev Med
Pharmacol Sci. 23:5456–5467. 2019.PubMed/NCBI
|