Introduction
Cervical cancer is one of the most common
gynecological malignancies, accounting for 10% of all gynecological
cancers, and its incidence accounts for approximately 5% of all
tumors (1). Worldwide, there are
nearly 500,000 new cases and approximately 270,000 patients die
from cervical cancer each year (2).
The majority of both the new and the fatal cases of cervical cancer
occur in developing countries (3).
The annual incidence of cervical cancer is increasing at a rate of
2–3% per year in China and the disease is increasingly seen in
younger patients. The number of new cases each year in China
accounts for 1/3 of the worldwide increase (4).
Surgery and radiotherapy are at present the main
treatments for cervical cancer, however, surgery is effective only
at early stages of the disease (5).
The most common pathological type of cervical cancer is squamous
cell carcinoma, accounting for 80–85% of cases (6). The main pathways for the metastasis of
cervical cancer are direct spread and lymphatic metastasis
(6). Distant metastasis largely
occurs through lymphatic metastasis (6). Multidrug resistance (MDR) is the main
cause of chemotherapy failure in cervical cancer patients, and MDR
has become one of the most difficult problems for cervical cancer
treatment (7). Once distant
metastasis and local recurrence of cervical cancer occur, the
patient survival rate is significantly reduced (8). It is therefore of great practical
significance to search for new treatment targets and prognostic
molecular markers for cervical cancer.
Aldehyde dehydrogenase 1 (ALDH1) is associated with
the occurrence and development of many human tumors. Increased
expression of ALDH1 usually suggests an insensitivity to
chemotherapy and poor prognosis (9,10). ALDH1
also plays a role in regulating apoptosis, but the mechanism
underlying this is not clear (11–13).
The expression of multiple microRNA (miR) molecules
and proteins in cervical cancer suggests that miRs may play
important roles in the regulation of disease-related proteins
(14). miR-222 is found to be
abnormally expressed in many tumor tissues (15–17), and
is associated with MDR (18,19). However, the regulatory relationship
between miR-222 and ALDH1 in cervical cancer has, to the best of
our knowledge, not yet been reported.
The current study aimed to investigate the
expression of miR-222 and ALDH1 in tumor tissues and blood from
cervical cancer patients, and to elucidate the relationship between
them.
Materials and methods
Patients
A total of 33 patients with cervical cancer who
received surgical treatment at Yuquan Hospital of Tsinghua
University (Beijing, China) between January 2015 and March 2018
were included in the present study. The age range of the patients
was 27–60 years, and the median age was 42.6 years.
Tumor tissues were collected from cervical cancer
patients as the experimental group and tumor-adjacent tissues were
collected as a control group. Peripheral blood was collected from
the same 33 patients and from 28 healthy female subjects who
underwent physical examination and had no history of use of
hormones, traditional Chinese medicine or chemotherapy or
radiotherapy. The age range of the healthy subjects was 26–63
years, and the median age was 43.5 years.
Cervical cancer was diagnosed by hospital
pathologists according to the European Society for Medical Oncology
Clinical Practice Guidelines for diagnosis, treatment and follow-up
of cervical cancer (20). All
patients had no prior diagnosis of cervical cancer, and had no
history of use of steroids, traditional Chinese medicine,
radiotherapy or chemotherapy before surgery.
All procedures used in the current study were
approved by the Ethics Committee of Tsinghua University. All
patients or their families signed written informed consent
forms.
Cells
Human HeLA cells (The Cell Bank of Type Culture
Collection of the Chinese Academy of Sciences) were cultured in
DMEM (cat. no. SH30022.01; Hyclone, Thermo Fisher Scientific, Inc.)
containing 10% fetal bovine serum (FBS; E600001; Sangon Biotech
Co., Ltd.) at 37°C and 5% CO2. The cells
(3×105) in logarithmic growth phase were seeded onto
24-well plates one day before transfection, and cultured in
antibiotic-free nutrient mixture F12/DMEM medium (cat. no.
11320-033; Thermo Fisher Scientific, Inc.) supplemented with 10%
FBS until they reached 70% confluency. In the first vial, 1 µl
agomiR-negative control (agomiR-NC; 20 pmol/µl;
5′-UUCUCCGAACGUGUCACGUTT-3′ and 3′-TTAAGAGGCUUGCACAGUGCA-5′; Hanbio
Biotechnology Co., Ltd.) or agomiR-222 (20 pmol/µl;
5′-CUCAGUAGCCAGUGUAGAUCCU-3′ and 3′-GAGUCAUCGGUCACAUCUAGGA-5′;
Hanbio Biotechnology Co., Ltd.) was mixed with 50 µl Opti MEM. In
the second vial, 1 µl Lipofectamine® 3000 (Thermo Fisher
Scientific, Inc.) was mixed with 50 µl Opti MEM medium. After 5
min, the two vials were mixed an incubated for 20 min at room
temperature. Cells in their respective groups were treated with the
mixtures. Six hours later, the medium was replaced with F12/DMEM
medium containing 10% FBS. The cells were collected for use after
48 h of incubation.
Reverse transcription-quantitative PCR
(RT-qPCR)
Tissue samples (100 mg) were ground into a powder in
liquid nitrogen and lysed with 1 ml TRIzol® reagent
(Thermo Fisher Scientific, Inc.) following the manufacturer's
instructions. Plasma (100 µl) or cells (3×106) were
directly lysed with 1 ml TRIzol reagent. Total RNA was extracted
using the phenol chloroform method (21). The quality of extracted mRNA and
miRNA was assessed by gel electrophoresis. To obtain cDNA, 1 µg RNA
was reverse-transcribed and the resulting cDNA was stored at −20°C
until use. The TIANScript II cDNA First Strand Synthesis kit
(Tiangen Biotech Co., Ltd.) was used for reverse transcription of
mRNA, while the miRcute miRNA cDNA First Strand Synthesis kit
(Tiangen Biotech Co., Ltd.) was employed for reverse transcription
of miRNA according to the manufacturer's manual. The temperature
protocol was 30 min of incubation at 55°C.
The SuperReal PreMix (SYBR Green) RT-qPCR kit
(Tiangen Biotech Co., Ltd.) was used to determine ALDH1 mRNA
expression levels, and GAPDH was used as the internal reference.
The sequences of ALDH1 and GAPDH primers are listed in Table I. The reaction mixture (20 µl) was
composed of 10 µl SYBR Premix EXTaq, 0.5 µl forward primer, 0.5 µl
reverse primer, 2 µl cDNA and 7 µl ddH2O. The following
thermocycling conditions were used: Initial denaturation at 95°C
for 30 sec; 39 cycles of denaturation at 95°C for 10 sec, annealing
at 60°C for 30 sec and elongation at 72°C for 15 sec; and a final
extension at 72°C for 5 min in an iQ5 thermal cycler (Bio-Rad
Laboratories, Inc.). The 2−ΔΔCq method (22) was used to determine relative
expression of ALDH1 mRNA against GAPDH. Each sample was tested in
triplicate.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
|
| Primer aequence
(5′-3′) |
|---|
|
|
|
|---|
| Target | Forward | Reverse |
|---|
| ALDH1 |
CCGTGGCGTACTATGGATGC |
CGCAATGTTTTGATGCAGCCT |
| GAPDH |
TGTTCGTCATGGGTGTGAACC |
ATGGACTGTGGTCATGAGTCC |
| miR-222 |
CGCAGCTACATCTGGCTACTG |
GTGCAGGGTCCGAGGT |
| U6 |
CGCTTCGGCAGCACATATAC |
CAGGGGCCATGCTAATCTT |
The level of miR-222 was determined using the
miRcute miRNA RT-PCR kit (Tiangen Biotech Co., Ltd.), and U6 was
used as the internal reference. The primer sequences of miR-222 and
U6 are listed in Table I. The
reaction mixture (20 µl) contained 10 µl RT-qPCR-Mix, 0.5 µl
forward primer, 0.5 µl reverse primer, 2 µl cDNA and 7 µl
ddH2O. The following thermocycling conditions were used:
Initial denaturation at 95°C for 5 min; 40 cycles of denaturation
at 95°C for 30 sec, annealing at 60°C for 35 sec and elongation at
72°C for 20 sec in an iQ5 thermal cycler. The 2−ΔΔCq
method was used to calculate the relative expression of miR-222
against U6. Each sample was tested in triplicate.
Western blotting
Before lysis, 100 mg tissue samples were first
frozen in liquid nitrogen and then ground into powder, and cells
(1×106) were trypsinized and collected. Then, tissue
samples or cells were lysed with 600 µl RIPA lysis buffer (Beyotime
Institute of Biotechnology) for 30 min on ice. After centrifugation
at 1,200 × g and 4°C for 10 min, supernatant was collected and used
to determine protein concentration (BCA protein concentration
determination kit; cat. no. RTP7102; Real-Times (Beijing)
Biotechnology Co., Ltd.). After mixing the samples with 5X sodium
dodecyl sulfate (Beyotime Institute of Biotechnology) loading
buffer, the samples were denatured in boiling water for 5 min.
Afterwards, the samples (20 µg) underwent 10% gel sodium dodecyl
sulfate-polyacrylamide gel electrophoresis at 100 V. Proteins were
then transferred to PVDF membranes on ice (100 V, 2 h) and blocked
with 5% non-fat milk at room temperature for 1 h. The membranes
were treated with monoclonal rabbit anti-human ALDH1 (1:2,000; cat.
no. ab52492; Abcam) or β-actin (1:5,000; cat. no. ab129348; Abcam)
primary antibodies at 4°C overnight. After washing (3 times, each
for 15 min), the membranes were incubated with goat anti-rabbit
horseradish peroxidase-conjugated secondary antibody (1:3,000; cat.
no. ab6721; Abcam) for 1 h at room temperature. After washing (3
times, each for 15 min), the membrane was developed with an ECL
Western Blotting Substrate kit (cat. no. ab65623; Abcam). Imaging
signals were acquired and analyzed using Image Lab software
(version 3.0; Bio-Rad Laboratories, Inc.). Relative amounts of
target proteins were expressed in comparison to β-actin.
Enzyme-linked immunosorbent assay
(ELISA)
An ALDH1 ELISA kit (cat. no. sE94824Hu; Shanghai
Wuhao Bio-tech Co., Ltd.) was used to measure ALDH1 concentrations.
Standards (50 µl) and samples (10 µl serum and 40 µl diluent) were
added into predefined wells of microplates, and blank wells were
left empty. Horseradish peroxidase-labelled conjugates (100 µl)
were added into the wells for standards and samples. The plates
were then sealed before incubation at 37°C for 1 h. After washing
the plates (5 times), 50 µl substrate A and 50 µl substrate B were
added into each well. After incubation at 37°C for 15 min, 50 µl
stop solution was added, and absorbance was determined at 450
nm.
Bioinformatics
miRanda (August 2010 Release; http://www.microrna.org/microrna/home.do), TargetScan
(v6.2; http://www.targetscan.org), PiTa (v6;
http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html),
RNAhybrid (September 18 2017; http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/)
and PICTA (March 26, 2007; http://pictar.mdc-berlin.de/) were used to predict
target genes that may be regulated by miR-222.
Dual-luciferase reporter assay
Wild-type and mutant seed regions of miR-222 in the
3′-UTR of the ALDH1 gene were chemically synthesized in
vitro, and then cloned into pMIR-REPORT luciferase reporter
plasmids (Ambion; Thermo Fisher Scientific, Inc.). Plasmids (0.8
µg) with wild-type or mutant 3′-UTR sequences were co-transfected
with agomiR-222 (100 nM; forward, 5′-AGCUACAUCUGGCUACUGGGU-3′;
reverse, 3′-UCGAUGUAGACCGAUGACCCA-5′; Sangon Biotech Co., Ltd.)
using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.) into 293T
cells (The Cell Bank of Type Culture Collection of the Chinese
Academy of Sciences). For control, 293T cells were transfected with
agomiR-negative control (NC; scrambled sequence; forward,
5′-UUCUCCGAACGUGUCACGUTT-3′; reverse, 3′-TTAAGAGGCUUGCACAGUGCA-5′;
Sangon Biotech Co., Ltd.). After 24 h incubation, the cells were
treated with a dual-luciferase reporter assay kit (Promega
Corporation) according to the manufacturer's manual, and
luminescence intensity was measured using a luminometer (GloMax
20/20; Promega Corporation). The luminescence values of each group
of cells were measured using Renilla luminescence activity
as an internal reference.
MTT assay
To examine proliferation, 20 µl MTT (5 g/l; cat. no.
JRDC000003, JRDUN Biotechnology Co., Ltd.) was added at 24, 48 and
72 h after transfection, followed by incubation at 37°C for 4 h.
After removing medium, dimethyl sulfoxide was added at a volume of
150 µl per well to dissolve purple crystals. The absorbance in each
well was measured at 490 nm with a microplate reader and cell
proliferation curves were plotted against time.
Statistical analysis
SPSS version 20.0 statistical software (IBM Corp.)
was used for statistical analysis. Data are presented as mean ±
standard deviation and were tested for normality. Measurement data
were analyzed using one-way ANOVA for multiple groups, with
Student-Newman-Keuls post-hoc tests subsequently used. Comparisons
between two groups were performed using a paired or unpaired
Student's t-test. P<0.05 indicated a statistically significant
difference.
Results
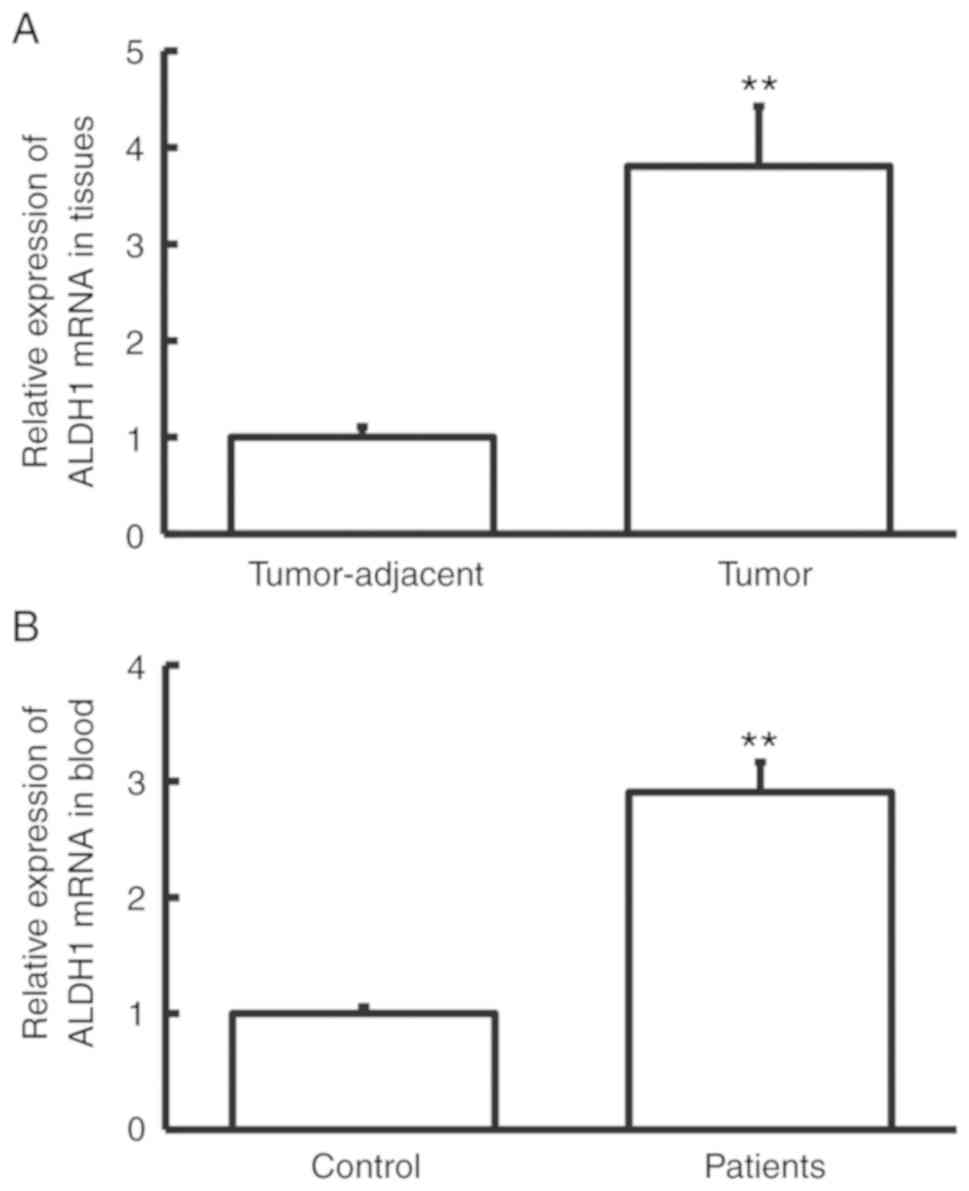
Expression of ALDH1 mRNA is elevated
in cervical cancer
RT-qPCR was performed to measure ALDH1 mRNA
expression. The level of ALDH1 mRNA in tumor tissues was
significantly higher than that in tumor-adjacent tissues
(P<0.01; Fig. 1A), and the level
of ALDH1 mRNA in peripheral blood from cervical cancer patients was
significantly higher than that from control subjects (P<0.01;
Fig. 1B). These results indicate
that the expression of ALDH1 mRNA was increased in cervical
cancer.
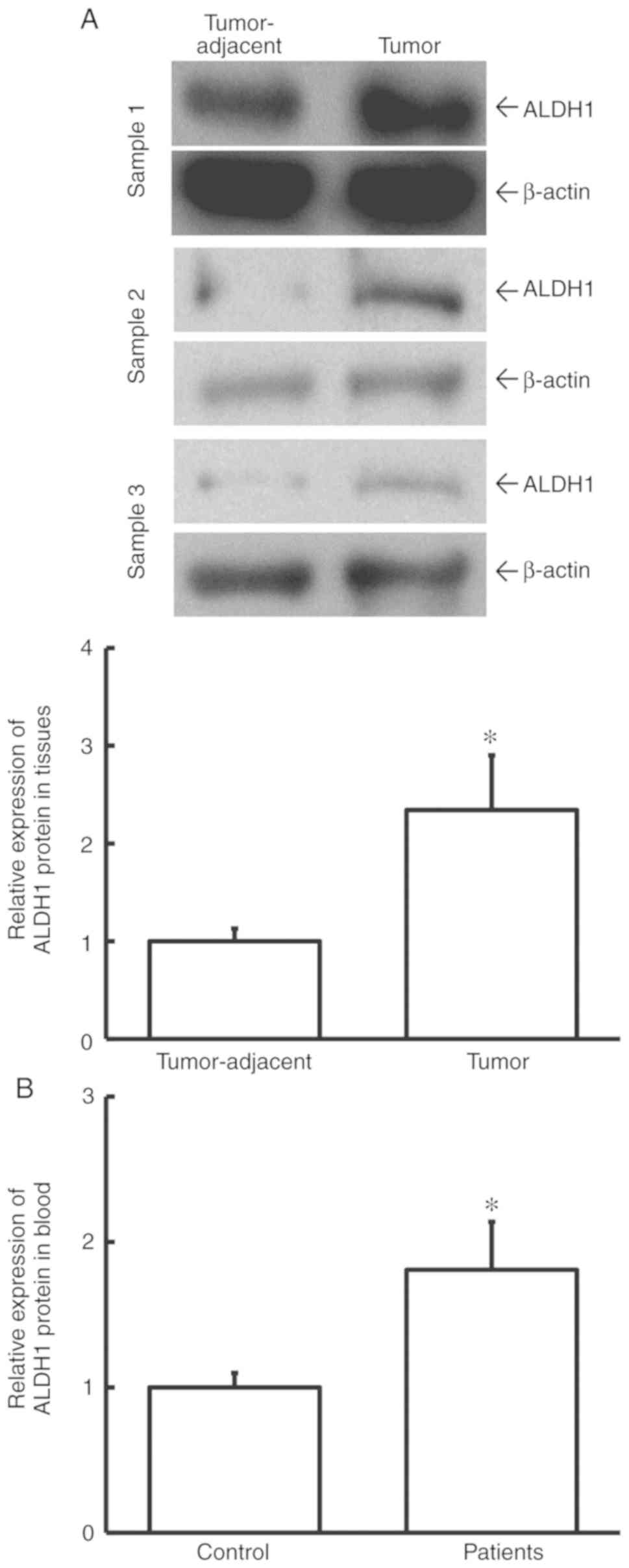
Expression of ALDH1 protein is
elevated in cervical cancer
To determine ALDH1 protein expression in tissues and
blood, western blotting and ELISA were used. The data showed that
the level of ALDH1 protein in tumor tissues from cervical cancer
patients was significantly higher than that in tumor-adjacent
tissues (P<0.05; Fig. 2A).
Additionally, the level of ALDH1 protein in peripheral blood from
cervical cancer patients was significantly elevated when compared
with healthy control subjects (P<0.05; Fig. 2B). This result indicated that ALDH1
protein level was increased in cervical cancer and is consistent
with the study findings regarding ALDH1 mRNA.
Expression of miR-222 in cervical
cancer is reduced
To study the expression of miR-222, RT-qPCR was
performed. The data showed that miR-222 expression in tumor tissues
was significantly lower than that in tumor-adjacent tissues
(P<0.01; Fig. 3A), and that
miR-222 levels in peripheral blood from cervical cancer patients
were significantly lower than those in a healthy control group
(P<0.05; Fig. 3B). These data
suggest that miR-222 expression was reduced in cervical cancer.
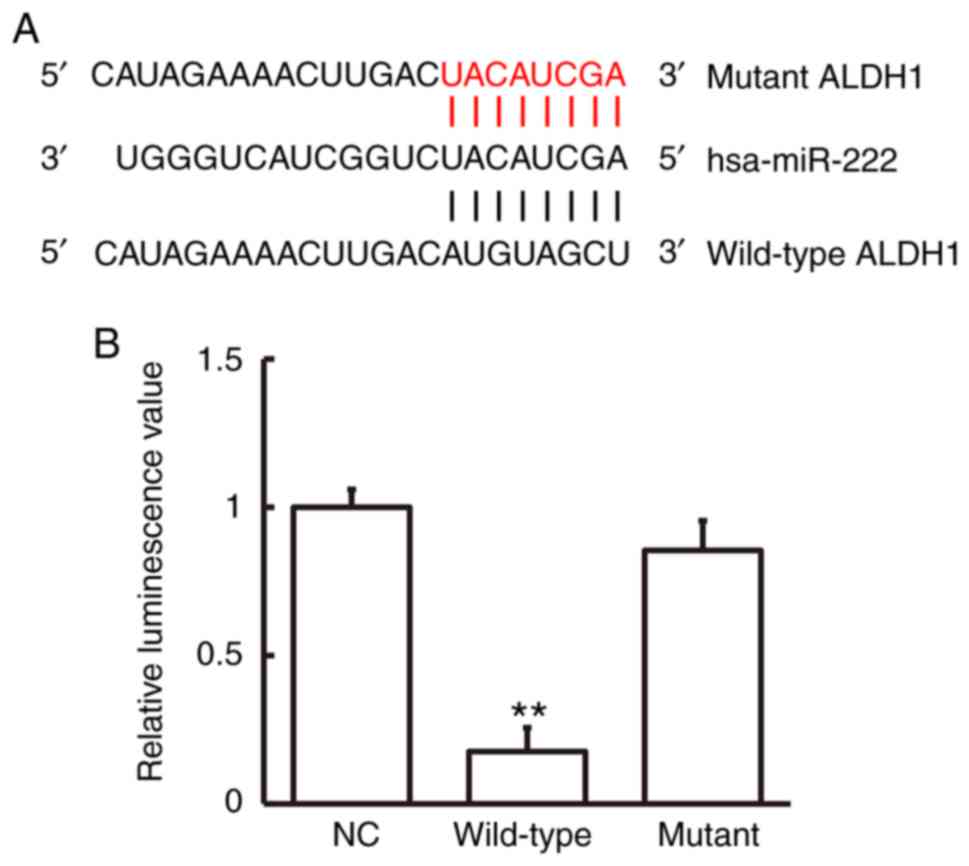
miR-222 regulates the expression of
ALDH1 mRNA by binding to the ALDH mRNA 3′-UTR seed region
Bioinformatics prediction showed that ALDH1 was a
potential target gene of miR-222 (Fig.
4A). To study miR-222 interaction with ALDH1 mRNA, a
dual-luciferase reporter assay was employed. Luminescence intensity
of cells in the wild-type group was significantly lower than that
in the negative control group (P<0.01), while luminescence
intensity in the mutant group was not significantly different from
that in the negative control group (Fig.
4B). These data indicate that miR-222 regulated expression of
ALDH1 mRNA by binding with its 3′-UTR seed region.
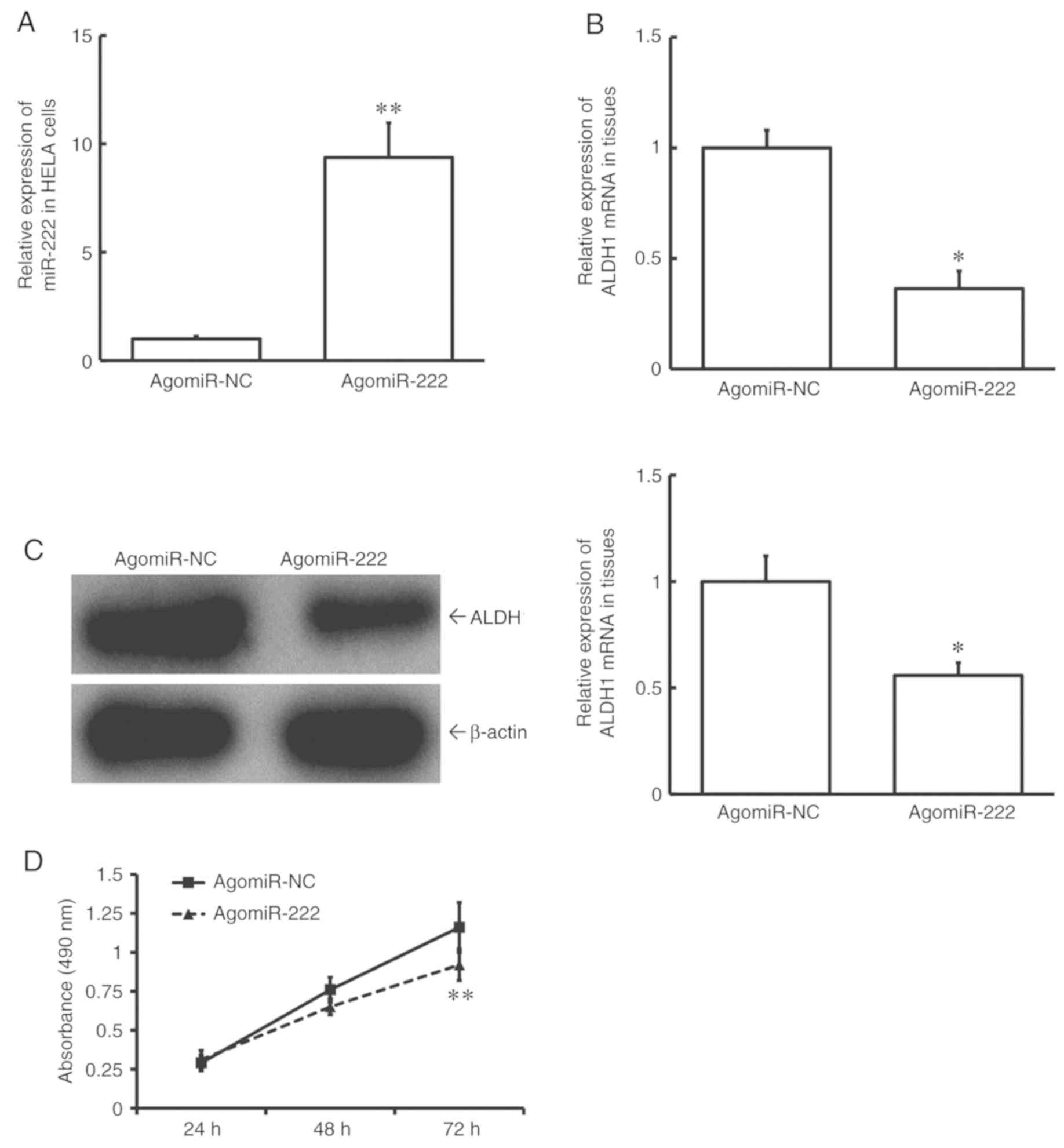
Overexpression of miR-222 reduces the
proliferation of HeLA cells possibly by a reduction in the
expression of ALDH1
To test how miR-222 affects cell proliferation, HeLA
cells were transfected with agomiR-NC or agomiR-222. The data
showed that the level of miR-222 in HeLA cells of the agomiR-222
group was significantly higher than that in the agomiR-NC group
(P<0.01; Fig. 5A). In addition,
expression levels of both ALDH1 mRNA and protein in HeLA cells of
the agomiR-222 group were significantly lower than those in the
agomiR-NC group (P<0.05; Fig. 5B and
C). MTT assay showed that the proliferation of HeLA cells of
the agomiR-222 group was significantly slower than that of the
agomiR-NC group (P<0.01 at 72 h; Fig.
5D). These data suggest that upregulation of miR-222 reduces
the proliferation of HeLA cells, possibly by reducing the
expression of ALDH1.
Discussion
Cervical cancer is the second most common female
malignancy in the world after breast cancer (23). A major challenge in the treatment of
cervical cancer is the invasion and metastasis. Lymphatic
metastasis is common in cervical cancer, and is closely related to
the prognosis of patients (23). It
is an important basis for judging the prognosis of patients and
treatment options (23).
Chemotherapy plays an important role in the treatment of cervical
cancer. However, the tolerance of cervical cancer cells to
chemotherapeutic drugs limits their efficacy. Forms of drug
resistance of tumor cells to chemotherapeutic drugs include natural
or primary resistance, acquired or secondary resistance and MDR or
cross resistance (24). ALDH1 is a
protein related to drug resistance and apoptosis. Proliferation,
colony-formation, adhesion, migration and invasion of breast cancer
cells are upregulated by ALDH1 overexpression in vitro
(25). Increased expression of ALDH1
in human breast cancer is associated with poor prognosis (11). It has also been discovered that the
percentage of African breast cancer patients with positive ALDH1
expression is about 48% (12). The
percentage of patients receiving cyclophosphamide-based
chemotherapy who have positive expression of ALDH1 in cancer
tissues is 61%, suggesting that ALDH1 may be a reference index
reflecting biological behavior of breast cancer (13). ALDH1 plays a role in regulating
apoptosis, though the underlying mechanism is not yet clear.
Several studies have shown that lowering the level of ALDH1 can
cause apoptosis (26,27). The expression of ALDH1 is also
associated with neurogenic locus notch homolog 3 (28), transforming growth factor-β (29) and nuclear factor-κB (30) signaling pathways. In cervical cancer,
the expression of ALDH1 is closely related to the Erk1/2 and Akt
signaling pathways (31). Based on
these data, it is hypothesized that high expression of ALDH1
contributes to drug resistance in cervical cancer. The results of
the present study show that expression levels of ALDH1 mRNA and
protein were elevated in cervical cancer tissues and peripheral
blood from cervical cancer patients when compared with
controls.
Using prediction by bioinformatics, it was
discovered that the miR-222 sequence is closely associated with
ALDH1, and may possibly be an upstream miRNA that regulates ALDH1
expression. miR-222 may cut the mRNA of ALDH1 to inhibit its
translation (32). Regulation by
miRNA promotes the up- or downregulation of mRNA and plays an
important role in the occurrence and development of diseases
(33). It has been reported that
miR-222 can target matrix metallopeptidase 1 to regulate the
biological functions of tongue squamous carcinoma cells (34). In addition, miR-222 can regulate dual
specificity mitogen-activated protein kinase and tumor necrosis
factor-related apoptosis-inducing ligand pathways of thyroid
follicular epithelial cells (35).
miR-222 can also affect the progression of gastric cancer by
regulating its target gene cyclin dependent kinase inhibitor 1B
(36). Abnormal expression of
miR-222 was also found in a transgenic mouse gastric cancer model
(37). miR-222 also has a unique
regulatory role in ovarian cancer-associated macrophages (38). These reports indicate that miRNA-222
is closely related to the occurrence and development of tumors.
In the present study reduced expression levels of
miR-222 and increased expression levels of ALDH1 were discovered in
tumor tissues and peripheral blood from cervical cancer patients.
These findings suggest that cleavage of ALDH1 by miR-222 may be
reduced by downregulation of miR-222. In order to further study the
underlying molecular mechanisms by which miR-222 and ALDH1 regulate
cervical cancer cells, HeLA cells were transfected with agomiR-222,
leading to overexpression of miR-222, and downregulation of ALDH1
mRNA and protein when compared with controls. The proliferation of
HeLA cells was also inhibited following transfection with
agomiR-222 compared with agomiR-NC. The results of a dual
luciferase reporter assay revealed that miR-222 may directly bind
with the 3′-UTR seed region of ALDH1 mRNA to regulate its
expression.
A limitation of the present study is that only one
type of cells was used in the in vitro experiments. This
work will be expanded on in future studies using a variety of
additional cell types to confirm these findings.
In conclusion, the present study demonstrated that
the expression of ALDH1 was elevated and that of miR-222 was
reduced in cervical cancer tissues and peripheral blood when
compared with controls. miR-222 may prevent the occurrence and
development of cervical cancer by regulating the proliferation of
tumor cells via ALDH1.
Acknowledgements
The authors wish to thank Professor Shuzhen Xu from
Friendship Hospital Affiliated to Capital Medical University
(Beijing, China) for her suggestions and instructions on the design
of the study.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
The final version of the manuscript was read and
approved by all authors, and each author believes that the
manuscript represents honest work. CL and YY collaborated to design
the study. CL, YZ and SL were responsible for performing
experiments. CL and YY analyzed the data. All authors collaborated
to interpret results and develop the manuscript.
Ethics approval and consent to
participate
All procedures performed in the current study were
approved by the Ethics Committee of Tsinghua University. Written
informed consent was obtained from all patients or their
families.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hovland S, Muller S, Skomedal H, Mints M,
Bergström J, Wallin KL, Karlsen F, Johansson B and Andersson S:
E6/E7 mRNA expression analysis: A test for the objective assessment
of cervical adenocarcinoma in clinical prognostic procedure. Int J
Oncol. 36:1533–1539. 2010.PubMed/NCBI
|
|
2
|
Wolfson IN: Letter: Blind defibrillation.
Am J Cardiol. 36:4121975. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sankaranarayanan R and Ferlay J: Worldwide
burden of gynaecological cancer: The size of the problem. Best
Pract Res Clin Obstet Gynaecol. 20:207–225. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Arbyn M, Castellsague X, de Sanjose S,
Bruni L, Saraiya M, Bray F and Ferlay J: Worldwide burden of
cervical cancer in 2008. Anna Oncol. 22:2675–2686. 2011. View Article : Google Scholar
|
|
5
|
Andrae B, Andersson TM, Lambert PC,
Kemetli L, Silfverdal L, Strander B, Ryd W, Dillner J, Törnberg S
and Sparén P: Screening and cervical cancer cure: Population based
cohort study. BMJ. 344:e9002012. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Voulgari A and Pintzas A:
Epithelial-mesenchymal transition in cancer metastasis: Mechanisms,
markers and strategies to overcome drug resistance in the clinic.
Biochim Biophys Acta. 1796:75–90. 2009.PubMed/NCBI
|
|
7
|
Huang R and Rofstad EK: Cancer stem cells
(CSCs), cervical CSCs and targeted therapies. Oncotarget.
8:35351–35367. 2017.PubMed/NCBI
|
|
8
|
Iden M, Fye S, Li K, Chowdhury T,
Ramchandran R and Rader JS: The lncRNA PVT1 contributes to the
cervical cancer phenotype and associates with poor patient
prognosis. PLoS One. 11:e01562742016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Canuto RA, Muzio G, Salvo RA, Maggiora M,
Trombetta A, Chantepie J, Fournet G, Reichert U and Quash G: The
effect of a novel irreversible inhibitor of aldehyde dehydrogenases
1 and 3 on tumour cell growth and death. Chem Biol Interact.
130-132:209–218. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang M, Shoeb M, Goswamy J, Liu P, Xiao
TL, Hogan D, Campbell GA and Ansari NH: Overexpression of aldehyde
dehydrogenase 1A1 reduces oxidation-induced toxicity in SH-SY5Y
neuroblastoma cells. J Neurosci Res. 88:686–694. 2010.PubMed/NCBI
|
|
11
|
Ginestier C, Hur MH, Charafe-Jauffret E,
Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG,
Liu S, et al: ALDH1 is a marker of normal and malignant human
mammary stem cells and a predictor of poor clinical outcome. Cell
Stem Cell. 1:555–567. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nalwoga H, Arnes JB, Wabinga H and Akslen
LA: Expression of aldehyde dehydrogenase 1 (ALDH1) is associated
with basal-like markers and features of aggressive tumours in
African breast cancer. Br J Cancer. 102:369–375. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sladek NE, Kollander R, Sreerama L and
Kiang DT: Cellular levels of aldehyde dehydrogenases (ALDH1A1 and
ALDH3A1) as predictors of therapeutic responses to
cyclophosphamide-based chemotherapy of breast cancer: A
retrospective study. Rational individualization of
oxazaphosphorine-based cancer chemotherapeutic regimens. Cancer
Chemother Pharmacol. 49:309–321. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao L, Zhang Z, Lou H, Liang J, Yan X, Li
W, Xu Y and Ou R: Exploration of the molecular mechanisms of
cervical cancer based on mRNA expression profiles and predicted
microRNA interactions. Oncol Lett. 15:8965–8972. 2018.PubMed/NCBI
|
|
15
|
Ding J, Xu Z, Zhang Y, Tan C, Hu W, Wang
M, Xu Y and Tang J: Exosome-mediated miR-222 transferring: An
insight into NF-kappaB-mediated breast cancer metastasis. Exp Cell
Res. 369:129–138. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Di Fazio P, Maass M, Roth S, Meyer C,
Grups J, Rexin P, Bartsch DK and Kirschbaum A: Expression of
hsa-let-7b-5p, hsa-let-7f-5p, and hsa-miR-222-3p and their putative
targets HMGA2 and CDKN1B in typical and atypical carcinoid tumors
of the lung. Tumour Biol. 39:10104283177284172017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li Y, Zhao L, Shi B, Ma S, Xu Z, Ge Y, Liu
Y, Zheng D and Shi J: Functions of miR-146a and miR-222 in
tumor-associated macrophages in breast cancer. Sci Rep.
5:186482015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu K, Liang X, Shen K, Sun L, Cui D, Zhao
Y, Tian J, Ni L and Liu J: MiR-222 modulates multidrug resistance
in human colorectal carcinoma by down-regulating ADAM-17. Exp Cell
Res. 318:2168–2177. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rao X, Di Leva G, Li M, Fang F, Devlin C,
Hartman-Frey C, Burow ME, Ivan M, Croce CM and Nephew KP:
MicroRNA-221/222 confers breast cancer fulvestrant resistance by
regulating multiple signaling pathways. Oncogene. 30:1082–1097.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Marth C, Landoni F, Mahner S, McCormack M,
Gonzalez- Martin A and Colombo N; ESMO Guidelines Committee, :
Cervical cancer: ESMO Clinical Practice Guidelines for diagnosis,
treatment and follow-up. Ann Oncol. 29 (Suppl 4):iv2622018.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yamaguchi M, Dieffenbach CW, Connolly R,
Cruess DF, Baur W and Sharefkin JB: Effect of different laboratory
techniques for guanidinium-phenol-chloroform RNA extraction on
A260/A280 and on accuracy of mRNA quantitation by reverse
transcriptase-PCR. PCR Methods Appl. 1:286–290. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Heid CA, Stevens J, Livak KJ and Williams
PM: Real time quantitative PCR. Genome Res. 6:986–994. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hakama M, Coleman MP, Alexe DM and Auvinen
A: Cancer screening: Evidence and practice in Europe 2008. Eur J
Cancer. 44:1404–1413. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nozaki Y, Tamori S, Inada M, Katayama R,
Nakane H, Minamishima O, Onodera Y, Abe M, Shiina S, Tamura K, et
al: Correlation between c-Met and ALDH1 contributes to the survival
and tumor-sphere formation of ALDH1 positive breast cancer stem
cells and predicts poor clinical outcome in breast cancer. Genes
Cancer. 8:628–639. 2017.PubMed/NCBI
|
|
26
|
Choudhary S, Xiao T, Vergara LA,
Srivastava S, Nees D, Piatigorsky J and Ansari NH: Role of aldehyde
dehydrogenase isozymes in the defense of rat lens and human lens
epithelial cells against oxidative stress. Invest Ophthalmol Vis
Sci. 46:259–267. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Quash G, Fournet G, Chantepie J, Gore J,
Ardiet C, Ardail D, Michal Y and Reichert U: Novel competitive
irreversible inhibitors of aldehyde dehydrogenase (ALDH1):
Restoration of chemosensitivity of L1210 cells overexpressing ALDH1
and induction of apoptosis in BAF(3) cells overexpressing bcl(2).
Biochem Pharmacol. 64:1279–1292. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim MJ, Kim AR, Jeong JY, Kim KI, Kim TH,
Lee C, Chung K, Ko YH and An HJ: Correlation of ALDH1 and notch3
expression: Clinical implication in ovarian carcinomas. J Cancer.
8:3331–3342. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zheng R, Wang J, Wu Q, Wang Z, Ou Y, Ma L,
Wang M, Wang J and Yang Y: Expression of ALDH1 and TGFβ2 in benign
and malignant breast tumors and their prognostic implications. Int
J Clin Exp Pathol. 7:4173–4183. 2014.PubMed/NCBI
|
|
30
|
House CD, Jordan E, Hernandez L, Ozaki M,
James JM, Kim M, Kruhlak MJ, Batchelor E, Elloumi F, Cam MC and
Annunziata CM: NFκB promotes ovarian tumorigenesis via classical
pathways that support proliferative cancer cells and alternative
pathways that support ALDH+ cancer stem-like cells. Cancer Res.
77:6927–6940. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang L, Liu Y, Zhou Y, Wang J, Tu L, Sun
Z, Wang X and Luo F: Zoledronic acid inhibits the growth of cancer
stem cell derived from cervical cancer cell by attenuating their
stemness phenotype and inducing apoptosis and cell cycle arrest
through the Erk1/2 and Akt pathways. J Exp Clin Cancer Res.
38:932019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kaehler M, Ruemenapp J, Gonnermann D,
Nagel I, Bruhn O, Haenisch S, Ammerpohl O, Wesch D, Cascorbi I and
Bruckmueller H: MicroRNA-212/ABCG2-axis contributes to development
of imatinib-resistance in leukemic cells. Oncotarget.
8:92018–92031. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu Q, Guo L, Jiang F, Li L, Li Z and Chen
F: Analysis of the miRNA-mRNA-lncRNA networks in ER+ and ER- breast
cancer cell lines. J Cell Mol Med. 19:2874–2887. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu X, Yu J, Jiang L, Wang A, Shi F, Ye H
and Zhou X: MicroRNA-222 regulates cell invasion by targeting
matrix metalloproteinase 1 (MMP1) and manganese superoxide
dismutase 2 (SOD2) in tongue squamous cell carcinoma cell lines.
Cancer Genomics Proteomics. 6:131–139. 2009.PubMed/NCBI
|
|
35
|
Aherne ST, Smyth P, Freeley M, Smith L,
Spillane C, O'Leary J and Sheils O: Altered expression of mir-222
and mir-25 influences diverse gene expression changes in
transformed normal and anaplastic thyroid cells, and impacts on MEK
and TRAIL protein expression. Int J Mol Med. 38:433–445. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lloyd KA, Moore AR, Parsons BN, O'Hara A,
Boyce M, Dockray GJ, Varro A and Pritchard DM: Gastrin-induced
miR-222 promotes gastric tumor development by suppressing p27kip1.
Oncotarget. 7:45462–45478. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Choi B, Yu J, Han TS, Kim YK, Hur K, Kang
BC, Kim WH, Kim DY, Lee HJ, Kim VN and Yang HK: Gastric
carcinogenesis in the miR-222/221 transgenic mouse model. Cancer
Res Treat. 49:150–160. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ying X, Wu Q, Wu X, Zhu Q and Wang X,
Jiang L, Chen X and Wang X: Epithelial ovarian cancer-secreted
exosomal miR-222-3p induces polarization of tumor-associated
macrophages. Oncotarget. 7:43076–43087. 2016. View Article : Google Scholar : PubMed/NCBI
|