Introduction
Cholangiocarcinoma (CCA) is a highly aggressive, malignant tumor arising from the ductal epithelium of the biliary tree. CCA represent the second most common type of primary hepatobiliary cancer worldwide, which account for 10-15% of all patients with liver cancer (1). Although CCA is considered a rare malignancy and accounts for a small proportion of total malignancies, in recent years, the incidence and mortality of CCA have rapidly increased worldwide according to epidemiological studies (2-4). CCA is classified into intrahepatic, extrahepatic or perihilar according to its anatomical locations (5). Both for intrahepatic and extrahepatic cancers, the incidence rate statistically increased from 1.5 to 2 per 100,000/year between 1999-2007 in Europe, and the prognosis remains poor (6). Radical surgical resection is the only available curative treatment for patients with CCA at the early stages of the disease; however, the majority of patients are diagnosed at advanced stages, as the early stages are commonly asymptomatic (7,8). Additionally, the postoperative recurrence and early metastasis rates are high, and the percentage of patients who survive 5 years after diagnosis remains at 10% (1). It is thus necessary to identify accurate prognostic predictors and molecular targeted therapies for the treatment of patients with CCA.
Micro (mi)RNAs are a type of endogenous small non-coding RNAs that regulate the expression of target genes post-transcriptionally by binding to the 3'-untranslated region (UTR) of its target Mrna (9). In recent years, miRNAs have been demonstrated to influence various biological processes, including cell proliferation, migration, invasion, differentiation and apoptosis, particularly in cancer cells (10-12). miR-96, a member of the miR-183 family, has been demonstrated to be involved in the tumorigenesis of various cancers. miR-96 typically serves an oncogenic role in multiple types of cancer, such as hepatocellular carcinoma, colorectal and breast cancer (13-15). Collins et al (16) have revealed that several differentially expressed miRNAs in CCA and pancreatic cancer tissues, including miR-96, distinguish CCA from pancreatic adenocarcinoma. However, the role of miR-96 in CCA has not yet been reported.
The present study aimed to investigate the expression level of miR-96 in CCA tissues and cell lines, as well as its prognostic value in patients with CCA. In addition, the biological function of miR-96 in CCA cells was explored.
Materials and methods
Patients and tissue specimens
A total of 115 patients with CCA (comprising 59 intrahepatic and 56 extrahepatic cases) admitted to Changhai Hospital (Shanghai, China) between March 2009 and February 2013 were recruited. These patients included 68 males and 47 females, aged 36-85 years, with a median age of 58 years. The inclusion criteria were as follows: i) All patients were initially diagnosed with CCA. ii) Pathologists confirmed the diagnosis of CCA. iii) Patients had normal liver and kidney function. The exclusion criteria were as follow: i) Patients who had received radiotherapy or chemotherapy prior to surgical treatment. ii) Patients had any other cancer history. Paired CCA and adjacent normal tissue specimens were obtained by resection from patients who underwent surgical treatment and immediately frozen in liquid nitrogen. The adjacent normal tissue specimens were para-cancerous tissues that were collected 2 cm from the tumor site. The clinical characteristics of the patients were collected and are presented in Table I. The 5-year survival information from the postoperative follow-up of all 115 CCA patients was collected via follow up phone calls. The study was approved by the Ethics Committee of Changhai Hospital (Shanghai, China) and all patients provided written informed consent. All specimens were anonymized according to the relevant ethical and legal standards.
 |
Table I
Correlation between the expression of miR-96 and clinical characteristics of patients with cholangiocarcinoma.
|
Table I
Correlation between the expression of miR-96 and clinical characteristics of patients with cholangiocarcinoma.
| |
|
miR-96 expression |
|
| Patient characteristics |
Total patients (n=115) |
Low (n=54) |
High (n=61) |
P-value |
| Sex |
|
|
|
0.724 |
| Male |
68 |
31 |
37 |
|
| Female |
47 |
23 |
24 |
|
| Age, years |
|
|
|
0.200 |
| <60 |
63 |
33 |
30 |
|
| ≥60 |
52 |
21 |
31 |
|
| Tumor size, cm |
|
|
|
0.810 |
| <2 |
54 |
26 |
28 |
|
| ≥2 |
61 |
28 |
33 |
|
| Lymph node metastasis |
|
|
|
0.023a |
| Negative |
66 |
37 |
29 |
|
| Positive |
49 |
17 |
32 |
|
| Differentiation |
|
|
|
0.036a |
| Well + moderate |
76 |
41 |
35 |
|
| Poor |
39 |
13 |
26 |
|
| TNM stage |
|
|
|
0.013a |
| I-II |
52 |
31 |
21 |
|
| III-IV |
63 |
23 |
40 |
|
Cell lines and transfection
Three human CCA cell lines (HuCCT1, HuH28, and RBE) and one normal human intrahepatic biliary epithelial cell line (HIBEC) were cultured in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) at 37˚C in a humidified chamber with 5% CO2. HuCCT1 and HuH28 cells were purchased from the Japanese Collection of Research Bioresources Cell Bank, RBE cells were obtained from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences, and HIBEC cells were purchased from the American Type Culture Collection.
The miR-96 mimic, inhibitor and respective corresponding negative controls (NC) were purchased from Shanghai GenePharma Co., Ltd. For cell transfection, CCA cells were inoculated in 6-well plates at a density of 5x104 cells/well. Then, miR-96 mimic (5'-UUUGGCACUAGCACAUUUUUGCU-3'), mimic NC (5'-UUCUCCGAACGUGUCACGUTT-3'), miR-96 inhibitor (5'-AGCAAAAAUGUGCUAGUGCCAAA-3'), or inhibitor NC (5'-CAGUACUUUUGUGUAGUACAA-3'), were transfected into the cells at a concentration of 50 nM using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Untreated cells were used as the transfection control and transfection efficiency was assessed after 48 h using reverse transcription-quantitative (RT-q)PCR.
RNA extraction and RT-qPCR
Total RNA was extracted from CCA tumor tissues and para-cancerous normal tissues, as well as CCA cell lines and normal HIBEC cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and cDNA was synthesized using a SuperScript III Reverse Transcriptase kit (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. qPCR was used to detect the expression of miR-96 in CCA tissues and cell lines using a SYBR Premix Ex Taq™ kit and an Applied Biosystems 7500 Sequence Detection System (both Applied Biosystems; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols. The relative expression levels of miR-96 and metastasis suppressor-1 (MTSS1) were calculated using the 2-ΔΔCq method (17) and normalized to those of U6 and GADPH, respectively. The primers used were as follows: MiR-96 forward, 5'-GCCCGCTTTGGCACTAGCACATT-3' and reverse, 5'-GTGCAGGGTCCGAGGT-3'; MTSS1 forward, 5'-TCAAGAACAGATGGAAGAATGG-3' and reverse, 5'-TGCGGTAGCGGTAATGTG-3'; U6 forward, 5'-CTCGCTTCGGCAGCACA-3' and reverse, 5'-AACGCTTCACGAATTTGCGT-3'; and GADPH forward, 5'-ATGATATCGCCGCGCTCGTC-3' and reverse, 5'-CGCTCGGTGAGGATCTTCA-3'. The thermocycling conditions were as follows: 95˚C for 5 min, 40 cycles of 95˚C for 10 sec, 60˚C for 20 sec, and 72˚C for 10 sec, and then final extension at 72˚C for 5 min.
Cell proliferation assay
Cell proliferation was measured using a Cell Counting Kit-8 (CCK-8; Beyotime Institute of Biotechnology) assay. A total of 100 µl cell suspension (2x103 cells/well) was inoculated in 96-well plates and incubated at 37˚C in a humidified chamber with 5% CO2. At 0, 24, 48 and 72 h, 10 µl CCK-8 reagent was added to the wells and further incubated for 4 h. A microplate reader (Bio-Rad Laboratories, Inc.) was used to measure the absorbance at 450 nm.
Cell migration and invasion assays
The migratory and invasive abilities of HuCCT1 and HuH28 cells that were transfected with miR-96 mimic, mimic NC, miR-96 inhibitor or inhibitor NC were investigated using Transwell assays. For the Transwell invasion assay, the upper chambers were precoated with 1 mg/ml Matrigel (BD Biosciences) for 4 h at 37˚C. For the migration assay, no Matrigel was used. Briefly, 1x105 transfected cells in serum-free RPMI-1640 medium were seeded into the upper chambers. RPMI-1640 medium supplemented with 10% FBS was added to the lower chambers. After 16 h of incubation at 37˚C, cells that migrated or invaded to the lower chambers were fixed with 4% paraformaldehyde for 20 min at room temperature and stained with crystal violet for 20 min at room temperature. Cells were counted under a light microscope (magnification, x200).
Bioinformatics analysis and dual-luciferase reporter assay
TargetScan (www.targetscan.org) and miRDB (www.mirdb.org) database were used to predict the target genes of miR-96, and MTSS1 was identified to be a potential target of miR-96. The binding site of miR-96 was at the MTSS1 3'-UTR. The MTSS1 wild-type (WT) or mutant (MUT) 3'-UTR was cloned into the pGL3 vector (Promega Corporation) for luciferase reporter experiments. The MTSS1-3'-UTR-WT or MTSS1-3'-UTR-MUT was co-transfected with the miR-96 mimic, NC mimic, miR-96 inhibitor or NC inhibitor into 5x104 HuCCT1 cells using Lipofectamine 2000 according to the manufacturer's instructions. Following 48 h, the luciferase activities of firefly luciferase reporters were measured with the dual-luciferase reporter gene assay system (Promega Corporation) according to the manufacturer's instructions. The relative firefly luciferase activities were normalized to Renilla luciferase activities.
Statistical analysis
Significant differences between two groups were identified using paired Student's t-test. One-way ANOVA followed by Tukey's post-hoc test was performed for comparisons among multiple groups. χ2 test was used to analyze the association between miR-96 expression and clinical characteristics of patients. Data are presented as the mean ± SD. The prognostic significance of miR-96 was assessed using a Kaplan-Meier curve and multivariate Cox regression analyses. Patients were divided into low- and high-miR-96 expression groups using the mean miR-96 expression level as a cutoff value for facilitating analyses. Statistical analyses were performed using SPSS 20.0 (IBM Corp.) and GraphPad Prism 6.0 (GraphPad Software, Inc.). P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of miR-96 is increased in CCA tissues and cell lines
The expression levels of miR-96 in CCA tissues and cell lines were determined. The RT-qPCR analysis results revealed that miR-96 expression was significantly upregulated in CCA tissues compared with adjacent normal tissues (P<0.001, Fig. 1A). Furthermore, the difference in miR-96 expression levels between intrahepatic and extrahepatic CCA tissues was also investigated. miR-96 expression was significantly higher in tumor tissues compared with normal tissues in the two subtypes (Fig. 1B). Considering the anatomical locations of intrahepatic and extrahepatic CCA, the expression levels of miR-96 in intrahepatic and extrahepatic CCA tissues were also compared (Fig. 1B), however, the expression level of miR-96 was not significantly different between intrahepatic and extrahepatic CCA tissues. miR-96 expression was also revealed to be upregulated in CCA cell lines compared with that in normal human intrahepatic biliary epithelial HIBEC cells (P<0.01, Fig. 1C).
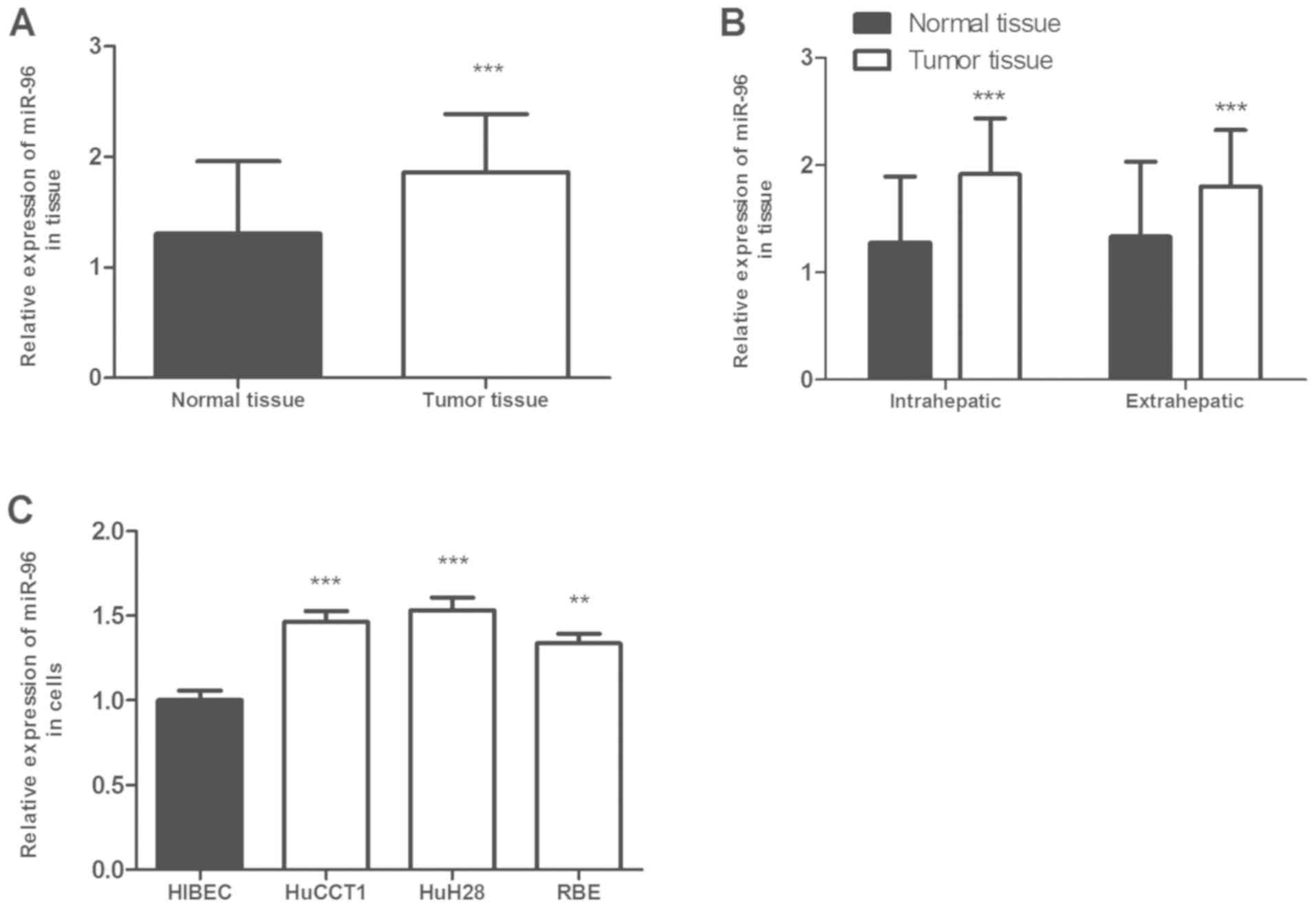 |
Figure 1.
miR-96 expression in CCA tissues and cells. (A) miR-96 expression was upregulated in CCA tissues compared with that in paired normal tissues. (B) miR-96 expression was upregulated in intrahepatic and extrahepatic CCA tissues compared with that in normal tissues. (C) miR-96 expression level was higher in CCA cell lines HUCCT1, HuH28 and RBE compared with a normal intrahepatic biliary epithelial cell line HIBEC. The data were analyzed using one-way ANOVA followed by Tukey's post-hoc test. **P<0.01, ***P<0.001 vs. normal tissues or HIBEC cells. miR, microRNA; CCA, cholangiocarcinoma.
|
Associations between miR-96 expression and clinical characteristics of patients with CCA
Patients were divided into a low-(n=54) and a high-miR-96 expression (n=61) groups using the mean expression level of miR-96 in CCA tissues as the cutoff value. Subsequently, the association between the expression levels of miR-96 with certain clinical characteristics of patients with CCA was analyzed. The results, which are presented in Table I, indicated that the expression level of miR-96 was significantly associated with lymph node metastasis (P=0.023), differentiation (P=0.036) and TNM stage (P=0.013). However, no significant associations were identified between the expression of miR-96 and other clinical features of patients with CCA, including sex, age and tumor size (P>0.05).
High miR-96 expression predicts a poor prognosis in patients with CCA
Considering that the expression of miR-96 was significantly associated with lymph node metastasis, differentiation and TNM stage, it was speculated that miR-96 expression level may influence the prognosis of CCA. Kaplan-Meier analysis indicated that high miR-96 expression was associated with a poor prognosis in patients with CCA (log-rank test; P=0.036; Fig. 2). Furthermore, the results of the multivariate Cox regression analysis revealed that high expression of miR-96 was an independent prognostic factor for patients with CCA (HR, 1.838; 95% CI, 1.060-3.187; P=0.030; Table II).
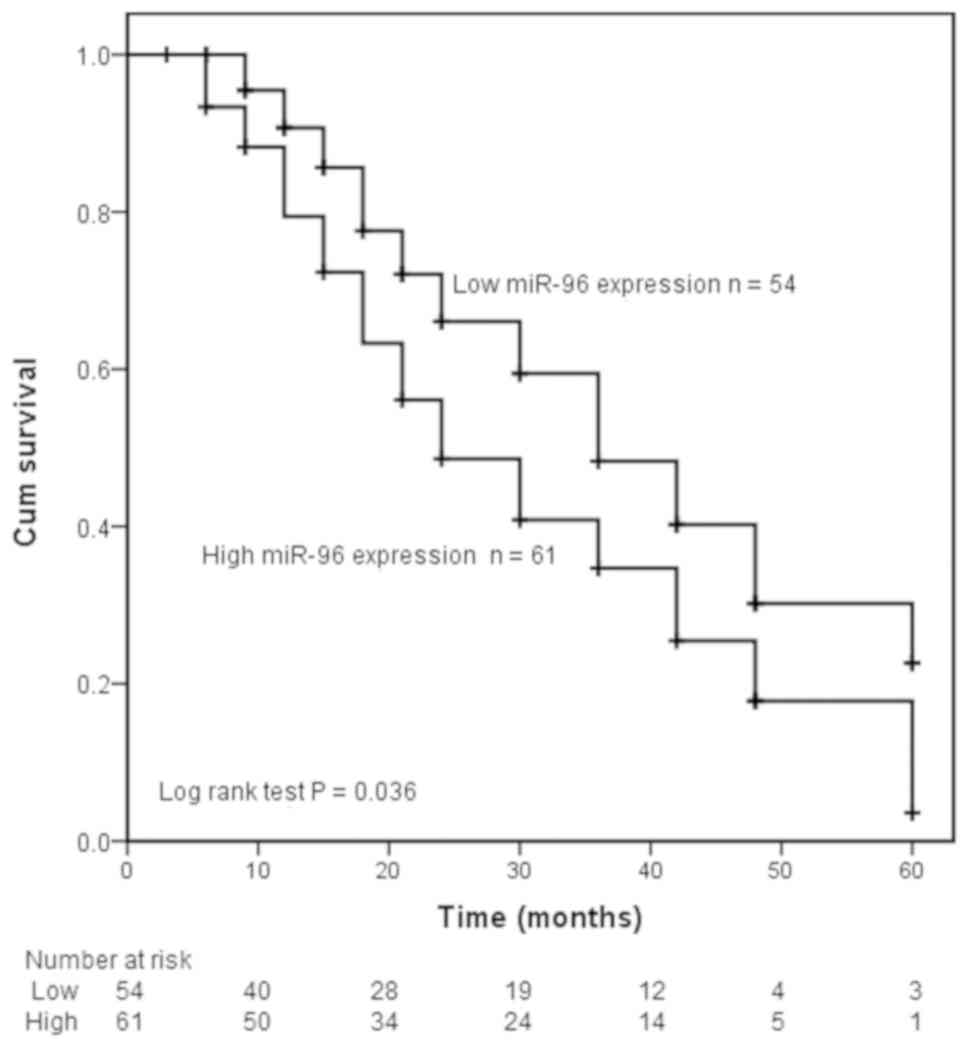 |
Figure 2.
Kaplan-Meier overall survival curve of patients with CCA. Patients with high expression of miR-96 exhibited lower overall survival time compared with those with low expression of miR-96. P=0.036. miR, microRNA; CCA, cholangiocarcinoma; Cum, cumulative.
|
 |
Table II
Multivariate Cox regression analyses of prognostic factors for overall survival in patients with cholangiocarcinoma.
|
Table II
Multivariate Cox regression analyses of prognostic factors for overall survival in patients with cholangiocarcinoma.
| |
Multivariate analysis |
| Patient characteristics |
HR |
95% CI |
P-value |
| miR-96 expression (High vs. low) |
1.838 |
1.060-3.187 |
0.030a |
| Sex (Female vs. Male) |
0.976 |
0.582-1.637 |
0.927 |
| Age (<60 vs. ≥60) |
0.983 |
0.598-1.615 |
0.945 |
| Tumor size (≥2 vs. <2) |
1.209 |
0.735-1.988 |
0.454 |
| Lymph node metastasis (Positive vs. Negative) |
1.575 |
0.966-2.567 |
0.068 |
| Differentiation (Well + moderate vs. poor) |
0.866 |
0.511-1.467 |
0.593 |
| TNM stage (I-II vs. III-IV) |
0.627 |
0.363-1.085 |
0.095 |
miR-96 promotes cell proliferation, migration and invasion
To investigate the role of miR-96 in CCA cells, the effect of miR-96 on CCA cell proliferation, migration and invasion was examined in HuCCT1 and HuH28 cells, which exhibited relatively high levels of miR-96 expression compared with other CCA cell lines (Fig. 1C). HuCCT1 and HuH28 cells were transfected with the miR-96 mimic or inhibitor, as presented in Fig. 3A. The expression levels of miR-96 were significantly higher in cells transfected with the miR-96 mimic, but lower in cells transfected with the miR-96 inhibitor compared with the respective controls (Fig. 3A; all P<0.001). A CCK-8 assay was used to determine the effects of miR-96 expression on cell proliferative ability. As indicated in Fig. 3B, transfection with the miR-96 mimic significantly promoted cell proliferation, whereas transfection with the miR-96 inhibitor significantly inhibited cell proliferation in the two CCA cell lines (all P<0.05). Transwell assays were used to assess the effects of miR-96 expression on the migration and invasion of CCA cells (Fig. 4). Compared with the respective control cells, transfection with the miR-96 mimic promoted the migratory and invasive abilities of CCA cells (P<0.001); in addition, the miR-96 inhibitor suppressed the migration and invasion of the cells (P<0.001). Taken together, these results indicated that high miR-96 expression was associated with the promotion of cell proliferation, migration and invasion of CCA cells.
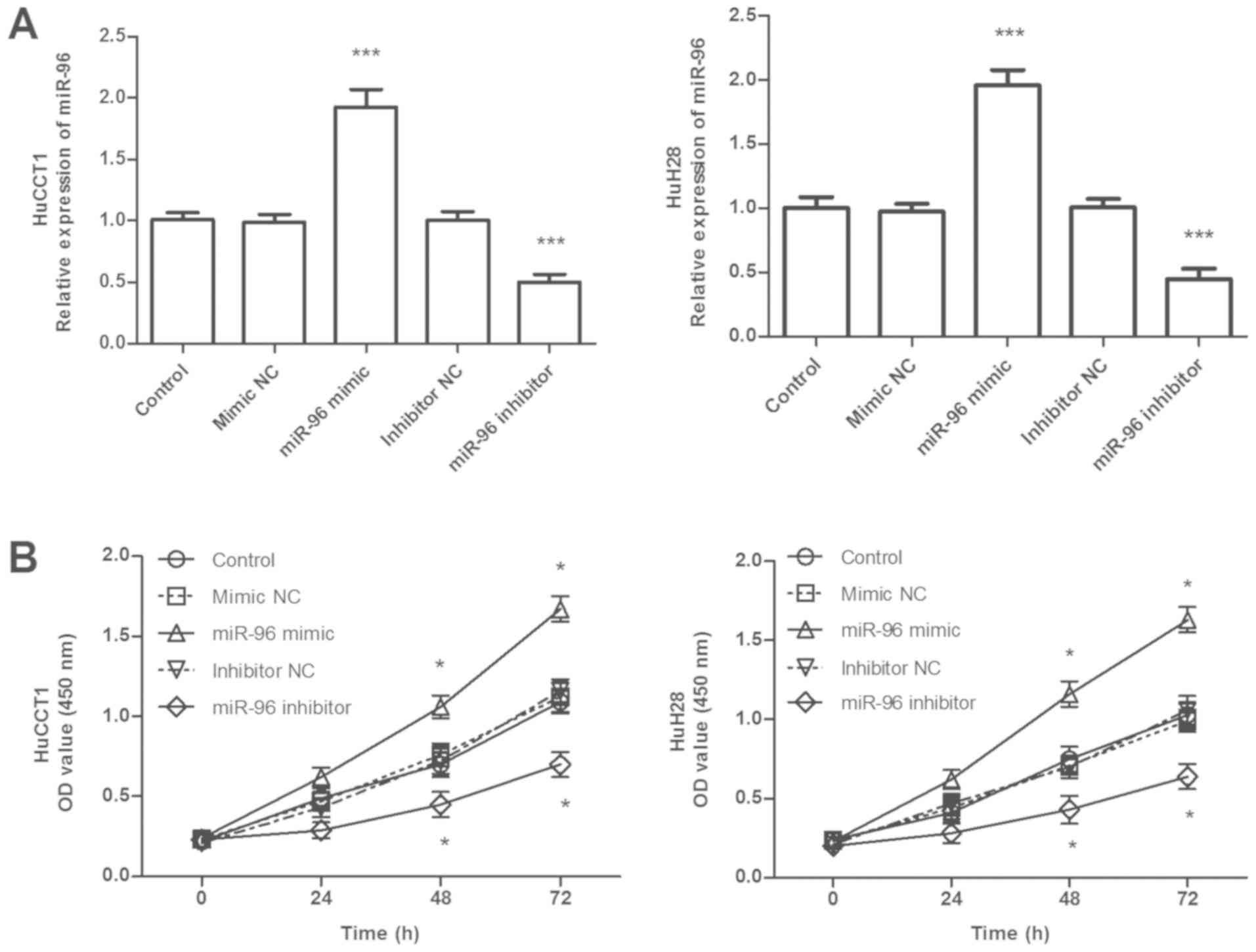 |
Figure 3.
Effects of miR-96 on CCA cell proliferation. (A) Efficiency of miR-96 mimics and miR-96 inhibitor in HUCCT1 and HuH28 cells confirmed by reverse transcription-quantitative PCR analysis. (B) Overexpression of miR-96 by miR-96 mimic promoted CCA cell proliferation, whereas downregulation of miR-96 by miR-96 inhibitor suppressed cell proliferation compared with the respective controls. *P<0.05, ***P<0.001 vs. control. miR, microRNA; CCA, cholangiocarcinoma; NC, negative control; OD, optical density.
|
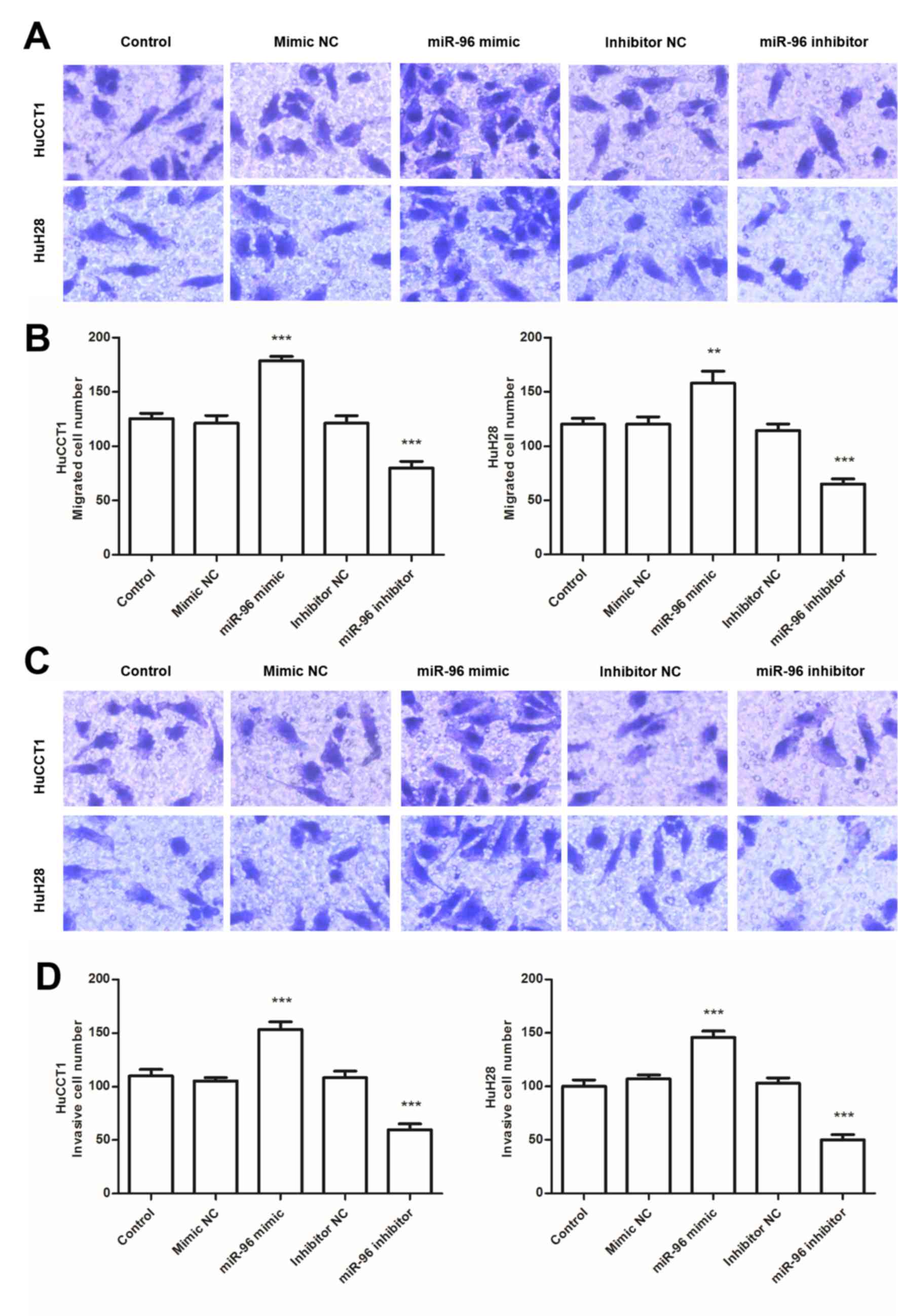 |
Figure 4.
Effects of miR-96 on CCA cell migration and invasion. (A) Representative images of the migration assay (magnification, x200). (B) Transwell migration assay results revealed that the migration of HUCCT1 and HuH28 cells was promoted by transfection with a miR-96 mimic and inhibited by transfection with a miR-96 inhibitor. (C) Representative images of the invasion assay (magnification, x200). (D) Transwell invasion assay results indicated that promotion of miR-96 expression enhanced the invasiveness of CCA cells, and knockdown of miR-96 expression inhibited the invasiveness of CCA cells. ***P<0.001 vs. control. miR, microRNA; CCA, cholangiocarcinoma.
|
MTSS1 is a direct target of miR-96
The potential downstream target genes of miR-96 were predicted by bioinformatics analysis using TargetScan and the miRDB database. Among the putative targets, MTSS1 was selected for further validation as it was downregulated in CCA tissue and associated with the recurrence and progression of CCA (18,19). A complementary binding site for miR-96 was identified in the MTSS1 3'-UTR. To validate whether miR-96 directly binds to the 3'-UTR of MTSS1, the mRNA expression of MTSS1 in CCA cells was determined, and MTSS1 was revealed to be downregulated in CCA cells compared with normal cells (P<0.001, Fig. 5B). Transfection efficiency was confirmed using RT-qPCR (P<0.001; Fig. 5C). As shown in Fig. 5C, the expression of miR-96 was upregulated by miR-96 mimic, while downregulated by miR-96 inhibitor, compared with that in control. In addition, the effects of miR-96 on MTSS1 expression were detected in CCA cells; the results demonstrated that MTSS1 expression was significantly decreased following overexpression of miR-96 and significantly increased by knockdown of miR-96 (P<0.01; Fig. 5D). To further confirm the bioinformatics prediction, WT- and MUT-MTSS1-3'-UTR were constructed and a luciferase reporter assay was conducted, which demonstrated that luciferase activity was suppressed in HuCCT1 cells co-transfected with the miR-96 mimic and pGL3-MTSS1-WT (P<0.05), whereas no significant changes were observed following co-transfection with the miR-96 mimic and pGL3-MTSS1-MUT (Fig. 5E). These results indicated that MTSS1 was a direct target of miR-96 in CCA cells.
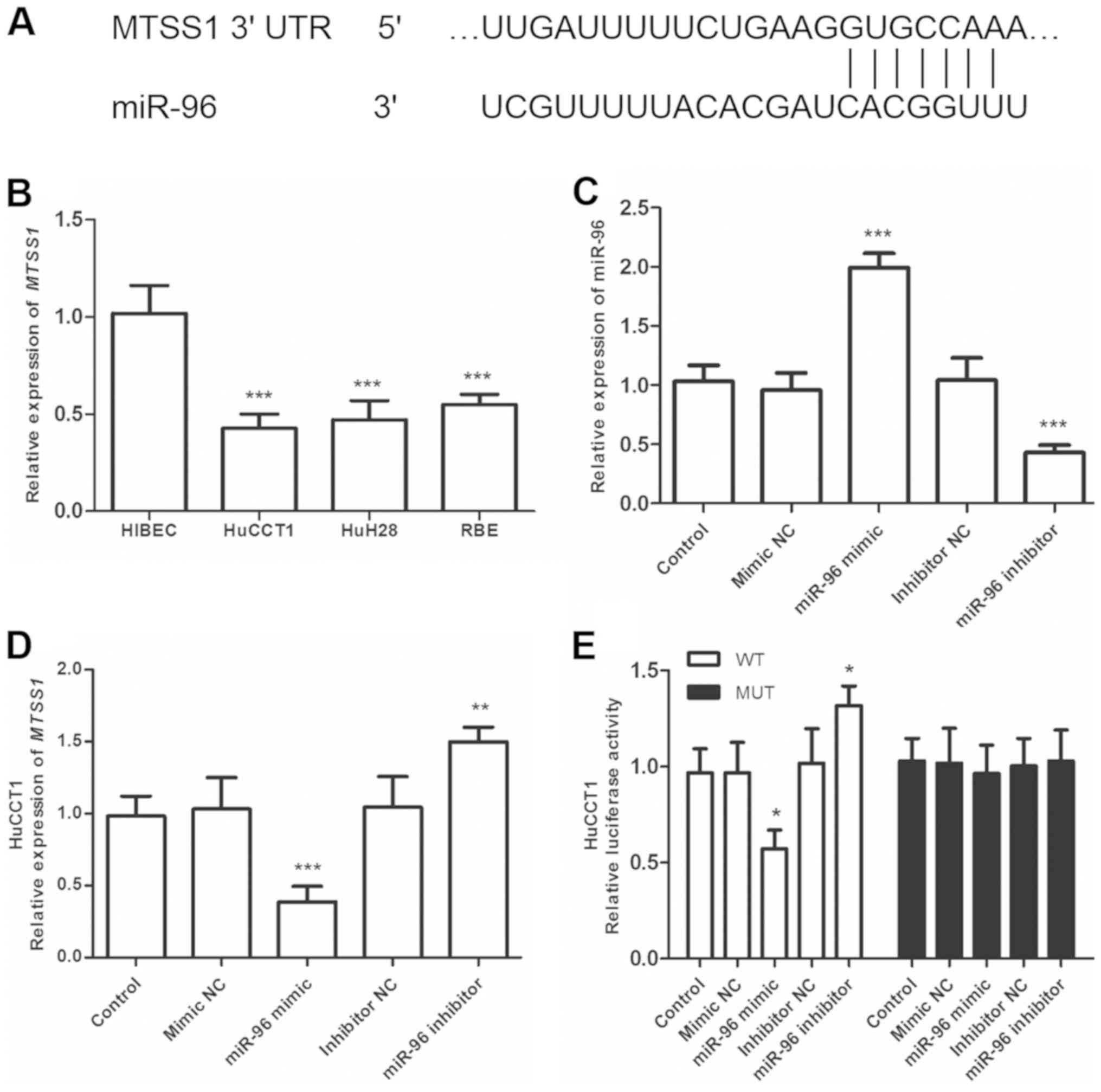 |
Figure 5.
MTSS1 is a direct target gene of miR-96. (A) Position of the miR-96 target site in the 3'-UTR of MTSS1 mRNA. (B) mRNA expression of MTSS1 was decreased in HuCCT1, HuH28 and RBE cells compared with normal hepatic HIBEC cells. (C) Transfection efficiency of miR-96 mimic and inhibitor in HuCCT1 cells was confirmed using RT-qPCR. (D) Effects of miR-96 on MTSS1 mRNA expression were determined using RT-qPCR. (E) The miR-96 mimic decreased the luciferase activity of the MTSS1-WT3'-UTR, but not the MTSS1-MUT-3'-UTR in HuCCT1 cells. The miR-96 inhibitor increased the luciferase activity of the MTSS1-WT-3'-UTR. *P<0.05, **P<0.01 and ***P<0.001 vs. HIBEC or control. miR, microRNA; CCA, cholangiocarcinoma; WT, wild type; MUT, mutant; RT-qPCR, reverse transcription-quantitative PCR; MTSS1, metastasis suppressor-1.
|
Discussion
CCA has a low prevalence, but a high mortality rate and a poor 5-year survival rate (20). CCA often develops chemotherapeutic resistance and, as a result, the only current curative treatment is surgical resection (21). The majority of patients are with CCA diagnosed in advanced stages due to the nonspecific presentation of CCA, and surgical resection may not be suitable for these patients (22). Considering the poor prognosis of advanced CCA, the present study aimed to improve CCA outcomes through the assessment of a miRNA considered to be an independent prognostic factor. A number of studies have demonstrated that miRNAs are abnormally expressed in various cancer types, such as non-small cell lung (23), bladder (24) and colorectal (25) cancer and osteosarcoma (26), and have broad potential for use as diagnostic or/and prognostic markers and therapeutic targets for cancer treatment (27-29).
The results of the present study revealed that miR-96 expression was significantly higher in CCA tissues compared with adjacent normal tissues. Furthermore, considering the anatomical locations of intrahepatic and extrahepatic CCA, the expression levels of miR-96 in intrahepatic and extrahepatic CCA tissues were also compared. Results showed miR-96 expression was significantly increased in intrahepatic and extrahepatic CCA tissues compared with the corresponding adjacent normal tissues. However, the expression levels of miR-96 between intrahepatic and extrahepatic CCA tissues showed no significant difference. Thus, we analyzed the role of miR-96 in all CCA samples for the subsequent analyses.
Except in CCA tissues, the expression of miR-96 was revealed to be upregulated in CCA cells (HuCCT1, HuH28 and RBE) compared with normal human intrahepatic biliary epithelial cells. The association between miR-96 expression and certain clinical characteristics of patients with CCA was also analyzed; the results demonstrated that high miR-96 expression was significantly associated with lymph node metastasis, poor differentiation and advanced TNM stage. Taken together, the results of the present study suggested that miR-96 may be an oncogene associated with CCA tumor development. Previous studies have also indicated that miR-96 functions as an oncogene in various types of cancer, including breast and non-small cell lung cancer, as well as cervical carcinoma (30-32). For instance, the expression of miR-96 is upregulated in human cervical carcinoma cells and may promote the proliferation and tumorigenicity of cervical cells by silencing PTPN9(32). CCA is divided into two major subtypes according to its anatomical location: Intrahepatic and extrahepatic CCA. In hepatocellular carcinoma, miR-96 has been demonstrated to be upregulated and to be a potential therapeutic target (13,33). In the current study, the expression pattern of miR-96 in CCA tissues was consistent with that in hepatocellular carcinoma.
A previous study by Yuan et al (34) suggested that members of the miR-183/182/96 gene cluster (which includes miR-96) may function as carcinogenic factors in kidney renal clear cell carcinoma and may therefore be utilized as prognostic predictors. Considering that in the present study, the expression of miR-96 was significantly associated with lymph node metastasis, differentiation and TNM stage in patients with CCA, it was hypothesized that miR-96 may also be associated with prognosis. Kaplan-Meier analysis and Cox regression analysis were performed to investigate the prognostic significance of miR-96 expression in patients with CCA; according to the 5-year follow-up data, patients with high miR-96 expression exhibited a shorter overall survival time compared with those with low-miR-96 expression. In addition, multivariate Cox regression analysis, combined with the other analyses, indicated that miR-96 may be an independent prognostic predictor in patients with CCA.
Numerous studies have reported that the expression levels and functions of miRNAs are associated with the genesis and progression of human cancers (35,36). In the present study, the functional role of miR-96 was investigated using HuCCT1 and HuH28 cell lines; miR-96 promoted the proliferation, migration and invasion of CCA cells. Several genes have been previously validated as direct targets of miR-96 (31,32,37). In hepatocellular carcinoma, the overexpression of miR-96 has been revealed to promote cell proliferation, migration and invasion by inhibiting the expression of SOX6(32). Fei et al (31) identified that miR-96 promoted lung cancer cell migration and invasion by targeting the GPC3 gene and that the miR-96/GPC3 axis may represent a therapeutic target for the treatment of non-small cell lung cancer. A recent study by Yang et al (37) indicated that miR-96 is upregulated in hepatocellular carcinoma and exerts a carcinogenic effect by activating the AKT/GSK-3β-catenin signaling pathway by targeting FOXO. In the present study, MTSS1 was identified as a novel target of miR-96. In addition, luciferase reporter assay demonstrated that miR-96 directly targeted the 3'-UTR of MTSS1, and overexpression of miR-96 inhibited MTSS1 mRNA expression in CCA cells. MTSS1, which is a metastasis suppressor gene, has been demonstrated to be downregulated in several types of cancer and is associated with tumor progression (38-40). In a recent study, MTSS1 was revealed to be downregulated in gastric cancer and suppressed the invasion, migration and epithelial-mesenchymal transition of gastric carcinoma cells through the inactivation of the PI3K/AKT signaling pathway (41). In CCA, MTSS1 was downregulated in CCA tissues; enhanced expression of MTSS1 inhibited the migration of QBC939 cells (18,19). Thus, it was speculated in the present study that miR-96 may serve an oncogenic role in CCA by targeting MTSS1. Further studies are required to assess whether miR-96 alters aggressive CCA progression through the PI3K/AKT signaling pathway.
In conclusion, the results of the present study demonstrated that miR-96 was upregulated and associated with lymph node metastasis, differentiation and TNM stage in patients with CCA, and may therefore represent a prognostic biomarker, potentially informing therapeutic strategy for patients with CCA. In addition, miR-96 may serve an oncogenic role, promoting proliferation, migration and invasion of CCA cells by targeting MTSS1.
Acknowledgements
The authors would like to thank Wei Hong (Changhai Hospital) for his support for helping analyzing the data during some experiments.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during the present study are included in this published article.
Authors' contributions
XY, ZC, and XS conceived the study, conducted the experiments and wrote the manuscript. JC and XW made substantial contributions to the recruitment of patients and collection of tissue specimens and contributed to the interpretation of the data. LY performed statistical analysis of the data and helped interpret the data. XZ performed bioinformatics analysis and dual-luciferase reporter assay. FL contributed to study design and supervision, as well as being involved in drafting the manuscript and critically revising the manuscript for important intellectual content. All authors approved the final article.
Ethics approval and consent to participate
The study protocol was approved by the Ethics Committee of Changhai Hospital, and all patients provided written informed consent. All specimens were anonymized according to the ethical and legal standards.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
1
|
Rizvi S and Gores GJ: Pathogenesis, diagnosis, and management of cholangiocarcinoma. Gastroenterology. 145:1215–1229. 2013.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Khan AS and Dageforde LA: Cholangiocarcinoma. Surg Clin North Am. 99:315–335. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Khan SA, Tavolari S and Brandi G: Cholangiocarcinoma: Epidemiology and risk factors. Liver Int. 39 (Suppl 1):S19–S31. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Razumilava N and Gores GJ: Cholangiocarcinoma. Lancet. 383:2168–2179. 2014.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Patel T: Cholangiocarcinoma-controversies and challenges. Nat Rev Gastroenterol Hepatol. 8:189–200. 2011.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Squadroni M, Tondulli L, Gatta G, Mosconi S, Beretta G and Labianca R: Cholangiocarcinoma. Crit Rev Oncol Hematol. 116:11–31. 2017.
|
|
7
|
Blechacz B: Cholangiocarcinoma: Current knowledge and new developments. Gut Liver. 11:13–26. 2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Ebata T, Mizuno T, Yokoyama Y, Igami T, Sugawara G and Nagino M: Surgical resection for Bismuth type IV perihilar cholangiocarcinoma. Br J Surg. 105:829–838. 2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Valencia-Sanchez MA, Liu J, Hannon GJ and Parker R: Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 20:515–524. 2006.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Wei R, Ye X, Zhao Y, Jia N, Liu T, Lian W, Wei H, Zhang G and Song L: MicroRNA-218 inhibits the cell proliferation and migration in clear cell renal cell carcinoma through targeting cancerous inhibitor of protein phosphatase 2A. Oncol Lett. 17:3211–3218. 2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Yao R, Zheng H, Wu L and Cai P: miRNA-641 inhibits the proliferation, migration, and invasion and induces apoptosis of cervical cancer cells by directly targeting ZEB1. Onco Targets Ther. 11:8965–8976. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Hrdlicka HC, Lee SK and Delany AM: MicroRNAs are critical regulators of osteoclast differentiation. Curr Mol Biol Rep. 5:65–74. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Iwai N, Yasui K, Tomie A, Gen Y, Terasaki K, Kitaichi T, Soda T, Yamada N, Dohi O, Seko Y, et al: Oncogenic miR-96-5p inhibits apoptosis by targeting the caspase-9 gene in hepatocellular carcinoma. Int J Oncol. 53:237–245. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Iseki Y, Shibutani M, Maeda K, Nagahara H, Fukuoka T, Matsutani S, Hirakawa K and Ohira M: MicroRNA-96 promotes tumor invasion in colorectal cancer via RECK. Anticancer Res. 38:2031–2035. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Xie W, Sun F, Chen L and Cao X: miR-96 promotes breast cancer metastasis by suppressing MTSS1. Oncol Lett. 15:3464–3471. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Collins AL, Wojcik S, Liu J, Frankel WL, Alder H, Yu L, Schmittgen TD, Croce CM and Bloomston M: A differential microRNA profile distinguishes cholangiocarcinoma from pancreatic adenocarcinoma. Ann Surg Oncol. 21:133–138. 2014.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Shi W, Hasimu G, Wang Y, Li N, Chen M and Zhang H: MTSS1 is an independent prognostic biomarker for survival in intrahepatic cholangiocarcinoma patients. Am J Transl Res. 7:1974–1983. 2015.PubMed/NCBI
|
|
19
|
Wang F, Liu Y and Zhang H: Loss of MTSS1 expression is an independent prognostic factor for Hilar cholangiocarcinoma. Pathol Oncol Res. 19:815–820. 2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Kirstein MM and Vogel A: Epidemiology and risk factors of cholangiocarcinoma. Visc Med. 32:395–400. 2016.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Puik JR, Meijer LL, Le Large TY, Prado MM, Frampton AE, Kazemier G and Giovannetti E: miRNA profiling for diagnosis, prognosis and stratification of cancer treatment in cholangiocarcinoma. Pharmacogenomics. 18:1343–1358. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Khan SA, Davidson BR, Goldin RD, Heaton N, Karani J, Pereira SP, Rosenberg WM, Tait P, Taylor-Robinson SD, Thillainayagam AV, et al: Guidelines for the diagnosis and treatment of cholangiocarcinoma: An update. Gut. 61:1657–1669. 2012.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Yang ZQ, Wu CA and Cheng YX: Prognostic value of microRNA-133a expression and its clinicopathologic significance in Non-small cell lung cancer: A comprehensive study based on Meta-analysis and the TCGA database. Oncol Res Treat. 41:762–768. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wang JR, Liu B, Zhou L and Huang YX: MicroRNA-124-3p suppresses cell migration and invasion by targeting ITGA3 signaling in bladder cancer. Cancer Biomark. 24:159–172. 2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Karimi N, Ali Hosseinpour Feizi M, Safaralizadeh R, Hashemzadeh S, Baradaran B, Shokouhi B and Teimourian S: Serum overexpression of miR-301a and miR-23a in patients with colorectal cancer. J Chin Med Assoc. 82:215–220. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Sekar D, Mani P, Biruntha M, Sivagurunathan P and Karthigeyan M: Dissecting the functional role of microRNA 21 in osteosarcoma. Cancer Gene Ther. 26:179–182. 2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Papadaki C, Stoupis G, Tsalikis L, Monastirioti A, Papadaki M, Maliotis N, Stratigos M, Mastrostamatis G, Mavroudis D and Agelaki S: Circulating miRNAs as a marker of metastatic disease and prognostic factor in metastatic breast cancer. Oncotarget. 10:966–981. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Bertoli G, Cava C and Castiglioni I: MicroRNAs: New biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 5:1122–1143. 2015.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Qin C, Huang RY and Wang ZX: Potential role of miR-100 in cancer diagnosis, prognosis, and therapy. Tumour Biol. 36:1403–1409. 2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Sun D, Zhong J, Wei W, Chen X, Liu J and Hu Z: Identification of microRNA expression in sentinel lymph nodes from patients with breast cancer via RNA sequencing for diagnostic accuracy. J Gene Med. 21(e3075)2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Fei X, Zhang J, Zhao Y, Sun M, Zhao H and Li S: MiR-96 promotes invasion and metastasis by targeting GPC3 in non-small cell lung cancer cells. Oncol Lett. 15:9081–9086. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Ma X, Shi W, Peng L, Qin X and Hui Y: MiR-96 enhances cellular proliferation and tumorigenicity of human cervical carcinoma cells through PTPN9. Saudi J Biol Sci. 25:863–867. 2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Li Z and Wang Y: miR-96 targets SOX6 and promotes proliferation, migration, and invasion of hepatocellular carcinoma. Biochem Cell Biol. 96:365–371. 2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Yuan J, Dong R, Liu F, Zhan L, Liu Y, Wei J and Wang N: The miR-183/182/96 cluster functions as a potential carcinogenic factor and prognostic factor in kidney renal clear cell carcinoma. Exp Ther Med. 17:2457–2464. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Sha HH, Wang DD, Chen D, Liu SW, Wang Z, Yan DL, Dong SC and Feng JF: MiR-138: A promising therapeutic target for cancer. Tumour Biol. 39(1010428317697575)2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Zhou MH, Zhou HW, Liu M and Sun JZ: The role of miR-92b in cholangiocarcinoma patients. Int J Biol Markers. 33:293–300. 2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Yang N, Zhou J, Li Q, Han F and Yu Z: miR-96 exerts carcinogenic effect by activating AKT/GSK-3β/β-catenin signaling pathway through targeting inhibition of FOXO1 in hepatocellular carcinoma. Cancer Cell Int. 19(38)2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Liu K, Jiao XD, Hao JL, Qin BD, Wu Y, Chen W, Liu J, He X and Zang YS: MTSS1 inhibits metastatic potential and induces G2/M phase cell cycle arrest in gastric cancer. Onco Targets Ther. 12:5143–5152. 2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Du P, Wang S, Tang X, An C, Yang Y and Jiang WG: Reduced expression of metastasis Suppressor-1 (MTSS1) accelerates progression of human bladder uroepithelium cell carcinoma. Anticancer Res. 37:4499–4505. 2017.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Taylor MD, Bollt O, Iyer SC and Robertson GP: Metastasis suppressor 1 (MTSS1) expression is associated with reduced in-vivo metastasis and enhanced patient survival in lung adenocarcinoma. Clin Exp Metastasis. 35:15–23. 2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Wang H, Zhao Y, Cao L, Zhang J, Wang Y and Xu M: Metastasis suppressor protein 1 regulated by PTEN suppresses invasion, migration, and EMT of gastric carcinoma by inactivating PI3K/AKT signaling. J Cell Biochem. 120:3447–3454. 2019.PubMed/NCBI View Article : Google Scholar
|



















