Introduction
With changes in diet and lifestyle, the incidence of
laryngeal cancer has increased in recent years. According to the
statistical data presented in GLOBOCAN in 2018, the number of new
cases of laryngeal cancer reached 177,422, accounting for 1% of all
new cancer cases worldwide (1-3). As
the most common type of malignant tumor in the head and neck,
laryngeal cancer has become a worldwide problem (4). Although there have been considerable
developments in treatment options in recent years, there remains a
number of problems and the prognosis of patients has not reached
what is desired (5,6). Therefore, it is of great significance
to investigate the potential molecular mechanisms and provide new
therapeutic targets for the clinical treatment of laryngeal
cancer.
Long non-coding RNAs (lncRNAs) have attracted much
attention due to their extensive biological activities (7). lncRNAs exhibit tissue specificity and
functional diversity, which makes them potentially effective
diagnostic and prognostic markers (8). Recent studies have revealed that lncRNA
SOX2-OT functions as an oncogenic factor and serves an important
role in numerous types of cancer, including osteosarcoma,
cholangiocarcinoma and lung squamous cell carcinoma (9-11).
Notably, using microarrays, Feng et al (12) identified that the expression of
SOX2-OT in cancer tissues was significantly higher compared with
that in adjacent non-neoplastic tissues in advanced laryngeal
squamous cell carcinoma (LSCC). Furthermore, Tai et al
(13) suggested that SOX2-OT
promotes the development of LSCC through silencing of phosphatase
and tensin homolog, which is induced by the methyltransferase EZH2.
These studies suggest that SOX2-OT is closely associated with the
development of laryngeal cancer. However, the underlying mechanism
by which SOX2-OT functions remains unclear in laryngeal cancer.
MicroRNAs (miRNAs) are composed of endogenous
non-coding small RNAs that can regulate mRNA stability and protein
translation (14). It has been
proved that miRNAs play take part in the development of various
cancer processes, such as proliferation, differentiation and
metastasis (15). miR-654 was found
to be abnormally expressed in many squamous cell carcinoma
including laryngeal squamous cell carcinoma (16). Nonetheless, the biological role of
miR-654 in laryngeal squamous cell carcinoma is still unclear.
The present study aimed to investigate whether
SOX2-OT is involved in the development of laryngeal cancer by
regulating microRNA (miR)-654. It was identified that the
expression of SOX2-OT is significantly increased in laryngeal
cancer cells. In order to evaluate the potential function of
SOX2-OT, RNA interference was applied to knockdown the expression
level of SOX2-OT, and further experiments were conducted to
identify the association between SOX2-OT and miR-654 in TU-177
cells.
Materials and methods
Cell culture and treatment
All cell lines, including the normal human
nasopharyngeal epithelial cell line NP69 and laryngeal cancer cell
lines TU-177, M4E, AMC-HN-8 and TU686, were purchased from the
American Type Culture Collection. Cells were cultured in RPMI-1640
medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with
10% FBS (Gibco; Thermo Fisher Scientific, Inc.) in an incubator
containing 95% air and 5% CO2 at a constant temperature
of 37˚C.
Cell transfection
The short hairpin RNA (shRNA) sequence targeting
SOX2-OT (shRNA-SOX2-OT-1/2), the negative control (shRNA-NC), the
miR-654 inhibitor, inhibitor NC (miR-NC), miR-654 mimic and mimic
NC (miR-654 NC) were designed and synthesized by Shanghai
GenePharma Co., Ltd. The shRNA-SOX2-OT-1 sequence was
GCACCGCTATACAGAGAAACCTTATCCTCGAGGATAAGGTTTCTCTGTATAGCTTTTTTG, the
shRNA-SOX2-OT-2 sequence was
GCACCGGAGCAAAGGTGCTGTCATTTCTCGAGAAATGACAGCACCTTTGCTC CTTTTTG, the
shRNA-NC sequence was
CGCGTCCCCCACCTTTCGGCACTCTCCCTTCAAGAGGGGAGAGTGCCGAAAGGTGTTTTTGGAAAT,
The miR-654 inhibitor sequence was 5' ACACAUGUUCUGCGGCCCACCA 3',
the negative control (miR-NC) sequence was 5' CAGUACUUUUGUGUAGUACAA
3', the miR-654 mimic sequence was 5' UGGUGGGCCGCAGAACAUGUGC 3' and
the miR-654 NC sequence was 5' UUGUACUACACAAAAGUACUG 3'. TU-177
cells were seeded in six-well plates at a density of
3x105/well and incubated for 24 h. Subsequently, TU-177
cells were transfected with 100 pmol shRNA-SOX2-OT-1/2 or shRNA-NC
with or without 100 nM miR-654 mimic, miR-654 inhibitor or
corresponding controls using Lipofectamine® 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). At 48 h after
transfection, cells were harvested for further experiments.
Reverse transcription-quantitative PCR
(RT-qPCR)
TU-177 cells were lysed and total RNA was extracted
using TRIzol reagent (Thermo Fisher Scientific, Inc.). For the
mRNAs, complementary DNA (cDNA) was synthesized using the
Transcriptor First Strand cDNA Synthesis kit (Roche Diagnostics).
For miR-654, cDNA was synthesized using specific stem-loop primers
combined with TaqMan MicroRNA Reverse Transcription kit (Takara
Bio, Inc.). The reverse transcription reaction was performed at
42˚C for 5 min and 95˚C for 10 sec. qPCR was then performed using
the SYBR Green kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.) in the ABI 7500 Real-time PCR system (Applied Biosystems).
The thermocycing conditions were as follows: One cycle of 95˚C for
10 min, followed by 40 cycles at 95˚C for 15 sec and 60˚C for 1
min. GAPDH and U6 were used as the internal controls for SOX2-OT
and miR-654, respectively. Primer sequences were exhibited at
Table I.
 | Table IPCR primer sequences. |
Table I
PCR primer sequences.
| | Primers (5'–3') |
|---|
| SOX2-OT forward |
5'-TTGGAAGGATGGCATACAC-3' |
| SOX2-OT reverse |
5'-CAATGAAGTTGACTGGACTC-3' |
| GAPDH forward |
5'-GGAGCGAGATCCCTCCAAAAT-3' |
| GAPDH reverse |
5'-GGCTGTTGTCATACTTCTCATGG-3' |
| miR-654 forward |
5'-TGGTGGGCCGCAGAACATGTGC-3' |
| miR-654 reverse |
5'-GCGAGCACAGAATTAATACGAC-3' |
| U6 forward |
5'-CGCTTCGGCAGCACATATACTA-3' |
| U6 reverse |
5'-CGCTTCACGAATTTGCGTGTCA-3' |
Cell Counting Kit-8 (CCK-8)
TU-177 cells were seeded in 96-well plates at a
density of 2x104/well and incubated for 24 h.
Subsequently, the shRNAs were transfected into TU-177 cells. At 24,
48 and 72 h after transfection, cell proliferation was evaluated
with a CCK-8 kit (catalog no. C0038; Beyotime Institute of
Biotechnology), according to manufacturer's instructions. The
absorbance value of each well was measured at 450 nm.
Western blot assay
The western blot assay was performed as previously
described (17). Briefly, total
protein was extracted from TU-177 cells using RIPA lysis buffer
(Beyotime Institute of Biotechnology). Following separation by 10%
SDS-PAGE, protein samples were transferred onto a PVDF membrane
(EMD Millipore). After blockage in 10% non-fat milk for 1 h at
37˚C, the membranes were incubated with primary antibodies (p21,
1:500, cat. no. 64016, Cell Signaling Technology; CDK2, 1:1,000,
cat. no. 2546, Cell Signaling Technology; cyclin E1, 1:500, cat.
no. sc-377100, Santa Cruz Biotechnology; MMP-7, 1:500, cat. no.
3801, Cell Signaling Technology; MMP-9, 1:1,000, cat. no. 3667,
Cell Signaling Technology; Bcl-2, 1:1,000, cat. no. 15071, Cell
Signaling Technology; Bax, 1:1,000, cat. no. 14796, Cell Signaling
Technology; cleaved caspase 3, 1:1,000, cat. no. 9661, Cell
Signaling Technology; GAPDH, 1:1,000, cat. no. MAB374, EMD
Millipore) overnight at 4˚C. Subsequently, the protein bands were
probed with rabbit anti-mouse IgG-HRP (1:10,000, cat. no.
sc-358914) or mouse anti-rabbit IgG-HRP secondary antibodies
(1:10,000, cat. no. sc-2357; both Santa Cruz Biotechnology) for 2 h
at room temperature. Finally, the ECL detection reagent (EMD
Millipore) was used to visualize the chemiluminescent signals.
ImageJ software (version 1.8, NIH) was used to quantify the protein
bands.
Transwell assay
Cell invasion was assessed using a Transwell assay.
During this assay, the inserts of the Transwell chamber (Corning
Inc.) were filled with Matrigel (BD Biosciences). At 48 h
post-transfection, TU-177 cells (5x103 cells/ml) were
plated in the upper chambers with serum-free RPMI-1640 medium. In
addition, complete DMEM containing 10% FBS was added to the lower
chamber as a chemoattractant. Following incubation for 24 h, cells
that migrated through the membrane were fixed with 4% formaldehyde
at room temperature. Finally, following staining with 0.1% crystal
violet, the invaded cells were counted with a light microscope
(Nikon Corporation) at x200 magnification.
Wound healing
TU-177 cells were seeded in six-well plates at a
density of 3x105/well and allowed to growth to 90%
confluence in DMEM with 10% FBS at 37˚C. A 10-µl sterile pipette
tip was used to generate a wound, and an inverted microscope was
used to monitor the migration of the cells at 0 and 24 h. The
widths of the wounds were measured to assess the cell
migration.
Cell apoptosis
At 48 h post-transfection, TU-177 cells were
harvested and an Annexin V-FITC/PI Apoptosis kit (catalog no.
KA3805; Abnova) was used to assess apoptosis. In brief, cells were
suspended in 100 µl 1X binding buffer. Following staining with 5 µl
Annexin V-FITC for 10 min, cells were stained with 5 µl PI for a
further 5 min in the dark at room temperature. Apoptotic cells were
analyzed using a flow cytometer (FACScan; BD Biosciences).
Dual-luciferase reporter assay
To verify the direct interactions between SOX2-OT
and miR-654, a dual-luciferase reporter assay was performed.
Briefly, the mutant type and wild type of SOX2-OT binding sequences
were cloned into a pGL3-promoter (Promega Corporation) to generate
the recombinant vectors pGL3-SOX2-OT-WT and pGL3-SOX2-OT-MUT.
Following seeding onto 24-well plates for 24 h, TU-177 cells were
co-transfected with 50 ng pGL3-SOX2-OT-WT/MUT and 20 µM miR-654
mimic/NC using Lipofectamine® 2000 reagent (Invitrogen;
Thermo Fisher Scientific, Inc.). At 48 h after transfection, the
luciferase activity of each well was determined using the
Dual-Luciferase Reporter assay system (Promega Corporation), and
the results were normalized to the Renilla luciferase
activity.
Statistical analysis
In the present study, each experiment was repeated a
minimum of three times and data are presented as the mean ±
standard deviation. Statistical analysis was performed using SPSS
version 20.0. Statistical analysis were performed using Student's
t-test and one-way analysis of variance, followed by a Dunnett's
post hoc test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Upregulation of SOX2-OT in laryngeal
cancer cell lines
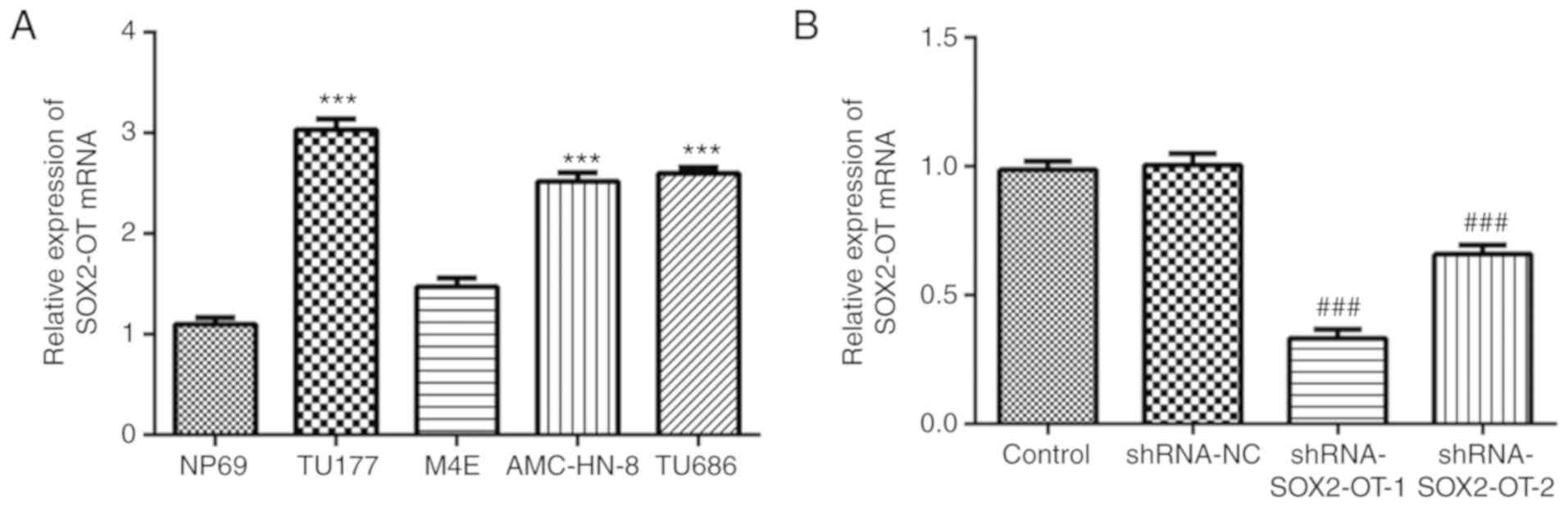
To evaluate the expression levels of SOX2-OT in
laryngeal cancer cell lines, an RT-qPCR assay was performed. As
presented in Fig. 1A, the levels of
SOX2-OT were significantly increased in the laryngeal cell lines
TU-177, AMC-HN-8 and TU686, especially in TU-177 cells, compared
with the normal cell line NP69. However, no significant difference
in SOX2-OT mRNA expression between NP69 and M4E was observed. With
this in mind, TU-177 cells were selected for the following
experiments. In order to further investigate the biological
function of SOX2-OT, shRNA-SOX2-OT-1/2 or shRNA-NC were transfected
into TU-177 cells. At 48 h after transfection, the interference
effect was detected, and the results demonstrated that expression
of SOX2-OT was markedly decreased by both shRNA-SOX2-OT-1 and -2
transfection (Fig. 1B). Moreover,
shRNA-SOX2-OT-1 was selected for subsequent experiments as it
exhibited a better interference effect.
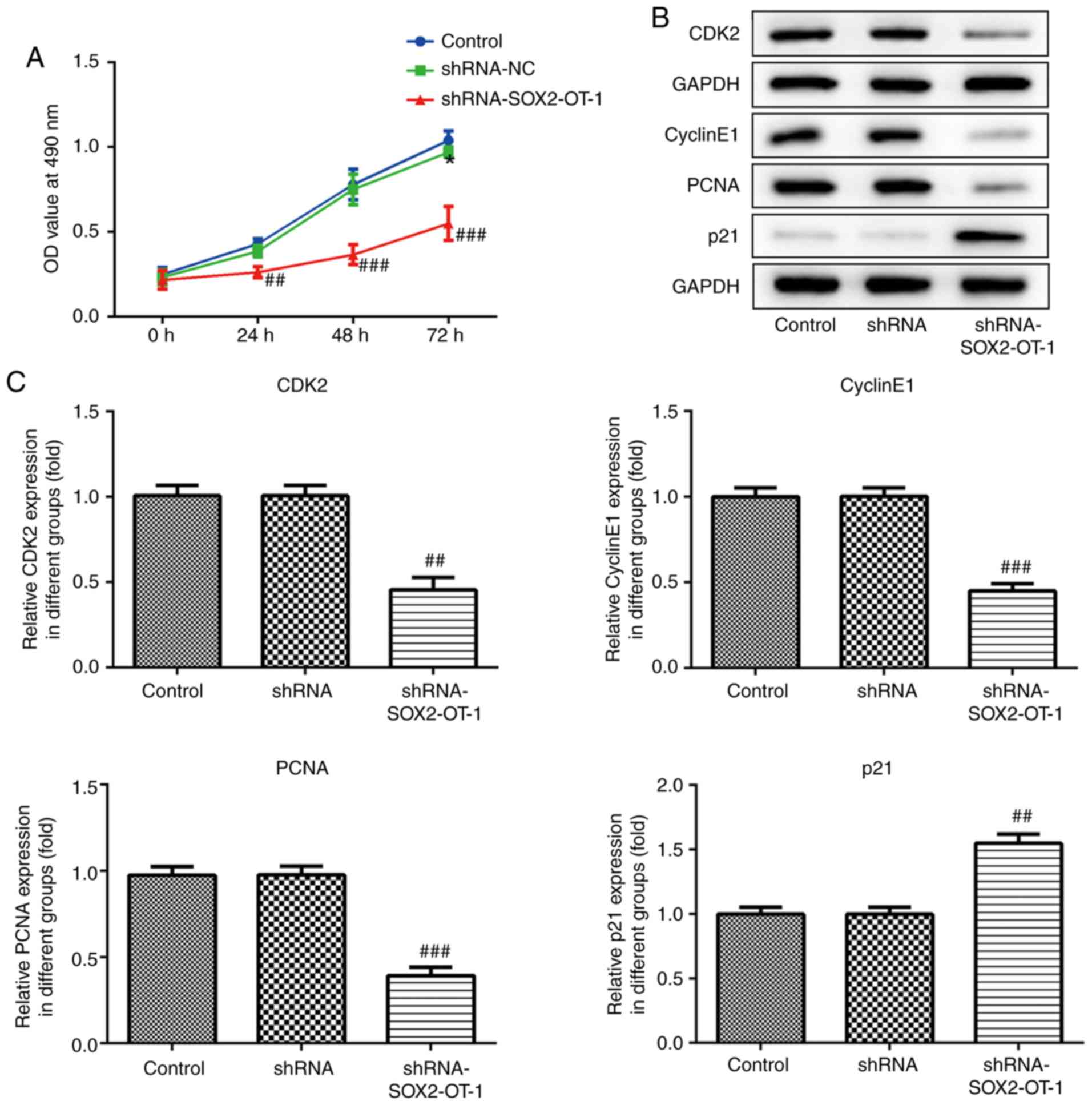
Knockdown of SOX2-OT suppresses the
proliferation of TU-177 cells
CCK-8 assay was performed to assess the effect of
SOX2-OT-knockdown on cell proliferation in TU-177 cells.
SOX2-OT-silencing markedly suppressed cell proliferation in TU-177
cells (Fig. 1A). Additionally, cell
proliferation following shRNA-SOX2-OT-1 transfection was assessed
using western blotting. The results demonstrated that
SOX2-OT-silencing significantly upregulated the expression of p21,
and downregulated the expression of CDK2, cyclin E1 and PCNA
(Fig. 2B and C).
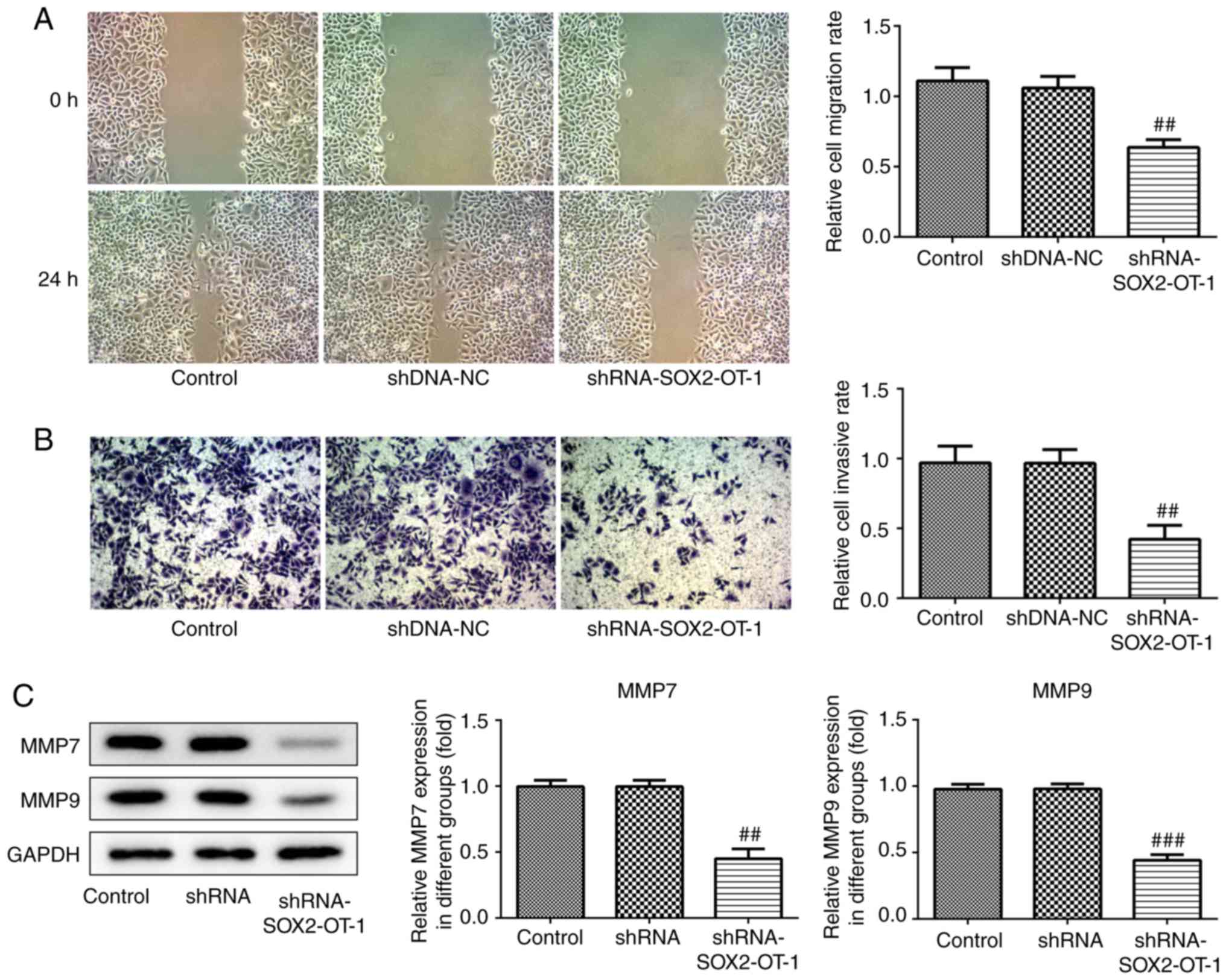
SOX2-OT-knockdown inhibits migration
and invasion in TU-177 cells
Migration and invasion are considered to be key
factors in the development of cancer. Therefore, wound healing and
transwell assays were used to measure the effects of SOX2-OT on
migration and invasion, respectively, in TU-177 cells. The results
indicated that both the migration and invasion of TU-177 cells were
inhibited following SOX2-OT-knockdown (Fig. 3A and B). Furthermore, western blot analysis was
used to determine the protein expression levels of MMP-7 and MMP-9.
As presented in Fig. 3C,
SOX2-OT-knockdown significantly reduced the protein levels of MMP-7
and MMP-9.
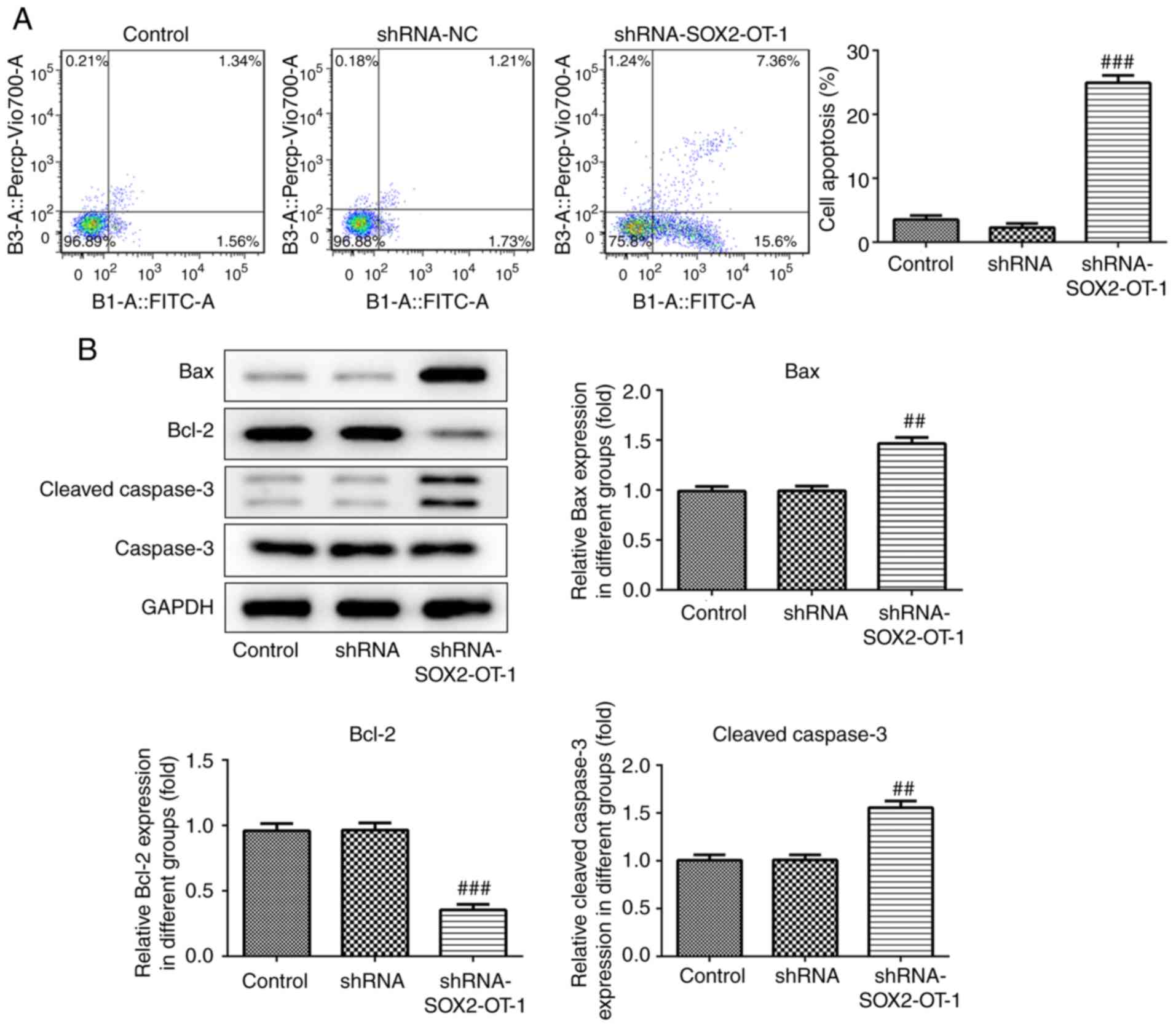
SOX2-OT-knockdown induces apoptosis in
TU-177 cells
To investigate whether the silencing of SOX2-OT has
an effect on the apoptosis of TU-177 cells, annexin V-FITC/PI
staining combined with flow cytometry was performed. According to
the results, the percentage of the apoptotic cell fraction was
significantly increased in shRNA-SOX2-OT-1-treated TU-177 cells
(Fig. 4A). In addition, the results
of western blot assay revealed that SOX2-OT-knockdown markedly
reduced the expression of Bcl-2 protein, and the protein levels of
Bax and cleaved caspase 3 were significantly increased in
shRNA-SOX2-OT-1-transfected TU-177 cells (Fig. 4B).
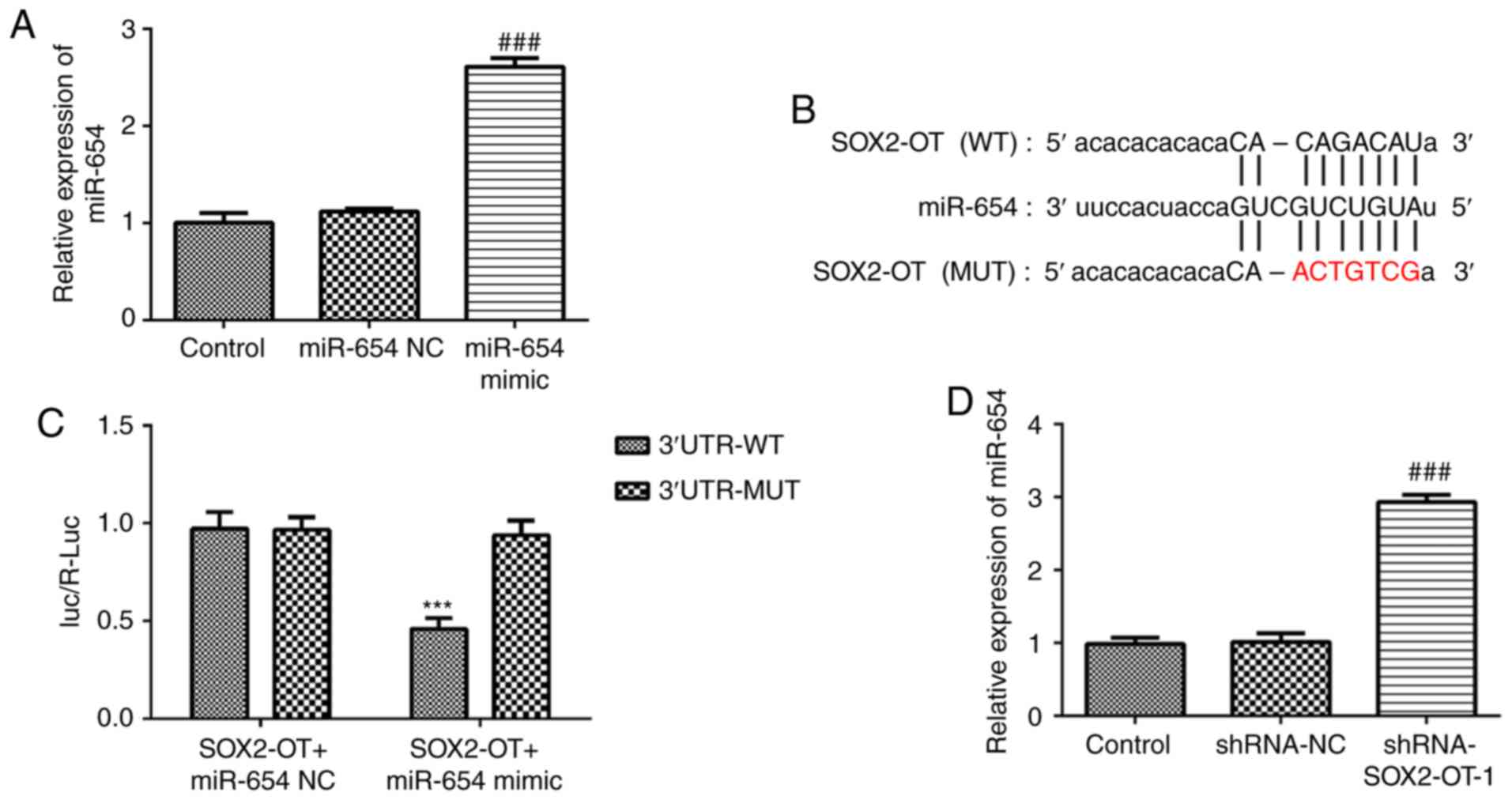
miR-654 serves as a direct target of
SOX2-OT
To assess the role of miR-654 in TU-177 cells, we
constructed miR-654 mimic and found that miR-654 expression in
TU-177 cells was significantly increased after transfection with
miR-654 mimic (Fig. 5A). As
presented in Fig. 5B and C, treatment with miR-654 mimic markedly
decreased the luciferase activity in pGL3-SOX2-OT-WT-transfected
cells. However, no significant effect was identified for the cells
transfected with pGL3-SOX2-OT-MUT. Furthermore, RT-qPCR results
demonstrated that SOX2-OT-silencing promoted the expression level
of miR-654 in TU-177 cells when compared with the control,
suggesting that miR-654 is a direct target of SOX2-OT (Fig. 5D).
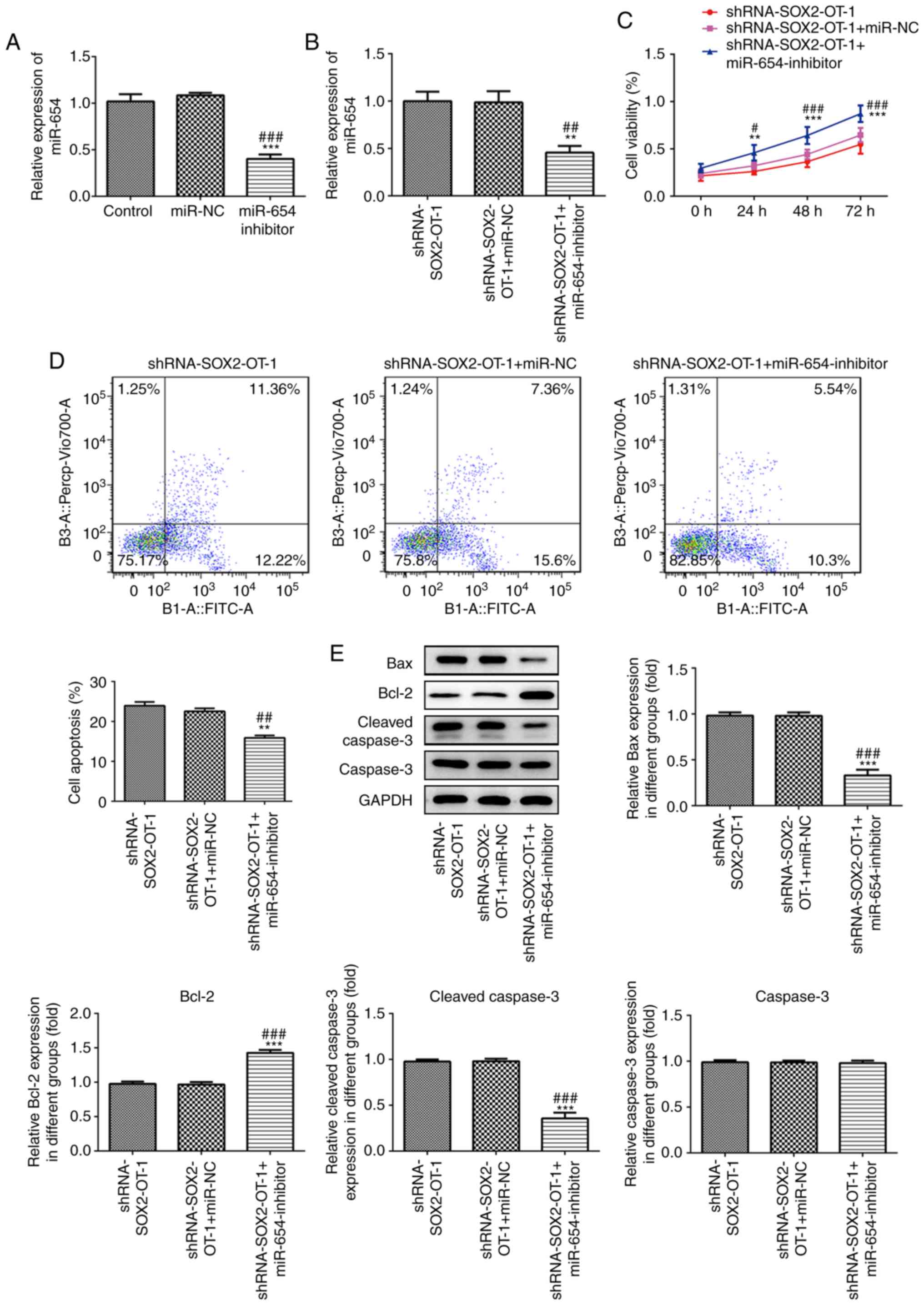
miR-654 is responsible for the effects
of SOX2-OT-silencing in TU-177 cells
To further investigate the role of miR-654 in
SOX2-OT-silenced laryngeal cancer cells, miR-654 inhibitor was
transfected to TU-177 cells and the effect was detected (Fig. 6A). As presented in Fig. 6B, the expression level of miR-654 was
significantly reduced in miR-654 inhibitor-transfected TU-177
cells. Moreover, the results of CCK-8 assay demonstrated that
downregulation of miR-654 significantly promoted cell proliferation
at 24, 48 and 72 h post-transfection (Fig. 6C). Additionally, downregulation of
miR-654 markedly inhibited the apoptosis induced by
SOX2-OT-knockdown in TU-177 cells (Fig.
6D). In addition, miR-654 silencing significantly reduced the
expression of Bax and cleaved caspase 3 while increased Bcl-2 level
compared with the shRNA-SOX2-OT-1-transfected TU-177 cells
(Fig. 6E).
Discussion
lncRNAs are involved in numerous physiological
processes, including cell growth, proliferation, apoptosis,
migration and differentiation (17-19).
The normal expression of lncRNA is necessary to maintain cell
homeostasis and function, and its aberrant expression is closely
associated with a variety of diseases, including cancer (20,21). The
present study reported that the expression level of SOX2-OT was
significantly increased in the human laryngeal cancer cell lines
TU-177, M4E, AMC-HN-8 and TU686 compared with the normal
nasopharyngeal epithelial cell line NP69. Consistent with the
present results, Feng et al (12) and Tai et al (13) revealed that the level of SOX2-OT is
markedly upregulated in laryngeal cancer tissues compared with the
adjacent tissues. These results suggest that SOX2-OT may play an
important role in the occurrence and development of laryngeal
carcinoma. In order to further investigate the potential role of
SOX2-OT in laryngeal cancer and to reveal the underlying molecular
mechanisms, shRNA-SOX2-OT-1 was transfected into TU-177 cells in
the present study. It was identified that SOX2-OT-knockdown
significantly suppressed cell proliferation, migration and
invasion, and induced apoptosis of TU-177 cells. Similarly, Tai
et al (13) suggested that
overexpression of SOX2-OT facilitates the tumorigenicity of
laryngeal squamous cell carcinoma Hep-2 cells), which indicates
that SOX2-OT is involved in the development of laryngeal cancer as
an oncogenic factor. Moreover, the present results demonstrated
that miR-654 serves as a direct target of SOX2-OT, and
downregulation of miR-654 reversed the effects of SOX2-OT-knockdown
in TU-177 cells, suggesting that SOX2-OT may facilitate laryngeal
cancer development through regulating miR-654.
A study involving oral squamous cell carcinoma
revealed that miR-654 expression is correlated with poor prognosis
of patients with OSCC, and miR-654-5p promotes the development of
OSCC via the GRAP/Ras/Erk signaling pathway (22). However, another study in glioma
reported that an upregulation of miR-654 markedly inhibits the
proliferation and invasion of the glioma cells U87 and U251 via the
IGF2BP3 signaling pathway (23).
These results suggest that miR-654 exhibits differences in
expression and function in different types of cancer. The present
results suggested that miR-654 serves as a tumor-suppressive
factor; downregulation of miR-654 significantly promoted cell
proliferation and inhibited apoptosis of TU-177 cells. However,
further studies are needed to explore the precise mechanism by
which miR-654 functions.
Mechanistically, the present study detected the
expression of key regulators of cell proliferation, migration,
invasion and apoptosis. It has been reported that p21 CDK2, cyclin
E1and PCNA are strongly implicated in cell proliferation (24,25). The
current results demonstrated that SOX2-OT-silencing significantly
upregulated the expression of p21 and downregulated the expression
of CDK2, cyclin E1and PCNA. Furthermore, Bcl-2 family members play
crucial roles in cell apoptosis by mediating the chronological
activation of caspases (26,27). SOX2-OT-knockdown markedly reduced the
expression of Bcl-2 protein, and increased the protein levels of
Bax and cleaved caspase 3. In addition, it has been reported that
increased expression of matrix metalloproteinases, particularly
MMP-7 and MMP-9, is associated with occurrence and development of
laryngeal carcinoma malignization (28-30).
The present data suggested that the expression levels of MMP-7 and
MMP-9 were markedly reduced by SOX2-OT-silencing. These results
suggest that the biological function of SOX2-OT is involved in the
regulation of multiple factors.
However, this study has several limitations. On one
hand, we detected the expression of SOX2-OT in several laryngeal
cancer cell lines but we investigated the potential roles of
SOX2-OT in only TU-177 cell lines in subsequent experiments. On the
other hand, all the experiments in this study were performed at the
cellular level thus we only proved the effects of SOX2-OT in
laryngeal cancer cells instead of animals or humans. Additionally,
10% FBS was supplemented to TU-177 cells in wound healing assay for
more confluence rate. In fact, >5% FBS might significantly
increase proliferative activity and it's hard to distinguish the
results caused by proliferation or migration. To distinguish
proliferation or migration is needed in the future research and the
roles of SOX2-OT should be observed in more several OSCC cell lines
and in animal experiments.
In conclusion, the present study identified SOX2-OT
as a crucial regulator in the development of laryngeal cancer by
targeting, at least partially, miR-654. The expression of SOX2-OT
was significantly upregulated in laryngeal cancer cell lines, and
silencing of SOX2-OT markedly suppressed cell proliferation,
migration and invasion, and induced apoptosis in TU-177 cells.
Therefore, SOX2-OT may serve as a potential therapeutic target in
the treatment of laryngeal cancer.
Acknowledgements
Not applicable.
Funding
This work was supported by Anhui Natural Science
Foundation (Grant no. 1808085QH248); the National Natural Sciences
Foundation of China (Grant no. 81800911); Fundamental Research
Funds for the Central Universities (Grant no. WK9110000053).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
GL and JWS designed the study, wrote and revised the
manuscript. GL, CP and JQS performed the experiments. GW analyzed
the data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Steuer CE, El-Deiry M, Parks JR, Higgins
KA and Saba NF: An update on larynx cancer. CA Cancer J Clin.
67:31–50. 2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Ferlito A, Devaney KO, Hunt JL and
Hellquist H: Some considerations on the WHO histological
classification of laryngeal neoplasms. Adv Ther. 36:1511–1517.
2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Wu CW, Dionigi G, Barczynski M, Chiang FY,
Dralle H, Schneider R, Al-Quaryshi Z, Angelos P, Brauckhoff K,
Brooks JA, et al: International neuromonitoring study group
guidelines 2018: Part II: Optimal recurrent laryngeal nerve
management for invasive thyroid cancer-incorporation of surgical,
laryngeal, and neural electrophysiologic data. Laryngoscope. 128
(Suppl 3):S18–S27. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Calkovsky V, Wallenfels P, Calkovska A and
Hajtman A: Laryngeal cancer: 12-year experience of a single center.
Adv Exp Med Biol. 911:9–16. 2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Laccourreye O, Bonfils P, Malinvaud D,
Ménard M and Giraud P: Survival and laryngeal preservation tradeoff
in advanced laryngeal cancer: From the otorhinolaryngology patient
to the managing physician. Head Neck. 39:1984–1989. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Yang G, Lu X and Yuan L: lncRNA: A link
between RNA and cancer. Biochim Biophys Acta. 1839:1097–1109.
2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Sang H, Liu H, Xiong P and Zhu M: Long
non-coding RNA functions in lung cancer. Tumour Biol. 36:4027–4037.
2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Teng Y, Kang H and Chu Y: Identification
of an exosomal long noncoding RNA SOX2-OT in plasma as a promising
biomarker for lung squamous cell carcinoma. Genet Test Mol
Biomarkers. 23:235–240. 2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Wang Z, Tan M, Chen G, Li Z and Lu X:
lncRNA SOX2-OT is a novel prognostic biomarker for osteosarcoma
patients and regulates osteosarcoma cells proliferation and
motility through modulating SOX2. IUBMB Life. 69:867–876.
2017.PubMed/NCBI View
Article : Google Scholar
|
|
11
|
Wei CX, Wong H, Xu F, Liu Z, Ran L and
Jiang RD: IRF4-induced upregulation of lncRNA SOX2-OT promotes cell
proliferation and metastasis in cholangiocarcinoma by regulating
SOX2 and PI3K/AKT signaling. Eur Rev Med Pharmacol Sci.
22:8169–8178. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Feng L, Wang R, Lian M, Ma H, He N, Liu H,
Wang H and Fang J: Integrated analysis of long noncoding RNA and
mRNA expression profile in advanced laryngeal squamous cell
carcinoma. PLoS One. 11(e0169232)2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Tai Y, Ji Y, Liu F, Zang Y, Xu D, Ma S,
Qin L and Ma J: Long noncoding RNA SOX2-OT facilitates laryngeal
squamous cell carcinoma development by epigenetically inhibiting
PTEN via methyltransferase EZH2. IUBMB Life. 71:1230–1239.
2019.PubMed/NCBI View
Article : Google Scholar
|
|
14
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714.
2009.PubMed/NCBI View
Article : Google Scholar
|
|
15
|
Zhang N, Lu C and Chen L: miR-217
regulates tumor growth and apoptosis by targeting the MAPK
signaling pathway in colorectal cancer. Oncol Lett. 12:4589–4597.
2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Cybula M, Wieteska L-,
Józefowicz-Korczyńska M, Karbownik MS, Grzelczyk WL and Szemraj J:
New miRNA expression abnormalities in laryngeal squamous cell
carcinoma. Cancer Biomark. 16:559–568. 2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wang FY, Jia J, Song HH, Jia CM, Chen CB
and Ma J: Icariin protects vascular endothelial cells from
oxidative stress through inhibiting endoplasmic reticulum stress. J
Integr Med. 17:205–212. 2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Shields EJ, Petracovici AF and Bonasio R:
lncRedibly versatile: Biochemical and biological functions of long
noncoding RNAs. Biochem J. 476:1083–1104. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Xie ZY, Wang P, Wu YF and Shen HY: Long
non-coding RNA: The functional regulator of mesenchymal stem cells.
World J Stem Cells. 11:167–179. 2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Xu J, Bai J, Zhang X, Lv Y, Gong Y, Liu L,
Zhao H, Yu F, Ping Y, Zhang G, et al: A comprehensive overview of
lncRNA annotation resources. Brief Bioinform. 18:236–249.
2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Sparber P, Filatova A, Khantemirova M and
Skoblov M: The role of long non-coding RNAs in the pathogenesis of
hereditary diseases. BMC Med Genomics. 12 (Suppl
2):S422019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Peng WX, Koirala P and Mo YY:
lncRNA-mediated regulation of cell signaling in cancer. Oncogene.
36:5661–5667. 2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Lu M, Wang C, Chen W, Mao C and Wang J:
miR-654-5p targets GRAP to promote proliferation, metastasis, and
chemoresistance of oral squamous cell carcinoma through Ras/MAPK
signaling. DNA Cell Biol. 37:381–388. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Jin P, Huang Y, Zhu P, Zou Y, Shao T and
Wang O: CircRNA circHIPK3 serves as a prognostic marker to promote
glioma progression by regulating miR-654/IGF2BP3 signaling. Biochem
Biophys Res Commun. 503:1570–1574. 2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Dhulipala VC, Welshons WV and Reddy CS:
Cell cycle proteins in normal and chemically induced abnormal
secondary palate development: A review. Hum Exp Toxicol.
25:675–682. 2006.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zatonski T, Ciesielska U, Nowinska K,
Ratajczak-Wielgomas K, Kobierzycki C, Pula B, Podhorska-Okolow M,
Krecicki T and Dziegiel P: Expression of cell cycle-related
proteins p16, p27, p53 and Ki-67 in HPV-positive and -negative
samples of papillomas of the upper respiratory tract. Anticancer
Res. 36:3917–3924. 2016.PubMed/NCBI
|
|
27
|
Lee EF, Harris TJ, Tran S, Evangelista M,
Arulananda S, John T, Ramnac C, Hobbs C, Zhu H, Gunasingh G, et al:
BCL-XL and MCL-1 are the key BCL-2 family proteins in melanoma cell
survival. Cell Death Dis. 10(342)2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Chrysovergis A, S Papanikolaou V, Tsiambas
E, Kikidis D, Maragoudakis P, Ragos V and Kyrodimos E: Caspase
complex in laryngeal squamous cell carcinoma. J BUON. 24:1–4.
2019.PubMed/NCBI
|
|
29
|
Lotfi A, Mohammadi G, Saniee L,
Mousaviagdas M, Chavoshi H and Tavassoli A: Serum level of matrix
metalloproteinase-2 and -9 in patients with laryngeal squamous cell
carcinoma and clinical significance. Asian Pac J Cancer Prev.
16:6749–6751. 2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Uloza V, Liutkevičius V, Pangonytė D,
Saferis V and Lesauskaitė V: Expression of matrix
metalloproteinases (MMP-2 and MMP-9) in recurrent respiratory
papillomas and laryngeal carcinoma: Clinical and morphological
parallels. Eur Arch Otorhinolaryngol. 268:871–878. 2011.PubMed/NCBI View Article : Google Scholar
|