Introduction
Enterococcus belongs to the Enterococcaceae
family, the members of which are symbiotic bacteria in the human
intestine (1). Enterococcus
infections may occur in the urinary tract and in meningitis,
diverticulitis, bacteremia and endocarditis infections with high
mortality rate of ~61% in Portugal (2). Enterococcus is used as a
probiotic to improve the intestinal environment when treating
bacterial diarrhea (3). There are
two main advantages to Enterococcus that make it the most
popular edible probiotic in animals and humans: i) In the
gastrointestinal (GI) tract, Enterococcus competes with
pathogens and thus decreases their virulence (2); ii) Enterococcus resists acid
stress and cannot be digested by GI-secreted digestive juice
(4). E. faecalis and E.
faecium are used as digestive agents for the treatment of
diarrhea caused by flatulence and indigestion (2). However, these two bacteria are
difficult to distinguish morphologically. Species identification
and genome characterization of these strains may elucidate
therapeutic strategies for bacterial infection and probiotic
treatment therapy.
The two bacteria were originally assigned to the
Streptococcus genus (2). In
1984, Schleifer et al (5)
indicated that they belonged to the Enterococcus genus via
DNA-DNA and DNA-rRNA hybridization. These changes were also
confirmed in ‘Bergey's Manual of Systematic Bacteriology Volume 3:
The Firmicutes’ in 2009, such that ‘Streptococcus faecalis’
was revised to Enterococcus faecalis, and ‘Streptococcus
faecium’ was revised to E. faecium (6). Due to previous changes in species
names, the bacterial standard used by a number of manufacturers is
inaccurate, which leads to incorrect or inconsistent identification
of bacteria. However, numerous species of Enterococci possess the
ability to transfer and carry antibiotic resistance genes (7). Therefore, the accurate definition and
identification of the different sources of the strains in
production is essential, as these issues may affect drug production
and safe use.
Full bacteria genome sequencing provides an
unprecedented method of investigating the biological processes and
evolutionary characteristics of bacteria. Using these data,
comparative analyses can be made to identify phylogenetic
relationships among species, obtain molecular markers for stains
and to investigate drug-resistant genes. To date, 527 E.
faecalis and 778 E. faecium draft genomes have been
submitted to GenBank (http://www.ncbi.nlm.nih.gov/genome; September 3,
2017). Although E. faecalis and E. faecium are
difficult to distinguish morphologically, different genome
structures have been identified between the two species. The gene
order of E. faecium is also significantly different from
that of E. faecalis according to complete genome data
analysis (8). In addition, using
competitive DNA hybridization, Shanks et al (9) demonstrated that a number of
cell-surface proteins were different between E. faecalis and
E. faecium, which may assist in developing biomarkers for
the identification of these species.
Shin Biofermin S is the most widely used commercial
medicine for the treatment of dyspepsia, abdominal distension and
diarrhea (10). Unfortunately, the
ingredients, including the species of strains, are yet to be fully
elucidated. The present study isolated the strain from Shin
Biofermin S. using the Illumina HiSeq 2000 platform, after which
the complete genomes of two E. faecium strains were
determined and annotated. The phylogenetic relationships between
these two strains and their genome characteristics were identified
and compared with those of other strains of E. faecalis and
E. faecium. This genomic information may provide a reference
genome data set of E. faecium strains to aid further
investigations into the ecological and functional diversity of
E. faecium. In addition, the results of the present
study identified the species of the two clinically applied strains,
140623 and SBS-1, which may guide the further production of edible
probiotics and the optimization of bacterial species.
Materials and methods
Isolation of strains and DNA
isolation
Strain 140623 was isolated from the National
Institute for Food and Drug Control as a control for the production
of Lactasin Tablets, and SBS-1 was isolated from Shin Biofermin S
(cat. no. X20000191, Biofermin Pharmaceutical Co., Ltd.; http://www.biofermin.co.jp/). The 140623 bacterial
powder was added to MRS medium (Tiangen Biotech Co., Ltd.) and
cultured for 48 h at 37°C. For the isolation of SBS-1, 1 g Shin
Biofermin S Tablet was first diluted in 9 ml 7.5% sodium chloride
solution and then cultured using 0.1 ml diluted test solution for
48 h at 37°C in MRS medium. Following culture, genomic DNA was
isolated using a Bacterial Genome DNA Extraction kit (Tiangen
Biotech Co., Ltd.) according to the manufacturer's protocol.
Biochemical identification of 140623
and SBS-1
The 140623 and SBS-1 strains were identified using
tests for the VITEK® 2 GP ID card in the VITEK 2 Compact
30 system (BioMérieux SA; http://www.biomerieux.com). The growth and biochemical
characteristics of these strains were assessed by the Bacteria
Preservation Center from the Institute of Microbiology, Chinese
Academy of Sciences (Beijing, China). The DuPont™
RiboPrinter® System (Hygiena, LLC) was used to identify
species by ribotyping (DuPont).
Analysis of fatty acid components by
gas chromatography-mass spectrometry (GC-MS)
In this experiment, the Enterococcus faecium
strain CGMCC1.131, provided by The China General Microbiological
Culture Collection Center (Institute of Microbiology; Chinese
Academy of Sciences) was used as control. Strains were grown in 100
ml MRS medium at 37°C for 48 h, centrifuged at 3000 × g for 10 min
at room temperature and washed three times with deionized water.
Collected bacteria were subsequently dried at 60°C for 3 h. The
fatty acid components were analyzed using an MS 5975C (Agilent
Technologies GmbH). The gas chromatography column was an HP-5MS (30
m × 0.25 mm × 0.25 µm) (Agilent Technologies GmbH). The carrier gas
was helium. The warming program was 120°C for 5 min, the
temperature was raised to 240°C (6°C/min) and maintained for 10
min. The temperature was then raised to 260°C (10°C/min) and
maintained for 2 min. The inlet temperature was 250°C, the split
ratio was 10:1, the flow was 1.0 and the injection volume was 1 µl.
The mass spectrometry conditions were an ion source with an
electron energy of 70 eV, a 3 min solvent delay, an electron
multiplier voltage gain factor (=1) mode with a full mass scan
range of 35–450 amu and a sampling frequency of 2. Multiple
reaction monitoring transitions was used to monitor entire eluate
with dwell time of 100 ms. The optimum voltage of the first
octupole was performed. To analyze the fatty acid methyl esters, 50
mg of samples or fatty acid standards was dissolved with 2 ml boron
trifluoride 14% in methanol solution and sealed by N2
(250°C), at a flow rate of 5 l/min and a nebulizer gas pressure of
20 psi, immediately following 10 min of ultrasonic shock. After
incubation in a 70°C water bath for 30 min, 2 ml N-hexane was added
and mixed. The supernatant was collected and washed with 1 ml
N-hexane following filtration with 0.22 µm filter membranes and
GC-MS analysis.
Genome sequencing and assembly
The genomes of 140623 and SBS-1 were sequenced using
the Illumina HiSeq 2000 platform (Illumina, Inc.). Genomic DNA (1
µg) from each strain was fragmented randomly and purified by
electrophoresis using 1.0% agarose gel (Tiangen Biotech Co., Ltd.).
The gel was then visualized by ethidium bromide staining under UV
light. The DNA fragments were connected to adaptors and sequenced
using the Illumina HiSeq 2000. The raw reads were used for de
novo assembly by SOAPdenovo software (version 2.04; http://soap.genomics.org.cn/soapdenovo.html) (11). The de novo assembly was
assessed by kmer and GC depth analysis. Kmer was calculated using
Meryl (https://github.com/marbl/meryl) with
parameter set to kmer=15. SOAP2 reads was used to map the genome
and calculate GC depth (11). E.
faecium T110 strain was used as control for the comparison of
sequences.
Genome annotation
The complete genome sequences of 140623 and SBS-1
were annotated using the Basic Local Alignment Search Tool (BLAST,
version: 2.2.27) (12) against the
Gene Ontology (GO) (13), Kyoto
Encyclopedia of Genes and Genomes (KEGG) (14–16),
Swiss-Prot (17), non-redundant
database and Clusters of Orthologous Groups (COG) (18,19)
databases (Expect (E) Value =1×10−100,000). The
colinearity analysis of genome was performed by mapping the
unigenes with high sequence similarity hits (threshold
value=1×10−30) (20). The
pathogenicity and drug resistance of animal pathogen analyses were
determined using type III secretion system (T3SS) prediction, which
was performed using EffectiveT3 (version 1.0.1) (21), Virulence Factors Database (version
20130128) (22) and Antibiotic
Resistance Genes Database (version 1.1) (23) database annotations. Repeated sequence
analyses, including small satellite sequences and microsatellite
sequences, were performed using Tandem Repeat Finder (24). Ribosomal RNA (rRNA) and bacterial
small RNA (sRNA) were predicted by searching the Rfam databases
(version 10.1) (25). The remainder
of the potential noncoding RNAs was predicted using RNAmmer
(version 1.2) (26) and tRNAscan
(version: 1.23) (27) software.
Potential virulence-associated genes were predicted by BLAST
queries against the pathogen-host interaction gene database
(http://www.phi-base.org). Genes of antibiotic
resistance were predicted using Antibiotic Resistance Genes
Database (ARDB) (https://ardb.cbcb.umd.edu/).
Results
Biochemical identification results of
140623 and SBS-1
Biochemical identification tests revealed that the
two tested strains were Gram-positive, spherical bacterium. No
spores were observed when culturing the bacteria. The catalase and
oxidase tests were negative in both strains. These bacteria could
grow in air and were able to survive and multiply at either 10 or
45°C. D-glucose, D-fructose, D-mannose, D-ribose, D-galactose,
L-arabinose, lactose, sucrose, maltose, trehalose, melibiose,
cellobiose, mannitol, sodium gluconate, esculin, salicin and
amygdalin tests were all positive, while the D-xylose, L-xylose,
D-arabinose, L-sorbose, L-rhamnose, melezitose, raffinose and
sorbitol tests exhibited negative results in the bacteria (Table I). These results indicated that the
two strains were E. faecium. The RiboPrinter ribotyping
results also indicated the two strains demonstrated high similarity
with E. faecium (Fig. 1).
 | Table I.Biochemical identification of two
strains. |
Table I.
Biochemical identification of two
strains.
| Test items | 140623 | SBS-1 |
|---|
| Gram staining | Positive | Positive |
| Cell shape | spherical | spherical |
| Spore | − | − |
| Catalase test | − | − |
| Oxidase test | − | − |
| Growth in air | + | + |
| Growth at 45°C | + | + |
| Growth at 10°C | + | + |
| 6.5% NaCl
growth | + | + |
| pH 9.6 growth | + | + |
| pH 4.5 growth | − | − |
| D-glucose | + | + |
| D-fructose | + | + |
| D-mannose | + | + |
| D-ribose | + | + |
| D-xylose | − | − |
| L-xylose | − | − |
| D-galactose | + | + |
| D-arabinose | − | − |
| L-arabinose | + | + |
| L-sorbose | − | − |
| L-rhamnose | − | − |
| Lactose | + | + |
| Sucrose | + | + |
| Maltose | + | + |
| Trehalose | + | + |
| Melibiose | + | + |
| Cellobiose | + | + |
| Melezitose | − | − |
| Raffinose | − | − |
| Sorbitol | − | − |
| Mannitol | + | + |
| Sodium
gluconate | + | + |
| Esculin | + | + |
| Salicin | + | + |
| Amygdalin | + | + |
| Species | Enterococcus
faecium | Enterococcus
faecium |
GC-MS analysis of fatty acid
components
The two strains revealed a GC-MS result of C14:0,
C16:1Δ9, C16:0, C18:1Δ9 and C18:0 peaks. A
specific 2-C8H17-C19:0 peak was observed in the tested strains. The
concentrations of C14:0, C16:1Δ9, C16:0,
C18:1Δ9, C18:0 and 2-C8H17-C19:0 were 6.60, 9.61, 17.50,
28.14, 3.93 and 36.29, respectively, in strain 140623. The
concentrations of C14:0, C16:1Δ9, C16:0,
C18:1Δ9, C18:0, and 2-C8H17-C19:0 were 6.29, 8.63,
17.58, 30.27, 3.95 and 32.47, respectively, in SBS-1. The two
strains had similar ratios of C14:0, C16:1Δ9, C16:0,
C18:1Δ9, C18:0, 2-C8H17-C19:0 (1.0:1.5:2.7:4.3:0.6:5.5
in 140623; 1.0:1.4:2.8:4.8:0.6:5.2 in SBS-1) with the control
(1.0:1.4:2.7:4.3:0.5:5.0 in control) E. faecium (Table II).
 | Table II.Relative content of standard strains
and the strains of lactasin tablets (%). |
Table II.
Relative content of standard strains
and the strains of lactasin tablets (%).
| Strains | C14:0 |
C16:1Δ9 | C16:0 |
C18:1Δ9 | C18:0 |
2-C8H17- C19:0 | Content ratio of
component |
|---|
| 140623 | 6.60 | 9.61 | 17.50 | 28.14 | 3.93 | 36.29 |
1.0:1.5:2.7:4.3:0.6:5.5 |
| SBS-1 | 6.29 | 8.63 | 17.58 | 30.27 | 3.95 | 32.47 |
1.0:1.4:2.8:4.8:0.6:5.2 |
|
Enterococcus | 6.68 | 9.66 | 17.85 | 28.56 | 3.43 | 33.5 |
1.0:1.4:2.7:4.3:0.5:5.0 |
Genome assembly of 140623 and
SBS-1
Illumina HiSeq 2000 generated 2,132 Mb and 2,413 Mb
raw data from 140623 and SBS-1, respectively. The sequence data of
which are available under NCBI BioProject number PRJNA549093. The
de novo assembly of 140623 generated 6 scaffolds (N50,
2,713,725 bp) and 63 contigs (N50, 125,144 bp), indicating that the
140623 genome had 2,812,926 bp nucleotides with 38.22% GC content
(Table III). The complete genome
of SBS-1 contained 2,797,745 bp nucleotides with 38.25% GC content,
which had 10 scaffolds (N50, 2,177,304 bp) and 38 contigs (N50,
208,101 bp; Table IV).
 | Table III.Summary of genome assembly of the two
strains. |
Table III.
Summary of genome assembly of the two
strains.
| A, 140623 |
|---|
|
|---|
| Characteristic | Scaffold | Contig |
|---|
| Total number,
n | 6 | 63 |
| Total length,
bp | 2,812,926 | 2,765,223 |
| N50, bp | 2,713,725 | 125,144 |
| N90, bp | 2,713,725 | 36,082 |
| Max length, bp | 2,713,725 | 460,559 |
| Min length, bp | 613 | 211 |
| GC content, % | 38.22 | 38.22 |
|
| B,
SBS-1 |
|
|
Characteristic |
Scaffold | Contig |
|
| Total number,
n | 10 | 38 |
| Total length,
bp | 2,797,745 | 2,754,045 |
| N50, bp | 2,177,304 | 208,101 |
| N90, bp | 415,863 | 49,841 |
| Max length, bp | 2,177,304 | 447,113 |
| Min length, bp | 1,148 | 232 |
| GC content, % | 38.25 | 38.25 |
 | Table IV.Genome component of the two
strains. |
Table IV.
Genome component of the two
strains.
|
| 140623 | SBS-1 |
|---|
| Genome size,
bp | 2,812,926 | 2,797,745 |
| GC content, % | 38.23 | 38.26 |
| Gene number | 2,766 | 2,734 |
| Gene length,
bp | 2,457,726 | 2,441,361 |
| Gene average
length, bp | 889 | 893 |
| Gene length/genome,
% | 87.37 | 87.26 |
| GC content in gene
region, % | 39.07 | 39.09 |
| Intergenic region
length, bp | 355,200 | 356,384 |
| GC content in
intergenic region, % | 32.42 | 32.57 |
| Intergenic region
length/genome, % | 12.63 | 12.74 |
| Tandem repeat
number | 70 | 70 |
| Tandem repeat
length, bp | 7,954 | 9,079 |
| Tandem repeat size,
bp | 5–676 | 5–692 |
| Tandem repeat
length/genome, % | 0.2828 | 0.3245 |
| Minisatellite DNA
Number | 39 | 38 |
| Microsatellite DNA
number | 2 | 1 |
| rRNA number | 0 | 3 |
| tRNA number | 41 | 54 |
| sRNA number | 4 | 4 |
| Genomic island
number | 0 | 0 |
| Prophage
number | 0 | 0 |
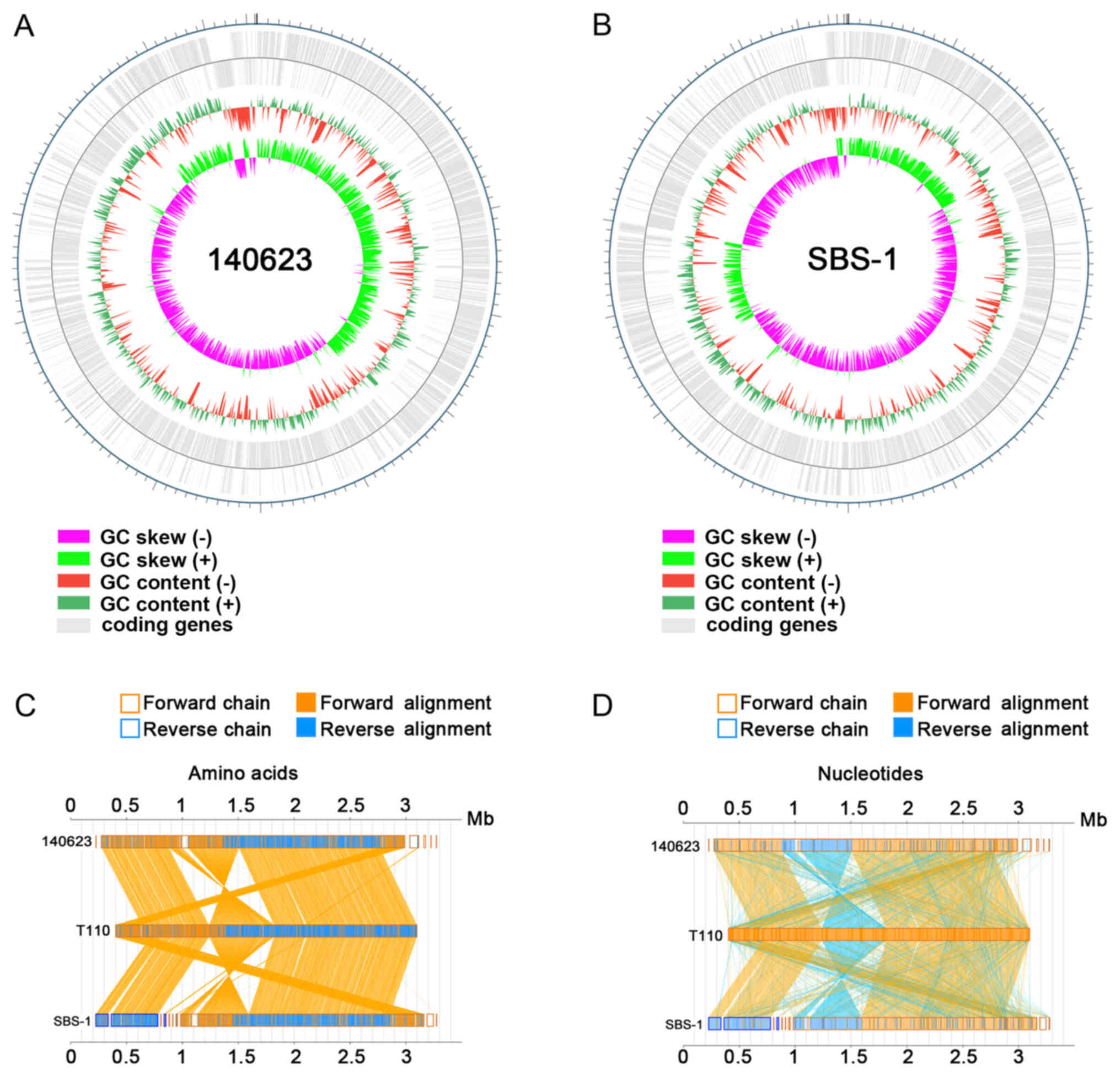
The genome characteristics, including gene
distribution and genome GC content distribution. A higher GC skew
(−) value was found in SBS-1, which may due to genome variation
between the two strains (Fig. 2A and
B). In the present study, the T110 strain was used as a control
to compare the genome sequences between the strains. The
collinearity analysis of the two strains and T110 demonstrated that
140623 and SBS-1 were most closely associated with sequences from
T110 (Fig. 2C and D). Similar
arrangement of sequences from the 140623 and SBS-1 genomes was
found by comparing them with sequences from the T110 genome. The
genome collinearity analysis of amino acid and nucleotide sequences
exhibited similar arrangements in 140623 and SBS-1 compared with
T110 (Fig. 2C and D).
Annotation of the 140623 and SBS-1
genomes
Gene annotations identified 2,766 genes (average
gene length, 889 bp) and 2,734 genes (average gene length, 893 bp)
from 140623 and SBS-1, respectively. Compared with the intergenic
region length (140623, 355,200 bp; SBS-1, 356,384 bp), larger gene
regions were identified in the 140623 genome (2,457,726 bp) and
SBS-1 genome (2,441,361 bp), representing 87.37 and 87.26% of the
complete genome sequences, respectively. Higher GC contents in gene
regions (140623, 39.07%; SBS-1, 39.09%) were observed when compared
with intergenic regions (140623, 32.42%; SBS-1, 32.57%). Both
genomes contained 70 tandem repeat sequences (7,954 bp in 140623,
9,079 bp in SBS-1), which accounted for 0.2828 and 0.3245% of the
genomes of 140623 and SBS-1, respectively. In 140623, 39
minisatellite DNA sequences, 2 microsatellite DNA sequences, 41
transfer RNAs (tRNAs) and 4 sRNAs were identified. The SBS-1 genome
contained 38 minisatellite DNA sequences, 1 microsatellite DNA
sequence, 3 rRNAs, 54 tRNAs and 4 sRNAs (Table IV).
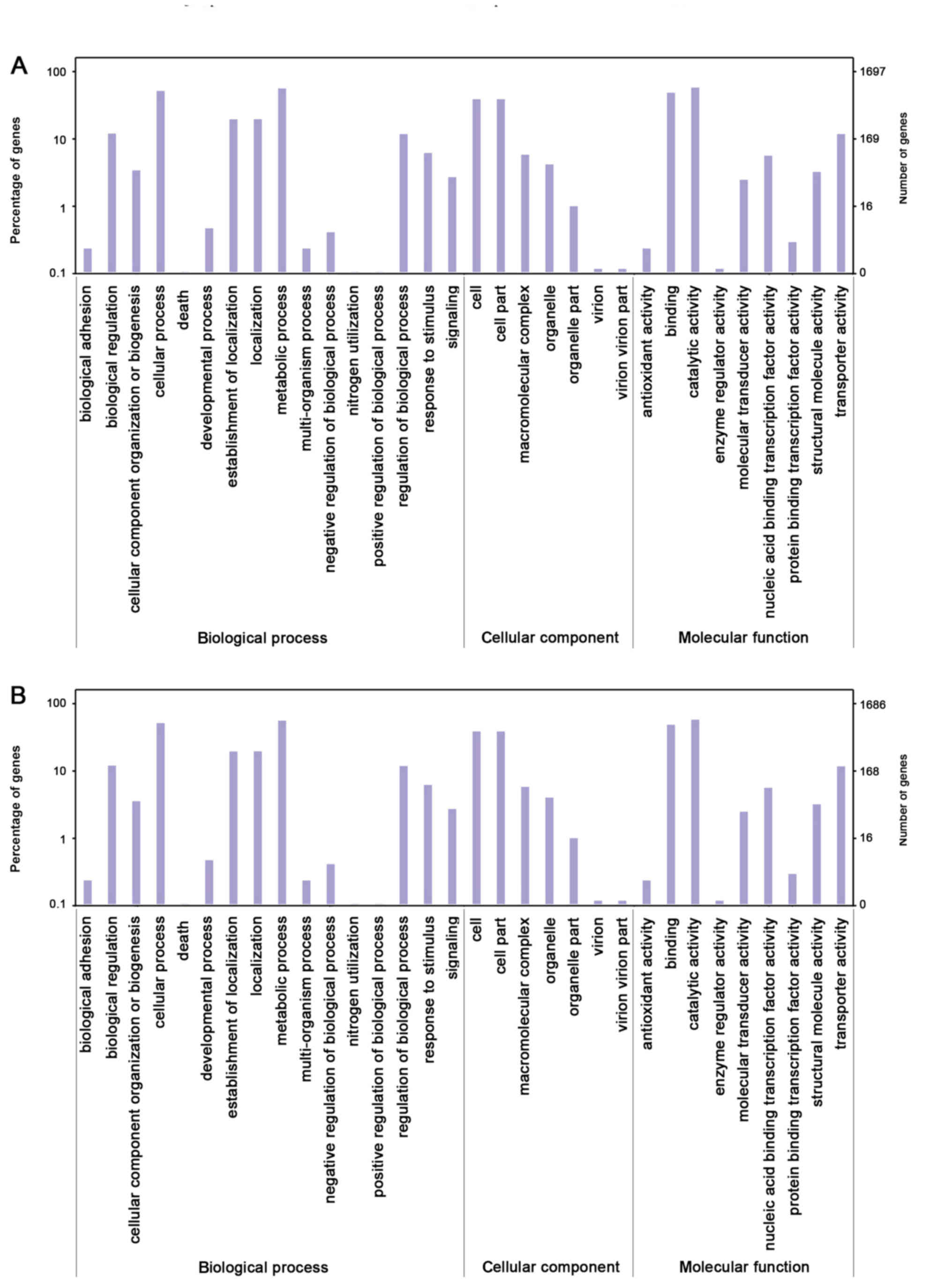
The distribution of GO categories was similar
between the 140623 and SBS-1 genomes. In 140623 and SBS-1, the
genes associated with ‘catalytic activity’, ‘metabolic process’,
‘cellular process’ and ‘binding’ were abundant (Fig. 3). The core genes were primarily
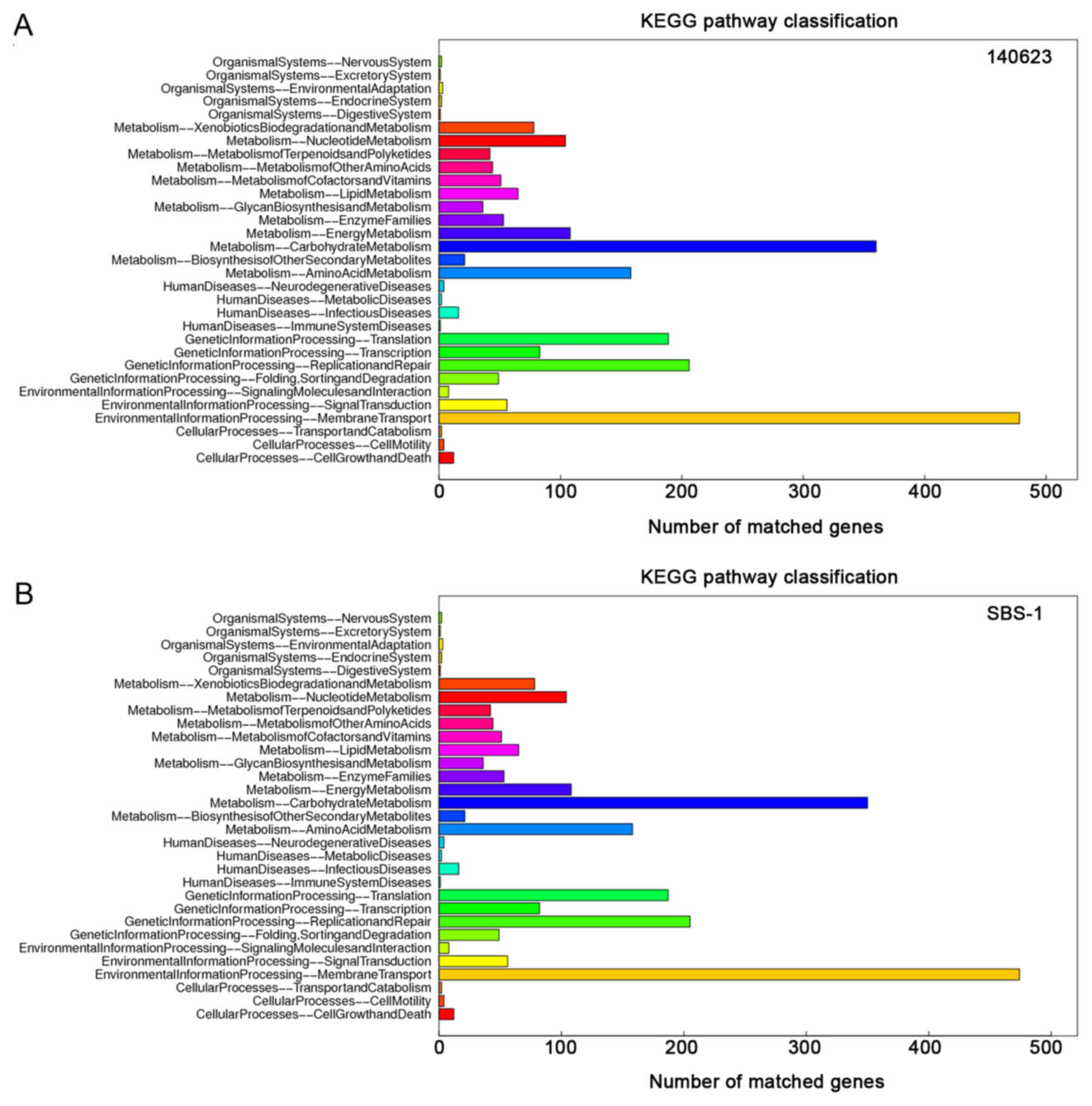
involved in the following KEGG categories: ‘Environmental
Information Processing-Membrane Transport’ (478 genes in 140623 and
474 genes in SBS-1) and ‘Metabolism-Carbohydrate Metabolism’ (360
genes in 140623 and 350 genes in SBS-1; Fig. 4). According to the COG
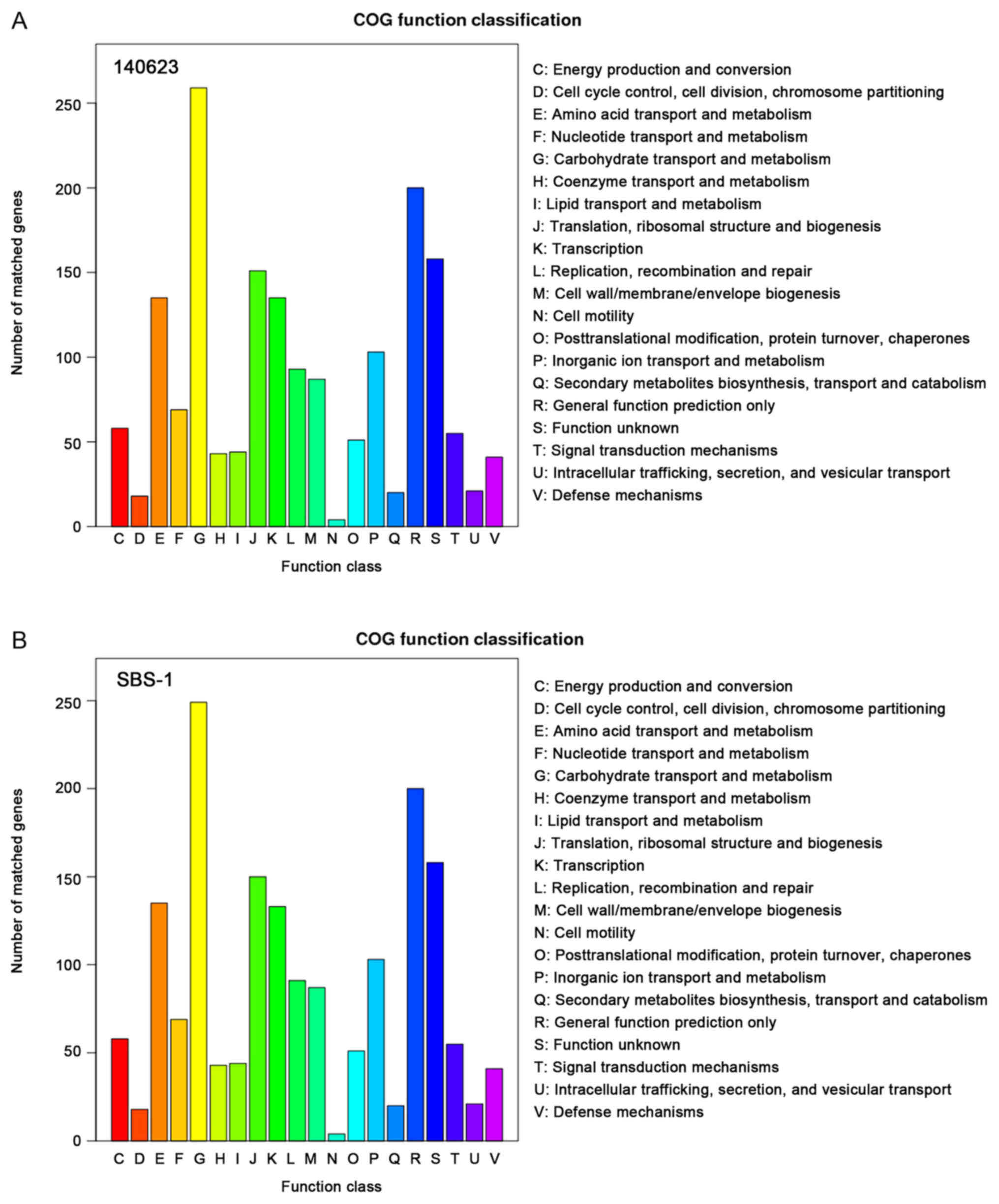
classification, 1,745 genes in 140623 and 1,730 genes in SBS-1 were
included. In the categories other than ‘Carbohydrate transport and
metabolism’, ‘Translation, ribosomal structure and biogenesis’,
‘Transcription’ and ‘Replication, recombination and repair’, the
two strains had the same gene numbers (Fig. 5).
Virulence- and drug
resistance-associated genes
A total of 6 putative virulence-associated genes
were identified from 140623 and SBS-1 genomes, including AphA,
CBL1, CPA1, GyrA, MGG_00383 and SOD2 (Table SI). Analysis using ARDB revealed
that there were only a few potential genes of antibiotic resistance
(15 genes in both 140623 and SBS-1 genomes; Table SII). The complete sequences of the
140623 and SBS-1 genomes each exhibited 31 genes associated with
bacterial toxins according to vfdb annotation (Table SIII).
Discussion
As a popular medicine for probiotic therapy to treat
with patients with diarrhea, the safety of Shin Biofermin S is
paramount. Therefore, in the present study, the species of the
SBS-1 strain isolated from a commercial medicine (Shin Biofermin S)
was determined via genome sequencing. Species identification is
important for evaluating safety and is favorable for formulating
therapeutic strategies. The scientific names of E. faecium
have been changed several times. Streptococcus faecalis and
Streptococcus faecium were separate until 1984, when DNA-DNA
and DNA-rRNA hybridization tests revealed that they belonged to
Enterococcus instead (5).
Bergey's Manual of Systematic Bacteriology Volume 3: The Firmicutes
also demonstrated this result (6),
such that Streptococcus faecalis was renamed Enterococcus
faecalis. In the food and health industry, E. faecium
and E. faecalis have been used to improve the intestinal
environment by regulating its inflammatory status and the gut
microflora (2). However, the
definition of strains is inaccurate among different companies. The
bacterial strain is the only active ingredient in probiotics. Thus,
if the identity of the original strain species is not assured, its
safety cannot be guaranteed. The present study identified two E.
faecium strains from the National Institute for Food and Drug
Control, and Shin Biofermin S. Biochemical identification, gene
fingerprinting, fatty acid component analysis and genome sequencing
were all used to clarify the species of these two strains.
Growth and sugar component tests classified the two
strains into E. faecium. The growth of E. faecium
requires high-quality culture medium, such as agar with human,
rabbit or horse blood. This species produced L-arabinose, in line
with a previous report (28), while
E. faecium could not generate L-arabinose. In addition, the
RiboPrinter automatic microbial genetic fingerprint identification
system, which is a tool designed for the identification of
bacteria, suggested that the two strains were E. faecium. A
number of studies have demonstrated that this method is a highly
targeted and effective approach to identify different species.
Previously, the two strains seemed to be E. faecalis
(2,5), while the present analyses strongly
supports E. faecium.
The fatty acid composition of the strains was
analyzed in the present study via GC-MS, which also supported the
classification of the two strains as E. faecium. The
characteristic peaks of C14:0, C16:1Δ9, C16:0,
C18:1Δ9, C18:0 and 2-C8H17-C19:0 were similar between
the two strains. This result demonstrated the high homology between
140623 and SBS-1. The fatty acid component of these two strains was
also similar to previously reported E. faecium (29).
The genome sizes of the two strains were 2,812,926
(140623) and 2,797,745 (SBS-1) bp. Zhong et al (30) reported five strains of E.
faecium genome sizes ranging from 2.64–2.99 Mb. The genome
sizes identified in the present study were also in this range.
Another report indicated that the genome of an E. faecium
strain isolated from the bloodstream of a patient in Melbourne,
Australia, contained 3 circular plasmids and a 2.9 Mb chromosome
(31). The chromosome of E.
faecium TX16 was 2,698,137 bp (8). In general, it appears that genome size
changes among different strains throughout evolution.
The genomes of 140623 and SBS-1 had 2,766 and 2,734
protein-coding open reading frames (ORFs), respectively. When three
E. faecium were isolated from bovine feces, it was revealed
that they contained 2,719, 2,665 and 2,659 genes, respectively
(32). In E. faecium TX16,
2,703 protein-coding ORFs were identified on the chromosome and the
three plasmids contained 43, 85 and 283 ORFs, respectively
(8). These previous reports indicate
that the genomes of E. faecium strains from different
sources have similar gene counts, indicating evolutionarily
conserved and similar functions. Similar gene annotations, such as
GO, KEGG and COG analyses, also supported this conclusion.
Virulence- and drug resistance-associated genes in
E. faecium were limited compared with the results of E.
faecalis. The present results demonstrated that AphA, CBL1,
CPA1, GyrA, MGG_00383 and SOD2 were exhibited by the two strains
and may be associated with pathogen-host interaction. CBL1 of
Fusarium graminearum has been reported to be associated with
pathogenicity, indicating a role in host infection (33). Waters et al (34) demonstrated the
ciprofloxacin-resistant function of GyrA. The SOD2 gene is
associated with the loss of pathogenicity in bacteria (35). The present study indicated that the
evolution of this species conferred resistance mechanisms in
natural environments and industrial environments. These resistance
mechanisms provide potential for clinical use in food and drug
safety.
The results of the present study indicated that the
species of 140623 and SBS-1 was E. faecium. The biochemical
assays and RiboPrinter ribotyping were comparable, both
characterizing the strains as E. faecium; GC-MS analysis
indicated specific peaks of fatty acid components that were similar
to other E. faecium strains; genome features of 140623 and
SBS-1 demonstrated high conservation of these strains as E.
faecium, which was also confirmed by phylogenetic analysis.
Taken together, the results of the present study
demonstrated that 140623 and SBS-1 from the National Institute for
Food and Drug Control and Shin Biofermin S were E. faecium
rather than E. faecalis. These data first elucidated the
genome information of SBS-1 strain from Shin Biofermin S, which is
a commercial medicine used to treat diarrhea (2). The present study demonstrated that the
strain is similar with 140623 and that the variations in the SBS-1
strain served potential roles in resistance. The virulence- and
drug resistance-associated genes identified in the present study,
which participate in antibiotic resistance, should be further
investigated. These could serve as key molecular factors for SBS-1
strain security during treatment. In conclusion, the results of the
present study may therefore be valuable for distinguishing
different species of the Enterococcus genus and guiding
edible probiotic usage.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
BZ designed the study. YQiG performed the
experiments and analyzed the data. YQiG and BZ wrote the article.
TZ, YQiangG and ZZ participated in the data processing and revision
of the manuscriptt
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rajilić-Stojanović M and de Vos WM: The
first 1000 cultured species of the human gastrointestinal
microbiota. FEMS Microbiol Rev. 38:996–1047. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fisher K and Phillips C: The ecology,
epidemiology and virulence of Enterococcus. Microbiology.
155:1749–1757. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Giannenas I, Papadopoulos E, Tsalie E,
Triantafillou E, Henikl S, Teichmann K and Tontis D: A
microbiological investigation on probiotic preparations used for
animal feeding. Microbi Res. 151:167–175. 1996. View Article : Google Scholar
|
|
4
|
Klein G: Taxonomy, ecology and antibiotic
resistance of Enterococci from food and the gastro-intestinal
tract. Int J Food Microbiol. 88:123–131. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schleifer KH and Kilpper-Bälz R: Transfer
of Streptococcus faecalis and Streptococcus faecium
to the Genus Enterococcus nom. rev. as Enterococcus
faecalis comb. nov. and E. faecium comb. nov. Int J Syst
Bacteriol. 34:31–34. 1984. View Article : Google Scholar
|
|
6
|
Vos P, Garrity G, Jones D, Krieg NR,
Ludwig W, Rainey FA, Schleifer KH and Whitman W: Bergey's Manual of
Systematic Bacteriology: Volume 3: The Firmicutes. 3. Springer
Science & Business Media; 2011
|
|
7
|
Salyers AA, Gupta A and Wang Y: Human
intestinal bacteria as reservoirs for antibiotic resistance genes.
Trends Microbiol. 12:412–416. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Qin X, Galloway-Peña JR, Sillanpaa J, Roh
JH, Nallapareddy SR, Chowdhury S, Bourgogne A, Choudhury T, Muzny
DM, Buhay CJ, et al: Complete genome sequence of Enterococcus
faecium strain TX16 and comparative genomic analysis of
Enterococcus faecium genomes. BMC Microbiol. 12:1352012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shanks OC, Santo Domingo JW and Graham JE:
Use of competitive DNA hybridization to identify differences in the
genomes of bacteria. J Microbiol Methods. 66:321–330. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Aiba Y, Ishikawa H, Shimizu K, Noda S,
Kitada Y, Sasaki M and Koga Y: Role of internalization in the
pathogenicity of Shiga Toxin-producing Escherichia coli infection
in a gnotobiotic murine model. Microbiol Immunol. 46:723–731. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan
J, He G, Chen Y, Pan Q, Liu Y, et al: SOAPdenovo2: An empirically
improved memory-efficient short-read de novo assembler.
Gigascience. 1:182012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Altschul SF, Madden TL, Schäffer AA, Zhang
J, Zhang Z, Miller W and Lipman DJ: Gapped BLAST and PSI-BLAST: A
new generation of protein database search programs. Nucleic Acids
Res. 25:3389–3402. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kanehisa M, Goto S, Kawashima S, Okuno Y
and Hattori M: The KEGG resource for deciphering the genome.
Nucleic Acids Res. 32:D277–D280. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kanehisa M: A database for post-genome
analysis. Trends Genet. 13:375–376. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kanehisa M, Goto S, Hattori M,
Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M and
Hirakawa M: From genomics to chemical genomics: New developments in
KEGG. Nucleic Acids Res. ((Database issue))34:D354–D357. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Magrane M and Consortium U: UniProt
Knowledgebase: A hub of integrated protein data. Database (Oxford).
2011:bar0092011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tatusov RL, Koonin EV and Lipman DJ: A
genomic perspective on protein families. Science. 278:631–637.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tatusov RL, Fedorova ND, Jackson JD,
Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov
SL, Nikolskaya AN, et al: The COG database: An updated version
includes eukaryotes. BMC Bioinformatics. 4:412003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kurtz S, Phillippy A, Delcher AL, Smoot M,
Shumway M, Antonescu C and Salzberg SL: Versatile and open software
for comparing large genomes. Genome Biol. 5:R122004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jehl MA, Arnold R and Rattei T:
Effective-a database of predicted secreted bacterial proteins.
Nucleic Acids Res (Database issue). 39:D591–D595. 2011. View Article : Google Scholar
|
|
22
|
Chen L, Xiong Z, Sun L, Yang J and Jin Q:
VFDB 2012 update: Toward the genetic diversity and molecular
evolution of bacterial virulence factors. Nucleic Acids Res.
40:D641–D645. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu B and Pop M: ARDB-Antibiotic
resistance genes database. Nucleic Acids Res. 37:D443–D447. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Benson G: Tandem repeats finder: A program
to analyze DNA sequences. Nucleic Acids Res. 27:573–580. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gardner PP, Daub J, Tate JG, Nawrocki EP,
Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S,
Eddy SR and Bateman A: Rfam: Updates to the RNA families database.
Nucleic Acids Res. 37:D136–D140. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lagesen K, Hallin P, Rødland EA,
Staerfeldt HH, Rognes T and Ussery DW: RNAmmer: Consistent and
rapid annotation of ribosomal RNA genes. Nucleic Acids Res.
35:3100–3108. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lowe TM and Eddy SR: tRNAscan-SE: A
program for improved detection of transfer RNA genes in genomic
sequence. Nucleic Acids Res. 25:955–964. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Manero A and Blanch AR: Identification of
Enterococcus spp. with a biochemical key. Appl Environ
Microbiol. 65:4425–4430. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sakayori Y, Muramatsu M, Hanada S,
Kamagata Y, Kawamoto S and Shima J: Characterization of E.
faecium mutants resistant to mundticin KS, a class IIa
bacteriocin. Microbiology. 149:2901–2908. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhong Z, Zhang W, Song Y, Liu W, Xu H, Xi
X, Menghe B, Zhang H and Sun Z: Comparative genomic analysis of the
genus Enterococcus. Microbiol Res. 196:95–105. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lam MM, Seemann T, Bulach DM, Gladman SL,
Chen H, Haring V, Moore RJ, Ballard S, Grayson ML, Johnson PD, et
al: Comparative analysis of the first complete E. faecium
genome. J Bacteriol. 194:2334–2341. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Beukers AG, Zaheer R, Goji N, Amoako KK,
Chaves AV, Ward MP and McAllister TA: Comparative genomics of
Enterococcus spp. isolated from bovine feces. BMC Microbiol.
17:522017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xie QN, Jia LJ, Wang YZ, Song RT and Tang
WH: High-resolution gene profiling of infection process indicates
serine metabolism adaptation of Fusarium graminearum in
host. Sci Bull. 62:758–760. 2017. View Article : Google Scholar
|
|
34
|
Waters B and Davies J: Amino acid
variation in the GyrA subunit of bacteria potentially associated
with natural resistance to fluoroquinolone antibiotics. Antimicrob
Agents Chemother. 41:2766–2769. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dimitrijevic L, Puppo A and Rigaud J:
Superoxide dismutase activities in Rhizobium phaseoli
bacteria and bacteroids. Arch Microbiol. 139:174–178. 1984.
View Article : Google Scholar
|