Introduction
In the past several decades, overuse and misuse of
antibiotics has led to increased antibiotic resistance which has
become a significant public health concern (1). There is no systematic surveillance of
antibiotic misuse and no data are available to quantify the problem
(2,3). Currently, antibiotic resistance
presents an increasing trend that leads to significant public
health issues and the increase in healthcare-associated infections
caused by antibiotic-resistant pathogens worldwide (4,5).
Resistance to different antimicrobial drugs is attributed to the
increased irregular employment of antibiotics that have resulted in
the drug resistance of expression of virulence factors, promotion
of intra-hospital cross infection and even increased challenges of
clinical treatments (6-8). A
previous report suggests that the major driving force for the
occurrence of antibiotic-resistant pathogens is the evolution of
metabolic function caused by the rapid antibiotic consumption
worldwide (9). Therefore, on the one
hand, health organizations are required to implement appropriate
policies to supervise antibiotic usage to reduce antibiotic
resistance. On the other hand, understanding the molecular
biological mechanisms underlying antimicrobial resistance is
imminent to evaluate the effects of intervention strategies.
Currently, antimicrobial resistance of gram-negative
bacilli is a persistent issue (10).
Antibiotic treatment is the most common therapeutic regiment for
pulmonary infections with Klebsiella pneumoniae in patients
with pneumonia (11). The current
antimicrobial resistance and susceptibility of Klebsiella
pneumoniae bacteria have been observed in clinical practice
(12). Previous research has shown
that the frequent outbreak of nosocomial infections is due to
extended-spectrum β-lactamase (ESBL) produced by Klebsiella
pneumoniae that is attributed to multiple mechanisms underlying
drug resistance (13). In addition,
strains of Klebsiella pneumoniae exhibit transferable
multiple drug resistance based on clinical sepsis observation
(14). Furthermore, antibacterial
drug susceptibility of Klebsiella pneumoniae has attracted
attention since pathogenic bacteria have acquired simultaneous
resistance to various antimicrobial classes mediated by the
production of ESBL. However, no precise molecular biological
mechanisms underlying the antimicrobial drug resistance of
Klebsiella pneumoniae have been reported (15).
The correlation between antimicrobial drug
resistance and biofilm formation along with ESBL lactamase produced
in Escherichia coli has been demonstrated in a previous
study (16). Recently, the increase
in drug resistance among Klebsiella pneumoniae has caused a
great problem in the treatment of pneumonia (17). The mechanisms involved when
β-lactamase hydrolyzes β-lactam antibiotics have been investigated
by performing different experiments (18). Previous research indicates that
ancient evolutionary relationships between β-lactamases and
antibiotic-producing bacteria are relatively conservative (19). In any way, antibiotic-resistance
genes originate in antibiotic-producing microorganisms and
subsequently integrate into the genome of other pathogens through
transduction and/or transformation (20,21).
Research has found that penicillin-binding proteins (PBPs),
membrane-associated macromolecules, play key roles in the cell wall
synthesis process (22). In
addition, zinc finger nuclease is a new approach to overcome
β-lactam antibiotic resistance (23).
In the present study, it was hypothesized that
interfering with ESBL synthesis could decrease antimicrobial drug
resistance leading to the control of nosocomial infections,
transmission and cross infection. The study also investigated the
association between the molecular biological mechanism underlying
the antibiotic resistance of Klebsiella pneumoniae and the
ESBL/PBP signaling pathway. The present study was designed to
elucidate β-lactam resistance and to understand the efficacy of
PBPs and zinc finger nuclease in increasing ESBL expression.
Materials and methods
Klebsiella pneumoniae culture and
reagents
Natural being Klebsiella pneumoniae (NB-K.p)
bacteria were purchased from American Type Culture Collection
(Klebsiella pneumoniae subsp. Pneumoniae; ATCC®
43816™). Klebsiella pneumoniae bacteria from patients with
pneumonia (PD-K.p) were isolated from a 56-year male patient with
pneumonia who suffered from the disease for approximately 30 years.
Cells of Klebsiella pneumoniae were grown in LBmedium at
37˚C for 24 h.
Growth potential assay
The Klebsiella pneumoniae bacteria were
cultured in 10 mg/ml penicillin medium with or without
penicillin-binding proteins (PBPs, 0.67 µg/ml, Sigma-Aldrich; Merck
KGaA) for 24 h. The number of Klebsiella pneumoniae cells
was calculated in the agar plating. The detailed procedures were
conducted according to a previous study (24).
Plasmid construction
To investigate the site of the zinc finger nuclease,
a recombinant plasmid expressing GFP and ZFN (rpGFP-ZFN) was
constructed. All plasmids were purchased from Invitrogen (Thermo
Fisher Scientific, Inc.). A full-length ZFN fragment was amplified
and subcloned into rpGFP-pET27b. The recombinant plasmid was named
rpGFP-ZFN. All expression plasmids were confirmed by sequencing.
Cells were transfected with rpGFP-ZFN or pET27b by using
electrotransfection according to the manufacturer's instructions.
After a 48-h transfection, the cells were captured using a Leica
DM5000 microscope equipped with Q-Imaging Retiga 4000RV camera
(Teledyne QImaging).
Antimicrobial susceptibility
testing
Antimicrobial susceptibility tests of Klebsiella
pneumoniae were performed by the disk diffusion method,
according to Clinical and Laboratory Standards Institute (CLSI)
recommendations (25). The final
results were performed according to the respective standards for
antimicrobial susceptibility testing.
Enzyme-linked immunosorbent assay
(ELISA)
This study analyzed the affinity of PBP with
penicillin by using ELISA Kit (cat. no. B21210; R&D Systems).
The ELISA assays were performed according to the manufacturer's
protocols. The results were measured at 450 nm in an ELISA
reader.
Real-time quantitative PCR (RT-qPCR)
analysis
Total RNA was obtained from Klebsiella
pneumoniae by using RNeasy Mini Kit (Qiagen) according to the
manufacturer's instructions. The cDNA was synthesized with
ReverTraAce (Toyobo Corp.). All forward and reverse primers were
synthesized by Invitrogen (Thermo Fisher Scientific, Inc.) and are
listed in Table I. PCR amplification
was followed by preliminary denaturation at 94˚C for 2 min,
followed by 45 cycles of 95˚C for 30 sec; annealing temperature was
reduced to 57˚C for 30 sec, and 72˚C for 30 sec. A volume of 20 µl
containing 50 ng of genomic DNA, 200 µM dNTP, 2.5 units of
Taq DNA polymerase, and 200 µM primers was used. Relative
mRNA expression level changes were calculated using the
2-ΔΔCq method (25). The results are expressed as n-fold
compared with the β-actin control.
 | Table IPrimers used for quantitative
real-time PCR. |
Table I
Primers used for quantitative
real-time PCR.
| | Sequence |
|---|
| Gene name | Reverse | Forward |
|---|
| BlaR1 |
5'-TCTAGAGGATCATATTACAATACCGAGCTC-3' |
5'-GAGCTCGGTATTGTAATATGATCCTCTAGA-3' |
| Bla1 |
5'-CGCTTAATTCAGCACTAAAC-3' |
5'-GAGCTCGGTATTGTAATATGATCCTCTAGA-3' |
| β-actin |
5'-CGGAGTCAACGGATTTGGTC-3' |
5'-AGCCTTCTCCATGGTCGTGA-3' |
Western blot analysis
Klebsiella pneumonia cells were homogenized
using RIPA buffer (M-PER reagent for the cells and T-PER reagent
for the tissues; Thermo Fisher Scientific, Inc.) and centrifuged at
6,000 x g at 4˚C for 15 min. The protein concentration was measured
using a BCA protein assay kit (Thermo Fisher Scientific, Inc.). A
total of 30 µg protein extracts was electrophoresed on 12%
polyacrylamide gradient gels and then transferred to polyvinylidene
fluoride (PVDF) membrane. The membranes were incubated in 5% BSA
(Sigma-Aldrich; Merck KGaA) for 2 h at 37˚C and then incubated with
primary antibodies (dilution 1:1,000, cat. no. ab11251; Abcam) or
β-actin (dilution 1:1,000, cat. no. ab8226; Abcam) for 2 h at 37˚C.
After washing with PBS three times, the membranes were then
incubating with secondary antibodies (dilution 1:1,000, cat. no.
SAB11045182; Sigma-Aldrich; Merck KGaA) for 12 h at 4˚C. After
three washings in PBST, the membranes were developed using a
chemiluminescence assay system (Roche) and exposed to Kodak
exposure film. Densitometric quantification of the immunoblot data
was performed by using Quantity-One software (version 1.2; Bio-Rad
Laboratories, Inc.).
Immunofluorescence
Klebsiella pneumoniae cells were prepared as
standard operation. Klebsiella pneumoniae cells were fixed
with formaldehyde solution (10%). Rehydrated slides or cells were
incubated with primary antibodies: Anti-PBP (dilution 1:500, cat.
no. ab226275, Abcam) or anti-zinc finger nuclease (dilution
1:1,000, cat. no, WH0284312M2-100UG; Sigma-Aldrich; Merck KGaA) for
2 h at 37˚C. Samples were washed with PBS three times and then
incubated with anti-IgG-FITC (dilution 1:1,000, cat. no. ab6785;
Abcam). Samples were washed and imaged using a fluorescence
microscopy (Nikon; magnification, x100).
Electrospray ionization-mass
spectrometric analysis
The mass spectrometry was recorded using a
quadrupole time-of-flight mass filter (Micromass, UK).
Spectrometric analysis was scanned over a range 100-2,000 units of
ratio m/z of mass to charge, with scan step 2 sec and interscan 0.1
sec/step. The quadrupole scan mode was performed under an
electrospray voltage 3 kV at the tip of a stainless-steel capillary
needle. The elution conditions were acetonitrile (50%) containing
formic acid (0.1%) at rate 2 µl/min.
Knockdown gene-coding PBPs
A total of 1x106 cells in 100 µl
H2O were transfected with control siRNA negative control
(control, 30 nM, 5'-UCACAACCUCCUAGAAAGAGUAGA-3') or siRNA targeting
PBPs (PBP, 30 nM, 5'-CUGAAGUGAUGUGUAACUGAUCAG-3') using Hiperfect
(Qiagen) transfection reagent. After 48 h of transfection, the
transfected cells were treated with zinc finger nuclease (0.5
µg/ml, Sigma-Aldrich; Merck KGaA) for 12 h at 37˚C.
Statistical methods
All presented data are reported as means ± SEM.
Unpaired data were analyzed by Student's t-test. Comparisons of
data between multiple groups were analyzed by analysis of variance
(ANOVA) followed by Tukey post hoc test. *P<0.05 and
**P<0.01 were assigned to indicate statistical
significance and are denoted with the relevant asterisks in the
figures.
Results
Identification of ESBL gene expression
and antimicrobial drug resistance of Klebsiella pneumoniae
In order to analysis the antimicrobial drug
resistance of Klebsiella pneumoniae for β-lactam, we first
determined ESBL gene expression and evaluated β-lactamase activity.
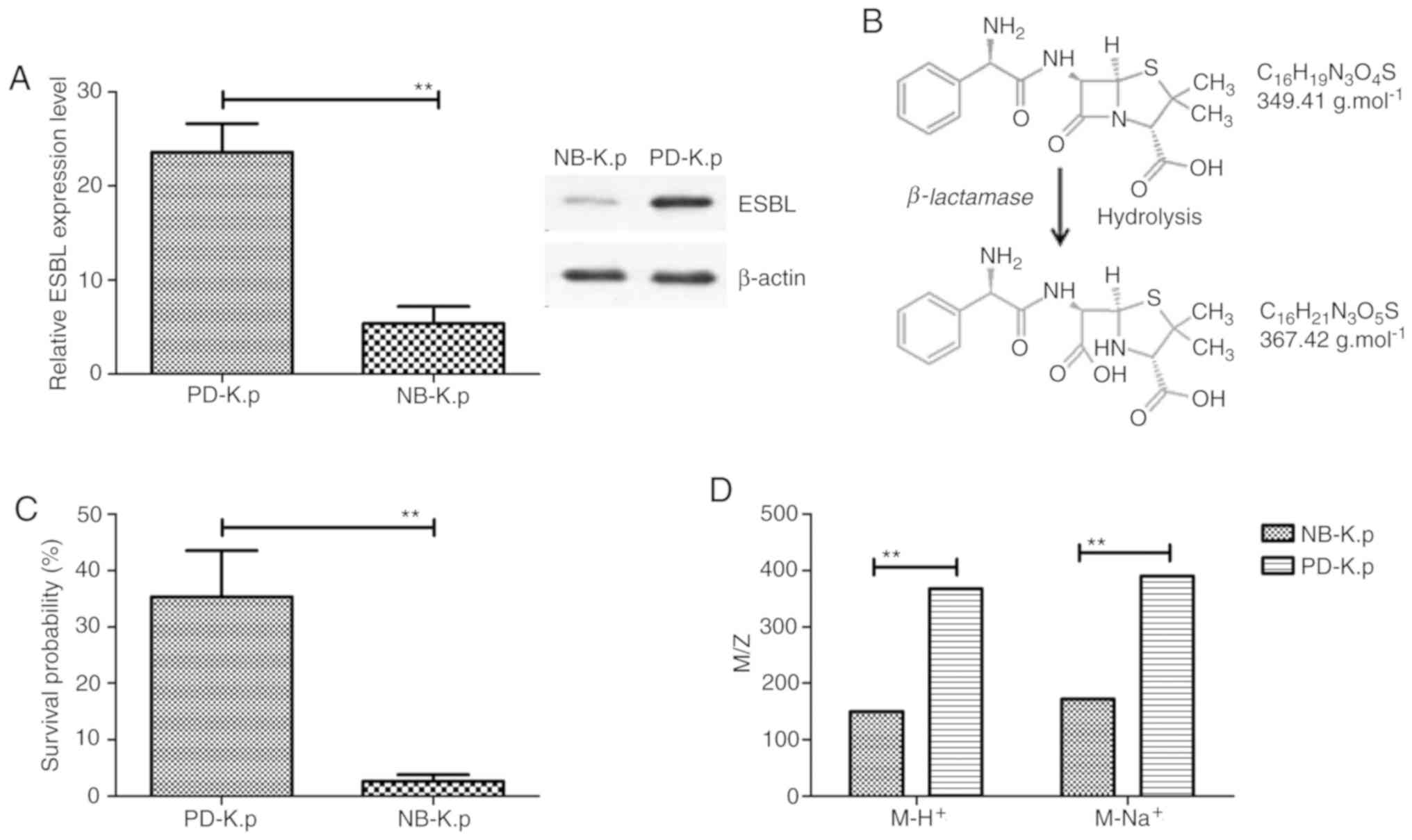
The results in Fig. 1A demonstrate
that both the mRNA and protein expression levels of ESBL were
higher in Klebsiella pneumoniae isolated from patients with
pneumonia (PD-K.p) than natural being Klebsiella pneumonia
(NB-K.p). A band at 42 kDa protein was identified in the cell
extract of Klebsiella pneumoniae. To determine the role of
ESBL in the antimicrobial resistance of Klebsiella
pneumoniae, the growth of Klebsiella
pneumoniae was assessed. Our data showed that β-lactamase
could hydrolyze the β-lactam ring, as determined by electrospray
ionization-mass spectrometric analysis (Fig. 1B). PD-K.p presented a higher survival
than NB-K.p in an environment with penicillin-based antibiotics
(Fig. 1C). This may be due to the
higher production of β-lactamase in PD-K.p than NB-K.p To determine
the β-lactamase activity in vivo, ampicillin was utilized as
a substrate to confirm the activity of β-lactamase. The data in
Fig. 1D illustrate that new signals
with m/z 368 units M-H+ and 390 units M-Na+
were observed in NB-K.p after incubation with β-lactamase protein
at 25˚C for 30 min (m/z of ampicillin is 150 units M-H+
and 172 units M-Na+ of PD-K.p). These data suggest that
β-lactamase hydrolyzed the amide bond of the β-lactam ring and
promoted antimicrobial drug resistance of PD-K.p.
Biological functions of PBPs in
Klebsiella pneumoniae growth
Penicillin-binding proteins (PBPs), bacterial
peripheral membrane enzymes, play an essential role in catalysis in
the final steps for the biosynthesis of the essential bacterial
cell wall peptidoglycan. PBPs are involved in antimicrobial drug
resistance In this study, we aimed to ascertain whether PBPs (25
mg/ml) could attenuate antimicrobial drug resistance of
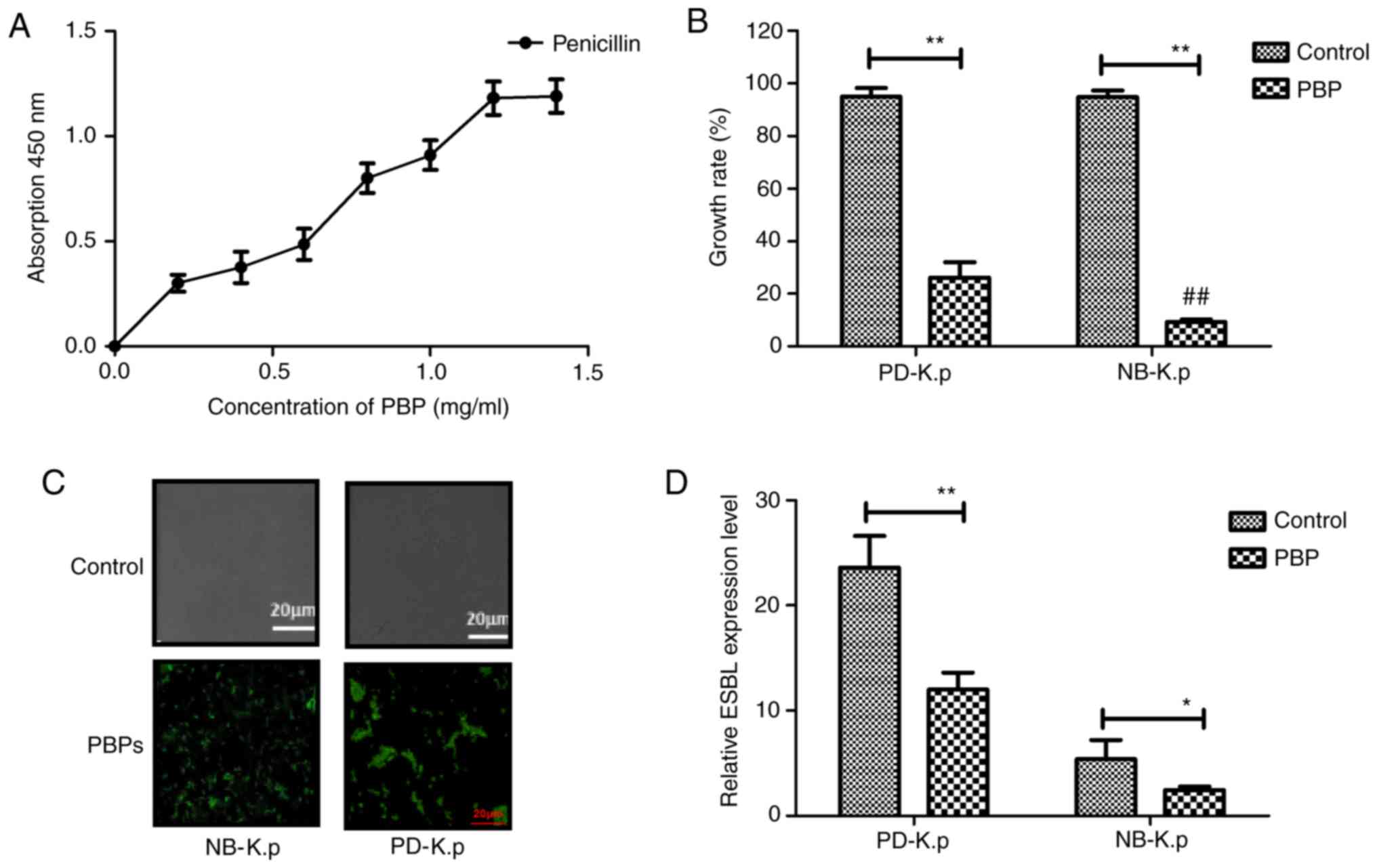
Klebsiella pneumoniae for β-lactam antibiotics. Our results
showed that PBPs could bind with β-lactam antibiotic (penicillin)
in vitro (Fig. 2A). It was
also found that PBPs inhibited Klebsiella pneumoniae growth
(PD-K.p and NB-K.p) compared to the control after incubation with
penicillin (Fig. 2B). This revealed
that PBPs significantly inhibited the growth of PD-K.p compared to
NB-K.p (P<0.01 vs. NB-K.p). In addition, our data in Fig. 2C showed that penicillin could bind
the FITC-labeled PBP-specific antibody, and these results suggested
that PBPs were expressed and presented at the cell surface of
Klebsiella pneumonia, as determined by confocal fluorescence
microscope. Furthermore, PBPs downregulated expression of
β-lactamase (Fig. 2D). These results
suggest that the binding of PBPs to β-lactam may interfere with the
synthesis of β-lactamases.
Efficacy of zinc finger nuclease for
growth of Klebsiella pneumoniae
A previous study indicates that zinc finger nuclease
technology is able to target and disrupt gene-encoded β-lactamase,
which prevents horizontal gene transfer-mediated evolution of
antibiotic resistant bacteria and antibiotic resistance genes
(23). Therefore, we analyzed the
function of zinc finger nuclease in Klebsiella pneumoniae.
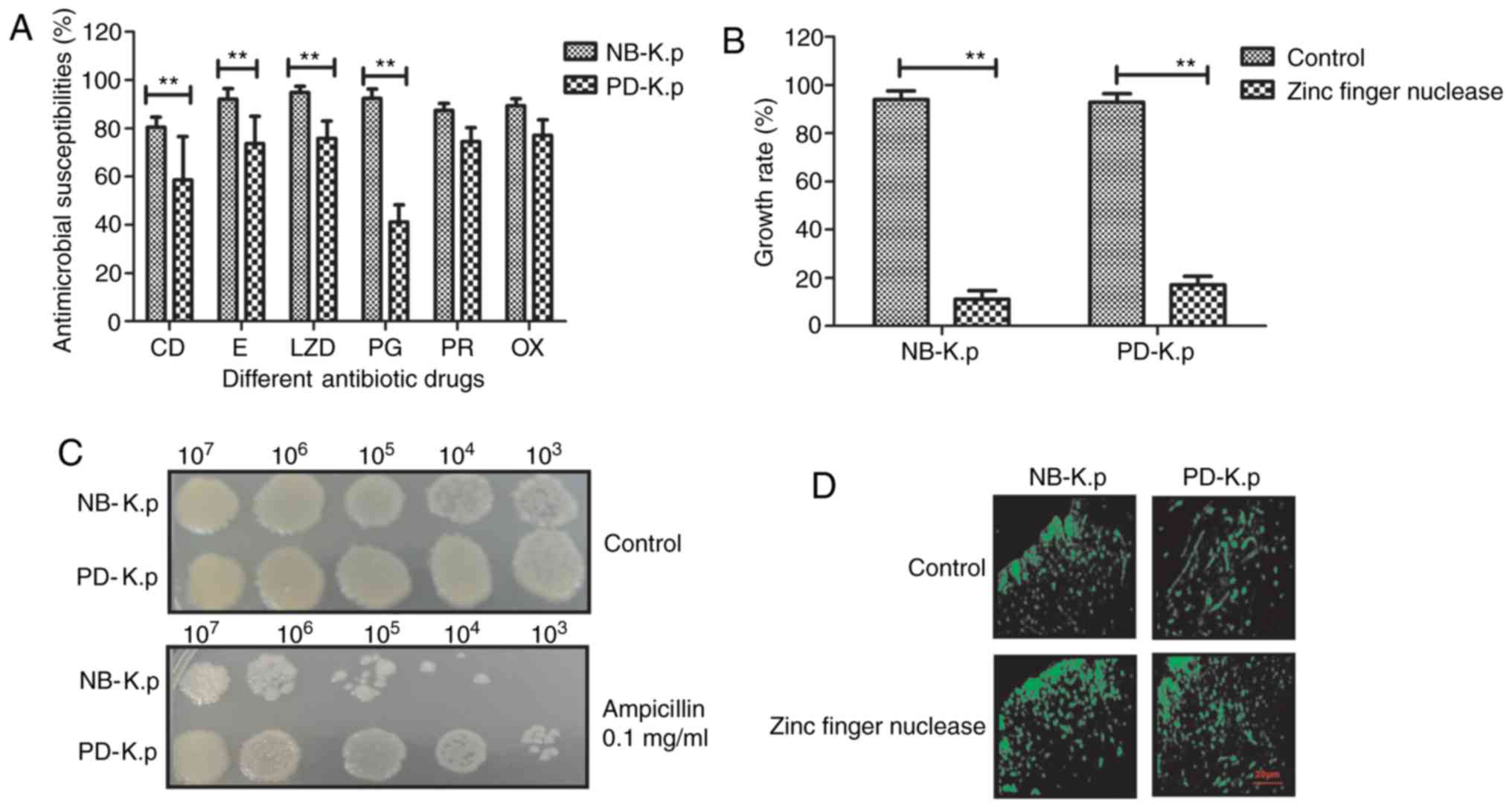
Fig. 3A shows the antimicrobial
susceptibility profile of Klebsiella pneumoniae. The results
revealed that Klebsiella pneumoniae bacteria exhibit varying
degrees of multi-drug resistance (CD, clindamycin; E, erythromycin;
LZD, linezolid; PG, penicillin G; RP, rifampin; OX, oxacillin;
concentrations of all the antibiotics were 1 mg/ml). However, zinc
finger nuclease treatment decreased drug resistance of
Klebsiella pneumoniae compared to control. Fig. 3B indicates that the growth of
Klebsiella pneumoniae was inhibited after transduction of
plasmids containing zinc finger nuclease. The optical density
experiment demonstrated a marked decrease in Klebsiella
pneumoniae growth after treatment with ampicillin compared with
the control groups (P<0.01) at the indicated temperature, which
was consistent with the results in Fig.
3A (Fig. 3C). In addition, the
kinetics of zinc finger nuclease in Klebsiella pneumoniae
was analyzed. The data showed that zinc finger nuclease markedly
increased the binding potential of PD-K.p to penicillin compared
with PBS (control) (Fig. 3D). The
data suggest that zinc finger nuclease inhibited Klebsiella
pneumoniae growth and enhanced the binding capacity of
penicillin to Klebsiella pneumoniae.
Relationship among β-lactamase, PBPs
and zinc finger nuclease in PD-K.p
To understand the mechanism of zinc finger nuclease
in inhibiting Klebsiella pneumoniae growth, we analyzed the
relationship among β-lactamase, PBPs and zinc finger nuclease.
First, we detected expression levels of β-lactamase and PBPs in
Klebsiella pneumoniae after incubation with zinc finger
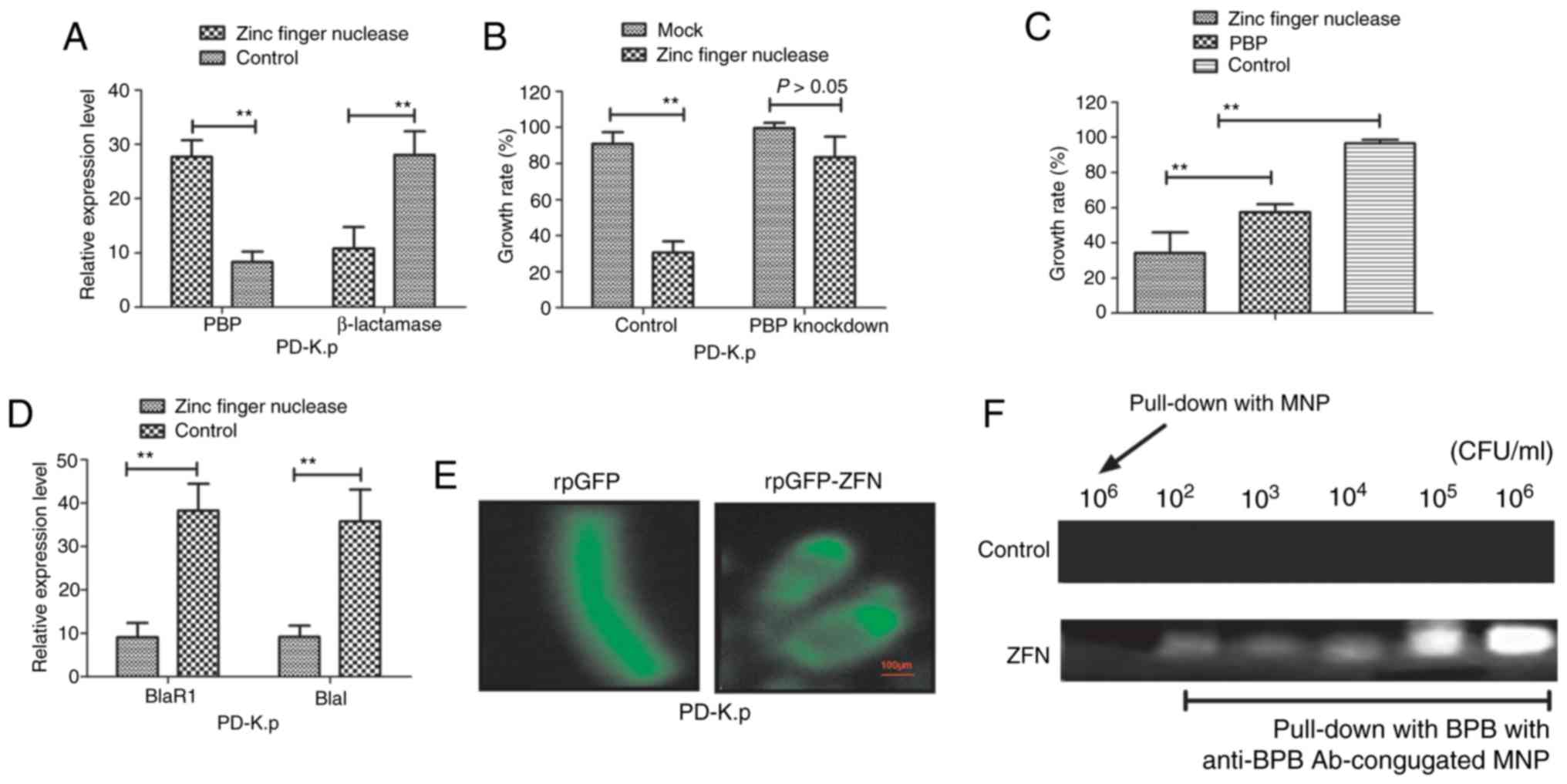
nuclease for 24 h. Fig. 4A shows
that zinc finger nuclease treatment upregulated PBP expression and
downregulated β-lactamase production. Knockdown of gene-coding PBPs
abrogated the inhibitory effects on zinc finger nuclease for PD-K.p
(Fig. 4B). Importantly, our data
showed that the inhibitory effects of zinc finger nuclease were
more superior to PBPs at the same concentration (10 mg/ml) in
regards to PD-K.p growth (Fig. 4C).
Notably, expression levels of two members (BlaR1 and Blal) of the
β-lactamase signaling pathway in PD-K.p were decreased after zinc
finger nuclease treatment (Fig. 4D).
To discover the action site of zinc finger nuclease, we transfected
plasmids of rpGFP-ZFN into Klebsiella pneumoniae with rpGFP
as control. As shown in Fig. 4E,
strong fluorescence was observed in surrounding Klebsiella
pneumoniae cells delivering rpGFP-ZFN plasmid, while
fluorescence was found in the cytoplasm carrying rpGFP, indicating
that the action site of zinc finger nuclease is associated with the
membrane of Klebsiella pneumoniae. Fig. 4F further confirmed that the action
site of zinc finger nuclease is located in the outer membrane of
Klebsiella pneumoniae. These results indicate that zinc
finger nuclease could regulate PBPs or zinc finger nuclease
expression, suggesting that it may represent a potential agent for
antibiotic resistance.
Discussion
The mean number of gram-positive bacilli isolated
from hospitalized patients has gradually revealed the different
degree of antibiotic resistance since 2000 (26,27).
Resistance to antimicrobial drugs is one of the greatest challenges
due to the overuse and misuse of antibiotics in modern medicine,
which significantly threatens public health (28,29). In
the present study, the potential molecular mechanism of antibiotic
resistance of Klebsiella pneumonia was investigated.
Although a previous study has proposed a primary theory of
antibiotic resistance, the main viewpoint focuses on the
β-lactamase signaling (30). In the
present study, extended-spectrum β-lactamase produced by
Klebsiella pneumoniae was confirmed by susceptibility
testing. Notably, our data indicated that penicillin-binding
proteins (PBPs) enhanced the affinity between Klebsiella
pneumoniae and penicillin, which led to the inhibition of the
growth of Klebsiella pneumoniae. Importantly, zinc finger
nuclease targeting the β-lactam resistance gene led to the
downregulation of β-lactamase synthesis. We also found that
expression of PBPs was decreased in Klebsiella pneumoniae
after treatment with zinc finger nuclease. We concluded that zinc
finger nuclease not only regulated the biological activity of
β-lactamase, but also promoted expression of PBPs in Klebsiella
pneumoniae.
Antibiotic resistance in Klebsiella
pneumoniae is associated with high rates of morbidity and
mortality in clinical patients (31). Therefore, understanding the
mechanisms involved in the antibiotic resistance of Klebsiella
pneumoniae are highly required to solve the antibiotic
resistance problem and decrease mortality of patients infected with
Klebsiella pneumoniae. β-lactam antibiotic resistance is
mainly caused by two mechanisms including extended-spectrum
β-lactamase (ESBL) and target mutation of gene-coding PBPs
(32,33). The results of the present study found
that ESBL expression was upregulated by the indiscriminate
application of antibiotics, which plays a hydrolytic role in
β-lactam antibiotic resistance. The expression level of
β-lactamases is responsible for β-lactam resistance in
Klebsiella pneumoniae by catalyzing the hydrolysis of
β-lactam antibiotics to generate antimicrobiologically inactive
compounds (34). In addition, a
recent study indicated that the genome of the β-lactamase
superfamily including RNA-metabolizing metallo-β-lactamases and
β-lactamase domain-containing proteins are expanded in
antibiotic-resistance Klebsiella pneumoniae (35). Furthermore, a study has showed that
various types of β-lactamases are distributed in a variety of
organisms (36). In the present
study, it was observed that the antibiotic resistance of
Klebsiella pneumoniae was associated with β-lactamase
expression (37). ESBL-producing
bacteria are resistant to most β-lactam antibiotics (38). ESBL-producing Klebsiella
pneumoniae with genetic diversity is characterized with
resistance genes, which is a significant threat for public health
(39). This study found that
patient-derived PD-K.p produced more ESBL than natural being
NB-K.p, which contributed to the increasing of drug resistance.
Future study should be specifically targeted towards the ESBL
signaling pathway as it is crucial to guide antibiotic therapy for
patients infected with PD-K.p.
A previous study has suggested that zinc finger
nuclease may be a potential agent to overcome β-lactam antibiotic
resistance (23). Antibiotic
resistance of Klebsiella pneumoniae has challenged the
therapeutic efficacy for patients with pneumonia (35). This preclinical study described the
efficacy of zinc finger nuclease technology via targeting and
disrupting the high expression of lactamase in Klebsiella
pneumoniae, which prevented horizontal gene transfer-mediated
antibiotic resistance. Our data showed that zinc finger nuclease
targeting the β-lactam resistance gene led to downregulation of the
β-lactamase signaling pathway and decreased β-lactamase synthesis
and therefore attenuated β-lactam resistance. The results also
found that zinc finger nuclease promoted PBP synthesis, which
enhanced the inhibitory effects of β-lactam on the growth of
Klebsiella pneumoniae. However, the present study only
analyzed the role of PBP synthesis in Klebsiella pneumoniae
growth, and further assessment of the function of PBPs need further
investigation. In addition, development of an advanced zinc finger
nuclease archive to target a broader range of DNA sequences would
allow zinc finger nuclease to target the catalytic domain encoding
region of drug resistance, which may result in more efficient
disruption of resistance. Furthermore, increasing zinc finger
nuclease expression may be a potential strategy to reduce
antibiotic resistance in Klebsiella pneumoniae.
In conclusion, the present study elucidated the
molecular mechanisms of the β-lactam resistance of Klebsiella
pneumoniae. PBPs and zinc finger nuclease were utilized to
design the drug resistance, while zinc finger nuclease was found to
be a potent regulator of PBP expression. Consistent with a previous
study, β-lactamases in antibiotic-resistance Klebsiella
pneumonia are various and PBP expression is variable in the
β-lactam system (40). In addition,
PBPs and β-lactamases are two important resistance mechanisms in
Klebsiella pneumoniae caused by the frequent use of
antibiotics in patients with pneumonia (41). Furthermore, the rate of emergence of
new antibiotic-resistant bacteria is rapidly increasing, that leads
to an increase in the number of patients presenting with tolerant
Klebsiella pneumonia. Taken together, the resistance of
Klebsiella pneumoniae bacteria to antimicrobial drugs acts
via the ESBL signaling pathway, which may be a potential target for
the treatment of patients infected with Klebsiella
pneumoniae through the β-lactamase signaling pathway.
Acknowledgements
Not applicable.
Funding
This study was supported by a grant from the
National Science Foundation (China).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WY performed experiments and data analysis. XZ, NW
and HR performed experiments, collected data and prepared the
diagrams. WJ designed the experiments and wrote the manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
Tianjin First Central Hospital (Tianjin, China). The patient
enrolled in this study signed a written consent form before
participation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lim MK, Lai PS, Ponnampalavanar SS, Syed
Omar SF, Taib NA, Yusof MY, Italiano CM, Kong DC and Kamarulzaman
A: Antibiotics in surgical wards: Use or misuse? A newly
industrialized country's perspective. J Infect Dev Countries.
9:1264–1271. 2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Kardas P, Devine S, Golembesky A and
Roberts C: A systematic review and meta-analysis of misuse of
antibiotic therapies in the community. Int J Antimicrob Agents.
26:106–113. 2005.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Heineman HS and Watt VS: All-inclusive
concurrent antibiotic usage review: A way to reduce misuse without
formal controls. Infect Control. 7:168–171. 1986.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Nõlvak H, Truu M, Kanger K, Tampere M,
Espenberg M, Loit E, Raave H and Truu J: Inorganic and organic
fertilizers impact the abundance and proportion of antibiotic
resistance and integron-integrase genes in agricultural grassland
soil. Sci Total Environ. 562:678–689. 2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Stogios PJ, Cox G, Spanogiannopoulos P,
Pillon MC, Waglechner N, Skarina T, Koteva K, Guarné A, Savchenko A
and Wright GD: Rifampin phosphotransferase is an unusual antibiotic
resistance kinase. Nat Commun. 7(11343)2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Ferro G, Guarino F, Castiglione S and
Rizzo L: Antibiotic resistance spread potential in urban wastewater
effluents disinfected by UV/H2O2 process. Sci
Total Environ. 560-561:29–35. 2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Ouwehand AC, Forssten S, Hibberd AA, Lyra
A and Stahl B: Probiotic approach to prevent antibiotic resistance.
Ann Med. 48:246–255. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Date KA, Newton AE, Medalla F, Blackstock
A, Richardson L, McCullough A, Mintz ED and Mahon BE: Changing
patterns in enteric fever incidence and increasing antibiotic
resistance of enteric fever isolates in the United States,
2008-2012. Clin Infect Dis. 63:322–329. 2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Çöl A, Dedeić-Ljubović A, Salimović-Bešić
I and Hukic M: Antibiotic resistance profiles and genetic
similarities within a new generation of carbapenem-resistant
Acinetobacter calcoaceticus-A. baumannii complex
resistotypes in Bosnia and Herzegovina. Microb Drug Resist.
22:655–661. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Barger A, Schaade L, Krause G and Kramer
M: Strategies for recognition, prevention and control of
antimicrobial resistances in Germany-a draft from the federal
german ministry of health. Gesundheitswesen. 70:631–635. 2008.(In
German). PubMed/NCBI View Article : Google Scholar
|
|
11
|
Oliva A, Gizzi F, Mascellino MT, Cipolla
A, D'Abramo A, D'Agostino C, Trinchieri V, Russo G, Tierno F,
Iannetta M, et al: Bactericidal and synergistic activity of
double-carbapenem regimen for infections caused by
carbapenemase-producing Klebsiella pneumoniae. Clin
Microbiol Infect. 22:147–153. 2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Gajul SV, Mohite ST, Mangalgi SS, Wavare
SM and Kakade SV: Klebsiella pneumoniae in septicemic
neonates with special reference to extended spectrum β-lactamase,
AmpC, metallo β-lactamase production and multiple drug resistance
in tertiary care hospital. J Lab Physicians. 7:32–37.
2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Rastogi V, Nirwan PS, Jain S and Kapil A:
Nosocomial outbreak of septicaemia in neonatal intensive care unit
due to extended spectrum β-lactamase producing Klebsiella
pneumoniae showing multiple mechanisms of drug resistance.
Indian J Med Microbiol. 28:380–384. 2010.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Babálová M, Blahová J, Králiková K,
Krcméry V Sr, Menkyna R, Bartoníková N and Skalicková R: Unexpected
reservoir of Klebsiella pneumoniae strains with transferable
multiple drug resistance causing clinical sepsis in newborns. J
Chemother. 17:454–455. 2005.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Sanchez GV, Master RN, Clark RB, Fyyaz M,
Duvvuri P, Ekta G and Bordon J: Klebsiellapneumoniae
antimicrobial drug resistance, United States, 1998-2010. Emerg
Infect Dis. 19:133–136. 2013.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Neupane S, Pant ND, Khatiwada S, Chaudhary
R and Banjara MR: Correlation between biofilm formation and
resistance toward different commonly used antibiotics along with
extended spectrum beta lactamase production in uropathogenic
Escherichia coli isolated from the patients suspected of
urinary tract infections visiting Shree Birendra Hospital, Chhauni,
Kathmandu, Nepal. Antimicrob Resist Infect Control.
5(5)2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Chen X, Che J, Zhao XF, Zhang LF and Li J:
Dissemination of insertion sequence common regions 1 and int1 gene
and drug resistance of 483 Escherichia coli and
Klebsiella pneumonia broiler isolates. Zhonghua Yu Fang Yi
Xue Za Zhi. 51:886–889. 2017.(In Chinese). PubMed/NCBI View Article : Google Scholar
|
|
18
|
Sekowska A, Janicka G, Klyszejko C, Wojda
M, Wroblewski M and Szymankiewicz M: Resistance of Klebsiella
pneumoniae strains producing and not producing ESBL
(extended-spectrum beta-lactamase) type enzymes to selected
non-beta-lactam antibiotics. Med Sci Monit. 8:BR100–BR104.
2002.PubMed/NCBI
|
|
19
|
Sanguinetti CM, De Benedetto F and
Miragliotta G: Bacterial agents of lower respiratory tract
infections (LRTIs), beta-lactamase production, and resistance to
antibiotics in elderly people. DEDALO study group. Int J Antimicrob
Agents. 16:467–471. 2000.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Boggio SB and Roveri OA: Catalytic
properties of an endogenous beta-lactamase responsible for the
resistance of Azospirillum lipoferum to beta-lactam antibiotics.
Microbiology. 149:445–450. 2003.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Uraz G, Simsek H and Celik B:
Beta-Lactamase activities and resistance to antibiotics of
Haemophilus influenzae, H. parainfluenzae and H. aphrophilus
strains identified in throat cultures from children. Drug Metabol
Drug Interact. 16:217–228. 2000.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Macheboeuf P, Contreras-Martel C, Job V,
Dideberg O and Dessen A: Penicillin binding proteins: Key players
in bacterial cell cycle and drug resistance processes. FEMS
Microbiol Rev. 30:673–691. 2006.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Shahbazi Dastjerdeh M, Kouhpayeh S,
Sabzehei F, Khanahmad H, Salehi M, Mohammadi Z, Shariati L, Hejazi
Z, Rabiei P and Manian M: Zinc finger nuclease: A new approach to
overcome beta-lactam antibiotic resistance. Jundishapur J
Microbiol. 9(e29384)2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zscheppang K, Kurth I, Wachtel N,
Dubrovska A, Kunz-Schughart LA and Cordes N: Efficacy of beta1
integrin and EGFR targeting in sphere-forming human head and neck
cancer cells. J Cancer. 7:736–745. 2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Hsueh PR, Ko WC, Wu JJ, Lu JJ, Wang FD, Wu
HY, Wu TL and Teng LJ: Consensus statement on the adherence to
clinical and laboratory standards institute (CLSI) antimicrobial
susceptibility testing guidelines (CLSI-2010 and CLSI-2010-update)
for enterobacteriaceae in clinical microbiology laboratories in
Taiwan. J Microbiol Immunol Infect. 43:452–455. 2010.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Baruah FK, Hussain AN, Kausalya and
Grover RK: Antibiotic resistance profile of non-fermenting
Gram-negative bacilli isolated from the blood cultures of cancer
patients. J Glob Infect Dis. 7:46–47. 2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Hill JT, Tran KD, Barton KL, Labreche MJ
and Sharp SE: Evaluation of the nanosphere Verigene BC-GN assay for
direct identification of gram-negative bacilli and antibiotic
resistance markers from positive blood cultures and potential
impact for more-rapid antibiotic interventions. J Clin Microbiol.
52:3805–3807. 2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Ares MA, Alcantar-Curiel MD,
Jimenez-Galicia C, Rios-Sarabia N, Pacheco S and De la Cruz MA:
Antibiotic resistance of gram-negative bacilli isolated from
pediatric patients with nosocomial bloodstream infections in a
mexican tertiary care hospital. Chemotherapy. 59:361–368.
2013.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Patel SJ, O'Toole D and Larson E: A new
metric of antibiotic class resistance in gram-negative bacilli
isolated from hospitalized children. Infect Control Hosp Epidemiol.
33:602–607. 2012.PubMed/NCBI View
Article : Google Scholar
|
|
30
|
Sasirekha B and Shivakumar S: Occurrence
of plasmid-mediated AmpC β-lactamases among Escherichia coli
and Klebsiella pneumoniae clinical isolates in a tertiary
care hospital in Bangalore. Indian J Microbiol. 52:174–179.
2012.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Koczura R, Mokracka J, Krzyminska S and
Kaznowski A: Virulence properties and integron-associated
antibiotic resistance of Klebsiella mobilis strains isolated
from clinical specimens. J Med Microbiol. 60:281–288.
2011.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Lee M, Dhar S, De Benedetti S, Hesek D,
Boggess B, Blázquez B, Mathee K and Mobashery S: Muropeptides in
pseudomonas aeruginosa and their role as elicitors of
β-lactam-antibiotic resistance. Angew Chem Int Ed Engl.
55:6882–6886. 2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Lee YD and Park JH: Phage conversion for
β-lactam antibiotic resistance of staphylococcus aureus from foods.
J Microbiol Biotechnol. 26:263–269. 2016.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Shafiq M, Rahman H, Qasim M, Ayub N,
Hussain S, Khan J and Naeem M: Prevalence of plasmid-mediated AmpC
β-lactamases in Escherichia coli and Klebsiella
pneumonia at tertiary care hospital of Islamabad, Pakistan. Eur
J Microbiol Immunol (Bp). 3:267–271. 2013.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Goessens WH, Mouton JW, Ten Kate MT,
Sorgel F, Kinzig M and Bakker-Woudenberg IA: The therapeutic effect
of tigecycline, unlike that of Ceftazidime, is not influenced by
whether the Klebsiella pneumoniae strain produces
extended-spectrum beta-lactamases in experimental pneumonia in
rats. Antimicrob Agents Chemother. 57:643–646. 2013.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Rivera A, Larrosa N, Mirelis B and Navarro
F: Importance of quality control for the detection of β-lactam
antibiotic resistance in enterobacteriaceae. Enferm Infecc
Microbiol Clin. 32 (Suppl 1):S30–S36. 2014.(In Spanish). PubMed/NCBI View Article : Google Scholar
|
|
37
|
Padilla E, Alonso D, Domenech-Sanchez A,
Gomez C, Pérez JL, Albertí S and Borrell N: Effect of porins and
plasmid-mediated AmpC beta-lactamases on the efficacy of
beta-lactams in rat pneumonia caused by Klebsiella
pneumoniae. Antimicrob Agents Chemother. 50:2258–2260.
2006.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Wragg R, Harris A, Patel M, Robb A,
Chandran H and McCarthy L: Extended spectrum beta lactamase (ESBL)
producing bacteria urinary tract infections and complex pediatric
urology. J Pediatr Surg. 52:286–288. 2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Charrouf FO, Hamze M, Mallat H, Achkar M
and Dabboussia F: Characterization of resistance genes in 68
ESBL-producing Klebsiella pneumonia in Lebanon. Med Mal
Infect. 44:535–538. 2014.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Ogawara H: Self-resistance in
streptomyces, with special reference to beta-lactam antibiotics.
Molecules. 21(E605)2016.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Navarro Jimenez C, Luque Marquez R, de
Torres Solis I and Aguilar Guisado M: Changes in empiric therapy
for nosocomial pneumonia due to the emergence of bacterial strains
with broad spectrum beta-lactamases. Med Clin (Barc).
122(679)2004.(In Spanish). PubMed/NCBI View Article : Google Scholar
|