Introduction
Osteosarcoma (Os) is one of the most common primary
malignant bone tumors in pediatrics and adolescents, and originates
from interstitial cells (1). Os
accounts for ~20% of primary malignant bone tumors and 0.5% of
malignant tumors (2). Os tends to
occur in the epiphyseal region where there is an abundant blood
supply, thus the incidence of hematogenous metastasis is high,
occurs early and progresses quickly (3). Previous studies have shown that, if not
treated in time, 80% of patients with Os will develop lung
metastases, which is often fatal (4–6).
Currently, the drugs used to treat Os include doxorubicin,
methotrexate, cisplatin and ifosfamide (7). Some patients have a poor prognosis or
drug resistance after chemotherapy, both of which are difficult to
improve despite alternative treatment combinations (8). In addition, high doses of
chemotherapeutic agents have side effects (9). Therefore, understanding the molecular
mechanism underlying the occurrence and development of Os could aid
in the discovery of novel targets for Os treatment (10–12).
Thyroid hormone receptor-interacting protein 6
(TRIP6), a member of the zyxin family of LIM proteins family, is an
adaptor protein that regulates a variety of cellular responses,
such as actin cytoskeletal reorganization and cell adhesion, and is
predominantly expressed in epithelial cells (13,14).
Many TRIP6 binding partners have been identified, including the
tyrosine phosphatase (PTP)-BL and the adaptor protein
reversion-induced LIM, indicating that these proteins have
regulatory effects on the actin cytoskeleton and cell viability
(15,16). Previous studies have reported that
TRIP6 is overexpressed in nasopharyngeal cancer, non-Hodgkin
lymphoma and Ewing sarcoma (17–19).
Miao et al (17) showed that
overexpression of TRIP6 can reverse the cell adhesion-mediated drug
resistance phenotype by decreasing the phosphorylation of P27 in
non-Hodgkin lymphoma. In addition, Lai et al (20) found that TRIP6 overexpression in
glioblastoma inhibits cell apoptosis and causes resistance to
Fas-mediated cell invasion by enhancing NF-κB activity. Therefore,
TRIP6 may play an important role in cancer progression and
development (21). However, the
clinical significance and biological role of TRIP6 in human Os
remains unknown. Whilst TRIP6 has been reported in other cancer
types, it has not been reported in Os; therefore, the present study
investigated the effect of TRIP6 on Os. In addition, TRIP6 has been
suggested to be involved in the regulation of the NF-κB signaling
pathway, but further investigation is required to understand
whether TRIP6 affects the occurrence and development of Os via the
NF-κB signaling pathway.
The NF-κB signaling pathway is activated by
extracellular stimulation (22).
Extracellular signaling factors bind to receptors on the cell
membrane and initiate a cascade of downstream pathways (23). Receptor proteins first activate IκB
kinase (IKK) upon stimulation (24).
IKK then phosphorylates serine at the regulatory site of the IκB
subunit on the intracellular NF-κB/IκB compound, which allows the
IκB subunit to be ubiquitinated and degraded by the proteasome to
release the NF-κB dimer (25–29).
With the degradation of IκB, free P65 is phosphorylated by protein
kinase A at serine 276 in the cytoplasm, and then phosphorylated
P65 enters the nucleus and binds to corresponding binding sites on
genes, which initiates transcription (30). NF-κB also activates the expression of
the inhibitor of κBα (IκBα) gene, and the newly formed IκBα
inhibits the activity of NF-κB, resulting in a spontaneous negative
feedback loop (31). IκB is an
inhibitory protein of NF-κB. The IκB family consists of eight
members, including P100, P105, IκBα, IκBβ, IκBγ, IκBε, Bcl-3 and
IκB-R (32). During resting state,
IκBα and the NF-κB subunits P65 and P50, exist in the cytoplasm in
an inactive state (33). When
upstream signaling factors activate IKK, IκBα is ubiquitinated,
phosphorylated and degraded, converting the two subunits of NF-κB
from the inactive to the active state and translocating the
subunits from the cytoplasm to the nucleus. NF-κB then binds to
corresponding inflammation-related genes, and initiates the
transcription of inflammatory cytokines and induces inflammation
(34).
A previous preliminary study found that TRIP6 was
overexpressed in a large number of human Os samples (data not
shown). The present results suggested that overexpression of TRIP6
significantly promoted cell proliferation, migration and invasion,
and inhibited apoptosis of Os cells. However, silencing TRIP6
inhibited proliferation, migration and invasion, and promoted
apoptosis in Os cells. The present results suggested that TRIP6 may
play a role as an oncoprotein in the progression of Os, providing
novel insights into the regulatory mechanism of the NF-κB signaling
pathway.
Materials and methods
Cell culture and transfection
Human Os cell lines U2OS and MG63, and the normal
osteoblast cell line hFOB1.19 were purchased from The Cell Bank of
The Chinese Academy of Sciences. U2OS and MG63 were cultured for 24
to 48 h in DMEM (Gibco; Thermo Fisher Scientific, Inc.) with 10%
FBS (Gibco; thermo Fisher Scientific, Inc.) at 37°C with 5%
CO2. hFOB1.19 cells were cultured for 72 to 96 h in DMEM
with F12, 0.3 mg/ml G418 and 10% FBS at 33.5°C with 5%
CO2. Culture media was changed regularly and cells were
split to maintain growth in the logarithmic growth phase. MG63 and
U2OS cells were harvested and seeded in 6-well plates
(2×105 cells/well). Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) was used to transfect
cells according to the manufacturer's instructions. The following
plasmids (Shanghai GenePharma Co., Ltd.; 2.5 ng/well) were used: i)
pcDNA Flag TRIP6 or shNC (non-coding shRNA)/pGPU6/GFP/Neo plasmids
as the negative control; ii) TRIP6-small interfering RNA (siRNA;
5′-GAAGCTGGTTCACGACATGAA-3′) or TRIP6-negative control (NC); iii)
pcDNA Flag P65 or shNC/pGPU6/GFP/Neo plasmids as the negative
control; and iv) P65-siRNA (5′-GAACACAATGGCCACTTGCC-3′) or P65-NC.
After transfection, the cells were cultured for 24–48 h before
further experiments were performed.
Patient information and tissue
specimens
The present study was conducted on a total of 55
paraffin-embedded archived Os samples which had been
histopathologically and clinically diagnosed in The Department of
Orthopedics at The Second Affiliated Hospital of Anhui Medical
University from June 2008 to June 2018. All the experiments
performed using human tissues were from the 55 paraffin-embedded
archived samples (males, 36; females 19; age range, 8–62 years;
mean age, 25±8.4 years). Enneking stages: 19 cases were in I stage,
in II stage were 17 cases and in III stage were 19 cases.
Osteosarcoma was found in the following sites in patients: In the
femur in 21 patients, in the tibia in 24 patients and in other
sites in ten patients. The tumor diameter was <5 cm in 19 cases
and ≥5 cm in 36 cases. There were 15 osteoblastoma cases, 12 cases
of chondroblastoma, 18 cases of fibroblastoma and ten cases
involving other factors. There were 28 cases with metastasis and 27
cases without metastasis. Inclusion criteria included patients who
were pathologically diagnosed with Os. Clinical and
clinicopathological stage was determined according to American
Joint Committee on Cancer criteria (35). For the use of clinical materials for
research, written informed consent was obtained from each patient
or their relatives prior to surgery. The present study was approved
by The Institutional Research Ethics Committee. The clinical
information for patient samples is summarized in Tables SI and SII.
Western blotting
Western blotting was performed according to standard
methods (26). Total protein from
cell lines was extracted using RIPA lysis buffer (Thermo Fisher
Scientific, Inc.) and quantified using the Bradford method
(36). According to
paraffin-embedded tissue protein extraction kit (BestBio; cat. no.
BB-3164-2) instructions, protein extracted from paraffin embedding
organization was used to analyze TRIP6 protein expression level in
the adjacent non-tumor tissue and Os tissue. Paraffin-embedded
adjacent non-tumor samples were also obtained from the same
patient. In total, 60 µg protein were separated by 10% SDS-PAGE and
transferred to a PVDF membrane (EMD Millipore). The membranes were
blocked using 5% milk at 37°C for 1 h. Membranes were incubated
overnight at 4°C with the following antibodies: Anti-TRIP6 (1:200;
ab70747; Abcam), anti-P65 (1:5,000; ab109458; Abcam), anti-PP65
(1:16,000; ab6503; Abcam), anti-IκBα (1:1,000; ab32518; Abcam),
anti-GAPDH (1:10,000; ab181602; Abcam). The membranes were then
incubated with the following secondary antibodies for 2 h at 37°C:
Horseradish peroxidase-conjugated goat anti-mouse (1:1,000; Abcam,
ab6789) and horseradish peroxidase-conjugated goat anti-rabbit
(1:1,000; Abcam, ab6721). Bound proteins were visualized using
enhanced chemiluminescent kit (Thermo Fisher Scientific, Inc.) and
detected using a BioImaging System (UVP, Inc.). Relative protein
levels were calculated based on the GAPDH (1:10,000; ab181602;
Abcam) expression levels, which was used as the loading control.
Experiments were performed in triplicate.
MTT assay
Cells were seeded in 96-well plates at a density of
2×103 cells/well in 200 µl of complete or serum-free
media (Gibco; Thermo Fisher Scientific, Inc.). At each time point
(0, 24, 48, 72 and 96 h) cells were stained with 200 µl sterile MTT
dye (0.5 mg/ml; Sigma-Aldrich; Merck KGaA) for 5 h at 37°C,
followed by removal of the culture medium and addition of 100 µl of
DMSO (Sigma-Aldrich; Merck KGaA). Absorbance was measured at 570
nm, with 655 nm used as a reference wavelength. Experiments were
performed in triplicate.
Colony formation assay
Cells were plated in 6-well plates at a density of
4×102 cells/well and cultured for 12 days at 37°C.
Colonies were stained with 1% crystal violet for 30 sec after
fixation with 4% formaldehyde for 5 min, 20–25°C. The number of
colonies of more than 10 cells (>0.1 mm diameter) was counted
under a light microscope (×400). Data are presented as the fold
change compared with the control group. Experiments were performed
in triplicate.
Flow cytometric analysis of
apoptosis
Apoptosis was measured using the Annexin V-FITC (50
mM Tris; 100 mM NaCl; 1% BSA; 0.02% sodium azide; pH 7.4) Apoptosis
Detection kit (Invitrogen; Thermo Fisher Scientific, Inc.). After
cells were harvested, washed twice with PBS and resuspended in 450
µl binding buffer (Annexin V-FITC cell apoptosis assay kit;
Beyotime Institute of Biotechnology), Annexin V-FITC was added (5
µl/well). Cells were stained for 15 min under light protection at
room temperature, followed by resuspension in 450 µl binding buffer
(Annexin V-FITC cell apoptosis assay kit; Beyotime Institute of
Biotechnology). Cells were then stained with propidium iodide (PI)
in the dark (10 µl, 20–25°C, 10–20 min). Cell apoptosis was
analyzed on a flow cytometer (Attune NxT; BD Biosciences) and the
percentage of apoptotic cells was analyzed using CellQuest Pro
analysis software (version 5.1; BD Biosciences; Attune NxT; FACS
Calibur). Experiments were performed in triplicate. The calculation
method of apoptotic rate was the sum of early and late apoptotic
rate.
Cell migration and invasion assay
Transwell inserts were used to detect cell migration
and invasion. After transfection, cells in logarithmic phase were
harvested according to the manufacturer's instructions for the 24
wells BD basement membrane matrix (dilution, 1:3; BD Biosciences)
invasion Transwell chamber. A cell suspension of 5×104
cells/ml was seeded in the upper chamber, with or without Matrigel,
for 24 h at 37°C). The upper chamber was filled with serum-free
DMEM culture media (Gibco; Thermo Fisher Scientific, Inc.), and the
lower chamber was filled with DMEM (Gibco; Thermo Fisher
Scientific, Inc.) with 10% FBS. Plates were then incubated at 37°C
with 5% CO2 for 24 h. After culturing the cells for 24
h, the non-invaded cells in the inner layer of the compartment
membrane were wiped with cotton swabs. The membranes were then
fixed for 10–20 min at 20–25°C, with 3% paraformaldehyde, stained
with 0.1% crystal violet for 5 min at 20–25°C, washed with water
and dried. The number of cells in the experimental group was
measured by placing the Transwell chamber (Costar; Corning, Inc.)
on a microscope slide and acquiring images using a light microscope
(×400; Olympus Corporation). Each experiment was performed in
triplicate.
Statistical analysis
Statistical analyses were performed using the SPSS
version 13.0 statistical software package (SPSS, Inc.). Statistical
tests for data analysis included log rank test, χ2 test,
Spearman-rank correlation test, Student's 2-tailed t-test and
one-way ANOVA followed by Newman-Keuls test. Multivariate
statistical analysis was performed using a Cox regression model.
Data are presented as the mean ± SD. P<0.05 was considered to
indicate a statistically significant difference.
Results
Overexpression of TRIP6 correlates
with the progression and poor prognosis of Os
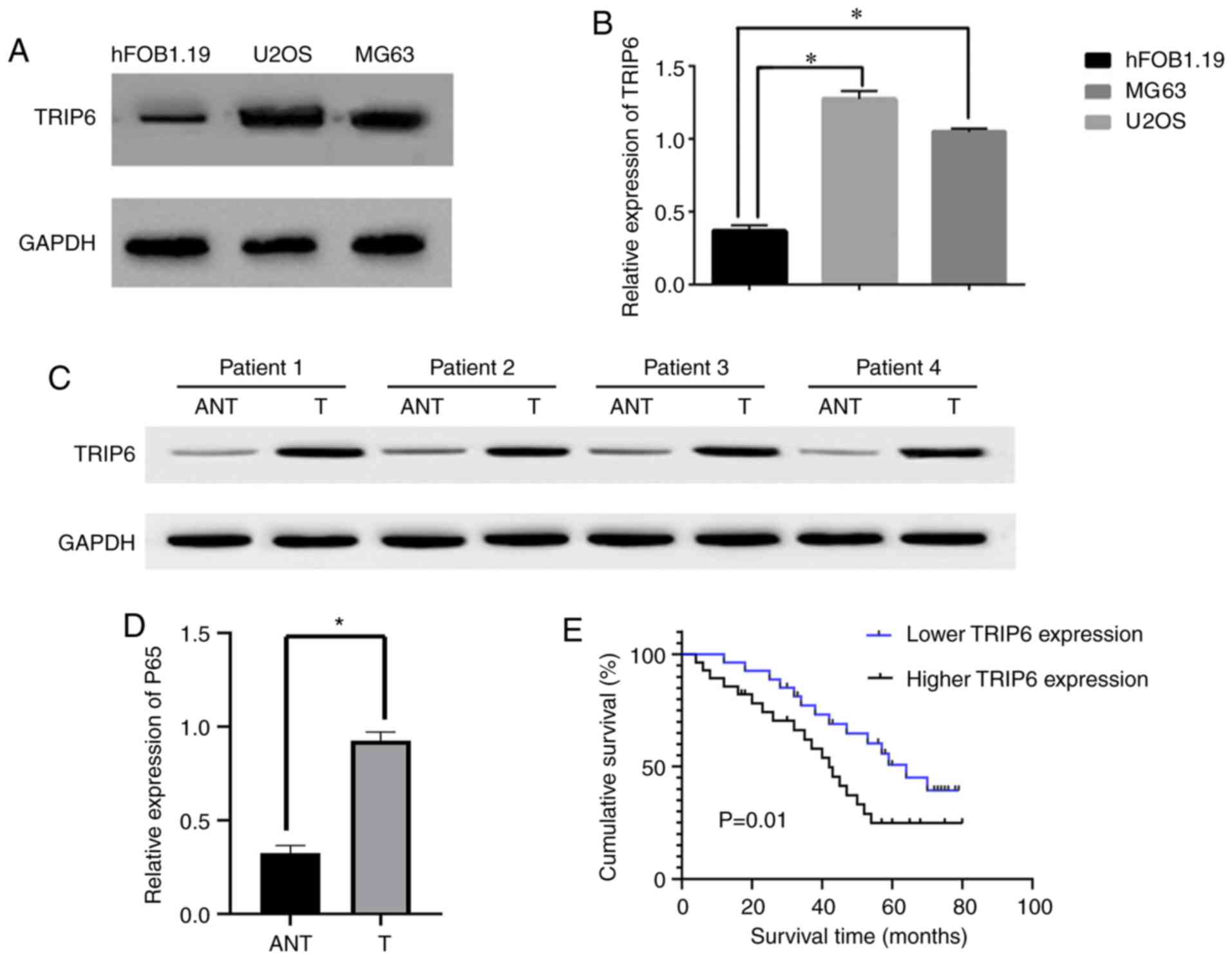
To investigate the carcinogenic effect of TRIP6 in
Os, the present study evaluated the protein expression level of
TRIP6 in Os cell lines and human Os tissue. TRIP6 was significantly
increased in U2OS and MG63 cells, compared with the normal
osteoblast cell line hFOB1.19 (Fig. 1A
and B). Comparative analysis results suggested that the
expression level of TRIP6 was significantly increased in the Os
samples compared with matched adjacent non-tumor tissues (Fig. 1C and D). The present results
suggested that that TRIP6 expression levels were significantly
increased in Os cells and human Os tissues compared with normal
osteoblasts and matched adjacent non-tumor tissues. Therefore, the
present results suggested that TRIP6 is overexpressed in Os.
Furthermore, the present results indicated that
TRIP6 overexpression was correlated with the Enneking stage and
metastasis (P<0.05), suggesting that TRIP6 overexpression is
associated with the clinical progression of human Os (Table SI). Moreover, based on the varying
expression levels of TRIP6 in different paraffin-embedded Os
samples, patients with Os could be separated into high- and
low-expression groups. The present results suggested that survival
time was significantly different between patients with Os with low
and high TRIP6 expression (P=0.01; Fig.
1E). In addition, univariate and multivariate analyses results
indicated that TRIP6 expression level may function as an
independent prognostic factor in Os (Table SII). Therefore, the present results
suggested that TRIP6 may be a potential prognostic indicator for
Os.
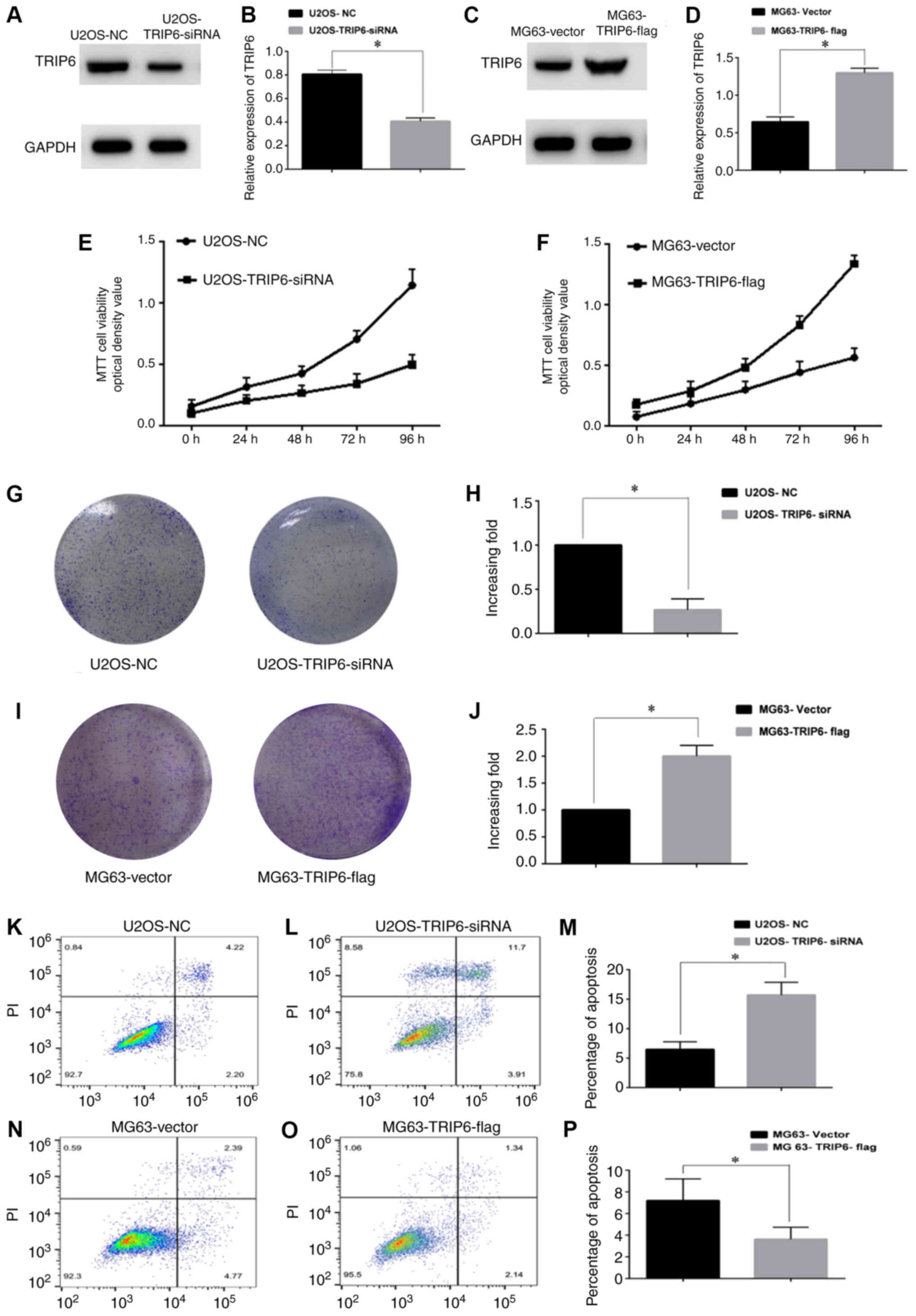
TRIP6 promotes Os cell proliferation
and apoptosis
The present study investigated the effect of TRIP6
on Os cell proliferation by overexpressing or silencing TRIP6.
Western blotting results suggested that TRIP6 was significantly
knocked down in the U2OS-TRIP6-siRNA cells compared with the
U2OS-NC cells (Fig. 2A and B). Cell
proliferation was significantly decreased in the U2OS-TRIP6-siRNA
group compared with the U2OS-NC group. After TROP6 overexpression,
TRIP6 expression level was significantly increased in the
MG63-TRIP6-flag group compared with the MG63-vector group (Fig. 2C and D). The MTT and colony formation
assay results suggested that, compared with the U2OS-NC group, cell
proliferation and colony formation in the U2OS-TRIP6-siRNA group
were significantly decreased (Fig. 2E, G
and H). In addition, compared with the MG63-Vector group, cell
proliferation and colony formation in the MG63-TRIP6-flag group
were significantly increased (Fig. 2F, I
and J). Flow cytometry using Annexin V-FITC and PI was
performed to quantify apoptosis after TRIP6 overexpression or
knockdown in U2OS and MG63 cells. The apoptotic rate was decreased
in U2OS-NC cells (6.42%) compared with U2OS-TRIP6-siRNA cells
(15.61%; Fig. 2K-M). The apoptotic
rate was 7.16% in the MG63-Vector group and 3.48% in the
MG63-TRIP6-flag group (Fig. 2N-P).
Overall, the present results suggested a potential role for TRIP6
in Os, as TRIP6 overexpression inhibited Os apoptosis and silencing
TRIP6 promoted Os apoptosis.
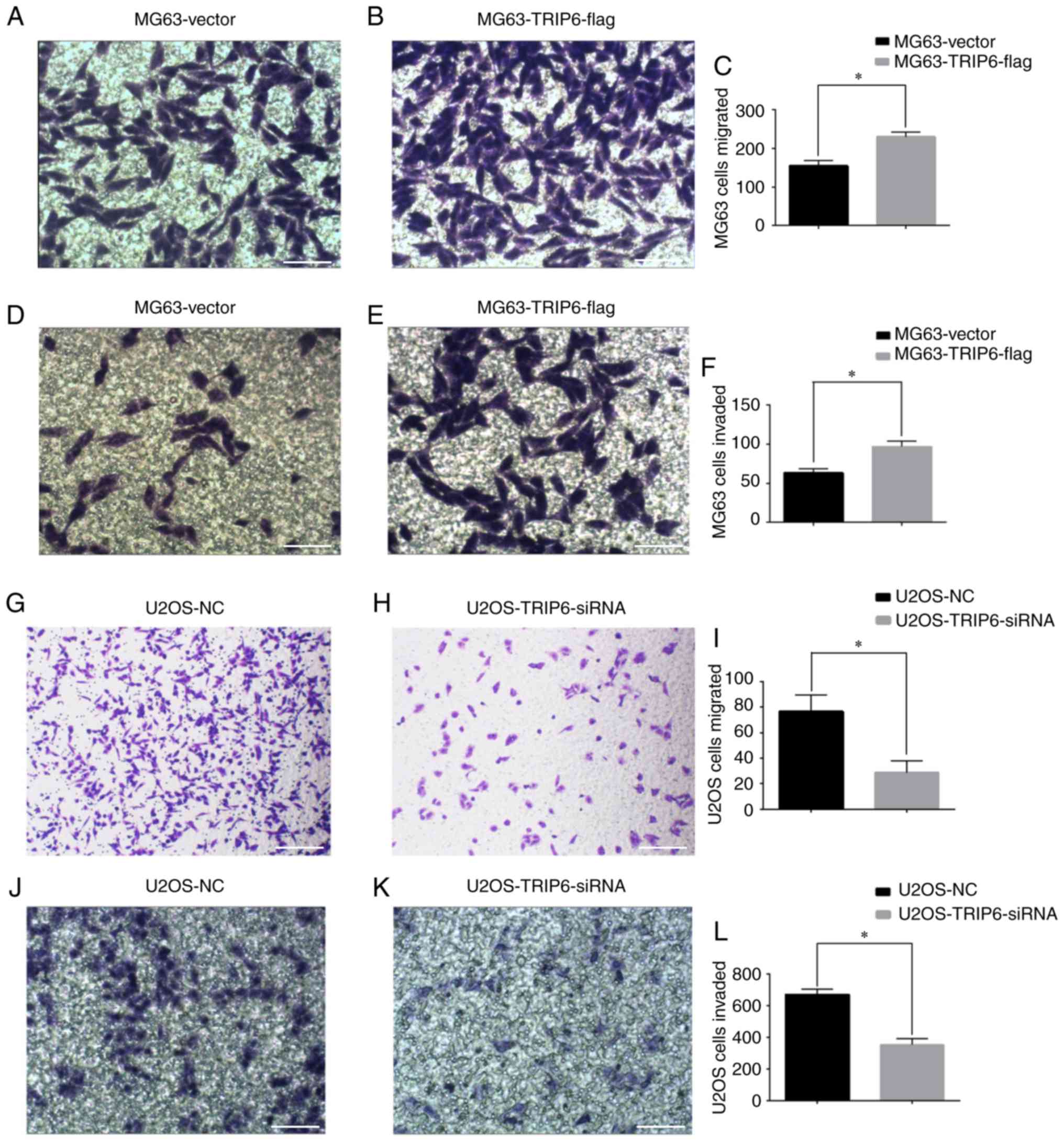
Effect of TRIP6 on the invasive and
migratory abilities of Os cells
TRIP6 was successfully transfected into U2OS and
MG63 cells, and Transwell assays were used to measure cell
migration and invasion. The present results suggested that the
migratory (Fig. 3A-C) and invasive
(Fig. 3D-F) ability of cells in the
MG63-TRIP6-flag group was significantly increased compared with the
MG63-Vector group. The migratory (Fig.
3G-I) and invasive (Fig. 3J-L)
ability of cells in the U2OS-TRIP6-siRNA group was significantly
lower compared with the U2OS-NC group. The present results
suggested that silencing TRIP6 inhibited, whereas overexpression of
TRIP6 promoted, the invasion and migration of Os cells.
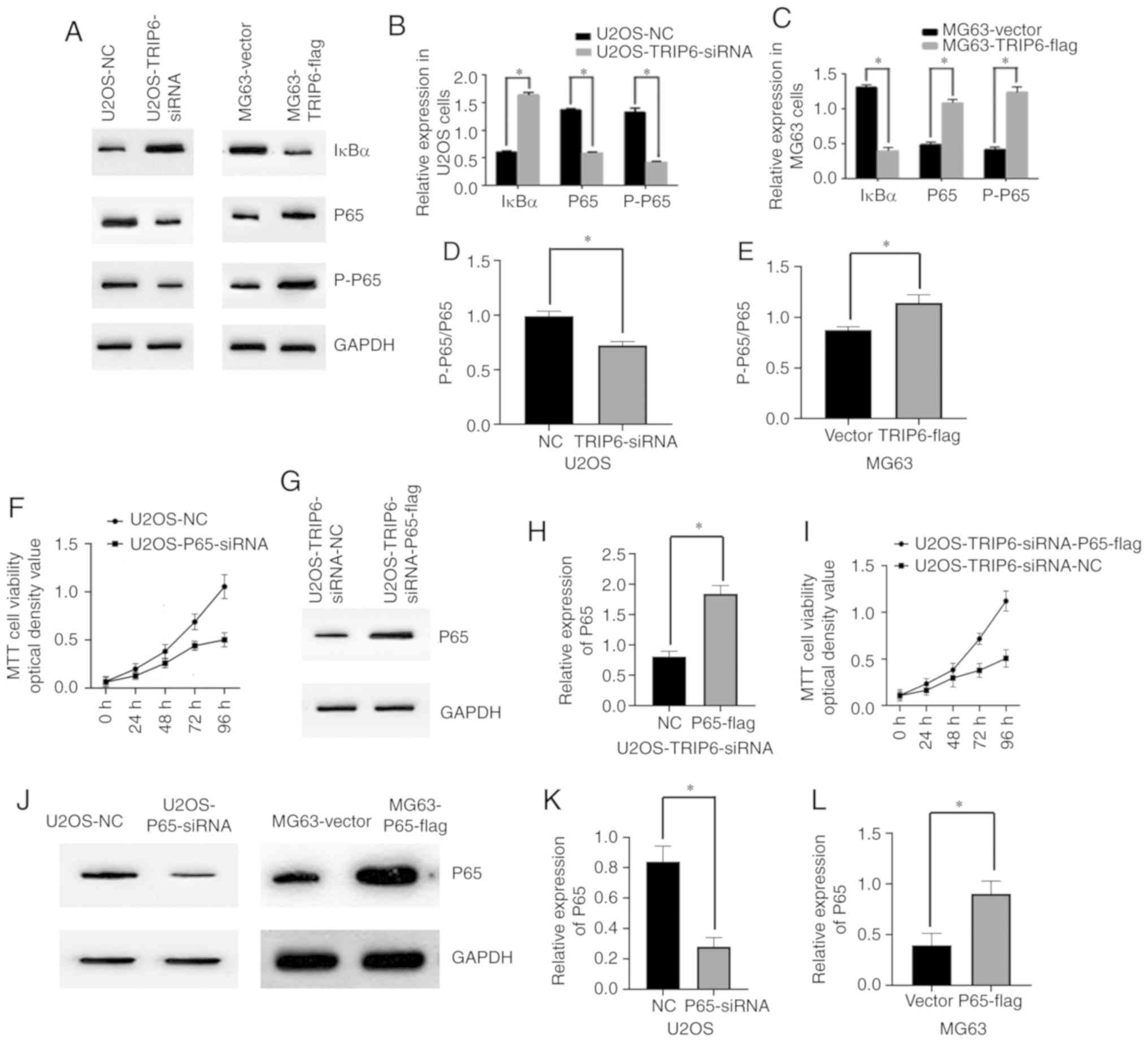
TRIP6 downregulates IκBα and activates
the NF-κB signaling pathway
Previous studies have shown that TRIP6 plays a role
in the NF-κB signaling pathway in non-Hodgkin lymphoma, Ewing
sarcoma, nasopharyngeal cancer and hepatocellular carcinoma
(18–20). Therefore, the present study
investigated whether TRIP6 was involved in the NF-κB signaling
pathway in OS cells. The present study knockdown TRIP6 using siRNA
in U2OS cells and overexpressed TRIP6 in MG63 cells. Knockdown of
TRIP6 in the U2OS-TRIP6-siRNA group, increased the expression level
of IκBα and decreased NF-κB P65 activation compared with the
control group (Fig. 4A and B).
Overexpression of TRIP6 in the MG63-TRIP6-flag group reduced the
expression level of IκBα and increased NF-κB P65 activation
compared with the MG63-Vector group (Fig. 4A and C). Therefore, the present
results suggested that TRIP6 may play a role in activating the
NF-κB signaling pathway in Os. The expression level of
phosphorylated (p)-P65 was related to the phosphorylation of this
protein (Fig. 4D and E). The present
study investigated the role of P65 in Os cell proliferation by
knocking down P65 in U2OS cells and overexpressing P65 in MG63
cells. MTT assay results suggested that the expression level of P65
in U2OS-P65-siRNA group was decreased compared with the U2OS-NC
group. In addition, the expression level of P65 in the
MG63-P65-flag group was significantly higher compared with the
MG63-Vector group (Fig. 4J-L). The
MTT assay results suggested that cell proliferation in the
U2OS-P65-siRNA group was decreased compared with the U2OS-NC group
(Fig. 4F). Furthermore, MTT assay
results suggested that overexpressing P65 in U2OS-TRIP6-siRNA cells
increased the growth rate of U2OS-TRIP6-siRNA cells (Fig. 4G-I). Collectively, the present
results suggested that P65 may play a role in the proliferative
effect of TRIP6 on Os cells.
Discussion
TRIP6 is an adaptor protein primarily expressed in
epithelial cells that can mediate several cellular responses, such
as actin cytoskeletal reorganization and cell adhesion (13,14).
However, the clinical significance and biological role of TRIP6 in
Os is not fully understood. Previous studies have shown that TRIP6
is involved in cell growth in Ewing sarcoma and non-Hodgkin
lymphoma (17,18). In line with a previous study, the
present results suggested that TRIP6 was highly expressed in human
Os tissue but weakly expressed in matched adjacent non-tumor
tissues (P<0.05) (18).
Therefore, the present results supported the hypothesis that TRIP6
may be an oncogenic protein that promotes cell proliferation,
migration and invasion, and inhibits apoptosis in Os cells.
The mechanism underlying the effect of TRIP6 in
promoting Os cell proliferation, migration and invasion, and
inhibiting apoptosis may be via the activation of the NF-κB
signaling pathway. The NF-κB signaling pathway is activated by IκB
phosphorylation and degradation (37). The present results suggested a novel
role for TRIP6 in Os, which may facilitate the development of novel
therapeutics for Os.
NF-κB is a family of transcription factor proteins
that includes the five subunits Rel, p65, RelB, p50 and p52
(38,39). The inhibitory protein IκBα, which is
downregulated in various types of cancer, such as breast cancer,
hepatocellular carcinoma and prostate cancer, plays vital roles in
multiple biological processes including proliferation, migration,
invasion and apoptosis (40–43). Previous studies have reported that
the target protein TRIP6 binds to receptors on the cell membrane
and initiates a cascade of downstream reactions (44,45).
Receptor proteins first activate IKK upon stimulation (46). Then, IKK phosphorylates serine at the
regulatory site of the IκB subunit of the intracellular NF-κB/IκB
compound, which allows the IκB subunit to be ubiquitinated and
degraded by the proteasome to release the NF-κB dimer (47). Then, P65, with the aid of P50, moves
from the cytoplasm to the nucleus and binds to specific DNA
sequences to regulate the transcription of target genes (48). Therefore, a role for NF-κB in
TRIP6-induced tumorigenesis has been suggested. However, the
molecular mechanism underlying TRIP6 signaling and NF-κB activation
are not fully understood. Activated IKK phosphorylates and degrades
IκBα, leading to activation and nuclear translocation of the two
subunits of NF-κB; especially the P65 subunit (49–54). The
present results suggested that TRIP6 overexpression increased IκBα
degradation and the phosphorylation of its downstream targets, such
as P65. Therefore, the present results indicated that IκBα may be
involved in TRIP6-induced cancer cell proliferation and survival,
and could regulate various characteristics of cancer cells,
including colony-formation, proliferation, migration and
invasion.
The present results supported the hypothesis that
TRIP6 may promote cell proliferation, migration and invasion, and
inhibit apoptosis of Os cells via activation of the NF-κB signaling
pathway. The present results may provide rationale for novel
potential therapeutic strategies for patients with Os. The present
study was limited to investigating TRIP6 function in Os in
vitro, and future in vivo experiments are critical for
the understanding of the role of TRIP6 in Os. Moreover, the effects
of TRIP6 on tumor formation in nude mouse models requires
investigation.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by The National Natural
Science Foundation of China (grant nos. 81702656 and 81671204) and
Natural Science Foundation of Anhui Province (grant no.
1708085QH215).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WL and LC performed the experiments. WL, LC and QL
collected and interpreted the data. WL and JJ drafted the
manuscript. WL, LC and JJ were responsible for the conception and
design of the study. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study protocol was approved by The Ethics
Committee of The Second Affiliated Hospital of Anhui Medical
University (SL-YX2019-042). Written informed consent was obtained
from each patient or their relatives prior to surgery.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhao Z, Lin X, Tong Y and Li W: Silencing
lncRNA ZFAS1 or elevated microRNA-135a represses proliferation,
migration, invasion and resistance to apoptosis of osteosarcoma
cells. Cancer Cell Int. 19:3262019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gill J, Ahluwalia MK, Geller D and Gorlick
R: New targets and approaches in osteosarcoma. Pharmacol Ther.
137:89–99. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang C, Ma K and Li WY: Cinobufagin
suppresses the characteristics of osteosarcoma cancer cells by
inhibiting the IL-6-OPN-STAT3 pathway. Drug Des Devel Ther.
13:4075–4090. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lindsey BA, Markel JE and Kleinerman ES:
Osteosarcoma overview. Rheumatol Ther. 4:25–43. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Friebele JC, Peck J, Pan X, Abdel-Rasoul M
and Mayerson JL: Osteosarcoma: A meta-analysis and review of the
literature. Am J Orthop (Belle Mead NJ). 44:547–553.
2015.PubMed/NCBI
|
|
6
|
Jour G, Wang L, Middha S, Zehir A, Chen W,
Sadowska J, Healey J, Agaram NP, Choi L, Nafa K and Hameed M: The
molecular landscape of extraskeletal osteosarcoma: A
clinicopathological and molecular biomarker study. J Pathol Clin
Res. 29:9–20. 2015.
|
|
7
|
Dong S, Xiao Y, Ma X, He W, Kang J, Peng
Z, Wang L and Li Z: miR-193b increases the chemosensitivity of
osteosarcoma cells by promoting FEN1-mediated autophagy. Onco
Targets Ther. 12:10089–10098. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yoshizawa M, Nakamura S, Sugiyama Y, Tamai
S, Ishida Y, Sueyoshi M, Toda Y, Hosogi S, Yano Y and Ashihara E:
6-Hydroxythiobinupharidine inhibits migration of LM8 osteosarcoma
cells by decreasing expression of LIM domain kinase 1. Anticancer
Res. 39:6507–6513. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Luetke A, Meyers PA, Lewis I and Juergens
H: Osteosarcoma treatment-Where do we stand? A state of the art
review. Cancer Treat Rev. 40:523–532. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xue M, Shen J, Cui J, Wu J, Qiao W, Ding
N, Song C and Shan B: MicroRNA-638 expression change in
osteosarcoma patients via PLD1 and VEGF expression. Exp Ther Med.
17:3899–3906. 2019.PubMed/NCBI
|
|
11
|
Cai W, Xu Y, Yin J, Zuo W and Su Z:
miR-552-5p facilitates osteosarcoma cell proliferation and
metastasis by targeting WIF1. Exp Ther Med. 17:3781–3788.
2019.PubMed/NCBI
|
|
12
|
Wu H, He Y, Chen H, Liu Y, Wei B, Chen G
and Lin H and Lin H: LncRNA THOR increases osteosarcoma cell
stemness and migration by enhancing SOX9 mRNA stability. FEBS Open
Bio. 9:781–790. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lin VT and Lin FT: TRIP6: An adaptor
protein that regulates cell motility, anti-apoptotic signaling and
transcriptional activity. Cell Signal. 23:1691–1697. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao MK, Wang Y, Murphy K, Yi J, Beckerle
MC and Gilmore TD: Gilmore TD LIM domain-containing protein trip6
can act as a coactivator for the v-Rel transcription factor. Gene
Expr. 8:207–217. 1999.PubMed/NCBI
|
|
15
|
Erdmann KS: The protein tyrosine
phosphatase PTP-Basophil/Basophil-like. Interacting proteins and
molecular functions. Eur J Biochem. 270:4789–4798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bashirova AA, Markelov ML, Shlykova TV,
Levshenkova EV, Alibaeva RA and Frolova EI: The human RIL gene:
Mapping to human chromosome 5q31.1, genomic organization and
alternative transcripts. Gene. 210:239–245. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Miao X, Xu X, Wu Y, Zhu X, Chen X, Li C,
Lu X, Chen Y, Liu Y, Huang J, et al: Overexpression of TRIP6
promotes tumor proliferation and reverses cell adhesion-mediated
drug resistance (CAM-DR) via regulating nuclear p27(Kip1)
expression in non-Hodgkin's lymphoma. Tumor Biol. 37:1369–1378.
2016. View Article : Google Scholar
|
|
18
|
Grunewald TG, Willier S, Janik D, Unland
R, Reiss C, Prazeres da Costa O, Buch T, Dirksen U, Richter GH,
Neff F, et al: The Zyxin-related protein thyroid receptor
interacting protein 6 (TRIP6) is overexpressed in Ewing's sarcoma
and promotes migration, invasion and cell growth. Biol Cell.
105:535–547. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fei J, Li J, Shen S and Zhou W:
Characterization of TRIP6-dependent nasopharyngeal cancer cell
migration. Tumor Biol. 34:2329–2335. 2013. View Article : Google Scholar
|
|
20
|
Lai YJ, Lin VT, Zheng Y, Benveniste EN and
Lin FT: The adaptor protein TRIP6 antagonizes fas-induced apoptosis
but promotes its effect on cell migration. Mol Cell Biol.
30:5582–5596. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhu L, Xu X, Tang Y and Zhu X: TRIP6
functions as a potential oncogene and facilitated proliferation and
metastasis of gastric cancer. Biologics. 13:101–110.
2019.PubMed/NCBI
|
|
22
|
Sun BQ, Sui YD, Huang H, Zou XB, Chen SC
and Yu ZK: Effect of lncRNA CRNDE on sepsis-related kidney injury
through the TLR3/NF-κB pathway. Eur Rev Med Pharmacol Sci.
23:10489–10497. 2019.PubMed/NCBI
|
|
23
|
Su XF, Li N, Meng FL, Chu YL, Li T and Gao
XZ: MiR-16 inhibits hepatocellular carcinoma progression by
targeting FEAT through NF-κB signaling pathway. Eur Rev Med
Pharmacol Sci. 23:10274–10282. 2019.PubMed/NCBI
|
|
24
|
Zhang Y, Zhou X, Zhang Q, Zhang Y, Wang X
and Cheng L: Involvement of NF-κB signaling pathway in the
regulation of PRKAA1-mediated tumorigenesis in gastric cancer.
Artif Cells Nanomed Biotechnol. 47:3677–3686. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li H, Wittwer T, Weber A, Schneider H,
Moreno R, Maine GN, Kracht M, Schmitz ML and Burstein E: Regulation
of NF-κB activity by competition between RelA acetylation and
ubiquitination. Oncogene. 31:611–623. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Schmitz ML, Mattioli I, Buss H and Kracht
M: Nf-kappaB: A multifaceted transcription factor regulated at
several levels. Chembiochem. 5:1348–1358. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wietek C and O'Neill LA: Diversity and
regulation in the NF-kappaB system. Trends Biochem Sci. 32:311–319.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pan D and Lin X: Epithelial growth factor
receptor-activated nuclear factor κB signaling and its role in
epithelial growth factor receptor-associated tumors. Cancer J.
19:461–467. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jiang T, Grabiner B, Zhu Y, Jiang C, Li H,
You Y, Lang J, Hung MC and Lin X: CARMA3 is crucial for
EGFR-Induced activation of NF-κB and tumor progression. Cancer Res.
71:2183–2192. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gu NJ, Wu MZ, He L, Wang XB, Wang S, Qiu
XS, Wang EH and Wu GP: HPV 16 E6/E7 up-regulate the expression of
both HIF-1α and GLUT1 by inhibition of RRAD and activation of NF-κB
in lung cancer cells. J Cancer. 10:6903–6909. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Natoli G, Saccani S, Bosisio D and Marazzi
I: Interactions of NF-kappaB with chromatin: The art of being at
the right place at the right time. Nat Immunol. 6:439–445. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Duan Y, An W, Wu H and Wu Y: Salvianolic
acid C attenuates LPS-induced inflammation and apoptosis in human
periodontal ligament stem cells via toll-like receptors 4
(TLR4)/nuclear factor kappa B (NF-κB) pathway. Med Sci Monit.
25:9499–9508. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Oliveira IDSDS, Colares AV, Cardoso FO,
Tellis CJM, Chagas MDSDS, Behrens MD, Calabrese KDS, Almeida-Souza
F and Abreu-Silva AL: Vernonia polysphaera Baker: Anti-inflammatory
activity in vivo and inhibitory effect in LPS-stimulated RAW 264.7
cells. PLoS One. 14:e02252752019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fei J, Ling YM, Zeng MJ and Zhang KW:
Shixiang plaster, a traditional Chinese medicine, promotes healing
in a rat model of diabetic ulcer through the receptor for advanced
glycation end products (RAGE)/nuclear factor kappa B (NF-κB) and
vascular endothelial growth factor (VEGF)/vascular cell adhesion
molecule-1 (VCAM-1)/endothelial nitric oxide synthase (eNOS)
signaling pathways. Med Sci Monit. 25:9446–9457. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pasquali S and Gronchi A: Neoadjuvant
chemotherapy in soft tissue sarcomas: Latest evidence and clinical
implications. Ther Adv Med Oncol. 9:415–429. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kouloulia S, Hallin EI, Simbriger K,
Amorim IS, Lach G, Amvrosiadis T, Chalkiadaki K, Kampaite A, Truong
VT, Hooshmandi M, et al: Raptor-mediated proteasomal degradation of
deamidated 4E-BP2 regulates postnatal neuronal translation and
NF-κB activity. Cell Rep. 29:3620–3635.e7. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sehgal K and Barbie DA: Clonal selection
drives NF-κB activation in recurrent nasopharyngeal carcinoma.
Cancer Res. 79:5915–5916. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Biswas DK and Iglehart JD: Linkage between
EGFR family receptors and nuclear factor kappaB (NF-kappaB)
signaling in breast cancer. J Cell Physiol. 209:645–652. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Biswas DK, Cruz AP, Gansberger E and
Pardee AB: Epidermal growth factor-induced nuclear factor kappa B
activation: A major pathway of cell-cycle progression in
estrogen-receptor negative breast cancer cells. Proc Natl Acad Sci
USA. 97:8542–8547. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang RK, Shao XM, Yang JP, Yan HL and Shao
Y: MicroRNA-145 inhibits proliferation and promotes apoptosis of
HepG2 cells by targeting ROCK1 through the ROCK1/NF-κB signaling
pathway. Eur Rev Med Pharmacol Sci. 23:2777–2785. 2019.PubMed/NCBI
|
|
41
|
Ma X and Ning S: Shikimic acid promotes
estrogen receptor (ER)-positive breast cancer cells proliferation
via activation of NF-κB signaling. Toxicol Lett. 312:65–71. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zou PL, Zhang QH, Zhou JF, Lin RW, Chen ZQ
and Xiang ST: Inhibitory effect of polyphyllin I on the
proliferation of prostate cancer PC3 cells via ERK1/2/P65/DNMT1 and
its molecular mechanism. Zhonghua Nan Ke Xue. 24:199–205. 2018.(In
Chinese). PubMed/NCBI
|
|
43
|
Zhou B, Yang Y and Li C: SIRT1 inhibits
hepatocellular carcinoma metastasis by promoting M1 macrophage
polarization via NF-κB pathway. Onco Targets Ther. 12:2519–2529.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li L, Bin LH, Li F, Liu Y, Chen D, Zhai Z
and Shu HB: TRIP6 is a RIP2-associated common signaling component
of multiple NF-kappaB activation pathways. J Cell Sci. 118:555–563.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kassel O, Schneider S, Heilbock C, Litfin
M, Göttlicher M and Herrlich P: A nuclear isoform of the focal
adhesion LIM-domain protein Trip6 integrates activating and
repressing signals at AP-1- and NF-kappaB-regulated promoters.
Genes Dev. 18:2518–2528. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li Q, Sun J, Mohammadtursun N, Wu J, Dong
J and Li L: Curcumin inhibits cigarette smoke-induced inflammation
via modulating the PPARγ-NF-κB signaling pathway. Food Funct.
10:7983–7994. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang P, Yin Y, Wang T, Li W, Li C, Zeng
X, Yang W, Zhang R, Tang Y, Shi L, et al: Maresin 1 mitigates
concanavalin A-induced acute liver injury in mice by inhibiting
ROS-mediated activation of NF-κB signaling. Free Radic Biol Med.
147:23–36. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ma X, Jiang Z, Li N, Jiang W, Gao P, Yang
M, Yu X, Wang G and Zhang Y: Ets2 suppresses inflammatory cytokines
through MAPK/NF-κB signaling and directly binds to the IL-6
promoter in macrophages. Aging (Albany NY). 11:10610–10625. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Guo G, Tian A, Lan X, Fu C, Yan Z and Wang
C: Nano hydroxyapatite induces glioma cell apoptosis by suppressing
NF-κB signaling pathway. Exp Ther Med. 17:4080–4088.
2019.PubMed/NCBI
|
|
50
|
Zhang R, Li X, Wei L, Qin Y and Fang J:
Lemur tyrosine kinase 2 acts as a positive regulator of NF-κB
activation and colon cancer cell proliferation. Cancer Lett.
454:70–77. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Fang W, Liao W, Zheng Y, Huang X, Weng X,
Fan S, Chen X, Zhang X, Chen J, Xiao S, et al: Neurotropin reduces
memory impairment and neuroinflammation via BDNF/NF-κB in a
transgenic mouse model of Alzheimer's disease. Am J Transl Res.
11:1541–1554. 2019.PubMed/NCBI
|
|
52
|
Yu Y, Feng S, Wei S, Zhong Y, Yi G, Chen
H, Liang L, Chen H and Lu X: Extracellular ATP activates
P2X7R-NF-κB (p65) pathway to promote the maturation of bone
marrow-derived dendritic cells of mice. Cytokine. 119:175–181.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Cai C, Zhu H, Ning X, Li L, Yang B, Chen
S, Wang L, Lu X and Gu D: LncRNA ENST00000602558.1 regulates ABCG1
expression and cholesterol efflux from vascular smooth muscle cells
through a p65-dependent pathway. Atherosclerosis. 285:31–39. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yamaguchi Y, Ayaki T, Li F, Tsujimura A,
Kamada M, Ito H, Maki T, Sawamoto N, Urushitani M and Takahashi R:
Phosphorylated NF-κB subunit p65 aggregates in granulovacuolar
degeneration and neurites in neurodegenerative diseases with
tauopathy. Neurosci Lett. 704:229–235. 2019. View Article : Google Scholar : PubMed/NCBI
|