Introduction
Oral cancer is one of the most common solid cancers
and an estimated 300,000 new cases were diagnosed and 145,000
related deaths were caused worldwide in 2012(1). Among the different types of oral
cancer, one of the most common malignancies that poses a threat to
human health and life is tongue squamous cell carcinoma (TSCC),
accounting for approximately 25-40% of cases of oral cancer
(2-4).
TSCC is characterized by high malignancy, invasive growth and early
lymph node metastasis (5). Current
therapeutic strategies for TSCC include traditional surgical
resection, radiotherapy and chemotherapy. Despite advances in these
treatments, there has been no significant improvement in the
prognosis and survival rate of patients with TSCC over the past 10
years, due to lymphatic metastasis, mortality and recurrence
(6,7). The rates of incidence and death remain
high (8,9) and the overall 5-year survival rate is
less than 60% (10,11). In addition, traditional treatments
may harm the functioning of surrounding organs leading to a decline
in patient quality of life, due to potential difficulties in
chewing, swallowing and speech (5).
In-depth research is urgently required to determine the precise
molecular mechanism underlying TSCC tumorigenesis, which may help
to more accurately locate and effectively inhibit cancer cell
proliferation during treatment, potentially an effective and
practical strategy to improve clinical outcomes. Targeted
therapeutic approaches are being developed to improve clinical
outcomes based on the identification of potential new biomarkers
and therapeutic targets for cancer (12-15).
The ectodysplasin-A receptor-associated adaptor
protein (EDARADD) gene encodes a protein containing a death
domain that interacts with the ectodysplasin-A receptor (EDAR),
which is part of the tumor necrosis factor (TNF) receptor family.
EDARADD was originally reported to be primarily associated with
anhidrotic ectodermal dysplasia, an ectodermal differentiation
disorder involving aberrant development of the exocrine sweat
glands, teeth and hair (16,17). Related studies have found that
EDARADD is not only associated with the occurrence of dental
caries, but also affects the development of tooth morphology and
tooth quantity (18,19). In the present study, EDARADD was
identified to be clinically relevant to head and neck squamous cell
carcinoma (HNSCC) through The Cancer Genome Atlas (TCGA) data
portal analysis. To assess the clinical significance of EDARADD in
TSCC and its potential molecular mechanisms, EDARADD was knocked
down in in vitro studies using interference technology to
identify whether EDARADD affects the tumorigenicity of TSCC
cells.
Materials and methods
Gene information
UALCAN database is a publicly available interactive
online portal used to perform in-depth analyses of TCGA gene
expression data (20), (http://ualcan.path.uab.edu/), TCGA analysis was used
to search for the expression of the EDARADD gene in the HNSCC and
generate a box-line diagram of EDARADD expression levels in 40
normal individuals and 520 patients with HNSCC.
Patients and tissue specimens
Specimens and histologically normal peri-tumor
tissues that had been collected from patients operated on for
primary TSCC between January 2016 and December 2018 at the
Department of Oral Medicine, Central Hospital of Xuzhou, The Xuzhou
Clinical College of Xuzhou Medical University (Xuzhou, China) were
analyzed in the current study after obtaining informed written
consent from each patient. The tissue samples were immediately
frozen in liquid nitrogen for further use. The present study was
approved by the Ethics Committee of the Central Hospital of Xuzhou,
The Xuzhou Clinical College of Xuzhou Medical University (reference
no. 2009XL002) and was conducted according to The Declaration of
Helsinki. A total of 33 patients were enrolled in the present study
(age, 28-87 years; mean age, 56.9 years), 22 (66.7%) were men and
11 (33.3%) were women. The tumor samples were evaluated according
to the World Health Organization stage and grading system (21).
Immunohistochemistry (IHC)
Paraffin-embedded sections of the tissue samples
(4-µm-thick) were deparaffinized and rehydrated before antigen
retrieval was performed in citrate buffer (pH 6.0; cat. no. G1202;
Wuhan Servicebio Technology Co., Ltd) for 10 min at 100˚C.
Thereafter, the preparations were incubated with 3%
H2O2 and 3% BSA (cat. no. G5001; Wuhan
Servicebio Technology Co., Ltd.) for 20 min at 25˚C, in order to
block endogenous peroxidase activity. The sections were then
incubated with primary rabbit anti-human monoclonal EDARADD
antibody overnight at 4˚C (1:200; cat. no. D123818; Sangon Biotech
(Shanghai) Co., Ltd.), followed by incubation with a horseradish
peroxidase-labeled goat anti-rabbit secondary antibody for 30 min
at 37˚C (1:200; cat. no. GB23204; Wuhan Servicebio Technology Co.,
Ltd.). The slides were then incubated with diaminobenzidine (cat.
no. G1211; Wuhan Servicebio Technology Co., Ltd.) for 20 min at
25˚C and finally counterstained with hematoxylin for 3 min at 25˚C
(cat. no. G1004; Wuhan Servicebio Technology Co., Ltd.). As a
negative control, PBS was used instead of the primary antibody for
one slide. All the stained slides were examined by two independent
pathologists, who were blinded to patient clinical information. A
total of three slices of each sample of TSCC tissues and adjacent
tissues were selected and five fields were observed under the
microscope (Olympus BX53; Olympus Corporation). The positive
staining area was processed quantitatively using Image-Pro Plus
Version 6.0 software (Media Cybernetics, Inc.). The immunostaining
intensity was classified into four categories: A score of 3
indicated strong staining, 2 indicated moderate staining, 1
indicated weak staining and 0 indicated no staining. Finally, the
data were divided into two categories: Low expression (weak or no
immunoreactivity) and high expression (strong or moderate
immunoreactivity).
Cell line and culture conditions
293T cells and the human TSCC cell lines CAL27,
SCC25 and SCC9 were used in this study. The cell lines were
purchased from the The Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences. The CAL27, SCC25, SCC9 and 293T cells
were grown in DMEM (Corning, Inc.) containing 10% FBS (Gibco;
Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin
(Gibco; Thermo Fisher Scientific, Inc.). 293T cells were grown in
DMEM without FBS 2 h prior to transduction. The cells were
maintained at 37˚C in a humidified incubator at 5% CO2
and 95% air (Sanyo; Panasonic Corporation).
Short hairpin (sh)RNA lentivirus
infection
shRNA for the inhibition of EDARADD expression was
purchased from Shanghai GeneChem Co., Ltd. The interference target
sequence of the EDARADD shRNA was 5'-GTACTTGTTCCTCCTGCTT-3'
(shEDARADD) and the sequence used for the negative control (shCtrl)
was 5'-TTCTCCGAACGTGTCACGT-3'. Lentiviral titers were determined by
reverse transcription-quantitative PCR (RT-qPCR; as described
below) and transfected into CAL27 cells using a lentiviral vector
(GV 112; Shanghai GeneChem Co., Ltd) with a multiplicity of
infection of 20. The cells were inoculated in six-well plates with
2 ml cell suspension at a density of 2x105 cells/ml and
incubated overnight; the spent medium was replaced with fresh
medium to continue the culture. After 72 h of incubation, the cells
were observed under a fluorescence microscope (Olympus Corporation)
and the successfully-infected cells were positive for green
fluorescent protein. The interference efficiency of the EDARADD
shRNA was measured by RT-qPCR and western blotting (as described
below).
Western blotting
Total protein was collected from the TSCC cell lines
using cell lysis buffer (P0013B; Beyotime Institute of
Biotechnology), and the protein concentration was determined using
a BCA protein detection kit (P0010S; Beyotime Institute of
Biotechnology). A total of 30 µg of each protein sample was
separated on an SDS-PAGE (10% gel) and transferred onto a PVDF
membrane at an electric current of 300 mA for 2.5 h. The membrane
was blocked for 1 h at 25˚C using 5% non-fat dry milk in TBST with
0.05% tween20 and incubated overnight at 4˚C with diluted primary
rabbit anti-human monoclonal antibodies against EDARADD (cat. no.
orb183301; 1:100; Biorbyt Ltd.), MYC (cat. no. ab32072; 1:1,000;
Abcam) and NF-κBp65 (cat. no. 8242; 1:500; Cell Signaling
Technology, Inc.) and primary mouse anti-human monoclonal
antibodies against Bcl-2 (cat. no. ab692; 1:500; Abcam) and GAPDH
(cat. no. sc-32233; 1:2,000; Santa Cruz Biotechnology, Inc.). After
washing three times with PBS, the membrane was incubated with
secondary anti-mouse IgG antibody (cat. no. 7076; 1:2,000; Cell
Signaling Technology, Inc.) or anti-rabbit IgG antibody (cat. no.
7074; 1:2,000; Cell Signaling Technology, Inc.) at room temperature
for 1.5 h. The protein levels were visualized with Pierce™ ECL
reagent (Thermo Fisher Scientific, Inc.) and developed onto X-ray
film.
RNA extraction and RT-qPCR
Total RNA was isolated from the TSCC cell lines and
purified using SuPerfecTRI™ reagent (Shanghai Pufei Biotechnology
Co., Ltd.) according to the manufacturer's instructions. The mRNA
fragments were used to generate cDNAs using M-MLV reverse
transcriptase (Promega Corp.) for 60 min at 37˚C according to the
manufacturer's protocol. The abundance of the EDARADD mRNA in the
CAL27, SCC25 and SCC9 cells was then detected by qPCR using the
Maxima SYBR Green qPCR Master Mix (Takara Bio, Inc.) by a two-step
method. The cycling conditions were as follows: 95˚C for 10 min,
then 40 cycles at 95˚C for 15 sec and 60˚C for 60 sec. The reverse
transcription primer and microRNA PCR primers were purchased from
Guangzhou Ruibo Biotechnology Co., Ltd. The relative change in the
mRNA expression levels was calculated by the comparative threshold
cycle method (2-ΔΔCq) (22) using the GAPDH as an internal
reference gene control. The experiment was repeated three times.
The sequences of the primers used are as follows: EDARADD forward,
5'-GACCAACCCAAAGAGGACAG-3' and reverse, 5'-CCAGAATGATGAGGCACCAT-3';
GAPDH forward, 5'-TGACTTCAACAGCGACACCCA-3' and reverse,
5'-CACCCTGTTGCTGTAGCCAAA-3'.
Cell counting assay
The TSCC cells (2x103 cells/well) were
plated in 96-well plates and cultured in 5% CO2 at 37˚C
for 24 h. After the incubation period, a Celigo cytometer (Beckman
Coulter, Inc.) was used to read the plates once a day for 5
consecutive days. The number of cells with cell viability was
accurately calculated and statistically analyzed based on the
quantity of green fluorescence protein. Using the cell count-fold
value, which indicated the cell count at each time point relative
to the mean of day 1, cell growth curves were drawn to represent
changes in cell proliferation.
MTT assay
The lentivirus-transduced TSCC cell lines were
incubated in 96-well plates at a density of 1,500 cells/well for 24
h. The cells were then cultured with 20 µl (5 mg/ml) MTT (Gen-view
Scientific, Inc.) at 37˚C and 5% CO2 for 4 h. After the
incubation period, the medium was removed and replaced with 100 µl
of dimethyl sulfoxide for 15 min at room temperature. The
absorbance was measured on a microplate reader (Tecan Group Ltd.)
at 490 nm. The assay was performed in triplicate.
Annexin V-allophycocyanin (APC)
apoptosis assay
A total of 2x105 cells/ml were cultured
in a 6-well plate after 3 days of transduction. The cell fusion
degree was detected to be ~85% on the 5th day following
transduction. The cells were trypsinized and centrifuged at 4˚C for
5 min at 300 x g, washed twice with D-Hanks solution (pH=7.2-7.4)
and once with the 1X Binding Buffer, then resuspended in 1X Binding
Buffer at a density of 1x106 cell/ml. Next, the 100 µl
of the cell suspension were incubated at 25˚C for 15 min in the
dark with the 5 µl of eBioscience™ Annexin V Apoptosis Detection
Kit APC (Thermo Fisher Scientific, Inc.) Finally, the samples were
analyzed in a flow cytometer using Guava easyCyte HT Version 8
system (EMD Millipore).
Colony formation assay
After 3 days of transfection, the cells were
disassociated and seeded in six-well plates at a density of
1.5x103 cells/well and cultured for ~10 days (medium
exchanged every 3 days). The cell clones were observed under a
fluorescence microscope. Next, the cells were washed once with PBS
before the end of the culture. They were then fixed for 30 min at
4˚C by adding 1 ml of 4% paraformaldehyde to each well, followed by
washing with PBS and staining with 1,000 µl Giemsa (Sigma-Aldrich;
Merck KGaA) for 15 min at 25˚C. The cells were then washed with
distilled deionized water several times and a digital camera was
used to capture images of the plates. The colonies were observed
with an inverted light microscope (Olympus BX53; Olympus
Corporation) and quantified using Image-Pro Plus Version 6.0
software (Media Cybernetics, Inc.). The assay was performed in
triplicate.
Statistical analysis
The results in this study are presented as the mean
± standard error of the mean from at least three experiments.
Student's t-test was used to analyze the data in two groups with
SPSS 21.0 software (IBM Corp). The clinical characteristics data
were analyzed by Fisher's exact test. P<0.05 was considered to
represent statistical significance.
Results
EDARADD is overexpressed in HNSCC
tissues
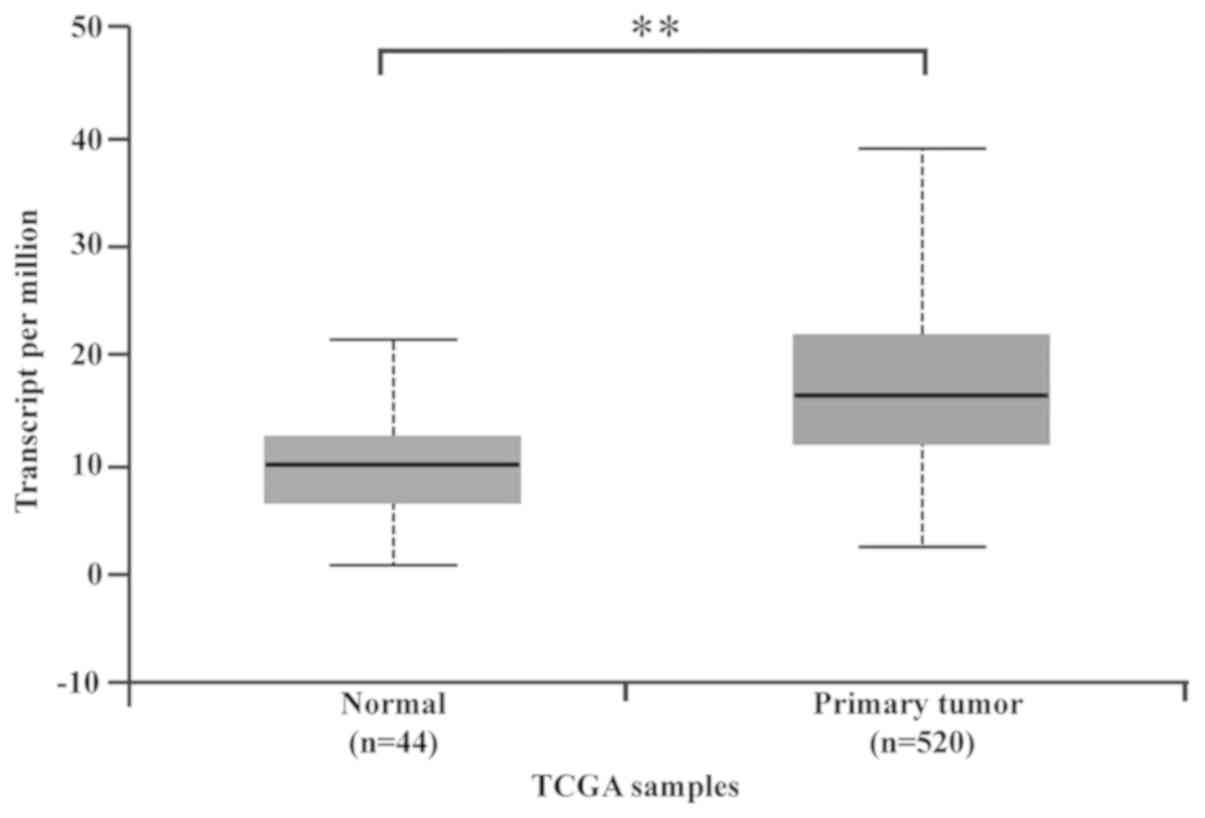
In the present study, EDARADD expression data were
obtained from the online UALCAN database. A box plot of EDARADD
expression showed that EDARADD was highly expressed in HNSCC
tissues compared to the levels of expression in non-tumor tissues
(P<0.01; Fig. 1).
EDARADD expression in TSCC
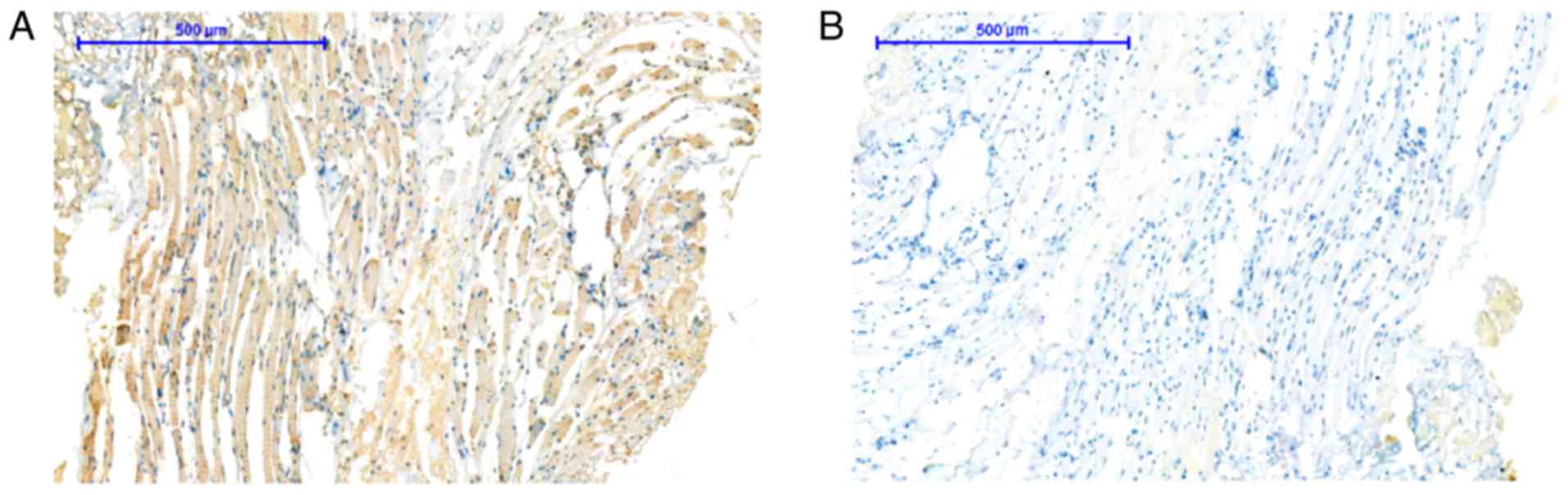
In the present study, the expression level of
EDARADD in TSCC tissues was markedly higher than that in normal
adjacent tissues (Fig. 2). The
results indicated that the expression level of EDARADD in the
normal adjacent tissues was 15.15% (5/33), while the expression
rate in TSCC was 60.61% (20/33). The difference was statistically
significant (P<0.001) and the results are shown in Table I.
 | Table IEDARADD expression in TSCC and
adjacent tissues. |
Table I
EDARADD expression in TSCC and
adjacent tissues.
| | | Expression of
EDARADD | |
|---|
| Characteristic | N | Low expression, n
(%) | High expression, n
(%) | P-value |
|---|
| TSCC tissue | 33 | 13 (39.39) | 20 (60.61) | <0.001 |
| Adjacent
tissue | 33 | 28 (84.85) | 5 (15.15) | |
Association between the expression of
EDARADD in tumor tissues and clinical and pathological parameters
of patients
The IHC results from tumor tissues were compared
with clinical data (sex and age) and pathological parameters (TNM
and clinical stages). The results showed that the expression levels
of EDARADD in TSCC were associated with the degree of tumor
differentiation (P=0.003) and local recurrence (P=0.027), as shown
in Table II. There were no
statistically significant differences in gender, age, TNM staging
and tumor size between patients with high or low levels of EDARADD
expression (P>0.05).
 | Table IIRelationship between the expression
of EDARADD and clinicopathological features of patients with
TSCC. |
Table II
Relationship between the expression
of EDARADD and clinicopathological features of patients with
TSCC.
| | | Expression of
EDARADD | |
|---|
| Characteristic | N (33) | Low expression, n
(%) | High expression, n
(%) | P-value |
|---|
| Sex | | | | 0.714 |
|
Male | 22 | 8 (36.36) | 14 (63.64) | |
|
Female | 11 | 5 (45.45) | 6 (54.55) | |
| Age (years) | | | | 0.485 |
|
≤60 | 13 | 4 (30.77) | 9 (69.23) | |
|
>60 | 20 | 9 (45.00) | 11 (55.00) | |
| Tumor size
(cm) | | | | 0.728 |
|
≤5 | 16 | 7 (43.75) | 9 (56.25) | |
|
>5 | 17 | 6 (35.29) | 11 (64.71) | |
| TNM stage | | | | 0.263 |
|
I+II | 24 | 11 (45.83) | 13 (54.17) | |
|
III+IV | 9 | 2 (22.22) | 7 (77.78) | |
| Tumor grade | | | | 0.003 |
|
G1 | 14 | 10 (71.43) | 4 (28.57) | |
|
G2/G3 | 19 | 3 (15.79) | 16 (84.21) | |
| Lymph node
metastasis | | | | 0.284 |
|
Yes | 18 | 9 (50.00) | 9 (50.00) | |
|
No | 15 | 4 (26.67) | 11 (73.33) | |
| Distant
metastasis | | | | 0.276 |
|
Yes | 13 | 7 (53.85) | 6 (46.15) | |
|
No | 20 | 6 (30.00) | 14 (70.00) | |
| Local
recurrence | | | | 0.027 |
|
Yes | 21 | 5 (23.81) | 16 (76.19) | |
|
No | 12 | 8 (66.67) | 4 (33.33) | |
EDARADD gene expression in TSCC cell
lines
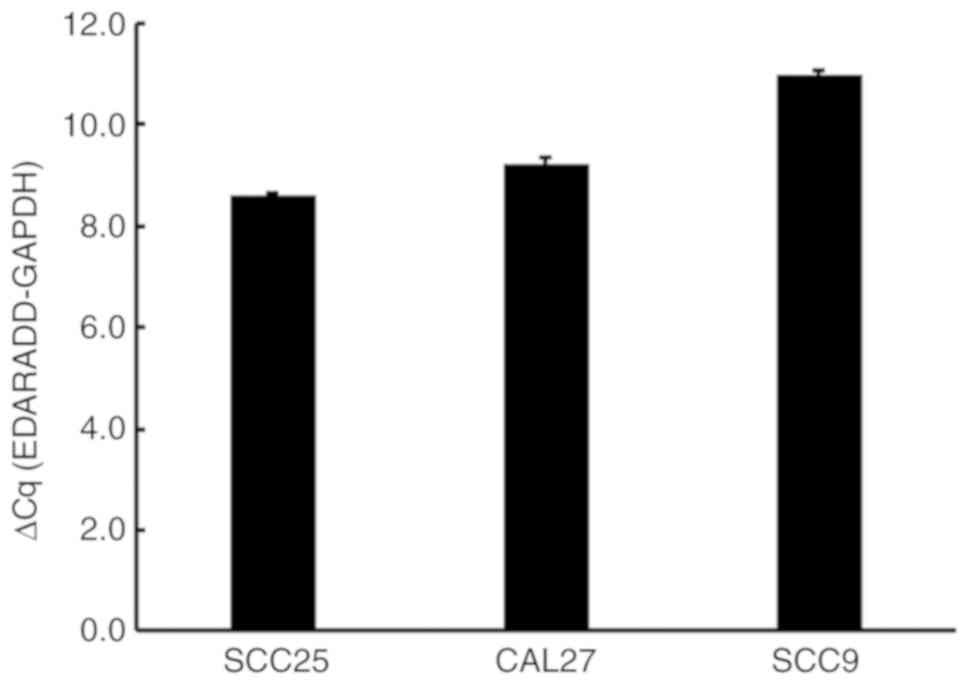
Using GAPDH as a standardized internal control, the
abundance of EDARADD mRNA in the CAL27, SCC25 and SCC9 cells was
evaluated by RT-qPCR (Fig. 3). The
results, (SCC25 cells, average ΔCt=8.58; CAL27 cells, average
ΔCt=9.21; SCC9 cells, average ΔCt=10.99), suggested there was
expression of EDARADD in all three cell lines.
EDARADD mRNA and protein expression in
TSCC cell lines
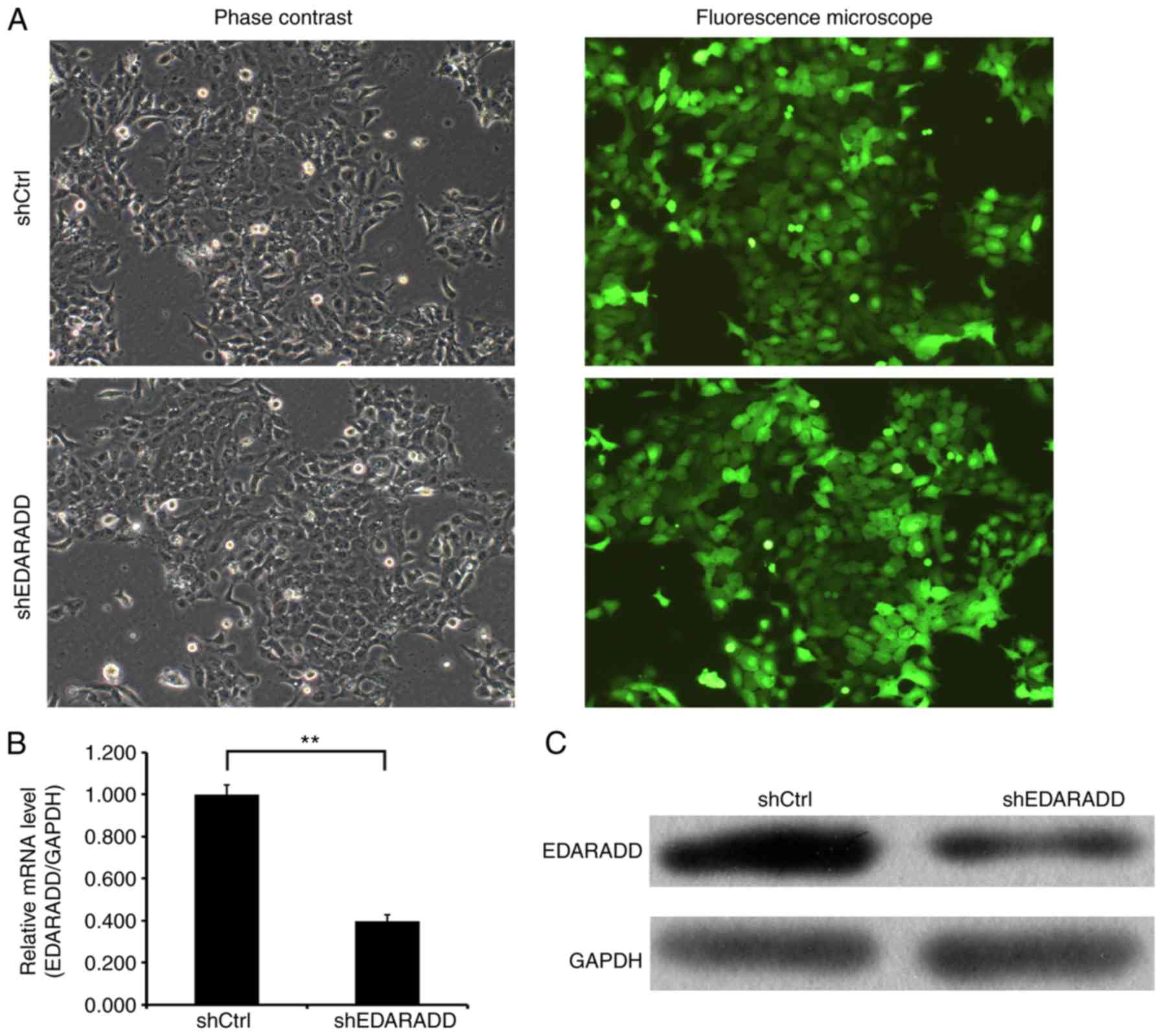
The stable knockdown of EDARADD expression in CAL27
cells was established to assess the potential effects of EDARADD.
In the present study, CAL27 cells were successfully infected with
the lentivirus expressing shEDARADD or shCtrl (Fig. 4A). Fluorescence microscopy (Olympus
IX71; Olympus Corporation) demonstrated that the rates of infection
and expression of green fluorescent protein were >80%. RT-qPCR
and western blotting were used to assess the interference
efficiency of shEDARADD. As indicated in Fig. 4B and C, there was an evident decrease in the
expression of EDARADD at both the mRNA (P<0.01) and protein
levels in cells transfected with shEDARADD compared with shCtrl.
These results indicated that the efficiency of EDARADD knockdown,
based on the mRNA expression levels, reached 60.4%, and the
endogenous expression of EDARADD gene was reduced at the protein
level.
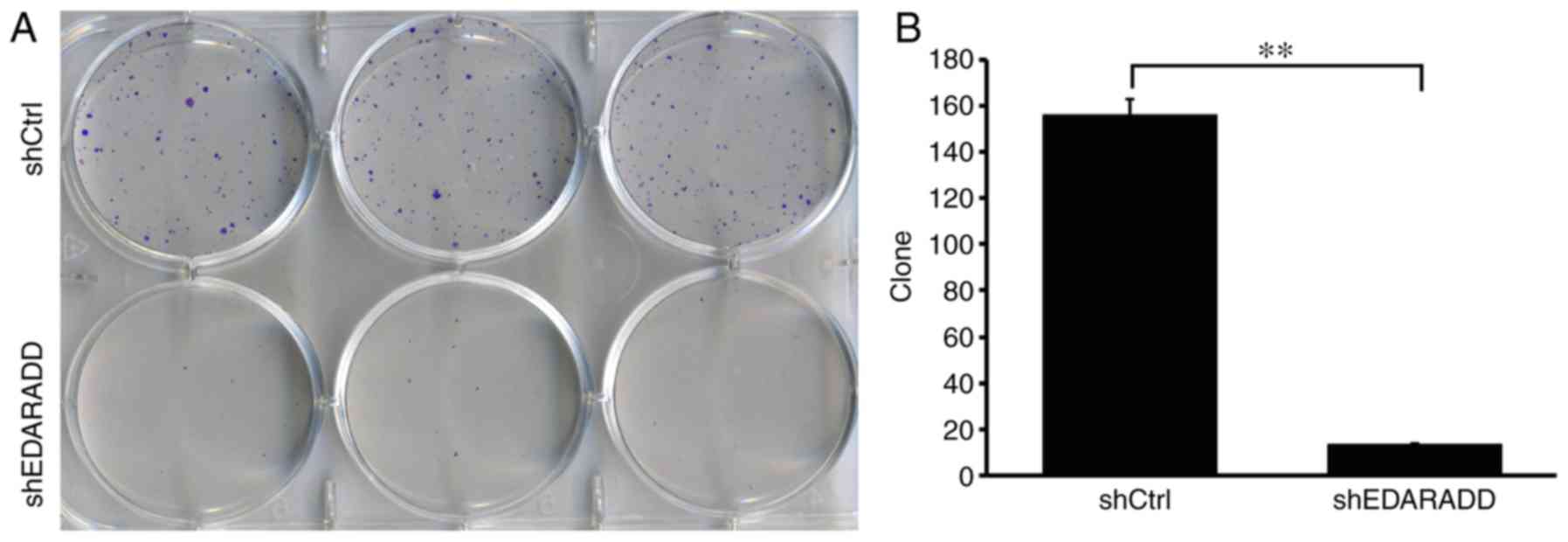
Knockdown of EDARADD affects the
cloning ability of TSCC cells
Following the knockdown of EDARADD expression in
CAL27 cells, cell counting assays were performed. The results
demonstrated that the number of CAL27 cell clones in the CAL27
culture infected with shEDARADD was significantly lower than that
in the shCtrl group (P<0.01; Fig.
5), implying that the EDARADD gene contributed to the the
cloning ability of the CAL27 cells.
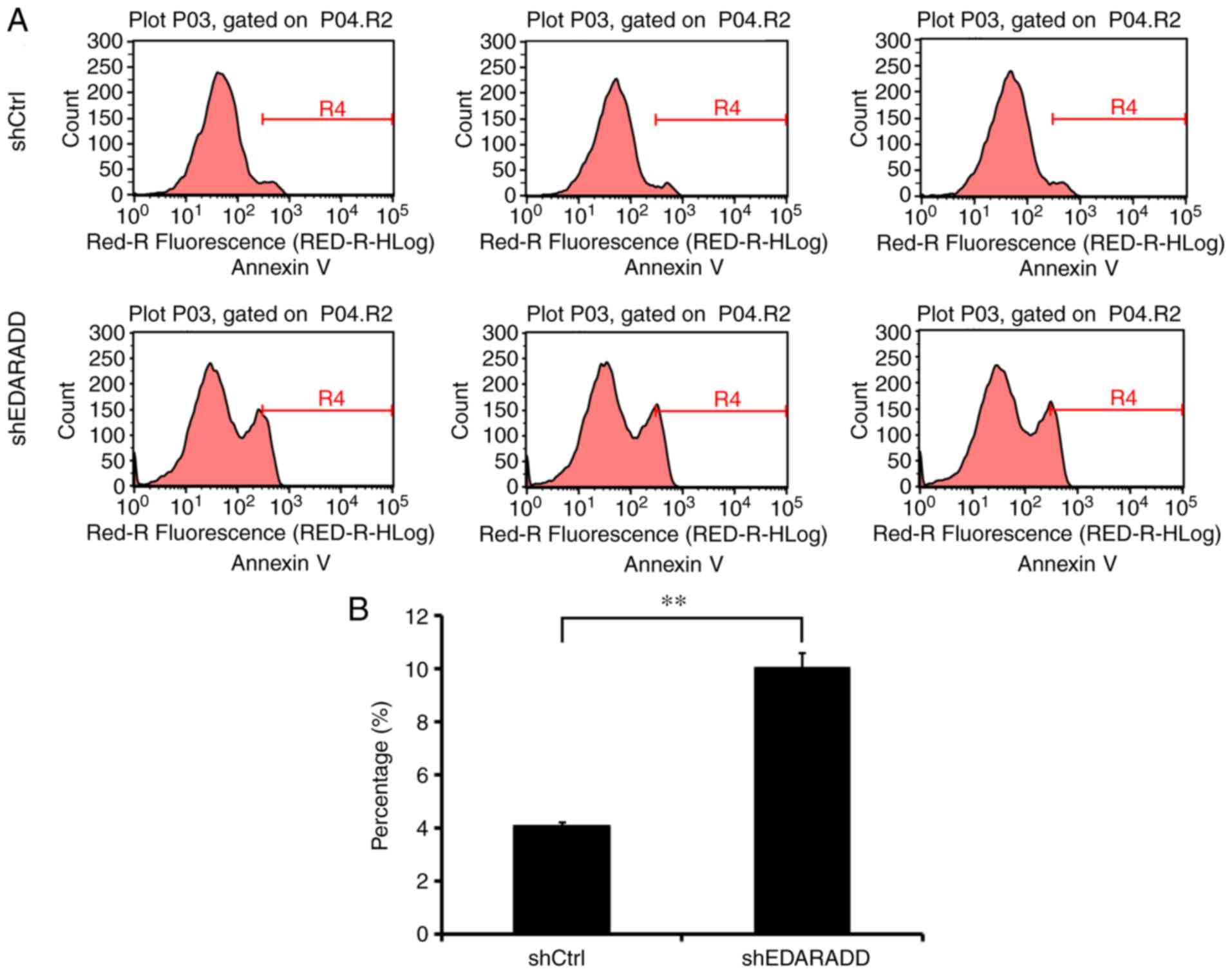
Knockdown of EDARADD induces apoptosis
in TSCC cell lines
To confirm whether EDARADD expression is associated
with the regulation of apoptosis in TSCC cells, flow cytometry
analysis was performed. The data indicated that shEDARADD
significantly enhanced the apoptotic ability of CAL27 cells when
compared with cells of the shCtrl group (P<0.01; Fig. 6), indicating that the EDARADD gene
may be involved in the regulation of apoptosis in CAL27 cells.
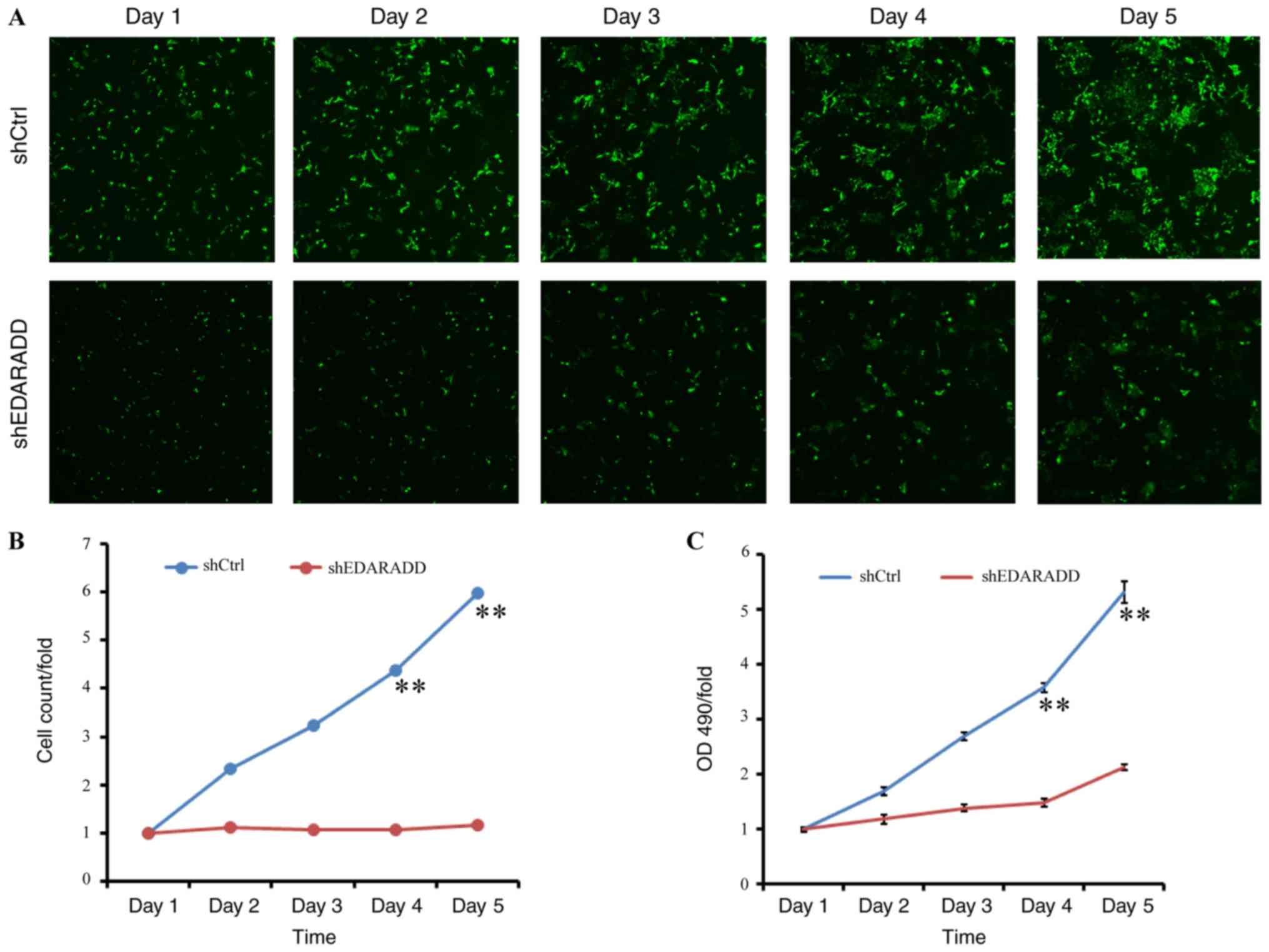
Knockdown of EDARADD suppresses the
proliferation of TSCC cells
To further elucidate the role of EDARADD in the
proliferation of TSCC cells, Celigo and MTT assays were performed
on CAL27 cells in the shEDARADD and shCtrl groups. The
cell-counting results showed that the rate of proliferation in the
shEDARADD group was markedly reduced compared to that in the shCtrl
group, especially on days 4 and 5 after transfection (P<0.01;
Fig. 7A and B). Similarly, the MTT assay indicated that
the OD 490-fold value of the shEDARADD group was markedly lower on
days 4 and 5 as compared to that in the shCtrl group (P<0.01;
Fig. 7C). These findings suggest
that knockdown of EDARADD may prevent the growth of TSCC cells
through inhibition of cell proliferation.
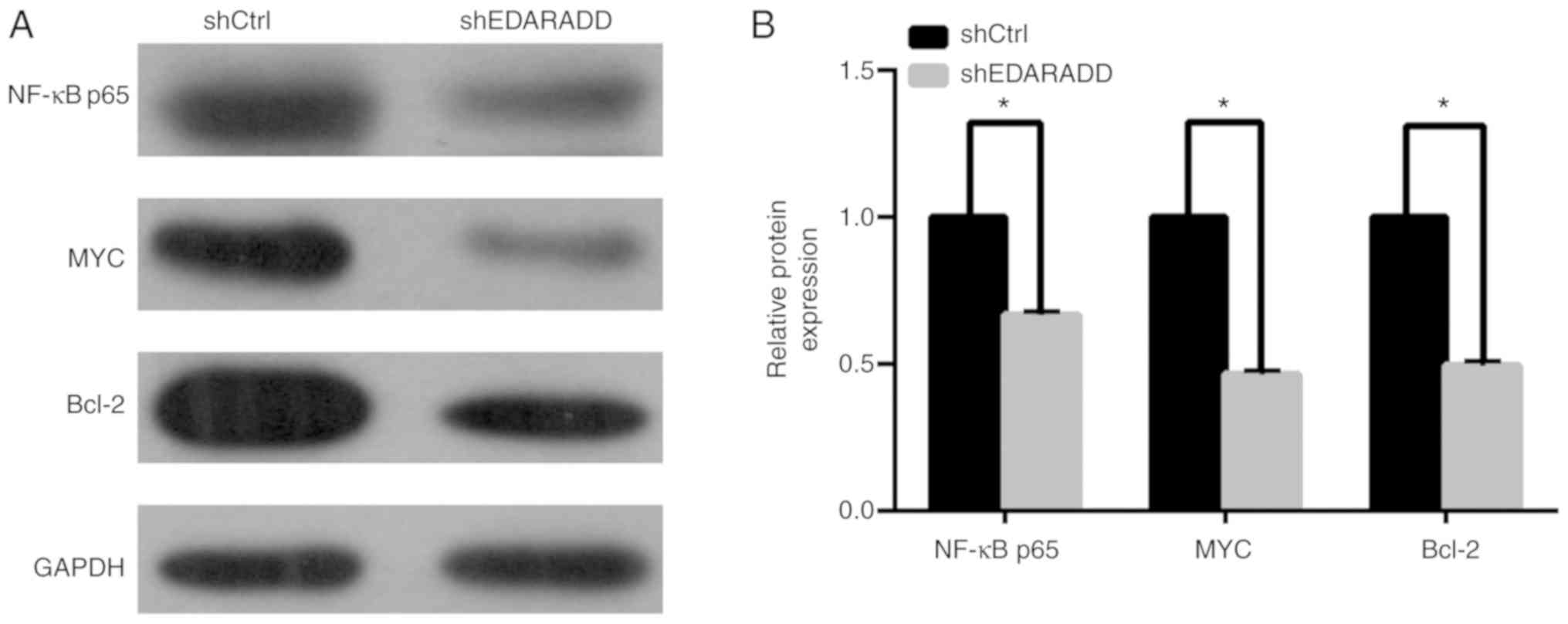
Knockdown of EDARADD inhibits the
expression of NF-κBp65, MYC and Bcl-2
To identify the molecular mechanism of EDARADD in
the regulation of proliferation and apoptosis of TSCC cells,
western blotting was performed to determine the expression of
NF-κBp65, MYC and Bcl-2. As illustrated in Fig. 8, the results suggested that knockdown
of the expression of EDARADD in TSCC cells reduced the protein
expression of NF-κBp65, MYC and Bcl-2 compared with the shCtrl
group (P<0.05).
Discussion
TSCC can be caused by a number of factors, such as
genetic alterations or environmental factors including tobacco use,
alcoholism, chronic inflammation and human papillomavirus infection
(23-25).
Recent reports have shown that the incidence of TSCC has increased
in young adults (<40 years old), especially in women, with
extremely high distant metastasis rate and poorer prognosis
(26,27). Therefore, different risk factors
leading to TSCC might have distinct molecular mechanism, including
the imbalance of cell proliferation and apoptosis (28-32),
and mucosal exophytic lesions (33).
To the best of our knowledge no previous studies have investigated
the biological function of EDARADD in TSCC and its underlying
molecular mechanisms.
In the present study, EDARADD expression was
identified in HNSCC using TCGA. TSCC is the most lethal and
high-incidence head neck squamous cell carcinoma worldwide
(34). Therefore, the expression of
EDARADD in TSCC was explored further. The expression of EDARADD in
TSCC tumor specimens was evidently higher than that in the adjacent
normal tissues. In addition, expression of EDARADD was observed in
TSCC cell lines. Subsequently, CAL27 cells were selected for shRNA
interference studies into the effect of EDARADD on the biological
progression of TSCC. Previous studies (35,36)
indicated that a lentivirus could be used to effectively infect the
CAL27 cells, leading to significant downregulation of gene
expression at both the mRNA and protein levels, providing a basis
for discovering the novel role of EDARADD in TSCC. Inhibition of
EDARADD effectively enhanced apoptosis and curbed the proliferation
of a TSCC cell line, providing evidence for the conclusion that
EDARADD has a tumorigenic effect in TSCC.
Few studies have been published that relate to the
possible function of EDARADD in tumors. Kumar et al
(37) reported that the presence of
death structures and cell death was observed during EDAR expression
and later studies (38) indicated
that EDAR was associated with the development of melanoma, and
observed that overexpression of EDAR in cells was able to induce
cell death. Furthermore, upon ligand binding, EDAR triggers the
activation of NF-κB. EDAR expression leads to apoptosis through an
EDARADD and caspase-8 dependent signaling pathway, and hence, it
can be defined as a potential tumor suppressor.
EDARADD mutations are associated with the
pathogenesis of multiple human disorders (39-43).
In mammals, there are two EDARADD isoforms exhibiting different
functions encoded by the EDARADD gene, isoforms A and B. Although
they exhibit differences in their dynamics, they both activate the
NF-κB pathway (44). Numerous
studies (45-47)
have shown that NF-κB transcription factors are adapted to adjust
the expression of genes that control cell survival and
proliferation. Moreover, deviated activation of the NF-κB signaling
is associated with the propagation of many cancers (48,49).
NF-κB has a dual role in cancer, it not only participates in the
immune defense, which can target and eliminate transformed cells,
but is also activated in many types of cancer, contributing to the
dysregulation of gene expression and prevention of apoptosis and
hence, it is responsible for various tumor-promoting functions
(50,51).
NF-κB plays a widespread role in cellular
proliferation and acts as a mediator of apoptosis during
tumorigenesis, particularly the RelA (p65) subunit (52,53). A
recent study proposed that the upregulation of miR-451 selectively
downregulated the expression of NF-κBp65 to inhibit the
inflammation and proliferation of human glomerular mesangial cells
(54). Furthermore, a study reported
that penta-acetyl geniposide reduced the protein level of NF-κBp65,
which has been shown to play a potent pro-apoptotic role in
fibroblast-like synoviocytes in adjuvant-induced arthritis in in
vitro studies (55).
MYC, located downstream of NF-κB, can stimulate the
growth and proliferation of cells, while Bcl-2 family proteins are
well known as regulators of apoptosis (56,57). MYC
is related to cell growth arrest and proliferation, thereby
contributing to tumorigenesis (58).
A previous study showed that knocking down MYC resulted in a
reduction in the viability of colon cancer cells (59). In addition, a study indicated that
the expression of MYC target genes was inhibited in response to the
downregulation of Gcn5 histone acetyltransferase activity,
contributing to apoptosis of lymphoma cells (60). c-MYC was also found to be repressed
in response to long stress-induced non-coding transcript 5
knockdown, which inhibited tumor cell proliferation, thereby
stimulating apoptosis of cells (61).
Most anticancer agents assist apoptosis by
downregulating the expression of Bcl-2 proteins. For example,
triptolide promotes the sensitivity of pancreatic cancer cell line,
PANC-1, to gemcitabine by reducing the rates of proliferation and
promoting apoptosis, following attenuation of NF-κB,
phosphorylated-p65 and Bcl-2 levels (62). The downregulation of DEAD-box
helicase 5 inhibits the expression of Bcl-2 of the
NF-κBp65-inducible anti-apoptotic factors through TNF-α stimulation
(63). Additionally, Wang et
al (64) demonstrated that the
tanshinone analog TC7 markedly induced prostate cancer cell
apoptosis by reducing the expression of Bcl-2.
As with most cancer cells, TSCC cells are
characterized by imbalanced cell proliferation and apoptosis
(65). It has also been reported
that the pro-inflammatory transcription factor NF-κB has a
significant effect on the initiation and progression of TSCC. Zhao
et al (66) reported that
c-MYC is likely to drive TSCC progression while Huang et al
(67) demonstrated that Iroquois
homeobox gene 5 can promote the aggressiveness of TSCC cells by
activating the NF-κB signaling pathway, leading to an increase in
the nuclear p65 levels and IκBα degradation. In addition, grape
seed proanthocyanidin markedly repressed the activity of NF-κB and
promoted apoptosis of TSCC cells by regulating the stability and
activity of Bax and downgrading the Bcl-2 and Bcl-xL proteins, and
additionally suppressed the aggressiveness and metastasis of TSCC
cells by restraining the secretion of MMP-2 and MMP-9 in the cells
(68). A previous study reported
that isobavachalcone may inhibit the expression of the
apoptosis-related protein Bcl-2(69).
Based on these previous studies and the decreased
expression of Bcl-2, MYC and NF-κBp65 in the present study after
EDARADD knockdown, the possible mechanism of action of EDARADD in
TSCC can be hypothesized. It can be speculated that knockdown of
EDARADD plays a role in the growth and survival of TSCC
cells by affecting the NF-κB and relevant pathway genes. Though a
significant biological effect of EDARADD has been detected at the
cellular level in in vitro studies, further in vivo
experiments are necessary to confirm whether the EDARADD
gene is a genuine target in TSCC and to explore its underlying
molecular mechanism.
In summary, it is speculated that knockdown of
EDARADD may influence the expression of NF-κB and relevant pathway
genes and thereby suppress cellular proliferation along with tumor
cell apoptosis in TSCC. These findings suggest that EDARADD may
serve as a novel therapeutic target and latent candidate for TSCC
treatment.
Acknowledgements
Not applicable.
Funding
The present study was supported by the grants from
Jiangsu Health and Family Planning Commission Project of Scientific
Research (grant no. H2017080) and Xuzhou Science and Technology
Project (grant no. KC17196).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. The data generated by TCGA Research Network (http://cancergenome.nih.gov/) has been used for UALCAN
development (http://ualcan.path.uab.edu/).
Authors' contributions
JM was involved in project administration and
supervision, playing a guiding role in the entire experimental
research process. ML and JM contributed to the conception of the
investigation. ML and YTB carried out the experiments. KH and XDL
were responsible for collecting experimental data and analyzing
data and all authors explained and discussed the results. ML wrote
the manuscript and YTB revised it. YTB contributed to supplementary
experiments. All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The present study was approved by The Ethical
Committee of the Central Hospital of Xuzhou and the Xuzhou Clinical
College of Xuzhou Medical University (reference no. 2009XL002) and
written informed consent was obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Casiglia J and Woo SB: A comprehensive
review of oral cancer. Gen Dent. 49:72–82. 2001.PubMed/NCBI
|
|
3
|
Rosebush MS, Rao SK, Samant S, Gu W,
Handorf CR, Pfeffer LM and Nosrat CA: Oral cancer: Enduring
characteristics and emerging trends. J Mich Dent Assoc. 94:64–68.
2012.PubMed/NCBI
|
|
4
|
Karatas OF, Oner M, Abay A and Diyapoglu
A: MicroRNAs in human tongue squamous cell carcinoma: From
pathogenesis to therapeutic implications. Oral Oncol. 67:124–130.
2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Xie N, Wang C, Liu X, Li R, Hou J, Chen X
and Huang H: Tumor budding correlates with occult cervical lymph
node metastasis and poor prognosis in clinical early-stage tongue
squamous cell carcinoma. J Oral Pathol Med. 44:266–272.
2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Nóbrega TD, Queiroz SI, Santos EM, Costa
AL, Pereira-Pinto L and de Souza LB: Clinicopathological evaluation
and survival of patients with squamous cell carcinoma of the
tongue. Med Oral Patol Oral Cir Bucal. 23:e579–e587.
2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Le Campion ACOV, Ribeiro CMB, Luiz RR, da
Silva Júnior FF, Barros HCS, Dos Santos KCB, Ferreira SJ, Gonçalves
LS and Ferreira SMS: Low survival rates of oral and oropharyngeal
squamous cell carcinoma. Int J Dent. 2017(5815493)2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Shakeel Uz Zaman, Adeel M and Suhail A:
Squamous cell carcinoma of oral tongue in young patients-a 10 years
tertiary care experience. J Pak Med Assoc. 66:155–158.
2016.PubMed/NCBI
|
|
9
|
Zhang YY, Wang DC, Su JZ, Jia LF, Peng X
and Yu GY: Clinicopathological characteristics and outcomes of
squamous cell carcinoma of the tongue in different age groups. Head
Neck. 39:2276–2282. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Tang Q, Cheng B, Xie M, Chen Y, Zhao J,
Zhou X and Chen L: Circadian clock gene bmal1 inhibits
tumorigenesis and increases paclitaxel sensitivity in tongue
squamous cell carcinoma. Cancer Res. 77:532–544. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Feng Z, Xu QS, Qin LZ, Li H and Han Z:
Predicting radiotherapy necessity in tongue cancer using lymph Node
Yield. J Oral Maxillofac Surg. 75:1062–1070. 2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Chen Y, Tian T, Mao MJ, Deng WY and Li H:
CRBP-1 over-expression is associated with poor prognosis in tongue
squamous cell carcinoma. BMC Cancer. 18(514)2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Daigo K, Takano A, Thang PM, Yoshitake Y,
Shinohara M, Tohnai I, Murakami Y, Maegawa J and Daigo Y:
Characterization of KIF11 as a novel prognostic biomarker and
therapeutic target for oral cancer. Int J Oncol. 52:155–165.
2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Shih CH, Chang YJ, Huang WC, Jang TH, Kung
HJ, Wang WC, Yang MH, Lin MC, Huang SF, Chou SW, et al:
EZH2-mediated upregulation of ROS1 oncogene promotes oral cancer
metastasis. Oncogene. 36:6542–6554. 2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Li TK, Yin K, Chen Z, Bao Y and Zhang SX:
MiR-214 regulates oral cancer KB cell apoptosis through targeting
RASSF5. Genet Mol Res. 16(gmr16019327)2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Bal E, Baala L, Cluzeau C, El Kerch F,
Ouldim K, Hadj-Rabia S, Bodemer C, Munnich A, Courtois G, Sefiani A
and Smahi A: Autosomal dominant anhidrotic ectodermal dysplasias at
the EDARADD locus. Hum Mutat. 28:703–709. 2007.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Podzus J, Kowalczyk-Quintas C,
Schuepbach-Mallepell S, Willen L, Staehlin G, Vigolo M, Tardivel A,
Headon D, Kirby N, Mikkola ML, et al: Ectodysplasin a in biological
fluids and diagnosis of ectodermal dysplasia. J Dent Res.
96:217–224. 2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Bergendal B, Klar J, Stecksén-Blicks C,
Norderyd J and Dahl N: Isolated oligodontia associated with
mutations in EDARADD, AXIN2, MSX1, and PAX9 genes. Am J Med Genet
A. 155A:1616–1622. 2011.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Arzoo PS, Klar J, Bergendal B, Norderyd J
and Dahl N: WNT10A mutations account for ¼ of population-based
isolated oligodontia and show phenotypic correlations. Am J Med
Genet A. 164A:353–359. 2014.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658.
2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Boeker M, França F, Bronsert P and Schulz
S: TNM-O: Ontology support for staging of malignant tumours. J
Biomed Semantics. 7(64)2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Hashibe M, Brennan P, Chuang SC, Boccia S,
Castellsague X, Chen C, Curado MP, Dal Maso L, Daudt AW, Fabianova
E, et al: Interaction between tobacco and alcohol use and the risk
of head and neck cancer: Pooled analysis in the international head
and neck cancer epidemiology consortium. Cancer Epidemiol
Biomarkers Prev. 18:541–550. 2009.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Cancer Genome Atlas Network: Comprehensive
genomic characterization of head and neck squamous cell carcinomas.
Nature 517: 576-582, 2015.
|
|
25
|
Tuna M, Amos CI and Mills GB: Genome-wide
analysis of head and neck squamous cell carcinomas reveals HPV,
TP53, smoking and alcohol-related allele-based acquired uniparental
disomy genomic alterations. Neoplasia. 21:197–205. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Mohideen K, Krithika C, Jeddy N, Bharathi
R, Thayumanavan B and Sankari SL: Meta-analysis on risk factors of
squamous cell carcinoma of the tongue in young adults. J Oral
Maxillofac Pathol. 23:450–457. 2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Jeon JH, Kim MG, Park JY, Lee JH, Kim MJ,
Myoung H and Choi SW: Analysis of the outcome of young age tongue
squamous cell carcinoma. Maxillofac Plast Reconstr Surg.
39(41)2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Zhou HY, Shu HY, Dai J, Li HC, Tang L,
Wang HW and Ni B: Maternal genetic backgrounds contribute to the
genetic susceptibility of tongue cancer patients in Hunan, central
of China. Mitochondrial DNA A DNA Mapp Seq Anal. 29:347–352.
2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Haeggblom L, Ramqvist T, Tommasino M,
Dalianis T and Näsman A: Time to change perspectives on HPV in
oropharyngeal cancer. A systematic review of HPV prevalence per
oropharyngeal sub-site the last 3 years. Papillomavirus Res.
4:1–11. 2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Ribeiro IP, Rodrigues JM, Mascarenhas A,
Kosyakova N, Caramelo F, Liehr T, Melo JB and Carreira IM:
Cytogenetic, genomic, and epigenetic characterization of the HSC-3
tongue cell line with lymph node metastasis. J Oral Sci. 60:70–81.
2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Bai Y, Cui X, Gao D, Wang Y, Wang B and
Wang W: Golgi integral membrane protein 4 manipulates cellular
proliferation, apoptosis, and cell cycle in human head and neck
cancer. Biosci Rep: Aug 31, 2018 (Epub ahead of print). doi:
10.1042/BSR20180454.
|
|
32
|
Chen G, Zhang Y, Liang J, Li W, Zhu Y,
Zhang M, Wang C and Hou J: Deregulation of hexokinase II is
associated with glycolysis, autophagy, and the
epithelial-mesenchymal transition in tongue squamous cell carcinoma
under hypoxia. Biomed Res Int. 2018(8480762)2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Ramos GO, Meyer GL, Visioli F, Manoela MD
and Oliveira MG: Carcinoma cuniculatum in the tongue of a patient
with oral lichen planus: Unusual presentation. Indian J Dent Res.
29:525–528. 2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Vigneswaran N and Williams MD:
Epidemiologic trends in head and neck cancer and aids in diagnosis.
Oral Maxillofac Surg Clin North Am. 26:123–141. 2014.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Zhang B, Li KY, Chen HY, Pan SD, Jiang LC,
Wu YP and Liu SW: Spindle and kinetochore associated complex
subunit 1 regulates the proliferation of oral adenosquamous
carcinoma CAL-27 cells in vitro. Cancer Cell Int.
13(83)2013.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Xu S, Ma D, Zhuang R, Sun W, Liu Y, Wen J
and Cui L: DJ-1 is upregulated in oral squamous cell carcinoma and
promotes oral cancer cell proliferation and invasion. J Cancer.
7:1020–1028. 2016.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Kumar A, Eby MT, Sinha S, Jasmin A and
Chaudhary PM: The ectodermal dysplasia receptor activates the
nuclear factor-kappaB, JNK, and cell death pathways and binds to
ectodysplasin A. J Biol Chem. 276:2668–2677. 2001.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Vial J, Royet A, Cassier P, Tortereau A,
Dinvaut S, Maillet D, Gratadou-Hupon L, Creveaux M, Sadier A,
Tondeur G, et al: The ectodysplasin receptor EDAR acts as a tumor
suppressor in melanoma by conditionally inducing cell death. Cell
Death Differ. 26:443–454. 2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Chen YT, Liu HC, Han D, Liu Y and Feng HL:
Association between EDAR polymorphisms and non-syndromic tooth
agenesis in the chinese han population. Chin J Dent Res.
20:153–159. 2017.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Suda N, Bazar A, Bold O, Jigjid B,
Garidkhuu A, Ganburged G and Moriyama K: A mongolian patient with
hypohidrotic ectodermal dysplasia with a novel P121S variant in
EDARADD. Orthod Craniofac Res. 13:114–117. 2010.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Chassaing N, Cluzeau C, Bal E, Guigue P,
Vincent MC, Viot G, Ginisty D, Munnich A, Smahi A and Calvas P:
Mutations in EDARADD account for a small proportion of hypohidrotic
ectodermal dysplasia cases. Br J Dermatol. 162:1044–1048.
2010.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Masui Y, Farooq M, Sato N, Fujimoto A,
Fujikawa H, Ito M and Shimomura Y: A missense mutation in the death
domain of EDAR abolishes the interaction with EDARADD and underlies
hypohidrotic ectodermal dysplasia. Dermatology. 223:74–79.
2011.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Wohlfart S, Söder S, Smahi A and Schneider
H: A novel missense mutation in the gene EDARADD associated with an
unusual phenotype of hypohidrotic ectodermal dysplasia. Am J Med
Genet A. 170A:249–253. 2016.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Sadier A, Lambert E, Chevret P, Décimo D,
Sémon M, Tohmé M, Ruggiero F, Ohlmann T, Pantalacci S and Laudet V:
Tinkering signaling pathways by gain and loss of protein isoforms:
The case of the EDA pathway regulator EDARADD. BMC Evol Biol.
15(129)2015.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Courtois G and Gilmore TD: Mutations in
the NF-kappaB signaling pathway: Implications for human disease.
Oncogene. 25:6831–6843. 2006.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Wang Y, Lin Z, Sun L, Fan S, Huang Z,
Zhang D, Yang Z, Li J and Chen W: Akt/Ezrin Tyr353/NF-κB pathway
regulates EGF-induced EMT and metastasis in tongue squamous cell
carcinoma. Br J Cancer. 110:695–705. 2014.PubMed/NCBI View Article : Google Scholar
|
|
47
|
He HJ, Bing H and Liu G: TSR2 induces
laryngeal cancer cell apoptosis through inhibiting NF-κB signaling
pathway. Laryngoscope. 128:E130–E134. 2018.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Shishodia S and Aggarwal BB: Nuclear
factor-kappaB: A friend or a foe in cancer? Biochem Pharmacol.
68:1071–1680. 2004.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Charbonneau B, Block MS, Bamlet WR,
Vierkant RA, Kalli KR, Fogarty Z, Rider DN, Sellers TA, Tworoger
SS, Poole E, et al: Risk of ovarian cancer and the NF-κB pathway:
Genetic association with IL1A and TNFSF10. Cancer Res. 74:852–861.
2014.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Hoesel B and Schmid JA: The complexity of
NF-κB signaling in inflammation and cancer. Mol Cancer.
12(86)2013.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Zeligs KP, Neuman MK and Annunziata CM:
Molecular pathways: The balance between cancer and the immune
system challenges the therapeutic specificity of targeting nuclear
factor-κB signaling for cancer treatment. Clin Cancer Res.
22:4302–4308. 2016.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Zeng S, Zhao X, Xu LS, Yang D, Chen L and
Xu MH: Apoptosis induction effect of Apocynum venetum polyphenol on
human U87 glioma cells via NF-κB pathway. Future Oncol.
15:3723–3738. 2019.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Giridharan S and Srinivasan M: Mechanisms
of NF-κB p65 and strategies for therapeutic manipulation. J Inflamm
Res. 11:407–419. 2018.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Wei H, Li J, Li Y and Song J: MicroRNA-451
inhibits inflammation and proliferation of glomerular mesangial
cells through down-regulating PSMD11 and NF-κB p65. Biosci Rep.
39(BSR20191455)2019.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Cai L, Li CM, Chen WN, Qiu YY, Guo YL and
Li R: Penta-acetyl geniposide induces apoptosis of fibroblast-like
synoviocytes from adjuvant-induced arthritis rats in vitro,
associated with inhibition of NF-κB activation. Pharmacol Rep.
71:1006–1013. 2019.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Adams JM and Cory S: The Bcl-2 protein
family: Arbiters of cell survival. Science. 281:1322–1326.
1998.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Zhao M, Zhang Y, Li J, Li X, Cheng N, Wang
Q, Cai W, Zhao C, He Y, Chang J and Zhou C: Histone deacetylation,
as opposed to promoter methylation, results in epigenetic BIM
silencing and resistance to EGFR TKI in NSCLC. Oncol Lett.
15:1089–1096. 2018.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Dang CV: c-Myc target genes involved in
cell growth, apoptosis, and metabolism. Mol Cell Biol. 19:1–11.
1999.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Hao YH, Lafita-Navarro MC, Zacharias L,
Borenstein-Auerbach N, Kim M, Barnes S, Kim J, Shay J, DeBerardinis
RJ and Conacci-Sorrell M: Induction of LEF1 by MYC activates the
WNT pathway and maintains cell proliferation. Cell Commun Signal.
17(129)2019.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Farria AT, Mustachio LM, Akdemir ZHC and
Dent SYR: GCN5 HAT inhibition reduces human Burkitt lymphoma cell
survival through reduction of MYC target gene expression and
impeding BCR signaling pathways. Oncotarget. 10:5847–5858.
2019.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Jiang H, Li Y, Li J, Zhang X, Niu G, Chen
S and Yao S: Long noncoding RNA LSINCT5 promotes endometrial
carcinoma cell proliferation, cycle, and invasion by promoting the
Wnt/β-catenin signaling pathway via HMGA2. Ther Adv Med Oncol.
11(1758835919874649)2019.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Ma JX, Sun YL, Yu Y, Zhang J, Wu HY and Yu
XF: Triptolide enhances the sensitivity of pancreatic cancer PANC-1
cells to gemcitabine by inhibiting TLR4/NF-κB signaling. Am J
Transl Res. 11:3750–3760. 2019.PubMed/NCBI
|
|
63
|
Tanaka K, Tanaka T, Nakano T, Hozumi Y,
Yanagida M, Araki Y, Iwazaki K, Takagi M and Goto K: Knockdown of
DEAD-box RNA helicase DDX5 selectively attenuates serine 311
phosphorylation of NF-κB p65 subunit and expression level of
anti-apoptotic factor Bcl-2. Cell Signal. 65(109428)2019.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Wang M, Zeng X, Li S, Sun Z, Yu J, Chen C,
Shen X, Pan W and Luo H: A novel tanshinone analog exerts
anti-cancer effects in prostate cancer by inducing cell apoptosis,
arresting cell cycle at G2 phase and blocking metastatic ability.
Int J Mol Sci. 20(E4459)2019.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Zhang S, Ma H, Zhang D, Xie S, Wang W, Li
Q, Lin Z and Wang Y: LncRNA KCNQ1OT1 regulates proliferation and
cisplatin resistance in tongue cancer via miR-211-5p mediated
Ezrin/Fak/Src signaling. Cell Death Dis. 9(742)2018.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Zhao L, Li P, Zhao L, Wang M, Tong D, Meng
Z, Zhang Q, Li Q and Zhang F: Expression and clinical value of
PD-L1 which is regulated by BRD4 in tongue squamous cell carcinoma.
J Cell Biochem. 121:1855–1869. 2020.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Huang L, Song F, Sun H, Zhang L and Huang
C: IRX5 promotes NF-κB signalling to increase proliferation,
migration and invasion via OPN in tongue squamous cell carcinoma. J
Cell Mol Med 2018 (Epub ahead of print).
|
|
68
|
Yang N, Gao J, Cheng X, Hou C, Yang Y, Qiu
Y, Xu M, Zhang Y and Huang S: Grape seed proanthocyanidins inhibit
the proliferation, migration and invasion of tongue squamous cell
carcinoma cells through suppressing the protein kinase B/nuclear
factor-κB signaling pathway. Int J Mol Med. 40:1881–1888.
2017.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Shi Y, Wu WZ, Huo A, Zhou W and Jin XH:
Isobavachalcone inhibits the proliferation and invasion of tongue
squamous cell carcinoma cells. Oncol Lett. 14:2852–2858.
2017.PubMed/NCBI View Article : Google Scholar
|