Introduction
Breast cancer (BC) is one of the most common
malignant tumors among women in the world that seriously threaten
women's health and quality of life (1-3). In
recent years, with the development of economy and the change of
lifestyle, the incidence of female BC in China has increased, and a
trend for younger age at diagnosis has emerged (4-6).
The occurrence of the majority of malignant tumors is associated
with abnormal changes in certain key genes, such as inactivation of
tumor suppressor genes, activation of oncogenes and abnormal
expression of certain apoptosis- or proliferation-associated
proteins (7,8). These changes lead to abnormal cell
proliferation, apoptosis and differentiation. At present, there are
numerous studies that have focused on the pathogenesis of BC, while
its specific mechanism remains unclear.
Ezrin-radixin-moesin (ERM) proteins are mainly
distributed on the surface of actin-rich cells and participate in
the regulation of cell proliferation, differentiation, adhesion and
movement; they serve an important role in maintaining cytoplasmic
stability and cell membrane structure (9-11).
ERM proteins participate in the development of malignant tumors and
serve a key role in tumor invasion and metastasis through
cytoskeleton and cell signal transduction (9,10,12).
Radixin is an important member of the ERM protein family involved
in the invasion, and migration of tumor cells (13-15).
Compared with ezrin and moesin, a limited number of studies have
focused on the role of radixin in tumor development.
microRNA (miR or miRNA) is an endogenous
single-stranded small non-coding RNA, 18-25 nucleotides in length.
miRNAs can bind to the 3'-untranslated region (UTR) of their target
gene mRNAs to affect the stability of the mRNA, resulting in
complete degradation or protein translation inhibition and
negatively regulating gene expression at the post-transcriptional
level (16). Numerous studies have
demonstrated that the abnormal expression of miRNAs serves
tumor-promoting or suppressing roles in the pathogenesis of
malignant tumors and can affect tumor invasion and metastasis by
regulating the expression of key genes (17-19).
miR-200b is closely associated with the occurrence and progression
of multiple types of tumors, including prostate, non-small cell
lung or cervical cancer (20-22).
Bioinformatics analysis has revealed that miR-200b has a
complementary binding in the 3'-UTR of radixin mRNA, indicating a
possible regulatory relationship. The present study aimed to
investigate the role of miR-200b in the regulation of radixin
expression, cell proliferation and invasion in BC.
Materials and methods
Reagents and materials.
The human normal breast cell line MCF-10A,
moderately metastatic BC cell line MCF-7 and highly metastatic BC
cell line MDA-MB-231 were purchased from the Cell Bank of Type
Culture Collection of the Chinese Academy of Sciences. DMEM and
fetal bovine serum (FBS) were purchased from Gibco; Thermo Fisher
Scientific, Inc. TRIzol® was purchased from Invitrogen;
Thermo Fisher Scientific, Inc. TransScript® First-Strand
cDNA Synthesis SuperMix was purchased from Beijing Transgen Biotech
Co., Ltd. miR-negative control (NC) (5'-UUCUCCGAACGUGUCACGUTT-3'),
miR-200b mimic (5'-UAAUACUGCCUGGUAAUGAUGA-3'), miR-200b inhibitor
(5'-UCAUCAUUACCAGGCAGUAUUA-3') and miR-200b NC inhibitor control
(5'-UUCUCCGAACGUGUCACGUTT-3') and riboFECT™ CP transfection reagent
were purchased from Guangzhou RiboBio Co., Ltd. Rabbit anti-human
N-cadherin (cat. no. 4061) and E-cadherin (cat. no. 3195)
antibodies were purchased from Cell Signaling Technology, Inc.
Rabbit anti-human β-actin (cat. no. ab16039) and radixin (cat. no.
ab227266) antibodies were purchased from Abcam. Horseradish
peroxidase (HRP)-conjugated goat anti-Rabbit IgG (H+L) secondary
antibody (cat. no. 31460) was purchased from Thermo Fisher
Scientific, Inc. The Transwell chamber was purchased from EMD
Millipore. Matrigel was obtained from BD Biosciences.
Dual-Luciferase assay kit was purchased from Promega Corporation.
pMIR luciferase reporter plasmid was purchased from Shaanxi Youbio
Technology Co., Ltd.
Clinical information.
A total of 36 patients with BC aged between 41 and
71 years (mean age, 53.69±14.59 years) were treated at Nantong
Traditional Chinese Medicine Hospital (Nantong, China) between
January and December 2017 were recruited for this study. All
patients were diagnosed by pathological examination and did not
receive radiotherapy or chemotherapy prior to the surgery. A total
of 12 cases of stage II, 14 cases of stage III and 10 cases of
stage IV were diagnosed. In addition, 14 cases aged between 39 and
70 years (mean age, 52.03±10.91 years) of normal breast tissue with
mammary gland hyperplasia were recruited at the same hospital
during the same time period as a control group.
The study was approved by the Research Ethics
Committee of Nantong Traditional Chinese Medicine Hospital, and all
patients provided written informed consent prior to the study.
Cell culture.
MCF-10A, MCF-7 and MDA-MB-231 cells were maintained
in DMEM containing 10% FBS and 1% penicillin/streptomycin (Thermo
Fisher Scientific, Inc.) and cultured in an incubator at 37˚C with
5% CO2. The cells were passaged every 3-4 days.
Dual luciferase reporter gene
assay.
The PCR product of the radixin 3'-UTR full-length
fragment was amplified from MCF-7 cells (Primer sequences: Forward,
5'-AGCTGAACCACCAACAGAGAA-3' and reverse,
5'-TGGAAAAGAGGCAATGGAAC-3') using the Titanium® Taq PCR
Kit according to manufacturer's protocol (Clontech Laboratories,
Inc.). The thermocycling conditions were as follows: Initial
denaturation at 94˚C for 5 min, followed by 25-30 cycles of
denaturation at 94˚C for 30 sec, annealing at 55˚C for 30 sec and
extension at 72˚C for 30 sec, with a final extension step at 72˚C
for 10 min. The PCR production was then double-digested by
HindIII and MluI and ligated into the pMIR plasmid.
Following connection by T4 DNA ligase, the plasmid was transformed
into DH5α-competent E. coli cells (Thermo Fisher Scientific,
Inc.) to screen a positive clone. Following sequencing,
pMIR-Radixin-wild-type (wt) and pMIR-Radixin-mutant (mut) plasmids
were selected. 293T cells (Thermo Fisher Scientific, Inc.) were
transfected with 1 µg pMIR-Radixin-wt or pMIR-Radixin-mut with the
miR-200b mimic, inhibitor or NC using riboFECT™ CP transfection
reagent. Following incubation for 48 h, luciferase activity was
detected using a Dual-Luciferase assay kit according to the
manufacturer's protocol. All luciferase activities were normalized
to that of Renilla luciferase.
Manipulation of miR-200b expression in
MDA-MB-231 cells.
MDA-MB-231 cells were divided into two groups,
inoculated into 10-cm culture dishes and cultured to 50-60%
confluency, followed by transfection with miR-NC or miR-200b mimic.
A total of 5 nM miR-NC-mimic or miR-200b mimic and miR-NC-inhibitor
or miR-200b inhibitor were diluted in 100 µl riboFECT™
CP Buffer at room temperature for 5 min and incubated with 10 µl
riboFECT™ CP Reagent at room temperature for 0-15 min.
The mixture was added to the cell culture medium and incubated for
72 h at 37˚C prior to further experiments.
Radixin siRNA transfection
MDA-MB-231 cells were divided into two groups,
inoculated into 10-cm culture dishes and cultured to 50-60%
confluency, followed by transfection with siRNA-NC or
siRNA-radixin. The transfection protocol was the same as that
aforementioned.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from MDA-MB-231 cells using
the miRNeasy FFPE kit (Qiagen China Co., Ltd.).
TransScript® Green One-Step qRT-PCR SuperMix (Beijing
Transgen Biotech Co., Ltd.) was used for one step RT-qPCR
detection. The PCR system comprised 1 µg template RNA, 0.3 µM
forward and reverse primers, 10 µl 2X TransStart Tip Green qPCR
SuperMix, 0.4 µl One-Step RT Enzyme mix and 0.4 µl Passive
Reference Dye II dissolved in RNase-free water. The reaction was
performed on an ABI ViiA™7 PCR system at 94˚C for 5 min, followed
by 40 cycles of 94˚C for 5 sec and 60˚C for 30 sec. U6 and GAPDH
was used as reference genes for miRNA and mRNA expression,
respectively. Quantitative analysis was performed using the
2-ΔΔCq method (23).
Primer sequences used for RT-qPCR were as follows: Radixin forward,
5'-CTCGAAAAGCTCTAGAACTGG-3' and reverse,
5'-GGTTCATTACCCCTTCATTTG-3'; miR-200b forward,
5'-ACAGTAATACTGCCTGGTAATG-3' and reverse,
5'-GGTCCAGTTTTTTTTTTTTTTTCATC-3'; U6 forward,
5'-CTCGCTTCGGCAGCACA-3' and reverse, 5'-ACGCTTCACGAATTTGCGT-3';
GAPDH forward, 5'-CAGCGACACCCACTCCTCCACCTT-3' and reverse,
5'-CATGAGGTCCACCACCCTGTTGCT-3'.
Western blotting.
Total protein was extracted from MDA-MB-231 cells by
SDS lysis and quantified by the BCA method. A total of 40 µg
protein/lane was separated by 8-10% SDS-PAGE and transferred to a
PVDF membrane at 300 mA for 1.5 h. Following blocking by 5% skimmed
milk at room temperature for 6 h, the membrane was incubated with
primary antibodies at 4˚C overnight (N-cadherin, 1:2,000;
E-cadherin, 1:2,000; radixin, 1:1,000; and β-actin, 1:10,000). The
membrane was washed with PBST and further incubated with the
HRP-conjugated secondary antibody (1:10,000) at room temperature
for 60 min. Finally, the membrane was treated with ECL
chemiluminescence reagent and developed. Image J software 1.52
(National Institutes of Health) was used for densitometric analysis
of band intensity.
Flow cytometry
MDA-MB-231 Cells (1x106/ml) were treated
with 10 µM 5-ethynyl-2'-deoxyuridine (EdU) solution in the
logarithmic phase. Following 48-h incubation, the cells were
digested with trypsin, collected, stained with Alexa Fluor-488
labeled reaction liquid (cat. no. C10337; Thermo Fisher Scientific,
Inc.) at room temperature for 30 min and detected using a Beckman
Coulter FC 500 MCL/MPL flow cytometer (Becton, Dickinson and
Company). FlowJo software (version 7.6.1; FlowJo LLC) was used for
the analysis of results.
Cell Counting Kit-8 (CCK-8) assay
CCK-8 (Shanghai Shenggong Biology Engineering
Technology Service) was used to evaluate cell proliferation. BC
cells were seeded in 96-well plates at a density of
1x105 cells per 200 µl for 24 h, followed by the
addition of a total of 10 µl CCK-8 solution into each well of the
plate and incubation for 2 h at 37˚C, following which absorbance at
450 nm was measured for each well using a microplate reader. A
fixed time point was set up for detection every day.
Transwell assay.
Matrigel (100 µl) was spread on the upper surface of
the Transwell chamber filter and incubated at 37˚C for 30 min to
allow polymerization. A total of 500 µl complete medium containing
10% FBS was added to a 24-well plate, and the Transwell chamber was
inserted. MDA-MB-231 cells (1x106/ml) were suspended in
200 µl serum-free DMEM and added to the upper chamber. Following
48-h incubation, the cells were fixed with 100% methanol for 15 min
at room temperature and stained with 0.2% crystal violet and
incubated for 10 min at room temperature. The number of invasive
cells was counted under an inverted TS2R-FL microscope
(Magnification, x40; Nikon Corporation).
Construction of pcDNA3.1-WT and
pcDNA3.1-Radixin overexpression vectors.
The microRNA.org web prediction software (http://www.microrna.org/microrna/home.do) was used to
predict the target of miR-200b. According to the requirements of
lentiviral packaging, the radixin sequence was inserted in the
front of the cPPT/CTS site, and the linearization of the vector was
achieved through SwaI or PacI single digestion. The
overlapping sequence size was 15-20 bp excluding the restriction
site. Using PacI single enzyme digestion as an example, the
upstream and downstream primers of radixin gene were designed as
follows: pcDNA3.1-WT forward, 5'-TCTGCCATAGCAAAACAAG-3' and
reverse, 5'-CTGGTCGAGCTGGACGGCGACG-3'; and pcDNA3.1-radixin
forward, 5'-TCTGCCATAGCAAAACAAGC-3' and reverse,
5'-CTGGTCGAGCTGGACGGCGACG-3'. The radixin gene was amplified by PCR
(94˚C for 1 min, 35 cycles of 94˚C for 15 sec, 55˚C for 20 sec and
72˚C for 30 sec, before 72˚C for 1 min), which was ligated into the
vector using the NEB® PCR Cloning Kit (New England
BioLabs, Inc.) according to the manufacturer's protocols. A single
clone was selected and verified to produce a positive clone. After
the plasmid was successfully constructed, the 293T cells were
transfected for 24 h at 37˚C using Lipofectamine® 2000
(Thermo Fisher Scientific, Inc.) followed by analysis; if stable
expression was achieved, the recombinant plasmid was used to
package the lentivirus using High-titer, one-step lentivirus
packaging systems (Lenti-X™ Packaging Single Shots; cat. no.
631278; Takara Bio, Inc.) according to the manufacturer's
protocols.
Colony formation experiments.
Cells (1x104) in logarithmic growth phase
were seeded onto 6 cm plates and cultured for 2-3 weeks. On
appearance of visible colonies cells were washed twice with PBS,
fixed in 100% methanol for 15 min at room temperature, stained with
0.2% crystal violet for 20 min at room temperature and air-dried
after further rinsing with water. The number of visible colonies
was counted in five random fields under an inverted light
microscope (magnification, x40; IX51; Olympus Corporation) using a
transparent film with grids.
Wound healing assay.
MDA-MB-231 cells (1x104 cells/well) were
inoculated into six-well plates and cultured routinely until
~70-80% confluence was reached following which scratches were
introduced using a 200 µl sterile pipette tip. The cells were
washed three times with sterile PBS to remove the cells from the
scratched region and were then cultured in serum-free culture
medium at 37˚C with 5% CO2. Images of the cells were
captured at 0 and 48 h following wound introduction under an
inverted light microscope (magnification, x40; IX51; Olympus
Corporation). The following formula was used to calculate cell
migration: Cell migration distance=distance at 0 h-distance at 48
h.
Statistical analysis.
SPSS 18.0 software was used for data analysis. Data
are presented as the mean ± standard deviation and were compared by
one-way ANOVA followed by Bonferroni post hoc test. The correlation
between miR-200b and radixin mRNA expression levels in BC tissue
was analyzed by Spearman's rank test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Regulatory relationship between
miR-200b and radixin.
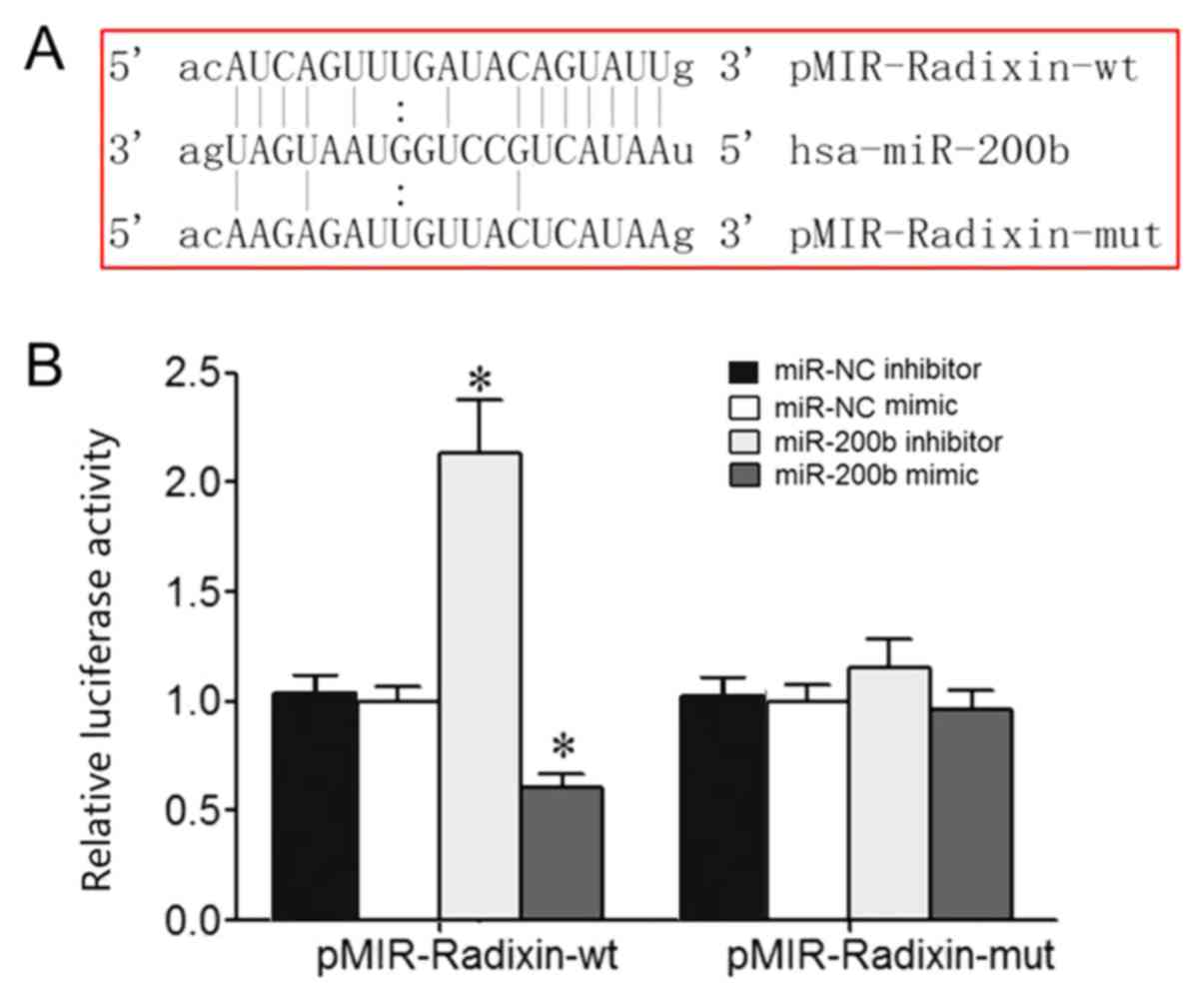
Bioinformatics analysis identified a potential
complementary binding site for miR-200b in the 3'-UTR of radixin
mRNA (Fig. 1A). Dual luciferase
reporter assay results demonstrated that miR-200b mimic or
inhibitor transfection significantly reduced or enhanced the
relative luciferase activity in 293T cells, respectively, which
suggested that radixin was the target gene of miR-200b (Fig. 1B).
miR-220b expression is downregulated,
whereas radixin level is elevated in BC tissue.
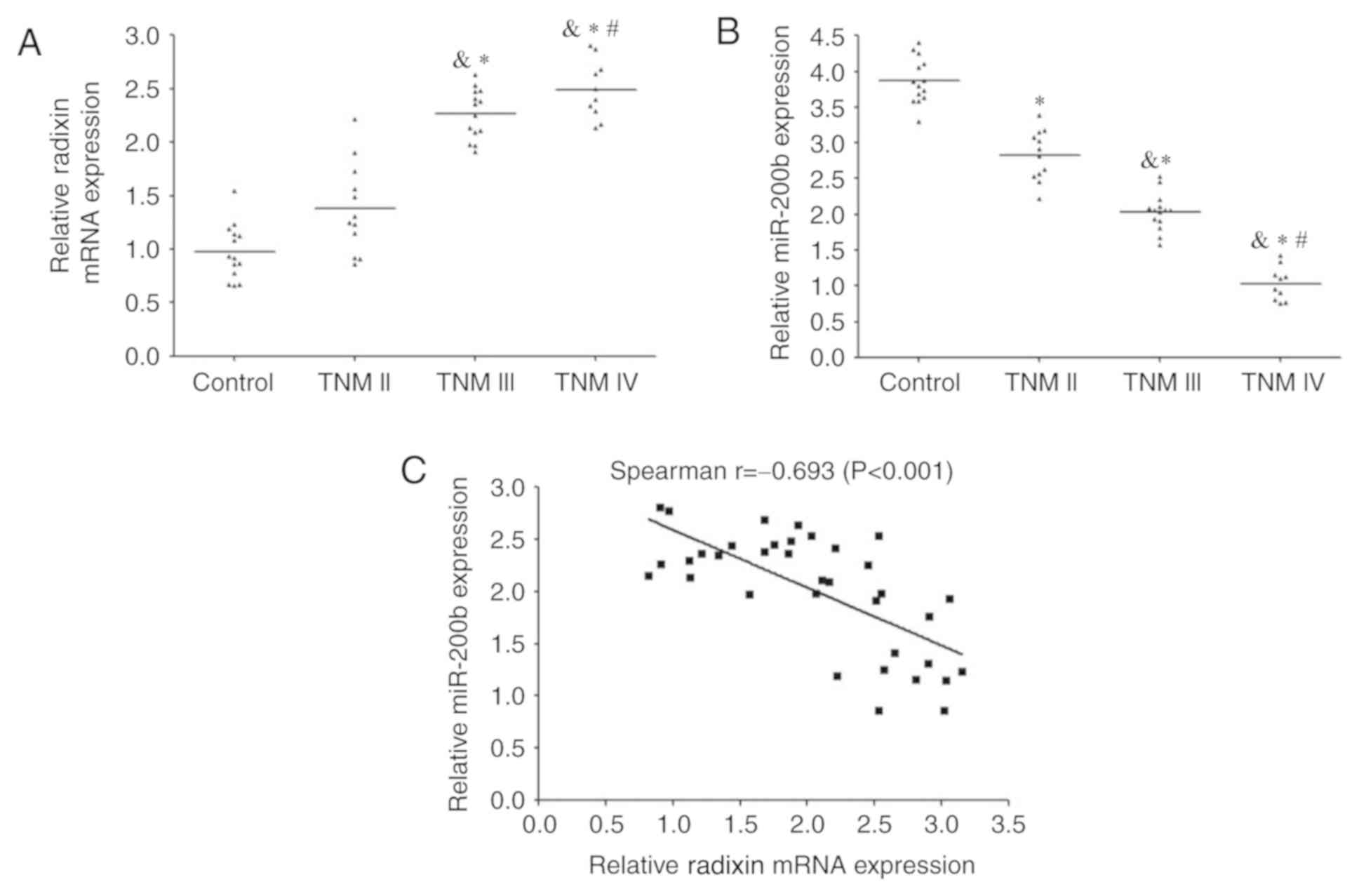
RT-qPCR analysis demonstrated that the expression of
radixin mRNA was significantly increased in patients with BC
compared with the control group; the levels increased with TNM
stage (Fig. 2A). By contrast, the
expression levels of miR-200b were reduced in BC tissue compared
with those in the control group and higher compared with lower TNM
stages (Fig. 2B). Spearman's rank
correlation analysis revealed that a significant moderate negative
correlation between miR-200b and radixin mRNA expression levels in
BC tissues (r=-0.693; P<0.001; Fig.
2C).
miR-220b and radixin expression levels
are associated with BC cell metastasis.
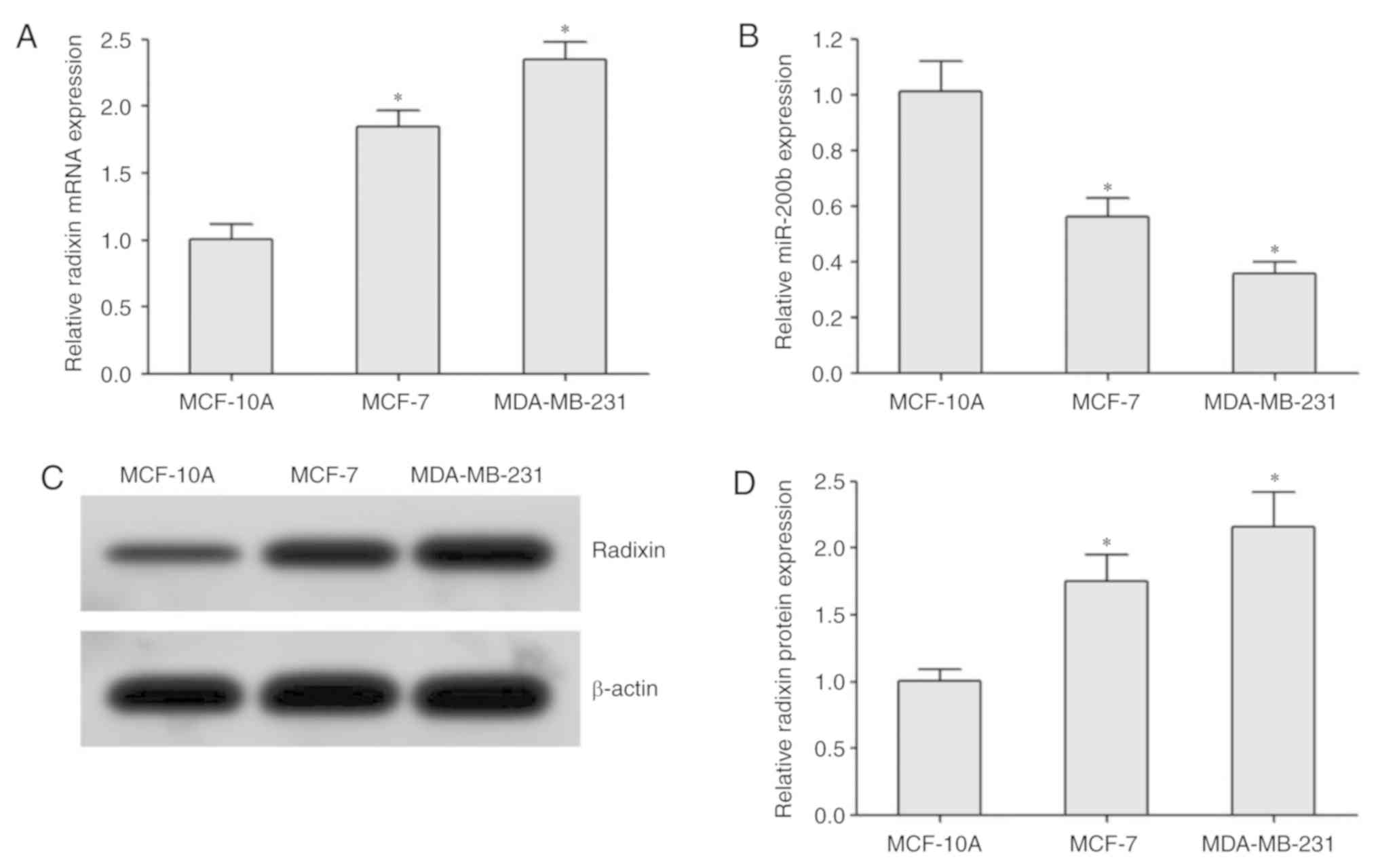
RT-qPCR analysis demonstrated that the expression of
radixin mRNA in BC cells was significantly higher compared with
that in normal breast MCF-10A cells; it was also higher in the
highly metastatic MDA-MB-231 cells compared with that in the
moderately metastatic MCF-7 cells (Fig.
3A). miR-200b expression was reduced in MDA-MB-231 cells
compared with that in MCF-7 and MCF-10A cells (Fig. 3B). Western blotting revealed that the
protein expression levels of radixin in BC cells were higher
compared with those in normal breast cells MCF-10A. In addition,
the protein expression levels of radixin in the highly metastatic
MDA-MB-231 cells was higher compared with that in MCF-7 cells
(Fig. 3C and D).
miR-200b overexpression reduces
radixin expression and cell invasion.
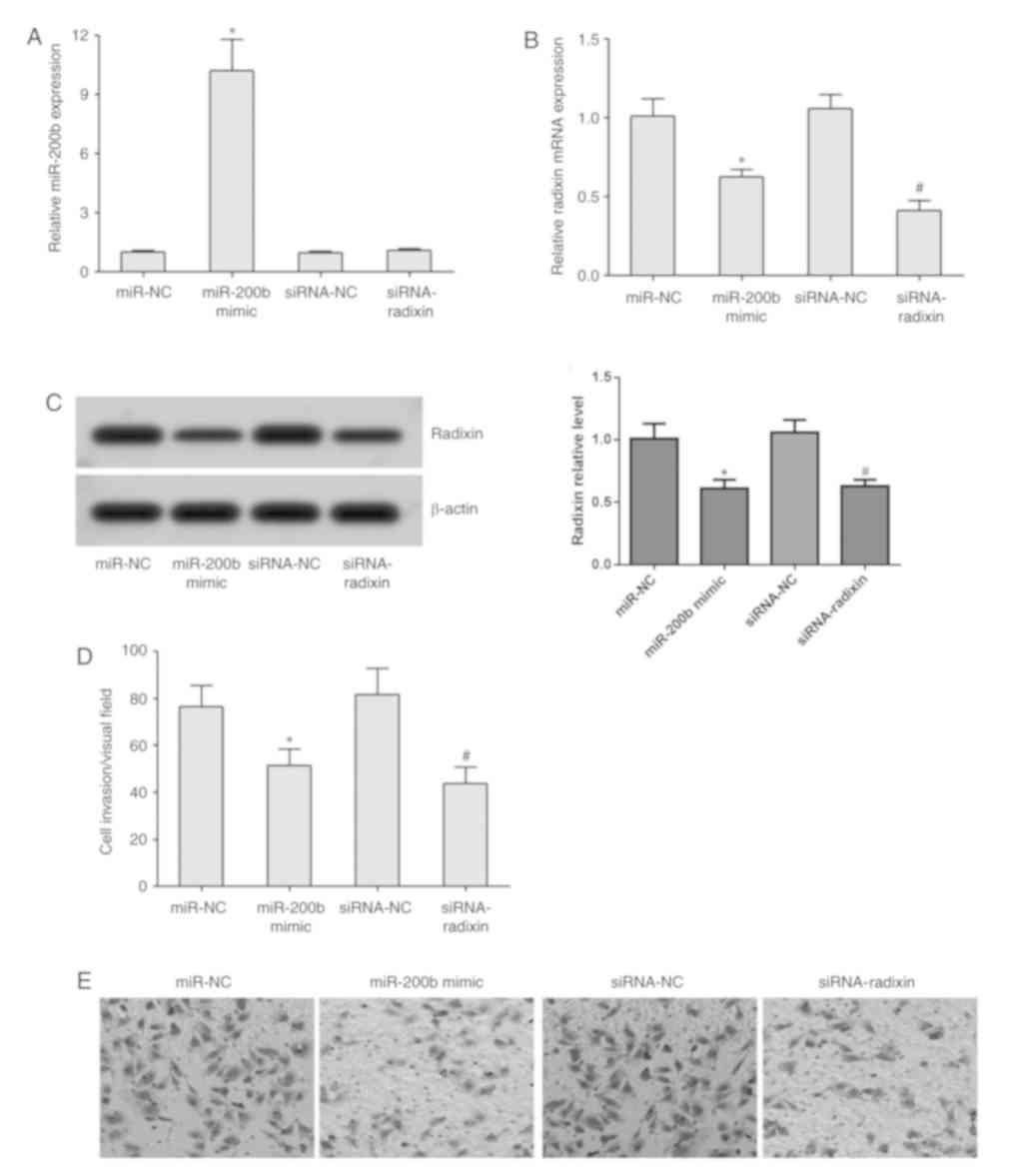
RT-qPCR analysis demonstrated that transfection with
the miR-200b mimic significantly increased miR-200b expression;
whilst transfection with the miR-200b mimic or siRNA-radixin
significantly reduced the expression of radixin mRNA in MDA-MB-231
cells (Fig. 4A and B). Western blotting revealed that compared
with the miR-NC group, the protein expression of radixin in
MDA-MB-231 cells was downregulated in the miR-200b mimic group; the
amount of intracellular radixin protein was significantly lower in
the siRNA-radixin group compared with that in the siRNA-NC group
(Fig. 4C). Transwell assay
demonstrated that transfection with the miR-200b mimic or
siRNA-radixin significantly reduced the invasive ability of
MDA-MB-231 cells (Fig. 4D and
E).
pcDNA3.1-radixin overexpression vector
increases cell proliferation and invasion.
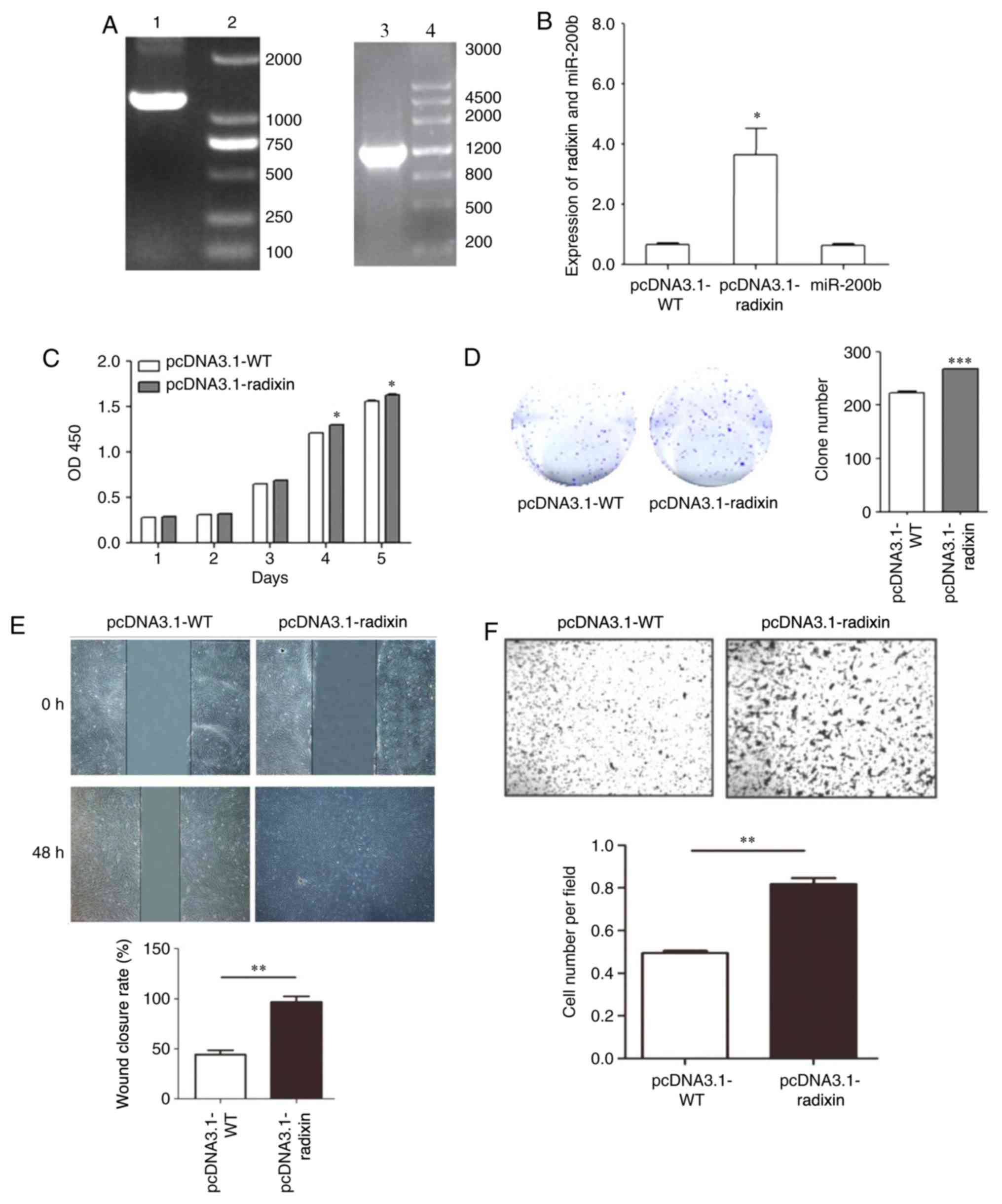
The radixin overexpression vector was constructed
and verified by PCR. The pcDNA3.1-WT was amplified by PCR to obtain
a fragment with a size of 1,771 bp. The pcDNA3.1-radixin
overexpression vector was also amplified by PCR to obtain a
fragment with a size of 1,771 bp. The expression of Radixin and
miR-200b was analyzed by RT-qPCR using the radixin overexpression
vector (Fig. 5A). RT-qPCR analysis
showed that the expression of radixin was significantly increased
following pcDNA3.1-radixin overexpression compared with cells
transfected with pcDNA3.1-WT (Fig.
5B). Radixin overexpression significantly promoted cell
proliferation (Fig. 5C and D), migration (Fig. 5E) and invasion (Fig. 5F).
miR-200b overexpression inhibits BC
cell proliferation.
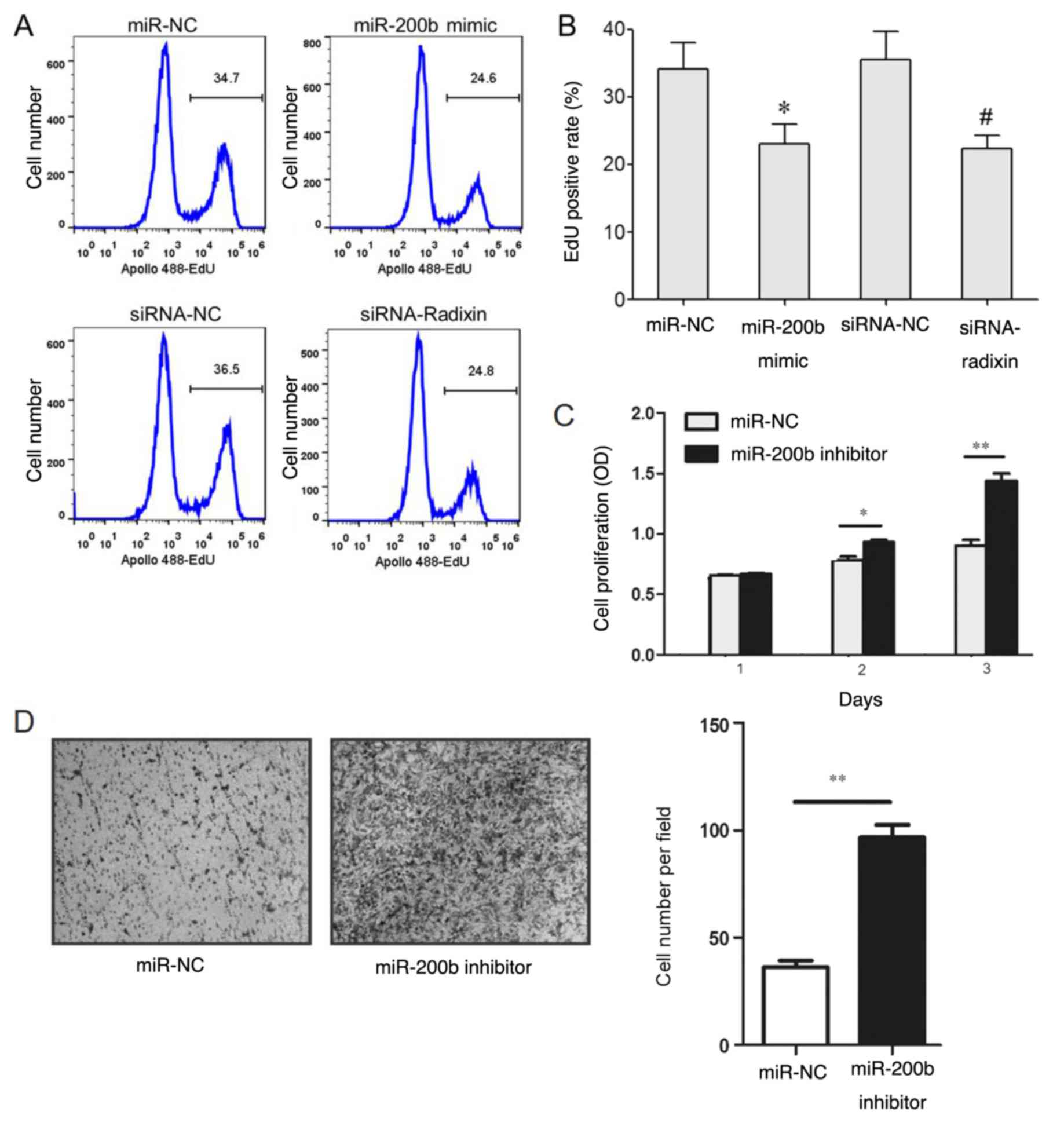
The results of the EdU staining revealed that the
EdU-positive staining rate in MDA-MB-231 cells transfected with the
miR-200b mimic was lower compared with that in cells transfected
with the miR-NC (Fig. 5A and
B). The EdU-positive staining rate
in the siRNA-radixin group was significantly lower compared with
that of the siRNA-NC group (Fig. 6A
and B). However, transfection with
the miR-200b inhibitor increased cell proliferation (Fig. 6C) and invasion (Fig. 6D).
Discussion
The ERM family comprises ezrin, radixin and moesin,
which exist in the microvilli and adherens junctions (24,25). ERM
proteins connect the cytoskeleton and the cell membrane through the
ERM domain and participate in cell morphogenesis, migration,
differentiation, adhesion and other functions (24,25).
Radixin is predominantly involved in physiological processes,
including intercellular adhesion, signal transduction and cell
movement (26). Abnormal changes in
radixin expression can cause the occurrence of a number of diseases
and may participate in the occurrence and progression of tumors by
regulating tumor-related signaling pathways. For example, Bartholow
et al (27) demonstrated that
the expression of radixin in prostate cancer tissues was
significantly lower compared with that of benign prostatic
hyperplasia and normal prostate tissues, which suggested that
radixin may be involved in the development of prostate cancer. Chen
et al (28) found altered
radixin expression in human glioma cells and implanted them into
nude mice; the results revealed that knockdown of radixin inhibited
tumor growth and invasion. The underlying molecular mechanism may
involve the expression of thrombospondin 1, E-cadherin, matrix
metallopeptidase 9 and other metastasis-inducing factors (22). Zhu et al (15) reported that silenced radixin
expression in the gastric cancer cell line SGC-7901 upregulated the
expression of E-cadherin through the NF-κB/Snail pathway, thus
inhibiting the metastasis of SGC-7901 cells. Chen et al
(28) demonstrated that silencing
radixin expression in pancreatic cancer cells significantly
inhibited cell proliferation in vitro and tumorigenicity
in vivo. Hua et al (29) reported that low expression of radixin
was associated with an enhanced invasive ability of glioma cells.
Knockdown of radixin expression inhibited the migration and
invasion of glioma cells (30).
miR-200b is associated with the development of various tumors, such
as prostate (20), lung (21) and cervical (22) cancer. Bioinformatics analysis in the
present study has identified a complementary binding site for
miR-200b in the 3'-UTR of radixin mRNA. Therefore, the present
study investigated the role of miR-200b in regulating radixin
expression, cell proliferation and invasion in BC.
In the present study, the results of the dual
luciferase gene reporter assay demonstrated that the miR-200b mimic
significantly reduced the relative luciferase activity in 293T
cells, whereas the miR-200b inhibitor enhanced the luciferase
activity. However, the miR-200b mimic and inhibitor exhibited no
significant effects on the relative luciferase activity in 293T
cells transfected with pMIR-Radixin-mut, indicating that miR-200b
directly targeted the 3'-UTR of radixin mRNA. The expression of
miR-200b was significantly decreased, whereas the expression of
Radixin was increased in patients with BC compared with the control
group, and the levels were associated with the TNM stage. In
addition, radixin expression was increased, whereas miR-200b
expression was reduced in BC cell lines compared with normal breast
cells, which was related to cell invasiveness. Li et al
(31) demonstrated that the
expression of miR-200b was significantly decreased in the highly
invasive MDA-MB-231 cells compared with the moderately invasive
MCF-7 cells. Yang et al (32)
reported that decreased expression of miR-200b was associated with
drug resistance and epithelial-mesenchymal transition (EMT) in BC
MCF-7 cells, suggesting that miR-200b downregulation was associated
with the enhancement of invasive characteristics in BC. Yao et
al (33) demonstrated that
compared with the normal breast epithelial HBL-10 cells, the
expression levels of miR-200b in BC MDA-MB-231, SK-BR-3 and
MDA-MB-468 cells were significantly reduced. Compared with normal
breast tissue, the expression of miR-200b in BC tissue was also
reduced, and was associated with TNM stage (33). In addition, miR-200b has been
reported to participate in BC cell migration and invasion by
regulating ERM in MCF-7 and MDA-MB-231(34). The results of these studies suggested
that miR-200b may serve a regulatory role in the pathogenesis and
progression of BC.
In the present study, overexpression of miR-200b or
knockdown of radixin significantly alleviated the invasive ability
of BC cells and inhibited cell proliferation. Humphries et
al (35) demonstrated that
overexpression of miR-200b in BC cells can inhibit the expression
of its target gene ARHGAP18 to suppress the migration and invasion
of BC cells. Li et al (31)
demonstrated that overexpressed miR-200b in MDA-MB-231 cells
alleviated the proliferation, migration and invasion of MDA-MB-231
cells through targeted inhibition of LIM domain kinase 1 gene
expression, indicating the tumor suppressor role of miR-200b in BC.
Yang et al (32) reported
that overexpression of miR-200b inhibited the expression of
fibronectin 1 gene, reduced the drug resistance and EMT process,
restrained cell migration and proliferation and weakened the
invasive ability of MCF-7 cells. Yao et al (33) reported that overexpression of
miR-200b induced apoptosis and reduced proliferation of BC cells.
Ye et al (36) also revealed
that the expression levels of miR-200b in BC tissues and cells were
abnormally decreased, whereas increased miR-200b expression
significantly inhibited proliferation, reduced cell invasive
ability and decreased malignant characteristics. Zheng et al
(37) demonstrated in xenograft
studies that low expression of miR-200b was associated with BC;
overexpression of miR-200b inhibited fucosyltransferase 4 and
reduced the clone formation, migration, invasion, tumorigenicity
and lung metastasis of breast cancer MCF-7 and MDA-MB-231 cells. In
the present study, miR-200b enhancement suppressed BC cell
proliferation and invasion, which was consistent with the previous
reports. Valastyan et al (38) reported that overexpression of radixin
significantly promoted the invasion and metastasis of BC cells.
Knockdown of radixin by siRNA reduced the invasion of BC cells
(38), suggesting that radixin may
serve a role in the regulation of BC cell invasion. The present
study revealed that low miR-200b expression may serve a role in
upregulating radixin and promoting the development of breast
cancer. Overexpression of miR-200b inhibited the expression of
radixin and reduced the proliferation and invasion of breast cancer
cells. However, the mechanism by which radixin affects the
proliferation and invasion of breast cancer cells has not been
elucidated.
In conclusion, low miR-200b and high radixin
expression levels may be associated with increased cell invasion in
BC. miR-200b overexpression inhibited BC cell proliferation and
invasion by targeting radixin expression.
Acknowledgments
Not applicable.
Funding
This work was supported by The Nantong City Social
Development Fund (grant No. MS12017017-4).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
JY, CX, HL and HY performed the majority of the
experiments and analyzed the data. HH, CG and ZW performed the cell
transfection, cell proliferation and invasion assays in addition to
analyzing the data. CX and HY designed the study and wrote the
manuscript.
Ethics approval and consent to
participate
This study was approved by the Research Ethics
Committee of Nantong Traditional Chinese Medicine Hospital
(Nantong, China), and all patients provided written informed
consent prior to the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Winters S, Martin C, Murphy D and Shokar
NK: Breast cancer epidemiology, prevention, and screening. Prog Mol
Biol Transl Sci. 151:1–32. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Fraser VJ, Nickel KB, Fox IK, Margenthaler
JA and Olsen MA: The epidemiology and outcomes of breast cancer
surgery. Trans Am Clin Climatol Assoc. 127:46–58. 2016.PubMed/NCBI
|
|
3
|
Rojas K and Stuckey A: Breast cancer
epidemiology and risk factors. Clin Obstet Gynecol. 59:651–672.
2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Sung H, Ren J, Li J, Pfeiffer RM, Wang Y,
Guida JL, Fang Y, Shi J, Zhang K and Li N: Breast cancer risk
factors and mammographic density among high-risk women in urban
China. NPJ Breast Cancer. 4(3)2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Li T, Tang L, Gandomkar Z, Heard R,
Mello-Thoms C, Shao Z and Brennan P: Mammographic density and other
risk factors for breast cancer among women in China. Breast J.
24:426–428. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Zuo TT, Zheng RS, Zeng HM, Zhang SW and
Chen WQ: Female breast cancer incidence and mortality in China,
2013. Thorac Cancer. 8:214–218. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Lee EY and Muller WJ: Oncogenes and tumor
suppressor genes. Cold Spring Harb Perspect Biol.
2(a003236)2010.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Wang LH, Wu CF, Rajasekaran N and Shin YK:
Loss of tumor suppressor gene function in human cancer: An
overview. Cell Physiol Biochem. 51:2647–2693. 2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Asp N, Kvalvaag A, Sandvig K and Pust S:
Regulation of ErbB2 localization and function in breast cancer
cells by ERM proteins. Oncotarget. 7:25443–25460. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Pokharel D, Padula MP, Lu JF, Jaiswal R,
Djordjevic SP and Bebawy M: The role of CD44 and ERM proteins in
expression and functionality of P-glycoprotein in breast cancer
cells. Molecules. 21(290)2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Clucas J and Valderrama F: ERM proteins in
cancer progression. J Cell Sci. 127:267–275. 2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Montt-Guevara MM, Shortrede JE, Giretti
MS, Giannini A, Mannella P, Russo E, Genazzani AD and Simoncini T:
Androgens regulate T47D cells motility and invasion through actin
cytoskeleton remodeling. Front Endocrinol (Lausanne).
7(136)2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Fernando H, Martin TA, Douglas-Jones A,
Kynaston HG, Mansel RE and Jiang WG: Expression of the ERM family
members (ezrin, radixin and moesin) in breast cancer. Exp Ther Med.
1:153–160. 2010.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Tsai MM, Wang CS, Tsai CY, Chen CY, Chi
HC, Tseng YH, Chung PJ, Lin YH, Chung IH, Chen CY and Lin KH:
MicroRNA-196a/-196b promote cell metastasis via negative regulation
of radixin in human gastric cancer. Cancer Lett. 351:222–231.
2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Zhu YW, Yan JK, Li JJ, Ou YM and Yang Q:
Knockdown of radixin suppresses gastric cancer metastasis in vitro
by up-regulation of E-Cadherin via NF-κB/Snail pathway. Cell
Physiol Biochem. 39:2509–2521. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
He J, Zhao J, Zhu W, Qi D, Wang L, Sun J,
Wang B, Ma X, Dai Q and Yu X: MicroRNA biogenesis pathway genes
polymorphisms and cancer risk: A systematic review and
meta-analysis. PeerJ. 4(e2706)2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Shrestha S, Hsu SD, Huang WY, Huang HY,
Chen W, Weng SL and Huang HD: A systematic review of microRNA
expression profiling studies in human gastric cancer. Cancer Med.
3:878–888. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
18
|
Catto JW, Alcaraz A, Bjartell AS, De Vere
White R, Evans CP, Fussel S, Hamdy FC, Kallioniemi O, Mengual L,
Schlomm T and Visakorpi T: MicroRNA in prostate, bladder, and
kidney cancer: A systematic review. Eur Urol. 59:671–681.
2011.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Li X, Abdel-Mageed AB, Mondal D and Kandil
E: MicroRNA expression profiles in differentiated thyroid cancer, a
review. Int J Clin Exp Med. 6:74–80. 2013.PubMed/NCBI
|
|
20
|
Janiak M, Paskal W, Rak B, Garbicz F,
Jarema R, Sikora K and Włodarski P: TIMP4 expression is regulated
by miR-200b-3p in prostate cancer cells. APMIS. 125:101–105.
2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Xiao P, Liu W and Zhou H: miR-200b
inhibits migration and invasion in non-small cell lung cancer cells
via targeting FSCN1. Mol Med Rep. 14:1835–1840. 2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zeng F, Xue M, Xiao T, Li Y, Xiao S, Jiang
B and Ren C: MiR-200b promotes the cell proliferation and
metastasis of cervical cancer by inhibiting FOXG1. Biomed
Pharmacother. 79:294–301. 2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Arpin M, Chirivino D, Naba A and
Zwaenepoel I: Emerging role for ERM proteins in cell adhesion and
migration. Cell Adh Migr. 5:199–206. 2011.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Fehon RG, McClatchey AI and Bretscher A:
Organizing the cell cortex: The role of ERM proteins. Nat Rev Mol
Cell Biol. 11:276–287. 2010.PubMed/NCBI View
Article : Google Scholar
|
|
26
|
Ivetic A and Ridley AJ:
Ezrin/radixin/moesin proteins and Rho GTPase signalling in
leucocytes. Immunology. 112:165–176. 2004.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Bartholow TL, Chandran UR, Becich MJ and
Parwani AV: Immunohistochemical staining of radixin and moesin in
prostatic adenocarcinoma. BMC Clin Pathol. 11(1)2011.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Chen SD, Song MM, Zhong ZQ, Li N, Wang PL,
Cheng S, Bai RX and Yuan H: Knockdown of radixin by RNA
interference suppresses the growth of human pancreatic cancer cells
in vitro and in vivo. Asian Pac J Cancer Prev. 13:753–759.
2012.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Hua D, Ding D, Han X, Zhang W, Zhao N,
Foltz G, Lan Q, Huang Q and Lin B: Human miR-31 targets radixin and
inhibits migration and invasion of glioma cells. Oncol Rep.
27:700–706. 2012.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Qin JJ, Wang JM, Du J, Zeng C, Han W, Li
ZD, Xie J and Li GL: Radixin knockdown by RNA interference
suppresses human glioblastoma cell growth in vitro and in vivo.
Asian Pac J Cancer Prev. 15:9805–9812. 2014.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Li D, Wang H, Song H, Xu H, Zhao B, Wu C,
Hu J, Wu T, Xie D, Zhao J, et al: The microRNAs miR-200b-3p and
miR-429-5p target the LIMK1/CFL1 pathway to inhibit growth and
motility of breast cancer cells. Oncotarget. 8:85276–85289.
2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Yang X, Hu Q, Hu LX, Lin XR, Liu JQ, Lin
X, Dinglin XX, Zeng JY, Hu H, Luo ML and Yao HR: miR-200b regulates
epithelial-mesenchymal transition of chemo-resistant breast cancer
cells by targeting FN1. Discov Med. 24:75–85. 2017.PubMed/NCBI
|
|
33
|
Yao Y, Hu J, Shen Z, Yao R, Liu S, Li Y,
Cong H, Wang X, Qiu W and Yue L: MiR-200b expression in breast
cancer: A prognostic marker and act on cell proliferation and
apoptosis by targeting Sp1. J Cell Mol Med. 19:760–769.
2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Hong H, Yu HZ, Yuan JF, Guo C, Cao H, Li W
and Xiao C: MicroRNA-200b impacts breast cancer cell migration and
invasion by regulating ezrin-radixin-moesin. Med Sci Monit.
22:1946–1952. 2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Humphries B, Wang Z, Li Y, Jhan JR, Jiang
Y and Yang C: ARHGAP18 downregulation by miR-200b suppresses
metastasis of triple-negative breast cancer by enhancing activation
of RhoA. Cancer Res. 77:4051–4064. 2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Ye F, Tang H, Liu Q and Xie X, Wu M, Liu
X, Chen B and Xie X: miR-200b as a prognostic factor in breast
cancer targets multiple members of RAB family. J Transl Med.
12(17)2014.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Zheng Q, Cui X, Zhang D, Yang Y, Yan X,
Liu M, Niang B, Aziz F, Liu S, Yan Q and Liu J: miR-200b inhibits
proliferation and metastasis of breast cancer by targeting
fucosyltransferase IV and α1,3-fucosylated glycans. Oncogenesis.
6(e358)2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Valastyan S, Benaich N, Chang A, Reinhardt
F and Weinberg RA: Concomitant suppression of three target genes
can explain the impact of a microRNA on metastasis. Genes Dev.
23:2592–2597. 2009.PubMed/NCBI View Article : Google Scholar
|