Introduction
Burn is a common type of traumatic disease. Heat,
chemicals, currents and radiation are the common causes of burn
injury. Among the numerous injury factors, heat is the most common
factor, and thermal damage is always combined with burns (1,2). The
injury factors of thermal burns are usually directly or indirectly
caused by individuals touching high-temperature solids, liquids or
gases (3). Cell morphology,
metabolism and function are altered in the tissues at the burn site
and the level of damage is related to many factors which are
associated with the ratio between the level of the heat energy and
the duration. The earliest cell histological changes after injury
are found in the epidermis which reveals the redistribution of
chromatin in the nucleus. With further damage, the cytoplasm of
basal cells and the nuclei of full-thickness cells begin to swell
or even become necrotic. If the heat of injury further increases,
the epidermis immediately undergoes coagulative necrosis and even
carbonization, eventually causing serious damage to the local
tissue and even to the entire body (4).
The skin is the largest organ, and healthy
conditions of the skin can aid humans to prevent attacks from the
external environment (5). Similar to
other stem cells, epidermal stem cells (EpSCs), derived from skin
tissue, display functions of self-proliferation and differentiation
and can maintain normal epidermal structure (6). The proliferation, differentiation and
apoptosis of EpSCs are coordinated by multiple gene pathways
(7), thus EpSCs play an important
role in promoting wound healing. Upregulation of the regeneration
process of EpSCs and inhibition of their apoptotic processes can
accelerate the repair of damaged wound tissue (8). EpSCs synthesize and secrete a variety
of biologically active substances such as immunomodulatory factors,
angiogenic factors, anti-apoptosis factors, antioxidants and cell
chemokines. Research has confirmed that cross-linking between
multiple signaling pathways forms a strict and orderly regulatory
network, and any regulatory changes in the network may interfere
with the wound healing process (8).
MicroRNAs (miRNAs) are important factors that
regulate the proliferation, differentiation and apoptosis of EpSCs
(9). miRNAs are small, non-coding
RNAs approximately 22 nucleotides (nt) in length that are encoded
by higher eukaryotic genomes. miRNAs can degrade to mRNAs and
inhibit mRNA transcription and enable post-transcriptional
regulation (10). Studies have shown
that miRNAs play an important role in skin tissue proliferation and
wound healing (5). There have been
studies confirming that miRNA-203 is an epithelial tissue-specific
miRNA that not only participates in skin development but also
induces the differentiation of EpSCs into various specialized cells
(11). In the present study, the
trypsin digestion method was used to isolate human EpSCs. The cells
were then incubated in a 51.5°C water tank for 35 sec to construct
a thermal injury model. The differentially expressed miRNAs were
identified using high-throughput sequencing technology, and
bioinformatic methods were used to predict their target genes and
signaling pathways that may be involved in wound repair.
Materials and methods
Sample collection
Normal skin samples were obtained from 10 male
patients, median age 25 years (range 18–35 years) who underwent
autologous grafting with epidermis at the Burn Center, The First
Affiliated Hospital of Nanchang University from January 2016 to
June 2016. The present study was conducted according to The
Declaration of Helsinki and was approved by the Ethics Committee of
Nanchang University. All patients provided written informed consent
prior to the study start.
Epidermal stem cell isolation and
culture
Following simple treatment, the specimens were
repeatedly rinsed three times with 1% penicillin-streptomycin in
phosphate-buffered saline (PBS), added to a concentration of 0.25%
trypsin + 0.02% ethylene diamine tetraacetic acid (EDTA), and
allowed to stand at 4°C for 8–10 h. The epidermal and dermal layers
of the epidermis were separated, and the separated epidermal layers
were crushed into tissue homogenates and subsequently 2 ml of 0.25%
trypsin + 0.02% EDTA were added and digestion was carried out at
37°C for 10 min. Defined K-SFM (2 ml) containing 10% fetal bovine
serum was added to stop the trypsin digestion, followed by 200 mesh
sieve filtration. The filtrate was collected in a 15-ml centrifuge
tube and centrifuged at 110 × g for 5 min at 37°C. The supernatant
was discarded. A total of 5 ml of 1% penicillin-streptomycin in PBS
was added, followed by centrifugation at 110 × g for 5 min at 37°C
and the supernatant was removed. Following centrifugation, 10 ml of
Defined K-SFM (Gibco; Thermo Fisher Scientific, Inc.) was added to
the cell layer to resuspend the cells, and after the count, the
concentration was adjusted to 3×106 cells/ml.
Subsequently, the cells were inoculated in a pre-plated type IV
collagen culture dish, and the culture dish was placed at 37°C.
Following incubation for 20 min in a CO2 incubator, the
upper unattached cell suspension was aspirated and washed twice
with PBS, and 5 ml of the culture medium was added to the culture
dish, which was then labeled and placed in an incubator.
Subsequently, IV collagen was used to prepare for adherence
screening, and the positive expression of ck19 and integrin β1 was
confirmed by immunochemical staining (12,13).
Human epidermal stem cell heat injury
treatment
The EpSCs were cultured at 5°C in a 5%
CO2 saturated humidity incubator and the culture fluid
was changed every other day. When the cell density reached 90% and
most of the cells grew well, the culture fluid was changed again. A
thermostatic water tank was preheated for 30 min in advance; the
temperature was set to 51.5°C, sealing material and timer were
prepared, and a Petri dish was sealed with sealing material. The
cells were randomly divided into two groups, control group (group
1) and experimental group (group 2). For the experimental group,
the sealed Petri dish was suspended in a 51.5°C water tank and
incubated for 35 sec. The timer was set. The control dish was
placed in a 37°C water tank and also incubated for 35 sec. The
procedure was the same as above and as previously described
(14). The treated cells were
observed under a fluorescent inverted phase contrast microscope
(magnification, ×100) to detect cell morphology and the number of
apoptotic cells. The samples were further collected after 6 h of
culture to extract RNA.
Total RNA extraction and
purification
Total RNA was isolated using Magzol reagent solution
(Thermo Fisher Scientific, Inc.) and purified using the RNeasy Mini
kit (Qiagen GmbH), according to the manufacturer's protocol. RNA
quality and quantity were measured using a NanoDrop
Spectrophotometer (ND-1000; NanoDrop; Thermo Fisher Scientific,
Inc.). RNA integrity was determined by electrophoresis on a
denatured agarose gel prepared in house. On the denaturing gels,
the 28S and 18S ribosomal RNA bands were visible, indicating that
the extracted total RNA was intact, the RNA degradation and
contamination were low, and the extracted total RNA showed high
purity levels. The A260/A280 ratio is a measure of RNA purity. A
ratio of <1.8 indicates sample contamination and a ratio of
>2.0 indicates RNA hydrolysis, between 1.8 and 2.1, indicating
that the RNA purity meets high-throughput sequencing
requirements.
High-throughput sequencing technology
library construction and quality inspection
Small RNA high-throughput sequencing was performed
by Guangzhou RiboBio Co., Ltd., and the library was constructed and
sequenced as follows: 1 µg of total RNA (≥50 ng small RNA) was used
as the initial amount of RNA sample. The sample was supplemented
with water to make the total reaction volume 7 µl. Subsequently,
the library was constructed using a small RNA sample preparation
kit (New England BioLabs, Inc.), according to the manufacturer's
protocol. After the library was constructed, the concentration of
the sample was determined by Qubit 2.0 (Thermo Fisher Scientific,
Inc.), and the sample in the library was diluted to a concentration
of 1 ng/µl. Library quality tests were performed using an Agilent
2200 TapeStation (Agilent Technologies, Inc.); cDNA fragments with
an insert size of ~18–40 nt were obtained by gel
electrophoresis.
Data analysis
We annotated clean sequences, as well as analyzed
the composition and expression difference of all types of miRNAs.
Bioinformatic methods were used for family analysis of the
differentially expressed miRNAs, and Target-Scan (www.targetscan.org), miRDB (www.mirdb.org), miRanda software (www.microrna.org) and high-throughput CLIP-seq data
(www.clipdb.ncrnalab.org) were used to
predict miRNA target genes, and miRNAs that were significantly
differentially expressed in this experiment were analyzed. Target
gene prediction was performed, and then the predicted target genes
were analyzed by Kyoto Encyclopedia of Genes and Genomes (KEGG)
(www.genome.jp/kegg) biological pathway
enrichment analysis and Gene Ontology (GO) (www.geneontology.org) gene function enrichment
analysis. Statistical analysis was performed using SPSS software
(version 22.0; IBM Corp.).
GO analysis
The gene list of the differentially expressed
upregulated and downregulated genes was prepared, and the data were
imported into the GO database, human species was selected, and then
calculation was carried out. The P-value related to the enrichment
of GO terms was calculated by the default statistical algorithm of
the GO analysis database. The smaller the P-value, the more notable
the entry of the GO term, and a term entry with P≤0.05 was
considered to be statistically significant. The base 10 logarithm
of the P-value was converted to a negative value, and an enrichment
score was obtained. The enrichment score value represents the
possibility that the differentially expressed mRNA is enriched in
the term entry. The higher the enrichment score more notable the
entry. Enrichment for significant features was sorted in descending
order, and the GO Enrichment histogram was plotted in Microsoft
Excel.
Pathway analysis
The data were imported into the KEGG database and
human species data were selected and investigations were performed.
The significance score of differential gene enrichment for each
pathway was calculated via the hypothesis testing to obtain
significant P-values. The smaller the P-value the more notable the
respective biological pathway. A biological pathway with P<0.05
was considered to be statistically significant. The base 10
logarithm of the P-value was converted to a base-negative
logarithmic scale, and the enrichment score value represents the
possibility that the differentially expressed mRNA is enriched in
this biological pathway. The higher the enrichment score the more
important this pathway was. Specific signal paths and data can be
exported in the system. P-values for significant pathways were
ranked in ascending order, and the Log p histogram of the KEGG
pathways were plotted by Microsoft Excel.
Results
Changes in epidermal stem cell growth
after heat injury
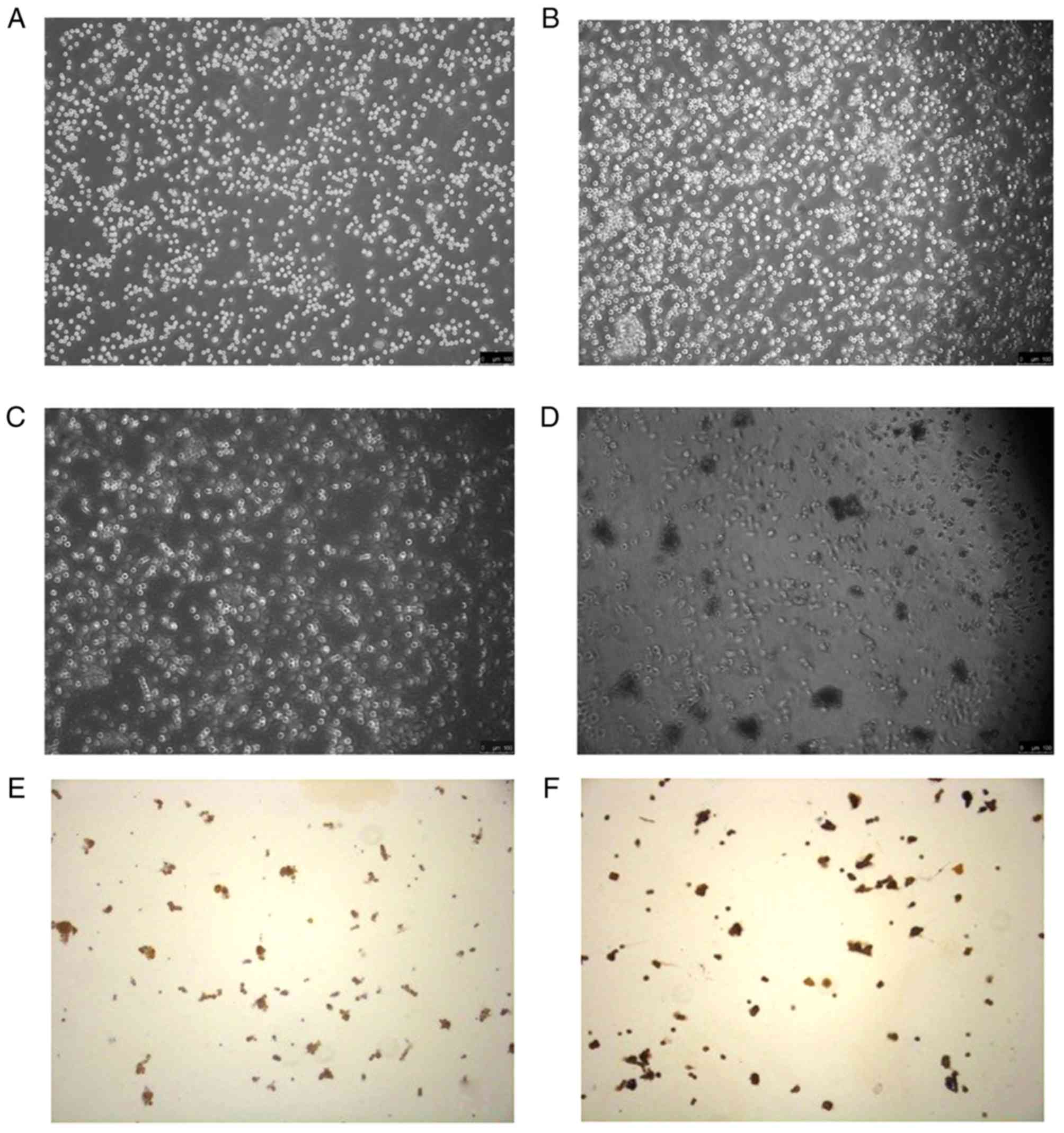
As a result of observation under an inverted
microscope, the EpSCs were found to be firmly attached, with a
small and rounded shape and well refraction was noted when they
were inoculated. After culturing for 2 days, the cells covered more
than 90% and the clones grew rapidly. The cells attached firmly.
The control group showed no obvious reduction in the number of
cells after a 37°C water bath. The cell shape was round, and cells
were firm. After the 51.5°C water bath exposure in the experimental
group, the number of cells was significantly decreased, the cells
showed irregular shapes, and were not attached firmly.
Immunochemical staining images showed that β1 integrin and CK19
were positively expressed which are characteristic of EpSCs
(Fig. 1).
Extraction and qualification of total
RNA
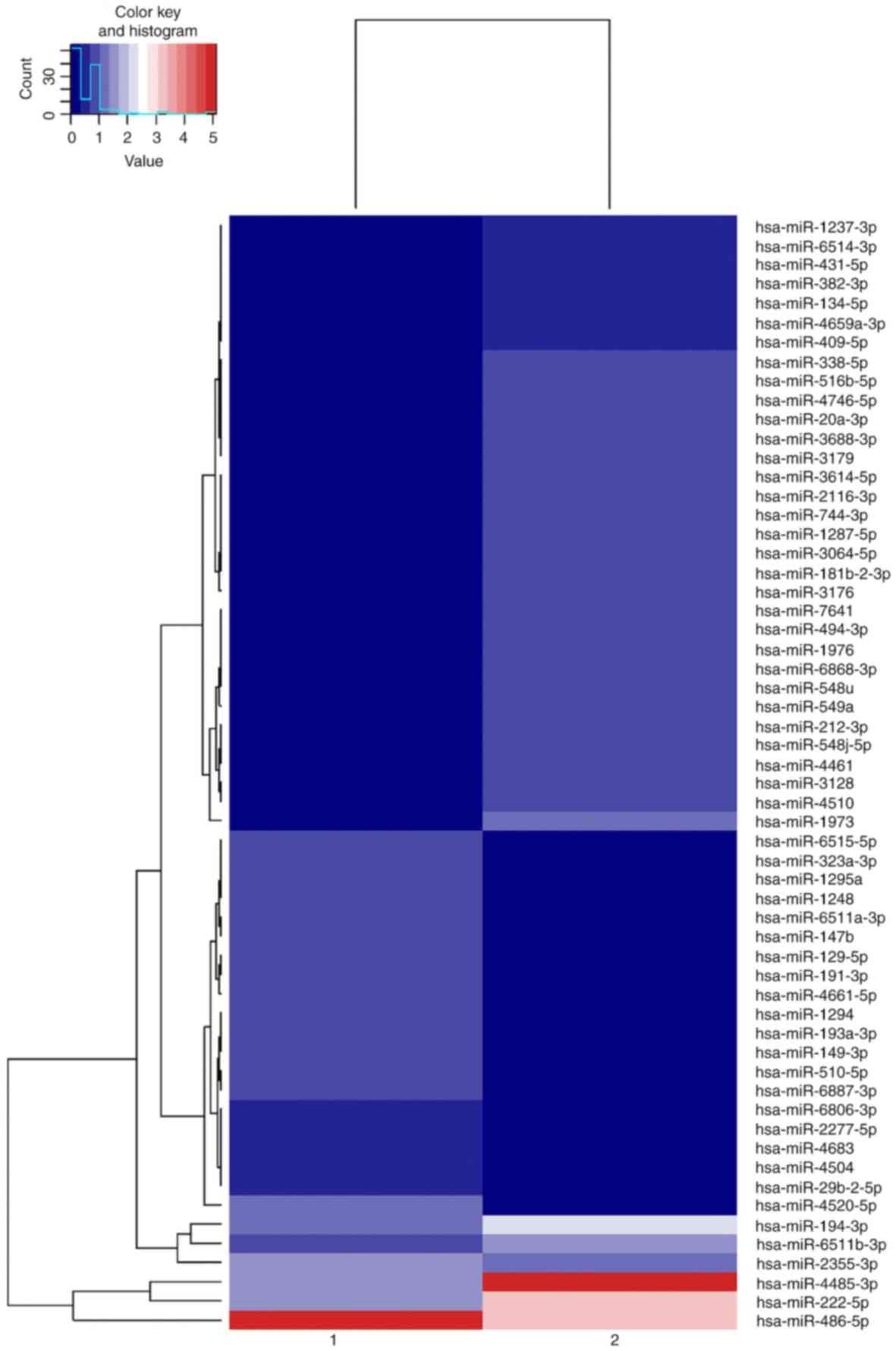
The quality test results of the two groups of total
RNA spectrophotometry met the experimental requirements, and the
miRNA cluster plots for experimental and control groups are
presented in Fig. 2.
Analysis of the differential
expression of the miRNAs between the two groups
Table I documents the
variations and patterns of miRNA expression between the two groups.
We analyzed the data in the results of the high-throughput
sequencing, statistical analysis of the two groups of samples for
the differential expression of miRNA was performed to determine
whether the difference was significant, and log2 ratio and scatter
plots were used to compare the differences in miRNA expression. In
Table IA, the experimental group has
a numerical value, while the control group is 0. As the negative
infinity cannot visually demonstrate the different relationships,
the control group takes 0.01 to calculate, and the log2 value is
obtained after the correction. The formula is log2 (experimental
group/control group). In Table IB,
the expression level of certain miRNAs in the experimental group
was 0, indicating that the sequence of the miRNA was not detected
in all samples of the group. This did not mean that there was no
expression, but the expression level was low. Low expression levels
are difficult to measure. Moreover, there were identical values due
to the low expression level of various miRNAs. In order to
accurately reflect the difference relationship, the system
calculated the corrected values as equal.
 | Table I.miRNAs with significantly upregulated
and downregulated expression in the experimental group when
compared with the control group. |
Table I.
miRNAs with significantly upregulated
and downregulated expression in the experimental group when
compared with the control group.
| A, Upregulated
miRNAs |
|---|
|
|---|
| miRNA_ID | Control group | Experimental
group | log2 (fold
change) | P-value |
|---|
|
hsa-miR-4485-3p | 1.7896 | 33.5333 | 4.2279 |
1.2×10−7 |
| hsa-miR-1973 | 0.0000 | 1.2268 | 6.9388 |
2.1×10−4 |
|
hsa-miR-548j-5p | 0.0000 | 0.9931 | 6.6339 |
7.2×10−4 |
| hsa-miR-212-3p | 0.0000 | 0.9931 | 6.6339 |
7.2×10−4 |
| hsa-miR-4461 | 0.0000 | 0.9931 | 6.6339 |
7.2×10−4 |
| hsa-miR-4510 | 0.0000 | 0.9347 | 6.5464 |
1.0×10−3 |
| hsa-miR-3128 | 0.0000 | 0.9347 | 6.5464 |
1.0×10−3 |
| hsa-miR-549a | 0.0000 | 0.8763 | 6.4534 |
1.4×10−3 |
| hsa-miR-494-3p | 0.0000 | 0.8179 | 6.3539 |
1.9×10−3 |
| hsa-miR-7641 | 0.0000 | 0.8179 | 6.3539 |
1.9×10−3 |
| hsa-miR-1976 | 0.0000 | 0.8179 | 6.3539 |
1.9×10−3 |
|
hsa-miR-6868-3p | 0.0000 | 0.8179 | 6.3539 |
1.9×10−3 |
| hsa-miR-548u | 0.0000 | 0.8179 | 6.3539 |
1.9×10−3 |
|
hsa-miR-2116-3p | 0.0000 | 0.7595 | 6.247 |
2.8×10−3 |
|
hsa-miR-3614-5p | 0.0000 | 0.7595 | 6.247 |
2.8×10−3 |
| hsa-miR-744-3p | 0.0000 | 0.7595 | 6.247 |
2.8×10−3 |
|
hsa-miR-1287-5p | 0.0000 | 0.7595 | 6.247 |
2.8×10−3 |
|
hsa-miR-3064-5p | 0.0000 | 0.7595 | 6.247 |
2.8×10−3 |
|
hsa-miR-181b-2-3p | 0.0000 | 0.7595 | 6.247 |
2.8×10−3 |
| hsa-miR-3176 | 0.0000 | 0.701 | 6.1313 |
4.1×10−3 |
|
hsa-miR-516b-5p | 0.0000 | 0.6426 | 6.0058 |
6.1×10−3 |
| hsa-miR-338-5p | 0.0000 | 0.6426 | 6.0058 |
6.1×10−3 |
|
hsa-miR-4746-5p | 0.0000 | 0.6426 | 6.0058 |
6.1×10−3 |
| hsa-miR-20a-3p | 0.0000 | 0.6426 | 6.0058 |
6.1×10−3 |
|
hsa-miR-3688-3p | 0.0000 | 0.6426 | 6.0058 |
6.1×10−3 |
| hsa-miR-3179 | 0.0000 | 0.6426 | 6.0058 |
6.1×10−3 |
|
hsa-miR-6514-3p | 0.0000 | 0.5842 | 5.8684 |
9.2×10−3 |
|
hsa-miR-1237-3p | 0.0000 | 0.5842 | 5.8684 |
9.2×10−3 |
| hsa-miR-431-5p | 0.0000 | 0.5842 | 5.8684 |
9.2×10−3 |
| hsa-miR-382-3p | 0.0000 | 0.5842 | 5.8684 |
9.2×10−3 |
| hsa-miR-134-5p | 0.0000 | 0.5842 | 5.8684 |
9.2×10−3 |
|
hsa-miR-4659a-3p | 0.0000 | 0.5842 | 5.8684 |
9.2×10−3 |
| hsa-miR-409-5p | 0.0000 | 0.5842 | 5.8684 |
9.2×10−3 |
|
| B, Downregulated
miRNAs |
|
|
miRNA_ID | Control
group | Experimental
group | log2 (fold
change) | P-value |
|
|
hsa-miR-4520-5p | 1.1185 | 0.0000 | −6.8054 |
2.9×10−4 |
|
hsa-miR-4661-5p | 0.8948 | 0.0000 | −6.4835 |
1.0×10−3 |
| hsa-miR-191-3p | 0.8389 | 0.0000 | −6.3904 |
1.4×10−3 |
| hsa-miR-129-5p | 0.8389 | 0.0000 | −6.3904 |
1.4×10−3 |
| hsa-miR-147b | 0.7829 | 0.0000 | −6.2908 |
1.9×10−3 |
|
hsa-miR-6868-3p | 0.7829 | 0.0000 | −6.2908 |
1.9×10−3 |
|
hsa-miR-323a-3p | 0.727 | 0.0000 | −6.1839 |
2.8×10−3 |
|
hsa-miR-6515-5p | 0.727 | 0.0000 | −6.1839 |
2.8×10−3 |
| hsa-miR-1295a | 0.727 | 0.0000 | −6.1839 |
2.8×10−3 |
| hsa-miR-1248 | 0.727 | 0.0000 | −6.1839 |
2.8×10−3 |
|
hsa-miR-193a-3p | 0.6711 | 0.0000 | −6.0685 |
4.1×10−3 |
| hsa-miR-1294 | 0.6711 | 0.0000 | −6.0685 |
4.1×10−3 |
| hsa-miR-149-3p | 0.6711 | 0.0000 | −6.0685 |
4.1×10−3 |
|
hsa-miR-6887-3p | 0.6152 | 0.0000 | −5.943 |
6.1×10−3 |
| hsa-miR-510-5p | 0.6152 | 0.0000 | −5.943 |
6.1×10−3 |
| hsa-miR-486-5p | 29.528 | 9.0552 | −1.7053 |
7.4×10−3 |
|
hsa-miR-2277-5p | 0.5592 | 0.0000 | −5.8053 |
9.2×10−3 |
|
hsa-miR-6806-3p | 0.5592 | 0.0000 | −5.8053 |
9.2×10−3 |
| hsa-miR-4683 | 0.5592 | 0.0000 | −5.8053 |
9.2×10−3 |
| hsa-miR-4504 | 0.5592 | 0.0000 | −5.8053 |
9.2×10−3 |
|
hsa-miR-29b-2-5p | 0.5592 | 0.0000 | −5.8053 |
9.2×10−3 |
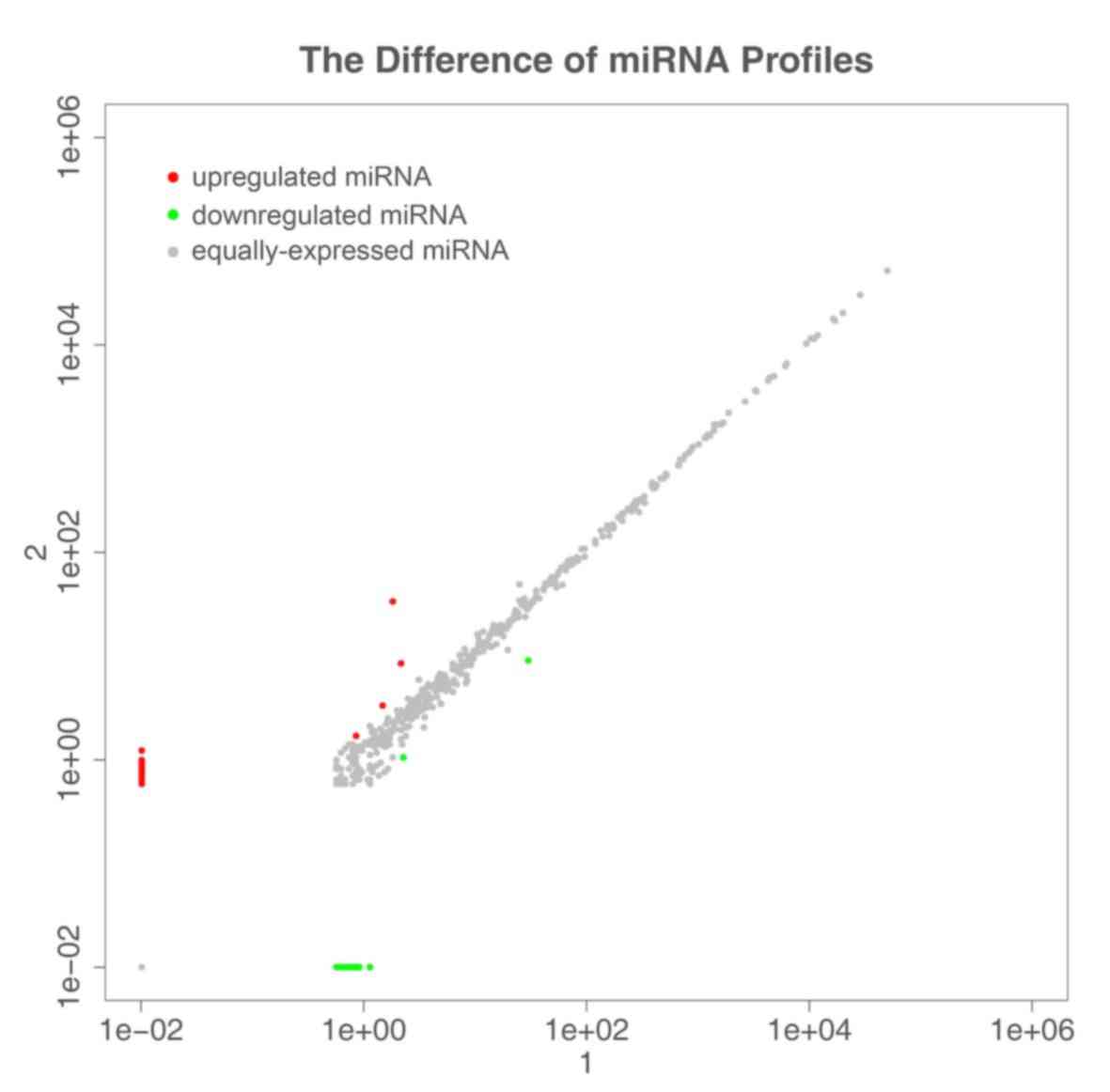
Differences in miRNA expression between samples
(Fig. 3) showed differences in the
expression of all miRNAs, with a log2 (fold change) ≥1 threshold.
Red represents miRNAs with upregulated expression, gray circles
indicate miRNAs with no significant difference in expression, and
green circles represent miRNAs with downregulated expression. log2
≥1 is the upregulation threshold, and −1 is the downregulation
threshold.
There were 33 significantly upregulated miRNAs and
21 significantly downregulated miRNAs. Among them, hsa-miR-1973
exhibited the most obvious upregulation and hsa-miR-4520-5p
exhibited the most significant downregulation.
Prediction of differentially expressed
miRNA target genes and KEGG pathway and GO analyses
Target genes were predicted for the miRNAs with
significant differential expression in the present study, using
Target-Scan, miRDB, miRanda and high-throughput CLIP-seq data
software. The prediction results were further screened and
organized, and the final results revealed a total of 391 predicted
target genes.
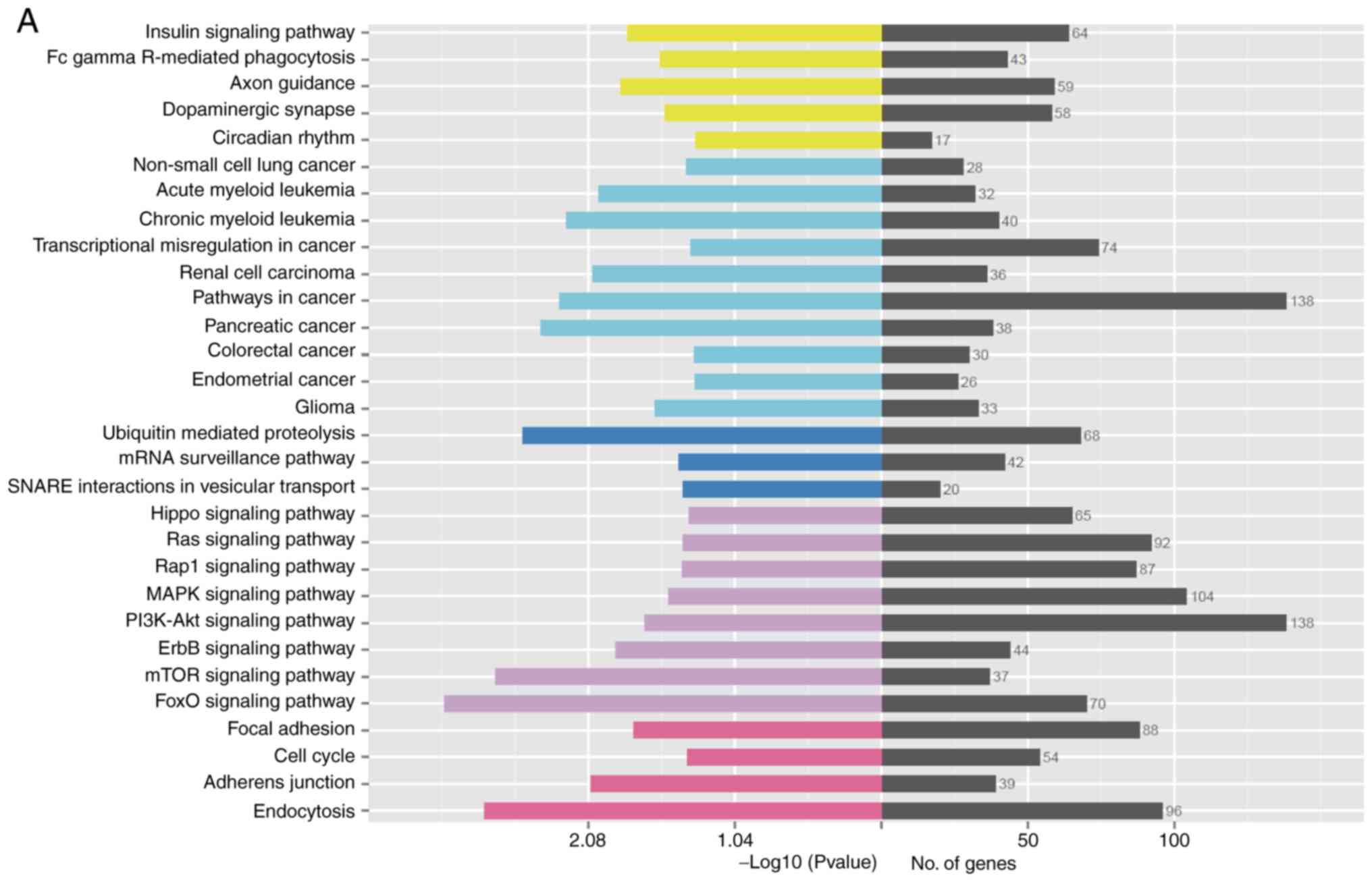
KEGG analysis of the target genes of the miRNAs,
with a P<0.05 KEGG enrichment pathway degree as a significant
threshold, resulted in significant enrichment of 32 KEGG pathways
(Table II). Among them, the
processes associated with wound healing include: FoxO signaling
pathway regulates cell cycle and apoptosis; mTOR signaling pathway
regulates cell metabolism and proliferation; ErbB signaling pathway
regulates cell proliferation, differentiation and migration;
insulin signaling pathway participates in the occurrence of
diabetes, participates in the metabolism of three major substances
and promotion of cell proliferation; focal adhesion signaling
pathway participates in adhesion of cell matrix, regulates cell
proliferation, differentiation, apoptosis, and migration; PI3K-Akt
signaling pathway regulates transcription and translation of genes,
and participates in cell biological metabolism, proliferation and
apoptosis process; MAPK signaling pathway is highly conserved and
involved in the regulation of cell proliferation, differentiation
and migration; cell cycle regulates cell mitosis; Activation of
Hippo signaling pathway leads to apoptosis, prevents cell
overgrowth, and restricts organ size; Wnt signaling pathway
regulates cells the final direction of growth involved in the
proliferation of stem cells and metabolism process. Fig. 4A is a statistical graph of the KEGG
enrichment pathways in the sample.
 | Table II.Results of the KEGG pathway analysis
of the differentially expressed miRNAs. |
Table II.
Results of the KEGG pathway analysis
of the differentially expressed miRNAs.
| Term | Sample number | Background
number | P-value |
|---|
| Axon guidance | 59 | 127 | 0.014083421 |
| FoxO signaling
pathway | 70 | 133 | 0.000780811 |
| Endocytosis | 96 | 203 | 0.001494435 |
| mTOR signaling
pathway | 37 | 60 | 0.001804281 |
| Ubiquitin mediated
proteolysis | 68 | 137 | 0.002811886 |
| Pancreatic
cancer | 38 | 66 | 0.003796989 |
| Pathways in
cancer | 138 | 327 | 0.005127192 |
| Chronic myeloid
leukemia | 40 | 73 | 0.005717632 |
| Adherens
junction | 39 | 73 | 0.008543298 |
| Renal cell
carcinoma | 36 | 66 | 0.008872749 |
| Acute myeloid
leukemia | 32 | 57 | 0.009730435 |
| ErbB signaling
pathway | 44 | 88 | 0.012823109 |
| Arrhythmogenic
right ventricular cardiomyopathy (ARVC) | 38 | 74 | 0.014654654 |
| Insulin signaling
pathway | 64 | 141 | 0.015666822 |
| Focal adhesion | 88 | 206 | 0.017299249 |
| PI3K-Akt signaling
pathway | 138 | 346 | 0.020570672 |
| Glioma | 33 | 65 | 0.024338625 |
| Fc gammaR-mediated
phagocytosis | 43 | 91 | 0.026594706 |
| Dopaminergic
synapse | 58 | 131 | 0.02879438 |
| MAPK signaling
pathway | 104 | 257 | 0.030410368 |
| mRNA surveillance
pathway | 42 | 91 | 0.036289303 |
| Rap1 signaling
pathway | 87 | 213 | 0.038127362 |
| SNARE interactions
in vesicular transport | 20 | 36 | 0.038478648 |
| Ras signaling
pathway | 92 | 227 | 0.038565721 |
| Non-small cell lung
cancer | 28 | 56 | 0.040725456 |
| Cell cycle | 54 | 124 | 0.041455661 |
| Hippo signaling
pathway | 65 | 154 | 0.042550777 |
| Thyroid hormone
signaling pathway | 52 | 119 | 0.04292765 |
| Transcriptional
misregulation in cancer | 74 | 179 | 0.043938096 |
| Colorectal
cancer | 30 | 62 | 0.046445737 |
| Endometrial
cancer | 26 | 52 | 0.047323945 |
| Circadian
rhythm | 17 | 30 | 0.047830477 |
The GO analysis of the cell composition demonstrated
each part of the cell and the cell internal and external
environments. GO analysis of the molecular function demonstrated
the activity of the target gene product at the molecular level,
such as gene transcription, translation and expression, and the
binding and catalysis in metabolic processes. Furthermore, GO
analysis of the biological processes revealed involvement of cell
proliferation, differentiation, signal transduction and apoptosis
processes (Fig. 4B). The above
results showed that they play a role in maintaining the stability
of the cell internal and external environment, gene transcription,
translation and expression, biochemical metabolic processes, cell
proliferation and differentiation, signal transduction, and
apoptosis.
Discussion
Wound healing is a complex physiological process and
is utilized to maintain the integrity of the skin. It is a complex
and dynamic process, requiring the well-orchestrated cooperation of
different types of cells (15).
Wound healing is often characterized as four sequential but
overlapping phases: Hemostasis, inflammation, proliferation and
remodeling (16). Current research
has confirmed that miRNAs play a pivotal role in the regulation of
wound repair networks (17). miRNAs
are important regulators of skin wound healing, especially in the
term of transition from the inflammatory to the proliferative phase
(17). Recent clinical trials have
demonstrated that modulation of miRNA expression by administration
of specific miRNA mimics or inhibitors exhibited beneficial effects
on a variety of diseases, such as cancer and viral infection
(18–20).
High-throughput sequencing enables the sequencing of
hundreds of thousands to millions of DNA molecules in parallel.
With the advancement in research, high-throughput sequencing
technology has been more widely used in the field of basic and
clinical medicine, and has become an important tool for elucidating
the physiological and pathological processes of the body from the
molecular level (21). The present
study sequenced the target genes and correlated bioinformatic
analysis to provide preliminary experimental support for further
research on gene-related targeted therapies.
A thermal injury model was constructed, and the
results demonstrated that cell morphology was significantly
altered. The total RNA of EpSCs was extracted, followed by
high-throughput sequencing, and related bioinformatic analysis was
performed. It was found that 33 of the differentially expressed
miRNAs in the experimental group were upregulated and 21 were
downregulated. hsa-miR-1973 exhibited the most significant increase
in expression, whereas hsa-miR-4520-5p exhibited the most
significant decrease. The significantly upregulated miRNAs also
included, hsa-miR-4485-3p, hsa-miR-548j-5p, hsa-miR-212-3p and
hsa-miR-4461, while the downregulated miRNAs included,
hsa-miR-4661-5p, hsa-miR-191-3p, hsa-miR-129-5p, hsa-miR-147b and
hsa-miR-6868-3p. Family analysis of the differentially expressed
miRNAs and prediction of target genes found that they are involved
in a series of signaling pathways that regulate biological
processes such as cell proliferation and differentiation, growth
and apoptosis, and cell migration. In terms of wound healing,
miR-155 can be involved in cell migration and transformation.
Overexpression of miR-155 at the wound edge can accelerate wound
healing mediated by enhanced keratinocyte migration (22). It has also been reported that miRNAs
are important regulators of inflammatory and tissue repair that act
through translation processing of target mRNAs (23). In terms of cell proliferation,
miR-1973 and miR-191-3p play an important role in breast cancer
proliferation and lymph node metastasis (24,25).
Tissue regeneration, tissue repair and regeneration depend on the
function of miRNAs, which are currently widely used in the tissue
engineering of cartilage, bone and skeletal muscle (26).
GO analysis of the differentially expressed miRNA
target genes showed that they play a role in maintaining cell
internal and external environment stability, gene transcription,
translation and expression, biochemical metabolic processes, cell
proliferation and differentiation, signal transduction and
apoptosis. Target gene prediction and KEGG function enrichment
analysis found that differentially expressed miRNA target
gene-related signaling pathways mainly include the ‘FoxO signaling
pathway’, ‘mTOR signaling pathway’, ‘ErbB signaling pathway’,
‘insulin signaling pathway’, ‘focal adhesion signaling pathway’,
‘PI3K-Akt signaling pathway’, ‘MAPK signaling pathway’, ‘cell
cycle’, ‘Hippo signaling pathway’ and ‘Wnt signaling pathway’. In
the FoxO signaling pathway, FoxO transcription factor changes play
an important role in cell proliferation, apoptosis, differentiation
and resistance to oxidative stress and other aspects. Among them,
the transcription factor FoxO1 may be closely related to
tumorigenesis. Low expression of miR-181a2/miR-181b2 can promote
tumor growth of cervical cancer through the PIK3R3/Akt/FoxO
signaling pathway (27). miR-486-5p
acts as a powerful prostate cancer driver which drives
tumorigenesis by directly targeting FoxO signaling (28).
Overactivation of the mTOR signaling pathway is
associated with tumorigenesis and is an important target for tumor
therapy. Regulation of the mTOR signaling pathway through miRNAs
and control of miRNA biogenesis through mTORs may be important for
the diagnosis and treatment of different types of human cancer.
Previous studies have shown that miR-126 targets the PI3K/AKT/mTOR
signaling pathway, maintains the stemness of leukemia stem cells
and promotes chemotherapy resistance (29,30). By
inhibiting hematopoietic pre-B cell leukemia transcription
factor-interacting protein-mediated mTOR signaling pathway,
miR-148a can reduce the growth, epithelial-to-mesenchymal
transition (EMT), invasion, and metastasis of HBx-expressing
hepatocarcinoma cells (31). Studies
have also shown that inhibition of miRNAs in the mTOR signaling
pathway may inhibit the growth of tumor cells. In addition,
miR-590-3p/MACC1 was found to inhibit the malignant biological
behavior of glioblastoma stem cells by inhibiting the PI3K/AKT/mTOR
pathways (32–35). There are also drugs that inhibit cell
proliferation and tumor growth of esophageal adenocarcinoma both
in vitro and in vivo via the AMPKα/mTOR signaling
pathway (36).
The focal adhesion pathway is closely related to
EMT, which directly or indirectly regulates EMT-related protein
expression and cytoskeletal remodeling, thereby affecting the EMT
process (37,38). It plays a role in the formation and
differentiation of bone and cartilage (39). The focal adhesion signaling pathway
mediates the involvement of miR-92a in cartilage formation and
chondrocyte response induced by IL-1β (40). In the field of cancer, its
involvement in miR-301/PTEN (phosphatase and tensin homolog)
promotes the progression of malignant melanoma (41).
Previous studies have shown that miR-29c-5p, miR-29b
and miR-193a-3p all participate in the MAPK signaling pathway, and
methylation-associated silencing of miR-193a-3p promotes ovarian
cancer aggressiveness by targeting MAPK/ERK pathways (42). miR-29c-5p inhibits gallbladder
carcinoma progression by directly targeting cytoplasmic
polyadenylation element binding protein 4 and inhibiting the MAPK
signaling pathway (43). miR-29b
negatively modulates the MAPK/ERK and PI3K/Akt signaling pathways
to inhibit angiogenesis in endometrial carcinomas by targeting
vascular endothelial growth factor A (44).
The Wnt signaling pathway plays a role in stem
cells. miR-214 is a key regulator of Wnt signaling pathway activity
and stem cell function during normal tissue homeostasis,
regeneration and aging (45). The
Wnt signaling pathway is an important signal transduction pathway
for the differentiation of mesenchymal stem cells into
cardiomyocytes, which is regulated by miR-1-2. Overexpression of
miR-1–2 in bone marrow-derived stem cells (BMSCs) of mice can
induce differentiation into cardiomyocytes by activating the
Wnt/β-catenin signaling pathway (46). At the same time, it also plays an
important role in the invasion and treatment of tumors. By
activating the Wnt/β-catenin signaling pathway, miR-106b-5p
promotes invasiveness of renal cell carcinoma and stem cell-like
phenotype (47). Low levels of
miR-600 are correlated with activation of the Wnt signaling pathway
and poor prognosis in breast cancer) (48).
EpSCs are the key source of skin damage repair. The
present study found that expression of miRNAs was significantly
altered after heat loss. If various strategies can be used to
activate endogenous stem cells, then it is beneficial to help the
wound healing direction of patients with extensive burns (49). At the same time, there is increasing
evidence that the recruitment of mesenchymal stem cells is
beneficial for tissue repair after injury. It has been reported
that migration of mesenchymal stem cells may contribute to tumor
angiogenesis (50); however, the
present study used the patient's own epidermal stem cells as this
eliminates the risk factors that may cause mesenchymal stem cells
to facilitate tumor angiogenesis, and have a good application
prospect.
Overall, through high-throughput sequencing
technology, the present study found that miRNAs in human EpSCs,
following heat injury were significantly differentially expressed
which may be related to the mechanism of the wound healing
signaling pathway. However, the current research is still at a
preliminary stage. Expression of miRNAs needs to be further
modified and intervened to clarify the specific regulatory
mechanisms that are involved during the wound healing process, as
well as provide more information on the safe and effective
application of targeted treatment of various types of wounds.
Acknowledgements
The authors would like to thank Mrs Meng-Yun Li, Mr
Shang-Feng Fu and Mr Long-Xiang Tu, all from The First Affiliated
Hospital of Nanchang University (Nanchang, China); Mr Ji Yan from
The First Clinical Medical College, Nanchang University (Nanchang,
China) and Mr Xing-Chao Liu from Beihang University (Beijing,
China) for their guidance and assistance.
Funding
The present study was funded by the National Natural
Science Foundation of China (grant no. 81460293).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
DWL designed the present study. HTR and DWL
performed the experiments, wrote the paper, and reviewed and edited
the initial manuscript. Both authors read and approved the final
published version of the manuscript.
Ethics approval and consent to
participate
The present study was performed according to The
Declaration of Helsinki and was approved by the Ethics Committee of
Nanchang University (Nanchang, China). Written informed consent was
provided by all the participating donors.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gurtner GC, Werner S, Barrandon Y and
Longaker MT: Wound repair and regeneration. Nature. 453:314–321.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bao P, Kodra A, Tomic-Canic M, Golinko MS,
Ehrlich HP and Brem H: The role of vascular endothelial growth
factor in wound healing. J Surg Res. 153:347–358. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xue M, Zhao R, Lin H and Jackson C:
Delivery systems of current biologicals for the treatment of
chronic cutaneous wounds and severe burns. Adv Drug Deliv Rev.
129:219–241. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Werner S and Grose R: Regulation of wound
healing by growth factors and cytokines. Physiol Rev. 83:835–870.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Horsburgh S, Fullard N, Roger M, Degnan A,
Todryk S, Przyborski S and O'Reilly S: MicroRNAs in the skin: Role
in development, homoeostasis and regeneration. Clin Sci (Lond).
131:1923–1940. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Brouard M and Barrandon Y: In-vivo
dedifferentiation of keratinocytes to epidermal stem cells. Lancet.
359:528–529. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Levy V, Lindon C, Zheng Y, Harfe BD and
Morgan BA: Epidermal stem cells arise from the hair follicle after
wounding. FASEB J. 21:1358–1366. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Blanpain C and Fuchs E: Epidermal stem
cells of the skin. Annu Rev Cell Dev Biol. 22:339–373. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Beck B and Blanpain C: Mechanisms
regulating epidermal stem cells. EMBO J. 31:2067–2075. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Grosshans H and Slack FJ: Micro-RNAs:
Small is plentiful. J Cell Biol. 156:17–21. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lewis CJ: Stem cell application in acute
burn care and reconstruction. J Wound Care. 22:7–8, 10, 12–16.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Song Z, Liu D, Peng Y, Li J, Zhang Z and
Ning P: Differential microRNA expression profile comparison between
epidermal stem cells and differentiated keratinocytes. Mol Med Rep.
11:2285–2291. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu Y, Zhong L, Liu D, Ye H, Mao Y and Hu
Y: Differential miRNA expression profiles in human keratinocytes in
response to protein kinase C inhibitor. Mol Med Rep. 16:6608–6619.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Qu M and Nourbakhsh M: Current
experimental models of burns. Discov Med. 23:95–103.
2017.PubMed/NCBI
|
|
15
|
Banerjee J and Sen CK: microRNA and wound
healing. Adv Exp Med Biol. 888:291–305. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Herter EK and Xu Landén N: Non-coding
RNAs: New players in skin wound healing. Adv Wound Care (New
Rochelle). 6:93–107. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Meng Z, Zhou D, Gao Y, Zeng M and Wang W:
miRNA delivery for skin wound healing. Adv Drug Deliv Rev.
129:308–318. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Berindan-Neagoe I, Monroig PC, Pasculli B
and Calin GA: MicroRNAome genome: A treasure for cancer diagnosis
and therapy. CA Cancer J Clin. 64:311–336. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tang L, Chen HY, Hao NB, Tang B, Guo H,
Yong X, Dong H and Yang SM: Microrna inhibitors: Natural and
artificial sequestration of microrna. Cancer Lett. 407:139–147.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Paul CP, Good PD, Li SX, Kleihauer A,
Rossi JJ and Engelke DR: Localized expression of small RNA
inhibitors in human cells. Mol Ther. 7:237–247. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mullard A: Oncology trials gear up for
high-throughput sequencing. Nat Rev Drug Discov. 11:339–340. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang L, Zheng Z, Zhou Q, Bai X, Fan L,
Yang C, Su L and Hu D: miR-155 promotes cutaneous wound healing
through enhanced keratinocytes migration by MMP-2. J Mol Histol.
48:147–155. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mori R, Tanaka K and Shimokawa I:
Identification and functional analysis of inflammation-related
miRNAs in skin wound repair. Dev Growth Differ. 60:306–315. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fomicheva KA, Knyazev EN and Mal'tseva DV:
hsa-miR-1973 MicroRNA is significantly and differentially expressed
in MDA-MB-231 cells of breast adenocarcinoma and xenografts derived
from the tumor. Bull Exp Biol Med. 163:660–662. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang B, Li J, Sun M, Sun L and Zhang X:
miRNA expression in breast cancer varies with lymph node metastasis
and other clinicopathologic features. IUBMB Life. 66:371–377. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sen CK and Ghatak S: miRNA control of
tissue repair and regeneration. Am J Pathol. 185:2629–2640. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mei Q, Li X, Zhang K, Wu Z, Li X, Meng Y,
Guo M, Luo G, Fu X and Han W: Genetic and methylation-induced loss
of miR-181a2/181b2 within chr9q33.3 facilitates tumor growth of
cervical cancer through the PIK3R3/Akt/FoxO signaling pathway. Clin
Cancer Res. 23:575–586. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang Y, Ji C, Guo S, Su X, Zhao X, Zhang
S, Liu G, Qiu X, Zhang Q, Guo H and Chen H: The miR-486-5p plays a
causative role in prostate cancer through negative regulation of
multiple tumor suppressor pathways. Oncotarget. 8:72835–72846.
2017.PubMed/NCBI
|
|
29
|
Lechman ER, Gentner B, Ng SWK, Schoof EM,
van Galen P, Kennedy JA, Nucera S, Ciceri F, Kaufmann KB, Takayama
N, et al: miR-126 regulates distinct self-renewal outcomes in
normal and malignant hematopoietic stem cells. Cancer Cell.
29:602–606. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Raffel S and Trumpp A: Mir-126 drives
quiescence and self-renewal in leukemic stem cells. Cancer Cell.
29:133–135. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu X, Fan Z, Kang L, Han J, Jiang C, Zheng
X, Zhu Z, Jiao H, Lin J, Jiang K, et al: Hepatitis B virus X
protein represses miRNA-148a to enhance tumorigenesis. J Clin
Invest. 123:630–645. 2013.PubMed/NCBI
|
|
32
|
Zhou W, Liu L, Xue Y, Zheng J, Liu X, Ma
J, Li Z and Liu Y: Combination of endothelial-monocyte-activating
polypeptide-II with temozolomide suppress malignant biological
behaviors of human glioblastoma stem cells via miR-590-3p/MACC1
Inhibiting PI3K/AKT/mTOR signal pathway. Front Mol Neurosci.
10:682017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang Y, Zhang X, Tang W, Lin Z, Xu L, Dong
R, Li Y, Li J, Zhang Z, Li X, et al: Mir-130a upregulates mTOR
pathway by targeting TSC1 and is transactivated by NF-κB in
high-grade serous ovarian carcinoma. Cell Death Differ.
24:2089–2100. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Matter MS, Decaens T, Andersen JB and
Thorgeirsson SS: Targeting the mTOR pathway in hepatocellular
carcinoma: Current state and future trends. J Hepatol. 60:855–865.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Minna E, Romeo P, Dugo M, De Cecco L,
Todoerti K, Pilotti S, Perrone F, Seregni E, Agnelli L, Neri A,
Greco A and Borrello MG: miR-451a is underexpressed and targets
AKT/mTOR pathway in papillary thyroid carcinoma. Oncotarget.
7:12731–12747. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fujihara S, Morishita A, Ogawa K, Tadokoro
T, Chiyo T, Kato K, Kobara H, Mori H, Iwama H and Masaki T: The
angiotensin II type 1 receptor antagonist telmisartan inhibits cell
proliferation and tumor growth of esophageal adenocarcinoma via the
AMPKα/mTOR pathway in vitro and in vivo. Oncotarget. 8:8536–8549.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wörthmüller J, Blum W, Pecze L, Salicio V
and Schwaller B: Calretinin promotes invasiveness and EMT in
malignant mesothelioma cells involving the activation of the FAK
signaling pathway. Oncotarget. 9:36256–36272. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jin H, He Y, Zhao P, Hu Y, Tao J, Chen J
and Huang Y: Targeting lipid metabolism to overcome EMT-associated
drug resistance via integrin β3/FAK pathway and tumor-associated
macrophage repolarization using legumain-activatable delivery.
Theranostics. 9:265–278. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Song J, Ye B, Liu H, Bi R, Zhang N, Hu J
and Luo E: Fak-Mapk, hippo and Wnt signalling pathway expression
and regulation in distraction osteogenesis. Cell Prolif.
51:e124532018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hou C, Zhang Z, Zhang Z, Wu P, Zhao X, Fu
M, Sheng P, Kang Y and Liao W: Presence and function of
microRNA-92a in chondrogenic ATDC5 and adipose-derived mesenchymal
stem cells. Mol Med Rep. 12:4877–4886. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cui L, Li Y, Lv X, Li J, Wang X, Lei Z and
Li X: Expression of nicroRNA-301a and its functional roles in
malignant melanoma. Cell Physiol Biochem. 40:230–244. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen K, Liu MX, Mak CS, Yung MM, Leung HY,
Xu D, Ngu SF, Chan KK, Yang H, Ngan HY and Chan DW:
Methylation-associated silencing of miR-193a-3p promotes ovarian
cancer aggressiveness by targeting GRB7 and MAPK/ERK pathways.
Theranostics. 8:423–436. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Shu YJ, Bao RF, Jiang L, Wang Z, Wang XA,
Zhang F, Liang HB, Li HF, Ye YY, Xiang SS, et al: MicroRNA-29c-5p
suppresses gallbladder carcinoma progression by directly targeting
CPEB4 and inhibiting the MAPK pathway. Cell Death Differ.
24:445–457. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chen HX, Xu XX, Tan BZ, Zhang Z and Zhou
XD: MicroRNA-29b Inhibits angiogenesis by targeting VEGFA through
the MAPK/ERK and PI3K/Akt signaling pathways in endometrial
carcinoma. Cell Physiol Biochem. 41:933–946. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ahmed MI, Alam M, Emelianov VU,
Poterlowicz K, Patel A, Sharov AA, Mardaryev AN and Botchkareva NV:
MicroRNA-214 controls skin and hair follicle development by
modulating the activity of the Wnt pathway. J Cell Biol.
207:549–567. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shen X, Pan B, Zhou H, Liu L, Lv T, Zhu J,
Huang X and Tian J: Differentiation of mesenchymal stem cells into
cardiomyocytes is regulated by miRNA-1-2 via WNT signaling pathway.
J Biomed Sci. 24:292017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lu J, Wei JH, Feng ZH, Chen ZH, Wang YQ,
Huang Y, Fang Y, Liang YP, Cen JJ, Pan YH, et al: miR-106b-5p
promotes renal cell carcinoma aggressiveness and stem-cell-like
phenotype by activating Wnt/β-catenin signalling. Oncotarget.
8:21461–21471. 2017.PubMed/NCBI
|
|
48
|
El Helou R, Pinna G, Cabaud O, Wicinski J,
Bhajun R, Guyon L, Rioualen C, Finetti P, Gros A, Mari B, et al:
miR-600 acts as a bimodal switch that regulates breast cancer stem
cell fate through WNT signaling. Cell Rep. 18:2256–2268. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Papaccio F, Paino F, Regad T, Papaccio G,
Desiderio V and Tirino V: Concise review: Cancer cells, cancer stem
cells, and mesenchymal stem cells: Influence in cancer development.
Stem Cells Transl Med. 6:2115–2125. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Mele L, Vitiello PP, Tirino V, Paino F, De
Rosa A, Liccardo D, Papaccio G and Desiderio V: Changing paradigms
in Cranio-facial regeneration: Current and new strategies for the
activation of endogenous stem cells. Front Physiol. 7:622016.
View Article : Google Scholar : PubMed/NCBI
|