Introduction
Cardiovascular diseases, including heart failure,
chronic ischemic heart disease and acute myocardial infarction,
remains a leading cause of morbidity and mortality worldwide
(1). The recent advent of the human
induced pluripotent stem cell (hiPSC) technology and an
increasingly refined capacity to differentiate hiPSCs into
cardiomyocytes (CMs), provides an unprecedented opportunity for
potential applications in disease modeling, drug discovery,
toxicity screening and novel cell-based therapies (2). Over the last two decades, a number of
protocols for cardiac differentiation have been established based
on embryonic body (EB) and monolayer methodologies (3–5).
Obtaining CMs of a high yield and purity by improving cell survival
may be invaluable for future applications in cell replacement
therapy and novel drug screening.
Rho-associated coiled-coil containing kinases
(ROCKs) are members of the serine/threonine protein kinase family
that are well documented to be involved in the regulation of a wide
range of fundamental cellular functions, including cell
proliferation, apoptosis, cell mitosis adhesion, cytoskeletal
adjustments and muscle cell contraction (6,7).
Evidence from a plethora of animal and clinical studies have
suggested ROCKs to be important mediators of a number of
pathophysiological characteristics associated with cardiovascular
diseases, including cardiac remodeling, fibrosis and hypertrophy
(6,8). Of note, Y27632, a pharmacological
inhibitor of ROCK and the calcium-sensitization pathway of smooth
muscle contraction, has been previously applied as a therapeutic
drug for the treatment of cardiovascular diseases (9). Apart from Y27632, fasudil is another
potent ROCK inhibitor and vasodilator which has been used for the
treatment of cerebral vasospasm (10). Indeed, a number of previous studies
have indicated that ROCK1 and ROCK2 contribute to pathological CM
apoptosis (11,12). Additionally, long-term fasudil and
Y27632 treatment has been demonstrated to attenuate CM apoptosis
and limit the progression of pathological cardiac fibrosis,
hypertrophy and remodeling, which lead to heart failure (13,14).
However, the effects of the ROCK signaling pathway on hiPSC-derived
CMs have not yet been thoroughly investigated.
Therefore, the aim of the present study was to
examine whether ROCK inhibitors can be used to increase the
survival of hiPSC-derived CMs, which may facilitate the development
of applications in stem cell therapy for the treatment of heart
disease. Specifically, the potential effects of the ROCK
inhibitors, Y27632 and fasudil, were evaluated on hiPSC-derived
CMs. The results indicated that ROCK inhibitors promoted hiPSC-CM
proliferation, increased the number of viable cells and inhibited
the apoptosis of hiPSC-CMs in suspension. The mechanism of these
physiological effects mediated by the ROCK inhibitor on hiPSC-CM
apoptosis may be via the suppression of caspase-3 expression.
Materials and methods
The differentiation of hiPSC into
cardiomyocytes using small molecules
HiPSCs (cat. no. DYR0100 Type Culture Collection of
the Chinese Academy of Sciences) were cultured at 37°C and 5%
CO2 in mTeSR™ 1 maintenance medium (Stemcell
Technologies, Inc.). Cells were seeded into 35 mm culture dishes
(2×105 cells/dish) coated with Matrigel (Corning, Inc.)
and passaged once every 2–3 days using ACCUTASE™ cell detachment
solution (Stemcell Technologies, Inc.).
For direct differentiation into cardiomyocytes
(5), hiPSC first were split at a 1:6
ratio using mTeSR media and cultured for 2 days to ~85% confluence.
On day 0, cells were first treated with 6 µM GSK-β inhibitor
CHIR99021 (MedChemExpress) diluted in in RPMI/B27 without insulin
for 48 h, followed by transferal to RPMI/B27 minus insulin
supplemented with 3 µg/ml Wnt signaling inhibitor IWP2
(MedChemExpress) for another 3 days. On day 5 following CHIR99021
treatment, the medium was changed to RPMI/B27 without insulin and
finally to RPMI/B27 with insulin medium on day 7 for further
maintenance until day 21.
To purify hiPSC-CMs, a biochemical purification
protocol was used using glucose-depleted DMEM (Invitrogen; Thermo
Fisher Scientific, Inc.) supplemented with 4 mM lactate
(Sigma-Aldrich; Merck KGaA) on day 15 (15). On day 15, the medium was changed to
the purification medium, consisting of glucose depleted DMEM, 500
µg/ml recombinant human albumin (Oryzogen; Wuhan Healthgen
Biotechnology, Corp.) and 213 µg/ml l-ascorbic acid 2-phosphate
supplemented (Sigma-Aldrich; Merck KGaA) with 4 mM l-lactic acid
(Sigma-Aldrich; Merck KGaA). Medium was refreshed every 2 days
during the purification process. On day 19, cells were maintained
in RPMI/B27 media and allowed to recover for 2 days.
To investigate the effects of Y27632 on hiPSC-CMs,
cells were first dissociated and seeded into gelatin-coated 12-well
plates (Corning, Inc.) at a density of 3×105 cells/well
under a number of different conditions. For controls, hiPSC-CMs
were cultured in DMEM supplemented with 20% FBS (Invitrogen; Thermo
Fisher Scientific, Inc.) in the absence of Y27632 (Selleck
Chemicals). For other treatment groups, cells were cultured in DMEM
containing 20% FBS supplemented with various concentrations of
Y27632 (5, 10 or 20 µM). After 24 and 48 h, cells were washed in
PBS, trypsinized and counted (n=3). Specifically, 10 µl cells mixed
with equal volumes of 0.4% trypan blue dye (Beijing Solarbio
Science & Technology Co., Ltd.) were counted using a
hemocytometer.
Immunostaining and fluorescent
microscopy
Cells were first fixed in 4% paraformaldehyde
(Beyotime Institute of Biotechnology) for 15 min at room
temperature (RT), permeabilized with 0.1% Triton X-100
(Sigma-Aldrich: Merck KGaA) for 10 min at RT and blocked with 10%
goat serum (Sigma-Aldrich; Merck KGaA) at RT for 1 h. Samples were
subsequently stained with primary antibodies against cardiac
troponin T (cTnT; 1:200; cat. no. 710635; Invitrogen; Thermo Fisher
Scientific, Inc.) and α-actinin (Invitrogen; 1:200; cat. no.
710947; Thermo Fisher Scientific, Inc.), dissolved in PBS at 4°C
for 12 h and further incubated with Alexa Fluor™ 555-conjugated
antibody (1:500; cat. no. A0460; Beyotime Institute of
Biotechnology) and Alexa Fluor™ 488-conjugated antibody (1:500;
cat. no. A0423; Beyotime Institute of Biotechnology) at RT for 2 h.
Nuclei were stained using 1 µg/ml DAPI (Sigma-Aldrich; Merck KGaA)
at RT for 5 min. An Optiphot-2 microscope (Nikon Corporation)
equipped with a CCD video camera system (Optronics Engineering,
Ltd.) and a computer interface (NIS-Elements; version 4.2.0; Nikon
Corporation) was used for imaging analysis (magnification, ×100).
In total, 40 images were taken for each well.
Crystal violet staining
Cells were first washed with PBS, fixed in 4%
paraformaldehyde at RT for 15 min and stained with 0.1% crystal
violet solution (Sigma-Aldrich; Merck KGaA) at RT for 30 min. After
the cells were washed with ddH2O and air-dried at RT,
images were obtained using a color digital camera (Optronics
Engineering, Ltd.). Absorbance was subsequently measured at 590 nm
using an automatic microplate reader (BioTek™ ELx800™; Omega
Bio-Tek, Inc.).
Cell counting kit-8 (CCK-8) assay
Cells were plated into 96-well plates in triplicate
at ~10,000 cells/well and subsequently cultured with or without 10
µM Y27632 at 37°C and 5% CO2 for 24 h. Cells were
analyzed using CCK-8 assay according to the manufacturer's protocol
(Beyotime Institute of Biotechnology). In total, 10 µl CCK-8 were
added per well and incubated for 2 h at 37°C. The absorbance value
at a wavelength of 450 nm was measured for each well using an
enzyme-linked immunosorbent assay reader (Thermo Lab Systems;
Thermo Fisher Scientific, Inc.).
5-bromo-2-deoxyuridine (BrdU)
incorporation assay
hiPSC-CM proliferation was evaluated using BrdU
incorporation assay. The same number of hiPSC-CMs
(1.5×105 cells) were cultured in DMEM supplemented with
20% FBS and 10 µM Y27632 for 24 h after passage on day 21,
following which the cells were treated with or without 10 µM Y27632
for 3 or 6 days, and incubation with or without 10 µM fasudil for 6
days (16). Cells were then treated
with 10 µM BrdU (Sigma-Aldrich; Merck KGaA) and incubated at 37°C
for 6 h. Samples were then fixed using 4% paraformaldehyde at RT
for 15 min, permeabilized by 0.1% Triton X-100 at RT for 15 min and
acid-washed by 1 M HCl on ice for 10 min. BrdU-positive cells were
subsequently detected by immunocytochemistry using mouse anti-BrdU
(1:1,000; cat. no. B2531; EMD Millipore) at 4°C overnight and goat
anti-mouse FITC-conjugated secondary antibodies (1:400; cat. no.
BA1101; Molecular Probes; Thermo fisher Scientific, Inc.) at RT for
1 h. Nuclei were visualized via 1 µg/ml DAPI staining at RT for 5
min. In total, each dish was divided into 4 regions, where 10
images were taken per region; ~2,000 cells were quantified and
normalized to the total cell number in each field using ImageJ
software (version 1.8.0; National Institutes of Health).
Annexin V-FITC/propidium iodide (PI)
apoptosis detection by fluorescence-activated cell sorting (FACS)
assay
Cell apoptosis was assessed using annexin-V-FITC and
PI staining kit (BD Biosciences). In total, 5×105 cells
were collected by trypsinization at 37°C for 5 min, centrifuged
under 200 × g at 4°C for 5 min and washed twice with PBS,
resuspended in 500 µl 1X binding buffer and incubated in 5 µl
Annexin V-FITC and 5 µl PI solution in the dark for 15 min at RT.
Cells were analyzed using an integrated BD Accuri™ C6 system (BD
Biosciences). The gates for flow cytometry was set using following
steps: Target cell population was circled by detecting unstained
cells. Annexin V-FITC single positive cells were detected,
following which fluorescence compensation was adjusted to ensure no
particles were in the upper left (UL) and upper right (UR)
quadrants. PI single positive cells were detected and fluorescence
compensation was adjusted to ensure no particles were present in
the upper right (UR) and lower right (LR) quadrants. A total of
four populations of cells were observed: i) Cells that were viable
and not undergoing apoptosis are both FITC-Annexin V and
PI-negative (LL); ii) cells undergoing apoptosis were FITC-Annexin
V-positive and PI-negative (LR); iii) the cells that are undergoing
late apoptosis or necrotic are both positive for FITC-Annexin V and
PI (UL and UR).
Reverse transcription-quantitative PCR
(RT-qPCR) or semi-quantitative PCR analysis (RT-PCR)
Total RNA was extracted and prepared using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. cDNA was
synthesized from 1 µg RNA using the AccuPower® RT Premix
(Bioneer Corporation). The temperature for the reverse
transcription reaction was 25°C for 5 min, 42°C for 60 min and 70°C
for 5 min, following which the product was used for RT-qPCR and
RT-PCR analyses.
RT-PCR was performed using ACTB mRNA as the
normalizing internal control and carried out using Taq DNA
Polymerase (Takara Bio, Inc.). The thermocycling conditions (total
volume per reaction, 20 µl) were as follows: Initial 5 min
denaturation at 95°C, followed by 30 cycles of 95°C for 30 sec,
60°C for 30 sec and 72°C for 30 sec and a final extension for 10
min at 72°C. PCR products were resolved by electrophoresis on a
1.5% agarose gel and stained using GelRed® Nucleic acid
gel stain (Biotium, Inc.), then visualized via UV transillumination
and photographed using a Gel Doc™ EZ System (Bio-Rad Laboratories,
Inc.).
qPCR reactions were set up in duplicate using
SYBR® Premix Ex Taq™ (Takara Bio, Inc.) and performed
using the 7500 Real-Time PCR system (Applied Biosystems; Thermo
Fisher Scientific, Inc.). qPCR reactions of 20 µl for eachsample
were made and consisted of cDNA (200 µg cDNA after dilution), 2X
SYBR Premix Ex Taq, 50X ROX Reference Dye, dH2O and 10
µM of each gene-specific primer. The thermocycling conditions were
as follows: Initial denaturation at 95°C for 30 sec, followed by 40
cycles of 95°C for 5 sec, annealing at 60°C for 34 sec, 72°C for 30
sec and a final extension at 72°C for 10 min. Relative mRNA
abundance was calculated using the 2−ΔΔCq method
(17) and normalized to the
expression levels of ACTB. The sequences of the primers used are
listed in Table I.
 | Table I.Primers used for PCR for the present
study. |
Table I.
Primers used for PCR for the present
study.
| Genes | Sequence
(5′-3′) | Size (bp) |
|---|
| ACTB | F:
AGCGAGCATCCCCCAAAGTT | 285 |
|
| R:
GGGCACGAAGGCTCATCATT |
|
| TNNT2 | F:
TTCACCAAAGATCTGCTCCTCGCT | 166 |
|
| R:
TTATTACTGGTGTGGAGTGGGTGTGG |
|
| TNNI3 | F:
CTGCAGATTGCAAAGCAAGA | 379 |
|
| R:
CCTCCTTCTTCACCTGCTTG |
|
| MYL7 | F:
GAGGAGAATGGCCAGCAGGAA | 450 |
|
| R:
GCGAACATCTGCTCCACCTCA |
|
| MYL2 | F:
ACATCATCACCCACGGAGAAGAGA | 247 |
|
| R:
ATTGGAACATGGCCTCTGGATGGA |
|
| Bax | F:
CCCCCGAGAGGTCTTTTTCC | 160 |
|
| R:
TGTCCAGCCCATGATGGTTC |
|
| Bcl-2 | F:
GAACTGGGGGAGGATTGTGG | 164 |
|
| R:
AAAGCCAGCCTCCGTTATCC |
|
| Caspase-3 | F:
GGGGATTGTTGTAGAAGTCTAACT | 178 |
|
| R:
AATAACCAGGTGCTGTGGAGTA |
|
| Caspase-8 | F:
ATTAGGGACAGGAATGGAACAC | 180 |
|
| R:
GGAGAGGATACAGCAGATGAAG |
|
Western blot analysis
Cells were prepared and lysed in ice-cold RIPA
buffer (0.5 M Tris-HCl, pH 7.4, 1.5 M NaCl, 2.5% deoxycholic acid,
10% NP-40 and 10 mM EDTA; EMD Millipore) containing freshly added
Halt Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific,
Inc.) and Protease Inhibitor Cocktail (Sigma-Aldrich; Merck KGaA).
Protein concentrations were determined using Bradford protein assay
(Bio-Rad Laboratories, Inc.). In total, 20 µg protein samples were
separated by 10% or 12% SDS-PAGE using an electrophoresis system
(Bio-Rad Laboratories, Inc.) and transferred onto nitrocellulose
membranes. The membranes were subsequently blocked with 5% non-fat
milk for 1 h at RT and probed with a number of different primary
antibodies (GAPDH, cat. no. 2118S; cleaved caspase-3, cat. no.
9661S; or caspase-8 cat. no. 4790S; all 1:1,000; Cell Signaling
Technology, Inc.) at 4°C overnight. Samples were then incubated
with horseradish peroxidase-conjugated goat anti-rabbit IgG (H+L)
(1:2,500; cat. no. SA00001-2; Proteintech Group, Inc.) at RT for 2
h. Blots were visualized using enhanced chemiluminescence (ECL)
reagent (Amersham; GE Healthcare Life Sciences), images were
captured using the ECL Tanon 5500 system (Tanon Science and
Technology Co., Ltd.) and analyzed using Tanon 5500 Multi automatic
chemiluminescence/fluorescent image analysis system (Tanon Science
and Technology Co., Ltd.).
FACS analysis
Following purification, 5×105 iPSC-CMs
were trypsinized under 37°C for 5 min, centrifugated at 200 × g at
RT for 5 min, fixed and permeabilized with 70% ethanol at 4°C
overnight and stained with phycoerythyrin-conjugated mouse
anti-cTnT antibodies (1:200; cat. no. 564767; BD Biosciences;) for
2 h at 4°C. Cells were washed three times with PBS and analyzed
using flow cytometry (BD Accuri™ C6, BD Biosciences).
Analysis of caspase-3 activity
Caspase-3 activity was detected using a caspase-3
Activity Assay kit (cat. no. C1116, Beyotime Institute of
Biotechnology). Cells (5×105) were cultured in the
presence or absence of 10 µM Y27632 for 3 days in a 6-well plate,
which were then washed with PBS, resuspended in the RIPA lysis
buffer (as part of the caspase-3 Activity Assay kit) and incubated
on ice for 15 min. For each well of a 96-well microplate, cell
lysates (50 µl/well), assay buffer and caspase-3 substrate
(Ac-DEVD-pNA, 10 µl/well) were combined together. Samples were then
incubated at 37°C for 2 h and quantified using microplate reader by
measuring absorbance at 405 nm.
Statistical analysis
All experiments of the current study were repeated
three times and the data are presented as the mean ± standard
deviation. GraphPad Prism 7 (GraphPad Software, Inc.) software was
used for the statistical analysis of all data. Statistical
differences between two groups were analyzed using Student's t-test
(two-tailed unpaired). Comparisons between multiple groups were
performed using one-way ANOVA followed by a Dunnett's post hoc
multiple comparison test. P<0.05 was considered to indicate a
statistically significant difference. To detect caspase-3 activity,
a linear regression line (Microsoft Corporation) was fitted to the
concentration: Optical density data. For comparative purposes,
caspase-3 activity values were generated for each group using
regression analyses.
Results
Cardiac differentiation and
characteristics of hiPSC-CMs
hiPSC-CMs were generated from hiPSC using CHIR9901
and IWP-2, inhibitors of GSK3β and Wnt respectively, in
insulin-free medium (Fig. 1A)
(5). Following ~8 days after
CHIR99021 treatment, spontaneously contracting cells were observed.
As presented in Fig. 1B, the
expression of cardiac markers, including troponin T type 2 (TNNT2),
troponin I type 3 (TNNI3), MYL7 and MYL2, were revealed to be
positive by RT-PCR analysis. In addition, hiPSC-CMs displayed
positive expression for cTnT and sarcomeric-α actinin on day 15
(Fig. 1C). FACS analysis
demonstrated that the purity of hiPSC-CMs isolated appeared to be
high, with >96% cells expressing cTnT on day 21 (Fig. 1D).
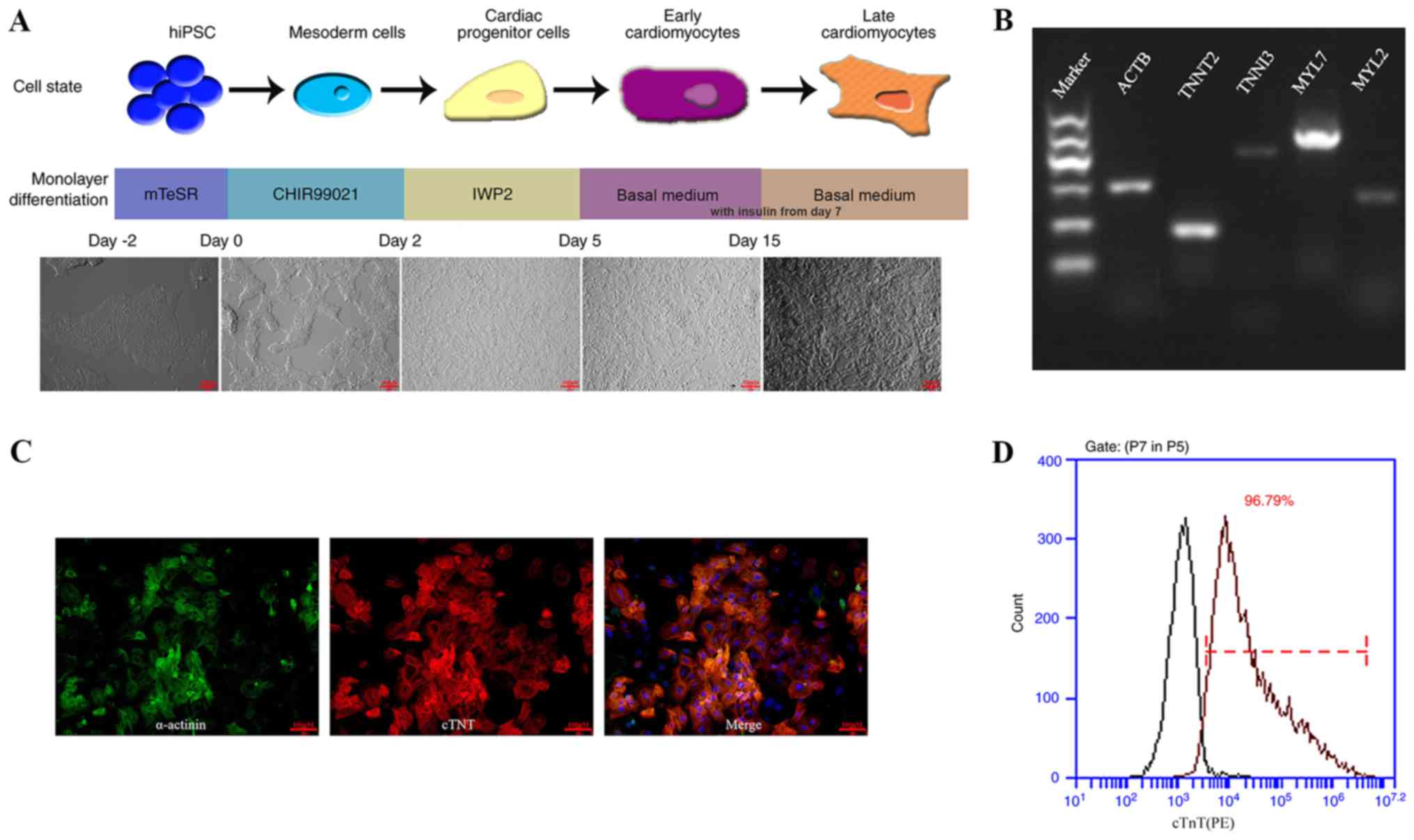 | Figure 1.hiPSC differentiation into hiPSC-CM
and characterization. (A) Schematic illustration of experimental
pipeline applied for the differentiation of human iPSC into
cardiomyocytes. The basal medium was supplemented with insulin from
day 7 onwards. (B) Reverse transcription-PCR analysis revealed that
cardiac gene expression was positive 30 days after the
differentiation process commenced. (C) On day 15, cells were
purified and re-plated onto 0.1% (wt/vol) gelatin-coated
coverslips. Immunostaining for α-actinin (green) and cTnT (red)
shows sarcomere organization. (Scale bar, 100 µm). (D)
Representative dot plots for cTnT populations. Left peaks represent
the isotype control. Numbers in plots indicate the percentage of
cTnT-positive cells after day 21 of differentiation. hiPSC, human
induced pluripotent stem cell; CM, cardiomyocites; ACTB, β-actin;
ACTB, β-actin; TNNT2, troponin T type 2; TNNI3, troponin I type 3;
MYL7, Myosin light chain 7; MYL2, Myosin light chain 2; hiPSC,
human induced pluripotent stem cells; CM, cardiomyocte; cTnT,
cardiac troponin T. |
ROCK inhibitor markedly promotes the
survival of dissociated of hiPSC-CMs
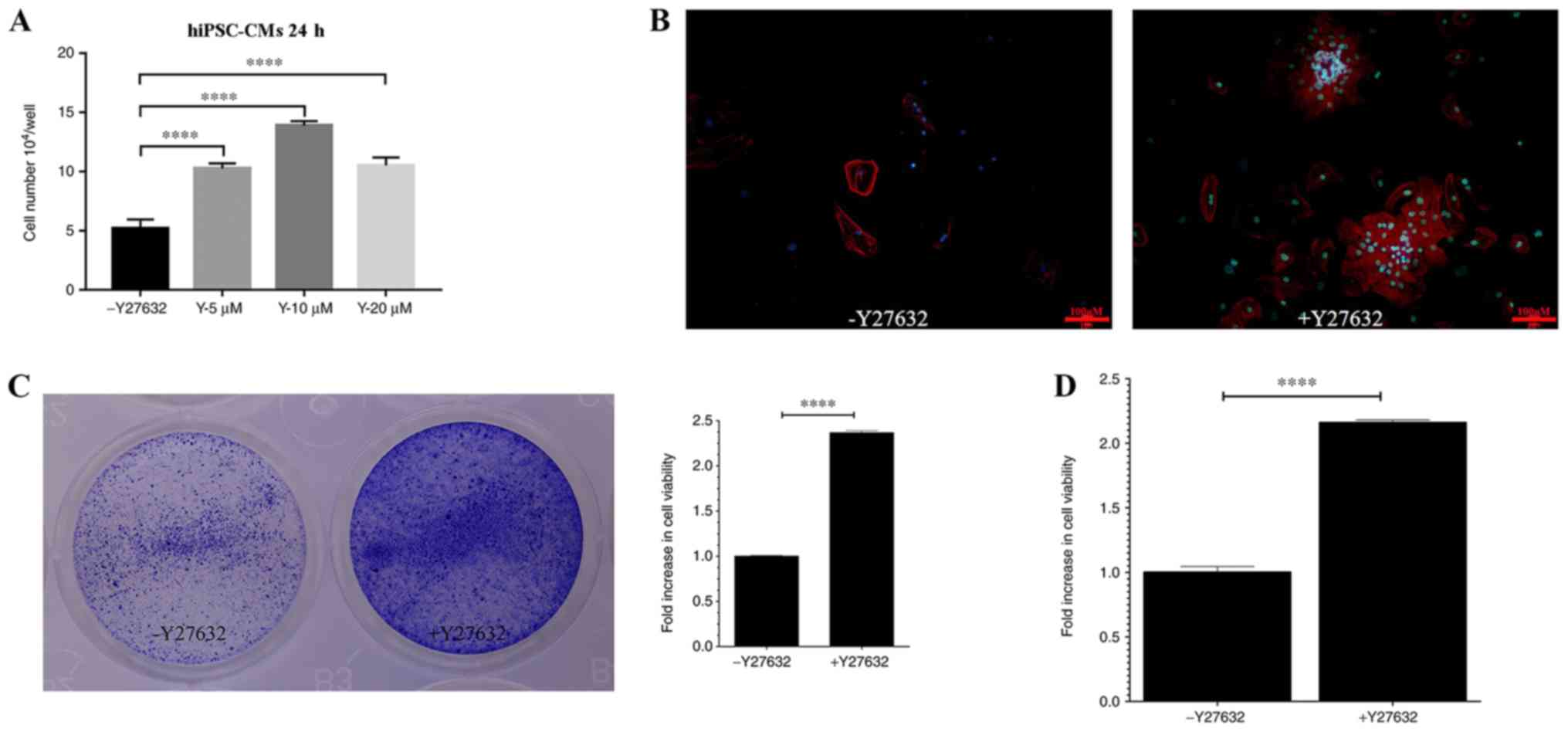
In the present study, the potential effects of
Y27632, a ROCK inhibitor, were investigated on hiPSC-CMs. To
examine if Y27632 exerts an effect on hiPSC-CM viability, cells
were cultured in the presence of 0, 5, 10 or 20 µM Y27632 for 24 h.
The number of viable cells increased in a dose-dependent manner, as
the concentration of Y27632 increased to 5 and 10 µM (Fig. 2A). However, at 20 µM, the number of
viable cells was decreased compared with 10 µM (Fig. 2A). Subsequently, the effects of
Y27632 on the viability of hiPSC-CMs, quantified as the number of
cells positively staining for the TNNT2, were examined. Y27632 (10
µM) markedly increased the plating efficiency of hiPSC-CMs
(Fig. 2B). The images and values
obtained from crystal violet staining demonstrated a significant
increase in relative cell viability following 10 µM Y27632
treatment compared with untreated cells (Fig. 2C). Quantitative analysis of the data
obtained from the CCK-8 assay also reported significant increases
in relative cell viability in Y27632 treated cells compared with
untreated cells (Fig. 2D). Taken
together, these data indicated that Y27632 treatment enhanced the
viability of hiPSC-CMs.
ROCK inhibitor enhances hiPSC-CMs
proliferation
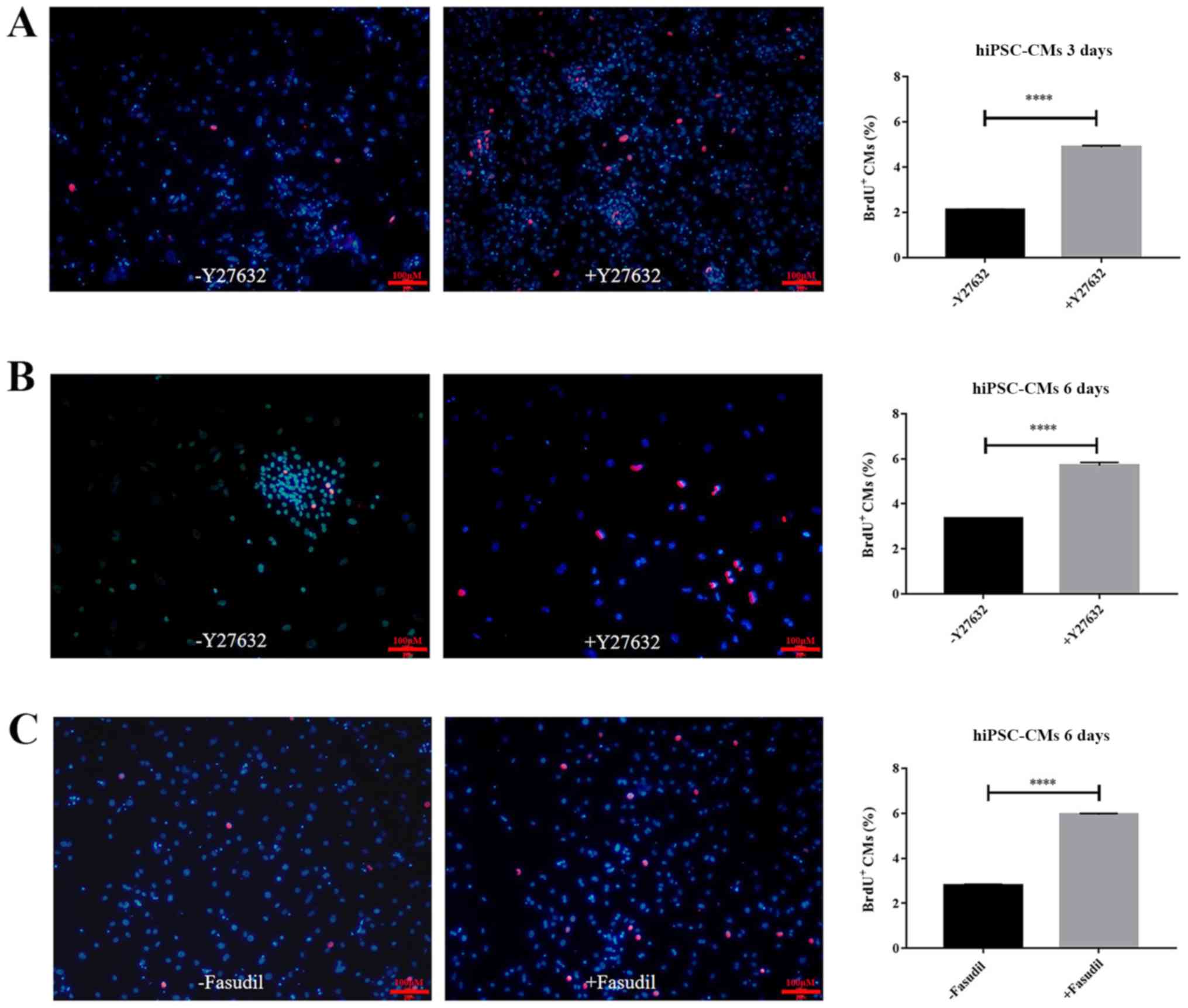
To determine whether the increased cell viability
was also due to increased cell proliferation, a BrdU incorporation
assay coupled with fluorescent microscopy was performed to measure
hiPSC-CM proliferation. After cells were cultured for 3 or 6 days
in the absence or presence of 10 µM Y27632, fluorescence images
were taken on day 3 (Fig. 3A) and
day 6 (Fig. 3B). Significant
increases in the numbers of BrdU-positive cells were observed
following Y27632 treatment for both days (P<0.0001; Fig. 3A and B). On day 3, the mean number of
BrdU+ cells counted in fluorescent images were 2.12±0.02
and 4.88±0.07% (Fig. 3A; n=3;
P<0.0001). After another 3 days, the proliferation rate of the
control was 3.34±0.01% of BrdU incorporation, compared with
5.7±0.12% for Y27632 treatment (Fig.
3B; n=3; P<0.0001). In addition, the results revealed that
BrdU-positive cells was also significantly increased following
treatment with fasudil, another ROCK inhibitor, for 6 days
(Fig. 3C; P<0.0001). These
findings indicated that treatment with ROCK inhibitors,
particularly Y27652, resulted in a 2.5-fold increase in hiPSC-CM
proliferation.
ROCK inhibitor inhibits the apoptosis
of dissociated hiPSC-CMs in serum-free medium
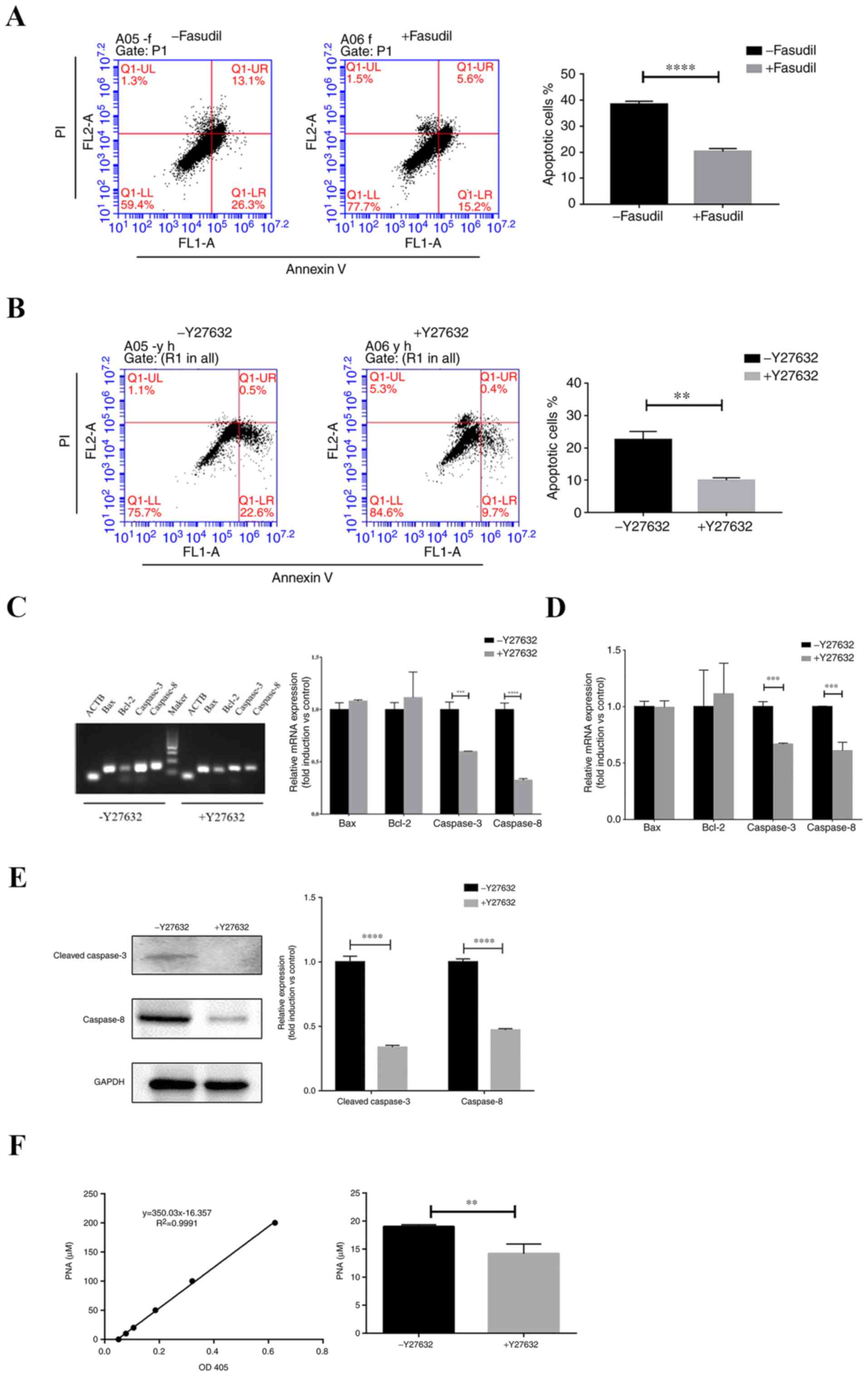
To assess the potential effects of Y27632 and
fasudil (both 10 µM) on hiPSC-CM apoptosis under serum starvation
and suspension conditions, hiPSC-CMs suspended in serum-free media
were seeded into non-adhesive dishes and cultured for 3 days.
Apoptosis was then evaluated using Annexin V/PI staining. Following
treatment with fasudil, number of apoptotic cells was significantly
decreased compared with control (P<0.0001; Fig. 4A). In addition, Y27632 treatment also
significantly reduced cell apoptosis compared with control
(P<0.001; Fig. 4B). Therefore,
treatment with ROCK inhibitors significantly reduced hiPSC-CM
apoptosis and necrosis, increasing survival under conditions of
stress.
To examine the potential mechanism of Y27632 on the
inhibition of apoptosis, mRNA levels of Bcl-2, Bax, caspases-3 and
−8 were detected after dissociated hiPSC-CMs were cultured in
suspension in the presence or absence of Y27632 for 72 h. The
expression of caspase-3 and −8 were significantly reduced by Y27632
(P<0.001), whilst levels of Bcl-2 and Bax did not exhibit
significant differences (Fig. 4C and
D).
Subsequent western blot analysis revealed that
Y27632 significantly reduced the protein expression of cleaved
caspase-3 and caspase-8 (P<0.0001; Fig. 4E). The colorimetric assay of
caspase-3 activity is based on the spectrophotometric detection of
pNA, which is produced by caspase-3-mediated cleavage of
acetyl-Asp-Glu-Val-Asp p-nitroanilide (Ac-DEVD-pNA). The level of
caspase-3 activity demonstrated that treatment with Y27632 reduced
caspase-3 activity (P<0.01; Fig.
4F). These results suggested that the preventive effects of
Y27632 on hiPSC-CMs apoptosis could be due to the suppression of
caspase-3 activation.
Discussion
Since being generated from a culture of human
fibroblasts using a cocktail of four transcription factors (Oct3/4,
Sox2, c-Myc and Klf4) in 2007, human iPSC technology has been
widely used for disease modeling, drug discovery and the
development of cell therapies (18,19).
Progress in hiPSC-derived cardiomyocytes has provided a platform
for personalized cardiovascular medicine (20). The development of effective
approaches in preventing cell death will facilitate the application
of hiPSC-derived CMs in multiple research areas. In the present
study, the effects of the ROCK inhibitor, Y27632, were evaluated in
hiPSC-CMs.
Significant improvements have been made in the
differentiation of pluripotent stem cells into cardiomyocytes over
the past two decades, which progressed from an original efficiency
of 5–10% to >90% (20,21). In the present study, a directed
cardiac differentiation method using small molecules (CHIR99021 and
IWP2) that modulate Wnt signaling were applied, which achieved an
efficiency of ~96.79%. Small molecules are advantageous compared
with growth factors in that they are more cost effective, easy to
store, more stable and more amenable to quality control (22).
Although structurally and functionally immature
hiPSC-CM limits its application in drug development and cell
therapy, their fetal characteristics may provide a possibility for
hiPSC-CMs to proliferate (23).
Uosaki et al (24) identified
four chemical compound groups: Inhibitors of glycogen synthase
kinase-3, p38 mitogen-activated protein kinase,
Ca2+/calmodulin-dependent protein kinase II and
activators of extra cellular signal-regulated kinase, all of which
synergistically enhanced the proliferation of cardiomyocytes
derived from both mouse and human PSCs. The present study
demonstrated that the ROCK inhibitor Y27632 exhibited proliferative
effects on hiPSC-CMs. Recently, Mohamed et al (25) screened a list of cell-cycle
regulators expressed in proliferating fetal cardiomyocytes. It was
demonstrated that the overexpression of cyclin-dependent kinase
(CDK) 1, CDK4, cyclin B1 and cyclin D1 efficiently induced cell
division in the post-mitotic mouse, rat and hiPSC-derived
cardiomyocytes (25). However, the
potential relationship between ROCK inhibition and cell-cycle
regulation requires further investigation.
Y27632 has been demonstrated to suppress cardiac
cell apoptosis in vivo and in vitro. Previous studies
have demonstrated that ROCK inhibitors enhanced post-ischemia
cardiac function, suppressed cardiac cell apoptosis and decreased
the accumulation of neutrophils in the heart following ischemia and
reperfusion injury in mice (26–29).
Bian et al (30) reported
that the pretreatment of Y27632 and 3-aminobenzamide reduced
myocardial infarction size and cardiomyocyte apoptosis via the poly
(ADP-ribose) polymerase/ERK signaling pathway. More recently, Dong
et al (31) evaluated the
cardioprotective effects of Y27632 and demonstrated that Y27632
attenuated myocardial injury by inhibiting the activation of the
MAPK and NF-κB signaling pathway, suppressing apoptosis and the
inflammatory response. The relationship between the Y27632-mediated
inhibition of apoptosis in the present study with the MAPK and
NF-κB signaling pathways merits further exploration.
ROCK inhibitors have been previously applied in
tissue engineering to improve cardiac cell engraftment for a number
of cardiovascular diseases based on cell therapy. Recently, Zhao
et al (32) revealed that
preconditioning with Y27632 increased the engraftment rate of
transplanted hiPSC-CMs in a murine myocardial infarction (MI) model
by a reduction in the number of apoptotic hiPSC-CMs but reported no
changes in proliferation. In the present study, the suppressive
effects of Y27632 on hiPSC-CM apoptosis was potentially caused by
the suppression of expression and activity of caspase-3 and
caspase-8. The results of proliferation were not consistent with
those of Zhao et al (32),
presumably due to different sources of hiPSC and differential
experimental conditions, including treatment time and culture
environments.
Additionally, previous studies have suggested that
ROCK kinase is involved in cell adhesion to the extracellular
matrix by modulating integrin avidity, which exerts further effects
on downstream physiological functions (7,33,34). In
particular, Martewicz et al (35) demonstrated the involvement of a
mechanotransduction pathway, RhoA/ROCK, in the structural
reorganization of hPSC-derived cardiomyocytes after adhesion.
Treatment with Y27632 prevented the structural reorganization of
the sarcoplasmic reticulum (SR) after attachment, modulating SERCA2
localization and promoting calcium cycling. They also suggested
that SR structural reorganization was observed in hPSC-CMs derived
from an embryonic bodies-based differentiation protocol that is
distinct from monolayer-based differentiation protocols (35). Recently, Yan et al (36) demonstrated that the application of
Y27632 promoted the gene expression of matrix metalloproteinases
2/3 and Notch-1 signaling to modulate Yes-associated protein-1
localization in the regulation of efficient patterning of
cardiovascular spheroids following mesoderm formation from hPSC.
Therefore, the possibility that ROCK kinase-associated integrin
signaling and receptors may be associated with cardiomyocyte
maturation, physiological function, proliferation and apoptosis,
which has not been examined in depth in the present study, cannot
be ruled out (37,38).
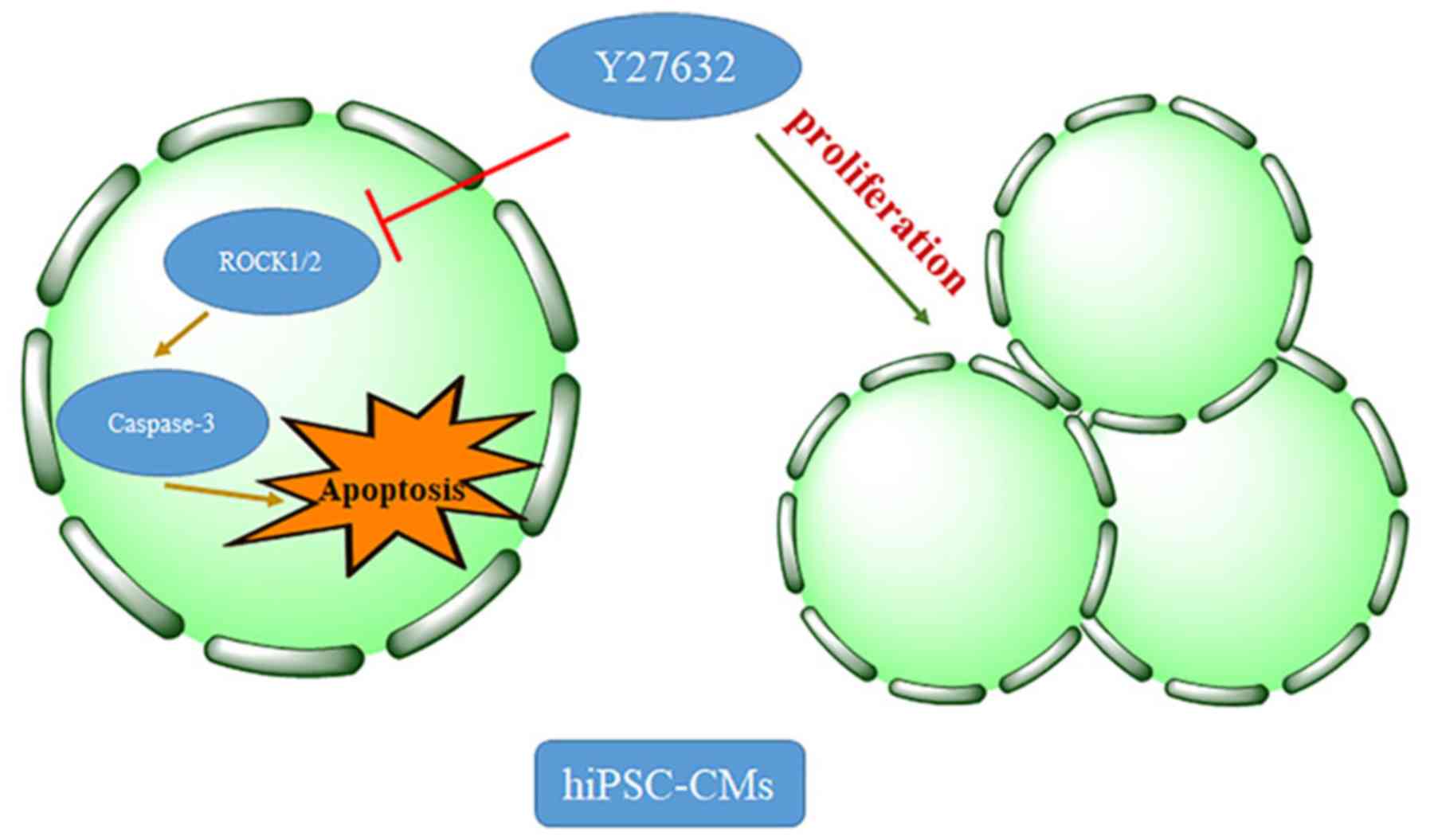
In conclusion, the present study demonstrated that
treatment with the ROCK inhibitor Y27632 significantly promoted the
survival of dissociated hiPSC-CMs by increasing proliferation and
attenuating apoptosis (Fig. 5). The
application of Rho-kinase inhibitors will be beneficial for the
culturing of CMs, which may facilitate the development of
applications of hiPSC-derived CMs for multiple research areas.
However, the associated mechanisms require further elucidation.
Acknowledgements
The authors would like to thank Dr Palecek
(Department of Chemical and Biological Engineering, University of
Wisconsin, Madison, WI, USA), for guidance with cardiac-directed
differentiation. The authors would also like to acknowledge Mr.
Ahmed Salah (Department of Biochemistry and Molecular Biology,
College of Life Science and Medicine, Zhejiang Sci-Tech University,
Hangzhou, Zhejiang, China) for the critical evaluation of the
manuscript and helpful comments.
Funding
The current study was supported the Enterprise
Commissioned R&D Project (grant no. 16040135-J).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
NQ and YW conceived and designed the experiments of
the current study. MK and MJ performed the experiments. MK wrote
the manuscript. HW and YY analyzed the data. All authors approved
the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ebert AD, Diecke S, Chen IY and Wu JC:
Reprogramming and transdifferentiation for cardiovascular
development and regenerative medicine: Where do we stand? EMBO Mol
Med. 7:1090–1103. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hartman ME, Dai DF and Laflamme MA: Human
pluripotent stem cells: Prospects and challenges as a source of
cardiomyocytes for in vitro modeling and cell-based cardiac repair.
Adv Drug Deliv Rev. 96:3–17. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kehat I, Kenyagin-Karsenti D, Snir M,
Segev H, Amit M, Gepstein A, Livne E, Binah O, Itskovitz-Eldor J
and Gepstein L: Human embryonic stem cells can differentiate into
myocytes with structural and functional properties of
cardiomyocytes. J Clin Invest. 108:407–414. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Burridge PW, Matsa E, Shukla P, Lin ZC,
Churko JM, Ebert AD, Lan F, Diecke S, Huber B, Mordwinkin NM, et
al: Chemically defined generation of human cardiomyocytes. Nat
Methods. 11:855–860. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lian X, Zhang J, Azarin SM, Zhu K,
Hazeltine LB, Bao X, Hsiao C, Kamp TJ and Palecek SP: Directed
cardiomyocyte differentiation from human pluripotent stem cells by
modulating Wnt/β-catenin signaling under fully defined conditions.
Nat Protoc. 8:162–175. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shimizu T and Liao JK: Rho kinases and
cardiac remodeling. Circ J. 80:1491–1498. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Narumiya S and Thumkeo D: Rho signaling
research: History, current status and future directions. FEBS Lett.
592:1763–1776. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shimizu T, Narang N, Chen P, Yu B, Knapp
M, Janardanan J, Blair J and Liao JK: Fibroblast deletion of ROCK2
attenuates cardiac hypertrophy, fibrosis, and diastolic
dysfunction. JCI Insight. 2(pii): 931872017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Surma M, Wei L and Shi J: Rho kinase as a
therapeutic target in cardiovascular disease. Future Cardiol.
7:657–671. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shibuya M and Suzuki Y: Treatment of
cerebral vasospasm by a protein kinase inhibitor AT 877. No To
Shinkei. 45:819–824. 1993.(In Japanese). PubMed/NCBI
|
|
11
|
Chang J, Xie M, Shah VR, Schneider MD,
Entman ML, Wei L and Schwartz RJ: Activation of Rho-associated
coiled-coil protein kinase 1 (ROCK-1) by caspase-3 cleavage plays
an essential role in cardiac myocyte apoptosis. Proc Natl Acad Sci
USA. 103:14495–14500. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hartmann S, Ridley AJ and Lutz S: The
function of Rho-associated kinases ROCK1 and ROCK2 in the
pathogenesis of cardiovascular disease. Front Pharmacol. 6:2762015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dong M, Ding W, Liao Y, Liu Y, Yan D,
Zhang Y, Wang R, Zheng N, Liu S and Liu J: Polydatin prevents
hypertrophy in phenylephrine induced neonatal mouse cardiomyocytes
and pressure-overload mouse models. Eur J Pharmacol. 746:186–197.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sawada N and Liao JK: Rho/Rho-associated
coiled-coil forming kinase pathway as therapeutic targets for
statins in atherosclerosis. Antioxid Redox Signal. 20:1251–1267.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tohyama S, Hattori F, Sano M, Hishiki T,
Nagahata Y, Matsuura T, Hashimoto H, Suzuki T, Yamashita H and
Satoh Y: Distinct metabolic flow enables large-scale purification
of mouse and human pluripotent stem cell-derived cardiomyocytes.
Cell Stem Cell. 12:127–137. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang L, Li Q, Li H, He Z, Cheng Z, Chen J
and Guo L: Inhibition of intracellular Ca2+ release by a
Rho-kinase inhibitor for the treatment of ischemic damage in
primary cultured rat hippocampal neurons. Eur J Pharmacol.
602:238–244. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Takahashi K, Tanabe K, Ohnuki M, Narita M,
Ichisaka T, Tomoda K and Yamanaka S: Induction of pluripotent stem
cells from adult human fibroblasts by defined factors. Cell.
131:861–872. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu J, Vodyanik MA, Smuga-Otto K,
Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA,
Ruotti V, Stewart R, et al: Induced pluripotent stem cell lines
derived from human somatic cells. Science. 318:1917–1920. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Matsa E, Ahrens JH and Wu JC: Human
induced pluripotent stem cells as a platform for personalized and
precision cardiovascular medicine. Physiol Rev. 96:1093–1126. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Qiu XX, Liu Y, Zhang YF, Guan YN, Jia QQ,
Wang C, Liang H, Li YQ, Yang HT and Qin YW: Rapamycin and CHIR99021
coordinate robust cardiomyocyte differentiation from human
pluripotent stem cells via reducing p53-dependent apoptosis. J Am
Heart Assoc. 6(pii): e0052952017.PubMed/NCBI
|
|
22
|
Kim WH, Jung DW and Williams DR: Making
cardiomyocytes with your chemistry set: Small molecule-induced
cardiogenesis in somatic cells. World J Cardiol. 7:125–133. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhu R, Blazeski A, Poon E, Costa KD, Tung
L and Boheler KR: Physical developmental cues for the maturation of
human pluripotent stem cell-derived cardiomyocytes. Stem Cell Res
Ther. 5:1172014. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Uosaki H, Magadum A, Seo K, Fukushima H,
Takeuchi A, Nakagawa Y, Moyes KW, Narazaki G, Kuwahara K, Laflamme
M, et al: Identification of chemicals inducing cardiomyocyte
proliferation in developmental stage-specific manner with
pluripotent stem cells. Circ Cardiovasc Genet. 6:624–633. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mohamed TMA, Ang YS, Radzinsky E, Zhou P,
Huang Y, Elfenbein A, Foley A, Magnitsky S and Srivastava D:
Regulation of cell cycle to stimulate adult cardiomyocyte
proliferation and cardiac regeneration. Cell. 173:104–116. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bao W, Hu E, Tao L, Boyce R, Mirabile R,
Thudium DT, Ma XL, Willette RN and Yue TL: Inhibition of Rho-kinase
protects the heart against ischemia/reperfusion injury. Cardiovasc
Res. 61:548–558. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dong M, Yan BP, Liao JK, Lam YY, Yip GW
and Yu CM: Rho-kinase inhibition: A novel therapeutic target for
the treatment of cardiovascular diseases. Drug Discov Today.
15:622–629. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Feng Y, LoGrasso PV, Defert O and Li R:
Rho kinase (ROCK) inhibitors and their therapeutic potential. J Med
Chem. 59:2269–2300. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li Y, Zhu W, Tao J, Xin P, Liu M, Li J and
Wei M: Fasudil protects the heart against ischemia-reperfusion
injury by attenuating endoplasmic reticulum stress and modulating
SERCA activity: The differential role for PI3K/Akt and JAK2/STAT3
signaling pathways. PLoS One. 7:e481152012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bian H, Zhou Y, Yu B, Shang D, Liu F, Li B
and Qi J: Rho-kinase signaling pathway promotes the expression of
PARP to accelerate cardiomyocyte apoptosis in ischemia/reperfusion.
Mol Med Rep. 16:2002–2008. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dong LY, Qiu XX, Zhuang Y and Xue S:
Y-27632, a Rho-kinase inhibitor, attenuates myocardial
ischemia-reperfusion injury in rats. Int J Mol Med. 43:1911–1919.
2019.PubMed/NCBI
|
|
32
|
Zhao M, Fan C, Ernst PJ, Tang Y, Zhu H,
Mattapally S, Oduk Y, Borovjagin AV, Zhou L, Zhang J and Zhu W:
Y-27632 preconditioning enhances transplantation of human-induced
pluripotent stem cell-derived cardiomyocytes in myocardial
infarction mice. Cardiovasc Res. 115:343–356. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Martinez-Rico C, Pincet F, Thiery JP and
Dufour S: Integrins stimulate E-cadherin-mediated intercellular
adhesion by regulating Src-kinase activation and actomyosin
contractility. J Cell Sci. 123:712–722. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Thuveson M, Gaengel K, Collu GM, Chin ML,
Singh J and Mlodzik M: Integrins are required for synchronous
ommatidial rotation in the Drosophila eye linking planar cell
polarity signalling to the extracellular matrix. Open Biol.
9:1901482019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Martewicz S, Serena E, Zatti S, Keller G
and Elvassore N: Substrate and mechanotransduction influence
SERCA2a localization in human pluripotent stem cell-derived
cardiomyocytes affecting functional performance. Stem Cell Res.
25:107–114. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yan Y, Bejoy J, Xia J, Griffin K, Guan J
and Li Y: Cell population balance of cardiovascular spheroids
derived from human induced pluripotent stem cells. Sci Rep.
9:12952019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Manzur MJ, Aguilera MO, Kotler ML, Berón W
and Ciuffo GM: Focal adhesion kinase, RhoA, and p38
mitogen-activated protein kinase modulates apoptosis mediated by
angiotensin II AT2 receptors. J Cell Biochem.
120:1835–1849. 2019. View Article : Google Scholar
|
|
38
|
Vitillo L, Baxter M, Iskender B, Whiting P
and Kimber SJ: Integrin-associated focal adhesion kinase protects
human embryonic stem cells from apoptosis, detachment, and
differentiation. Stem Cell Reports. 7:167–176. 2016. View Article : Google Scholar : PubMed/NCBI
|