Introduction
Lung cancer is the leading cause of
cancer-associated mortality in men and women and is responsible for
26% of all cancer-associated deaths worldwide (1). Non-small cell lung cancer (NSCLC) is
the predominant type of lung cancer, accounting for ~80% of all
lung cancers (1). Furthermore,
>50% patients with NSCLC present with locally advanced or
distantly metastatic cancer at the time of diagnosis (1,2).
Although targeted therapy has improved the management and outcome
of patients with NSCLC, the overall 5-year survival rates of
patients with NSCLC and advanced NSCLC are only 18 and 4%,
respectively (1).
Recently, long non-coding RNAs (lncRNAs) have
attracted great attention in the field of cancer. They serve
crucial roles in the development and progression of cancer and
might therefore represent a potential therapeutic target to treat
various types of cancer, including lung cancer and colon cancer
(3). Selective inhibition of target
lncRNA by RNA interference technology represents the most suitable
approach. Small interfering (si)RNAs can significantly inhibit the
expression of lncRNAs in tumor cells. For example, HOX transcript
antisense RNA (HOTAIR) knockdown by siRNA can reduce lung cancer
cell proliferation and invasive capacity in vitro (4). In addition, HOTAIR silencing by siRNA
can reverse cisplatin resistance in lung adenocarcinoma cells via
p21 downregulation (5). Cheng et
al (6) reported that the
expression of urothelial cancer associated 1 (UCA1) lncRNA is
significantly increased in patients with NSCLC and that UCA1
knockdown can partly improve the gefitinib sensitivity of
gefitinib-resistant NSCLC cell lines. In vivo experiments
also demonstrated that high expression of UCA1 might present a
novel mechanism for the acquired resistance of gefitinib-resistant
NSCLC cell lines (6). A previous
study also demonstrated that metastasis associated lung
adenocarcinoma transcript 1 (MALAT1) inhibition using a siRNA can
reduce NSCLC cell migratory and invasive ability (7). In addition, it was demonstrated in
vitro that Pvt1 oncogene knockdown can inhibit the
proliferation and induce apoptosis of NSCLC cells by sponge-like
adsorption of microRNA-195 therefore, increasing sensitivity to
radiotherapy of these cells (8).
LncRNAs may therefore be used to develop novel targeted therapy to
treat NSCLC.
The present study determined the expression of
LINC00210 in NSCLC tumor tissues and cells and investigated its
effects on NSCLC progression. Moreover, a previous study identified
that LINC00210 sponges miR-328-5p (9). Thus, this present study explored
whether LINC00210 also sponges miR-328-5p in NSCLC. Through
luciferase reporter assays, it was demonstrated that LINC00210
targeted miR-328-5p to promote NSCLC progression. The findings from
the present study may provide a novel therapeutic target for the
diagnosis and treatment of NSCLC.
Materials and methods
Ethical statement
The present study was approved by the Ethics
Committee of China-Japan Union Hospital of Jilin University and
patients provided written informed consent.
Patients and samples
A total of 39 patients who were diagnosed for the
first time with NSCLC, according to the grading system of the
American Joint Committee on Cancer (10), at the China-Japan Union Hospital of
Jilin University, between March 2010 and July 2012, were included
in the present study. Patients treated with radiotherapy or
chemotherapy before surgery were excluded. The 39 patients with
NSCLC included 29 men and 10 females, and their mean age was
58.7±9.2 years. The clinicopathological characteristics of all
patients are presented in Table I.
Patients' carcinoma tissues and corresponding adjacent normal
tissues (at least 3 cm from tumor tissues) were collected during
resection and immediately stored at -20˚C. Following surgery, all
patients were followed-up every 3 months for 5 years through
telephone consultations, to analyze their 5-year survival rate
using Kaplan-Meier survival analysis.
 | Table IAssociation between LINC00210
expression and the clinicopathological characteristics of patients
with non-small cell lung cancer. |
Table I
Association between LINC00210
expression and the clinicopathological characteristics of patients
with non-small cell lung cancer.
| Characteristics | LINC00210 low
expression (n=18) | LINC00210 high
expression (n=21) | P-value |
|---|
| Sex | | | 0.6508 |
| Male | 14 | 15 | |
| Female | 4 | 6 | |
| Age, years | | | 0.5189 |
| ≤60 | 13 | 17 | |
| >60 | 5 | 4 | |
| Tumor size, cm | | | 0.0071a |
| ≤3 | 12 | 5 | |
| >3 | 6 | 16 | |
| TNM stage | | | 0.0160a |
| I and II | 9 | 3 | |
| III and IV | 9 | 18 | |
| Lymph node | | | 0.0174a |
| metastasis | | | |
| Negative | 12 | 6 | |
| Positive | 6 | 15 | |
Cell culture
The SK-MES-1 and A549 NSCLC, and the human 16-HBE
bronchial epithelial cell line were all provided by The Cell Bank
of Type Culture Collection of the Chinese Academy of Sciences. All
cells were maintained separately in Dulbecco's Modified Eagle
medium (DMEM) supplemented with 10% FBS (Invitrogen; Thermo Fisher
Scientific, Inc.), 100 µg/ml streptomycin and 100 U/ml penicillin
and cultured at 37˚C in a humidified incubator containing 5%
CO2.
Transfection
A total of two siRNAs against LINC00210 (siRNA#1:
5'-GGUUCUCAUUCUCAUUUAAUU-3' and siRNA#2:
5'-CGGUAUUAUGACCACUACUUU-3') as well as LINC00210 scramble siRNA
negative control (NC) (5'-AAUUCUCCGAACGUGUCACGU-3'), miR-328-5p
mimics (5'-GGGGGGGCAGGAGGGGCUCAGGG-3'), miR-328-5p NC (scrambled;
5'-UCACAACCUCCUAGAAAGAGUAGA-3') and miR-328-5p inhibitors
(5'-CCCTGAGCCCCTCCTGCCCCCCC-3') were all synthesized by Shanghai
GenePharma Co., Ltd. A549 cells cultured in serum free DMEM were
transfected with 100 nM of the siRNA/miRNA/miRNA inhibitor, using
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.).
After transfection, A549 cells were grouped as follows:
siLINC00210#1 group (transfected with LINC00210 siRNA#1),
siLINC00210#2 group (transfected with LINC00210 siRNA#2), scramble
group (transfected with LINC00210 siRNA NC), miR-NC group
(transfected with miR-328-5p NC), miR-328-5p group (transfected
with miR-328-5p mimics), siLINC00210#1 + miR-328-5p inhibitor group
(co-transfected with LINC00210 siRNA#1 and miR-328-5p inhibitors).
The transfection efficiency was confirmed using reverse
transcription-quantitative PCR (RT-qPCR), 24 h after transfection
and transfected cells were used for following experiments.
Cell proliferation assay
Cell proliferation was measured using Cell Counting
Kit-8 (CCK8) assay (Sigma-Aldrich; Merck KGaA). Transfected A549
cells were seeded into 96-well plates (2x103 cells/well)
and cultured for 24, 48, 72 and 96 h. CCK8 solution (20 µl; 0.5
mg/ml; Sigma-Aldrich; Merck KGaA) was added into each well and
incubated for 2 h at 37˚C. Absorbance was measured at 450 nm using
a microplate reader (Thermo Labsystems).
Transwell assay
To investigate cell invasion, 24-well Transwell
chambers were pre-coated with Matrigel (BD Biosciences) for 30 min
at 37˚C and were inserted into 24-well plates containing 1 ml DMEM
supplemented with 10% FBS. A total of 100 µl of A549 cells in serum
free-suspension (1x105 cells/ml) was added to 24-well
Transwell chambers and incubated in a humidified incubator at 37˚C
with 5% CO2. After 2 days cells that had not penetrated
the membrane were removed with a cotton swab. Cells that penetrated
and adhered to the lower side of the membrane were fixed with 4%
paraformaldehyde for 30 min at room temperature and stained with
crystal violet (0.1%) for 10 min at room temperature. The number of
invading cells was counted in five random fields under the light
microscope at 100x magnification. Cell number was counted using
ImageJ (version 1.41; National Institutes of Health). For the
assessment of cell migration, the same method was performed,
although the 24-well Transwell chambers, were not pre-coated with
Matrigel.
Luciferase reporter gene assay
To determine if LINC0020 is a target of miR-328-5p
the target gene prediction software miRcode 11 (http://www.mircode.org/) was used. Mutant (Mut) and
wild-type (WT) sequences of the LINC00210 containing the
3'untranslated region were designed and amplified using PCR. A549
cells from the miR-NC and miR-328-5p groups were seeded in 24-well
plates (1x104 cells per well), and transfected with
pGL3-LINC00210-Mut and pGL3-LINC00210-WT plasmids (Promega
Corporation), respectively using Lipofectamine® 2000,
and cultured for 24 h at 37˚C. Cells were collected and firefly and
Renilla luciferase activity was detected using a
Dual-Luciferase Reporter Assay System kit (Promega Corporation)
according to the manufacturer's protocol. Firefly luciferase
activity was normalized to Renilla luciferase activity.
RT-qPCR
Total RNA was extracted from tissue samples and
cells using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. Total
RNA was reverse transcribed into cDNA at 37˚C for 15 min using a
Reverse Transcription kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. A
similar amount of cDNA product in each sample was subjected to PCR
amplification using the Applied Biosystems 7300 Real Time PCR
system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The
qPCR were performed using a SYBR Green I Master Mix kit (Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
as follows: Initial denaturation for 5 min at 95˚C, followed by 39
cycles of denaturation at 95˚C for 30 sec and annealing at 60˚C for
45 sec. The sequences of the primers (Sangon Biotech Co., Ltd.)
were as follows: LINC00210 forward, 5'-AACACGTTAGCGGGTTCTCA-3' and
reverse, 5'-TCAAAAACCACCGAGGGAGG-3'; miR-328-5p forward,
5'-AACGAGACGACGACAGAC-3' and reverse,
5'-GGGGGGGCAGGAGGGGCTCAGGG-3'; GAPDH forward,
5'-ATGTTGCAACCGGGAAGGAA-3', reverse 5'-AGGAAAAGCATCACCCGGAG-3'. and
U6 forward, 5'-AACGAGACGACGACAGAC-3' and reverse
5'-GCAAATTCGTGAAGCGTTCCATA-3'. The relative expressions levels of
LINC00210 and miR-328-5p were normalized to the endogenous control
U6 and expressed as 2-ΔΔCq
(11).
LINC00210 distribution detection
For A549 cells, nuclear and cytoplasmic RNA were
separated using a Cytoplasmic and Nuclear RNA Purification kit
(Norgen Biotek Corp.) according to the manufacturer's instructions.
The LINC00210 expression level in the nucleus and cytoplasm was
determined using RT-qPCR. U6 was used as endogenous control for
LINC00210 expression level in nucleus, whereas GAPDH used as the
endogenous control for cytoplasmic LINC00210 expression level.
Statistical analysis
All data are presented as the mean ± standard
deviation and were analyzed using SPSS v19.0 software (IBM Corp.)
or GraphPad Prism v6 (GraphPad Software, Inc.). The correlation
between LINC00210 and miR-328-5p expression level was determined
using Pearson's correlation analysis. Comparison between two groups
was performed using unpaired two tailed Student's t-test. One-way
analysis of variance followed by Tukey's post hoc test was used for
comparison of multiple groups. The Kaplan-Meier method was used to
create survival curves and the log-rank test was used to determine
statistical significance. χ2 test was used to determine
the association between LINC00210 expression level and the
clinicopathological characteristics of the 39 patients with NSCLC.
P<0.05 was considered to indicate a statistically significant
difference. Each experiment was repeated at least three times.
Results
LINC00210 is overexpressed in
NSCLC
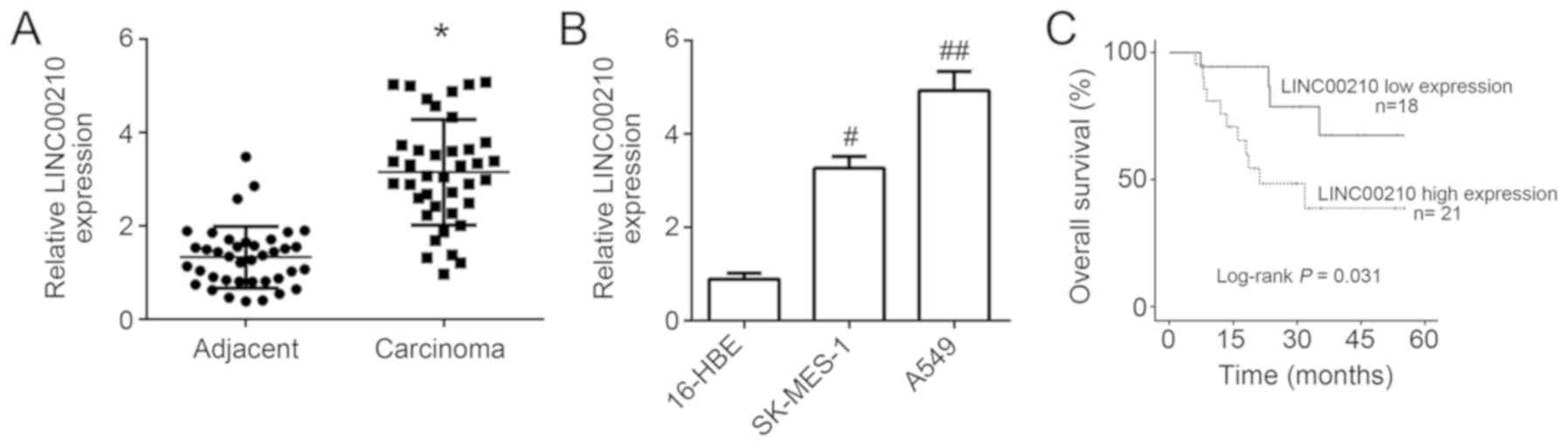
The results from the RT-qPCR revealed that LINC00210
expression level was significantly increased in NSCLC tissues
compared with that in adjacent tissues (P<0.05; Fig. 1A). Similarly, LINC00210 expression
level was significantly increased in the SK-MES-1 and A549 NSCLC
cell lines compared with that in the human 16-HBE bronchial
epithelial cell line (P<0.05; Fig.
1B). These results suggested that LINC00210 may by
overexpressed in NSCLC.
According to the follow-up study, patients with high
LINC00210 expression levels, using the median value as the cut-off
(n=21), had significantly lower overall 60-month survival compared
with patients with low LINC00210 expression level (n=18; P<0.05;
Fig. 1C; Log Rank χ2
value, 4.6). Analysis of the association between LINC00210
expression level and the clinicopathological characteristics of
patients with NSCLC revealed that LINC00210 expression level was
significantly associated with tumor size, TNM stage and lymph node
metastasis (Table I; P<0.05). High LINC00210 expression level
may therefore be used to predict larger tumor size, advanced tumor
stage and positive lymph node metastasis. The upregulation of
LINC00210 in patients with NSCLC indicated a poor prognosis.
LINC00210 knockdown inhibits A549 cell
proliferation, and migratory and invasive abilities
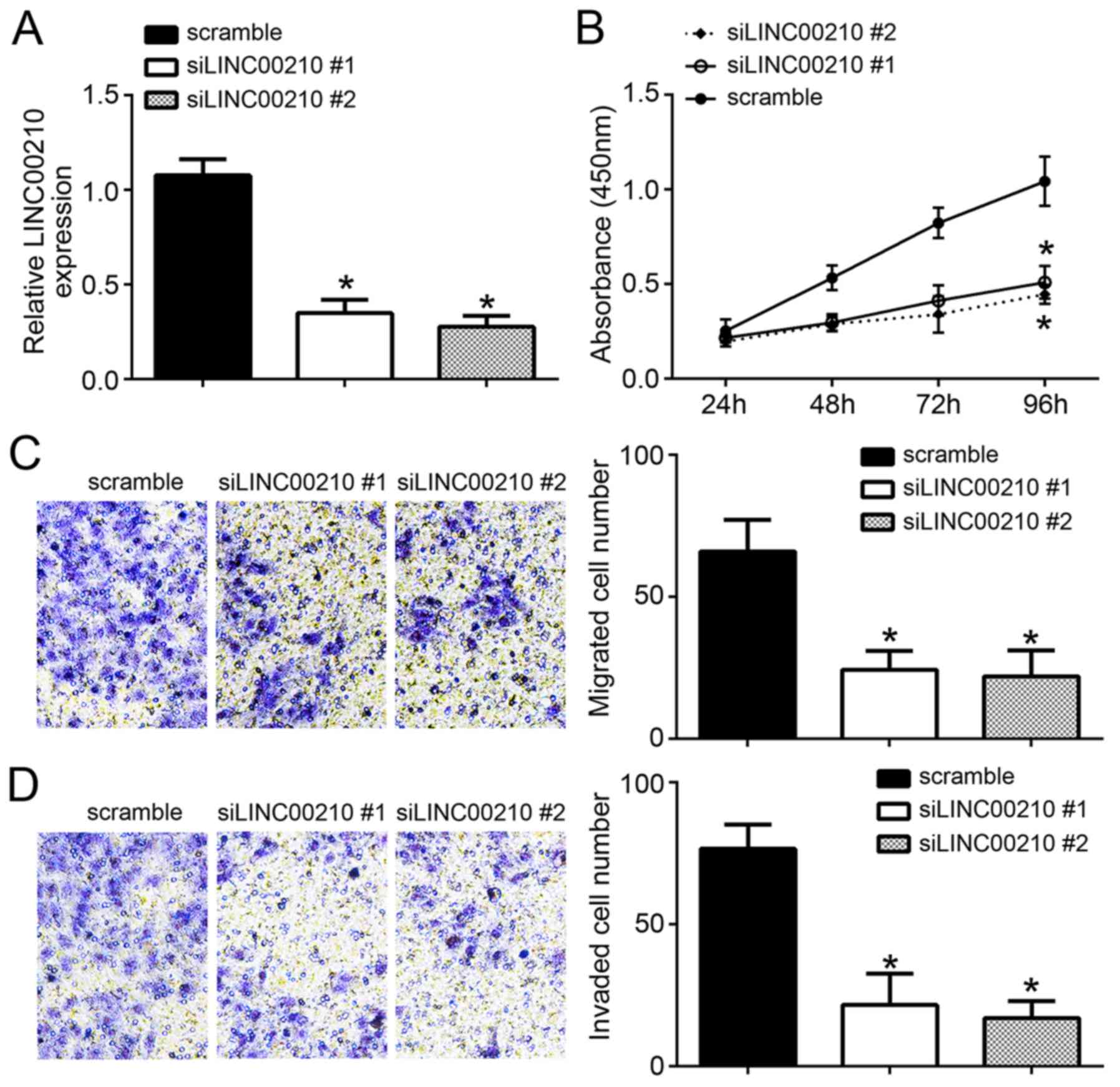
A549 cells in the siLINC00210#1 and siLINC00210#2
groups exhibited a significantly decreased LINC00210 expression
level compared with that in A549 cells from the scramble NC group
(P<0.05; Fig. 2A), indicating
that LINC00210 was successfully downregulated in A549 cells
following transfection. The results from the CCK8 assay revealed
that the absorbance was significantly reduced in A549 cells in the
siLINC00210#1 and siLINC00210#2 groups compared with that in A549
cells from the scramble NC group (P<0.05; Fig. 2B). In addition, transfection with
siLINC00210#1 and siLINC00210#2 induced a significant decrease in
the migratory and invasive ability of A549 cells compared with that
in A549 cells from the scramble NC group (P<0.05; Fig. 2C and D).
LINC00210 knockdown directly promotes
miR-328-5p expression
The distribution of LINC00210 expression level in
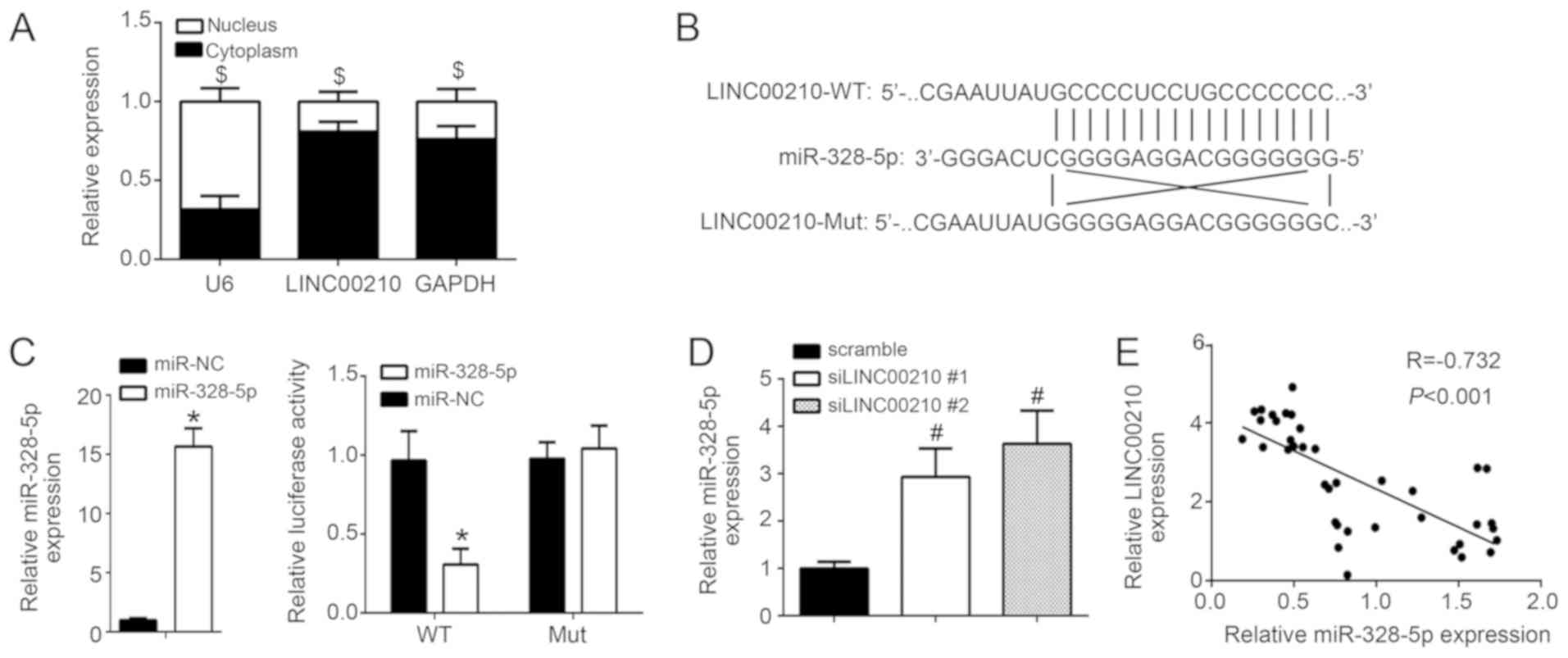
A549 cells was investigated. As presented in Fig. 3A, LINC00210 was mainly distributed in
the cytoplasm. A previous study reported that LINC00210 can
interact with miR-328-5p (9).
Through bioinformatics analysis, miR-328-5p was also identified as
a potential target of LINC00210, therefore miR-328-5p was
investigated in the present study. The WT and Mut sequences of
LINC00210 were designed separately, and their binding sites to
miR-328-5p were presented in Fig.
3B. A luciferase reporter assay was performed following A549
cell transfection with miR-328-5p mimics and WT- or Mut-LINC00210
reporter plasmid. The results revealed that miR-328-5p mimics
significantly inhibited the relative luciferase activity of
WT-LINC00210, but not Mut-LINC00210 (Fig. 3C; P<0.05) compared with that in
miR-NC group. Furthermore, A549 cells from the siLINC00210#1 and
siLINC00210#2 groups exhibited a significant increase in miR-328-5p
expression level compared with that in A549 cells from the scramble
NC group (P<0.05; Fig. 3D). The
results from the Pearson's correlation analysis revealed a negative
correlation between LINC00210 and miR-328-5p expression levels in
NSCLC tissues (Fig. 3E). These
findings suggest that LINC00210 knockdown may directly promote
miR-328-5p expression.
LINC00210 knockdown inhibits A549 cell
proliferation, and migratory and invasive ability by promoting
miR-328-5p expression
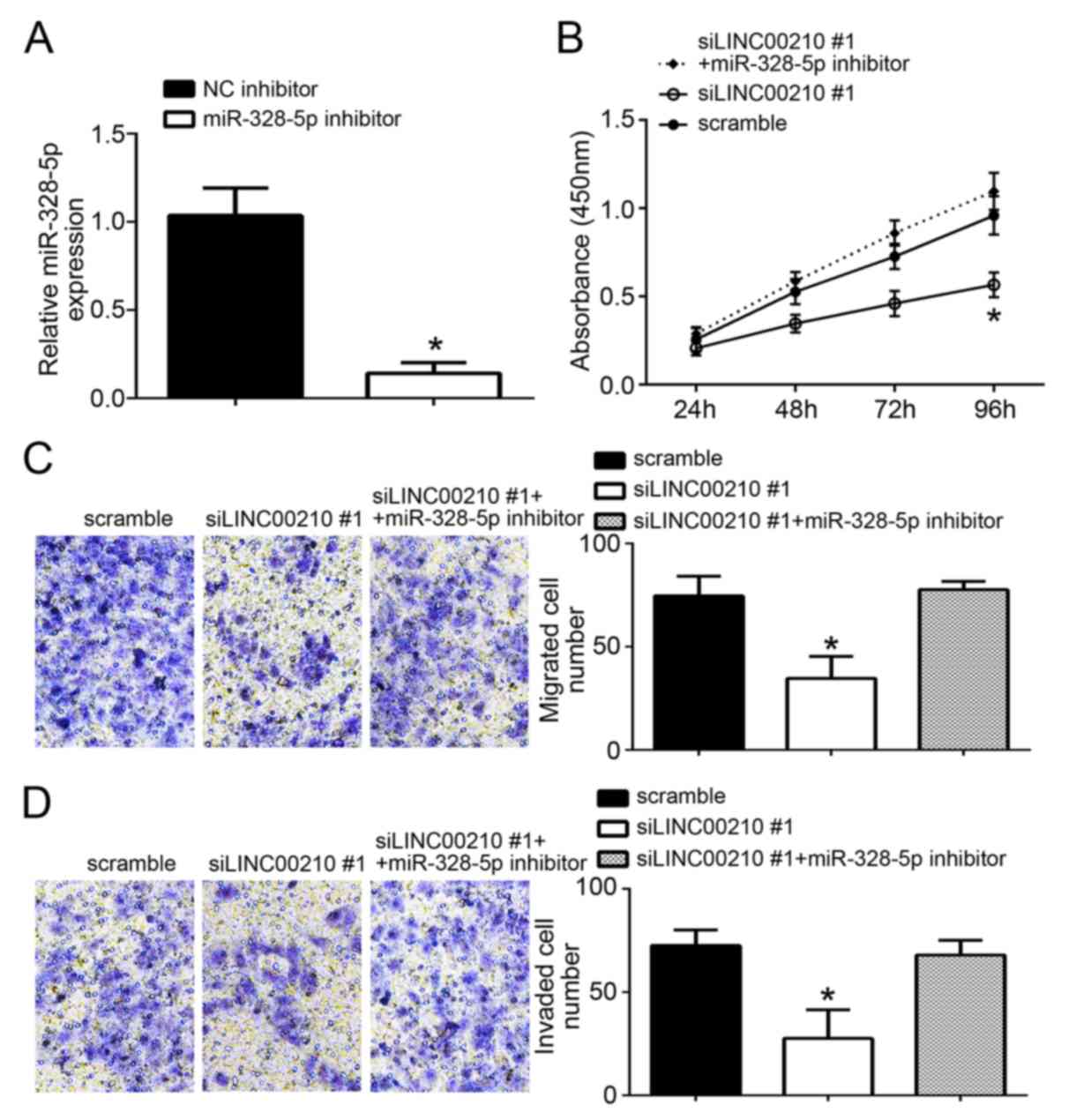
miR-328-5p inhibitor significantly inhibited the
expression of miR-328-5p (Fig. 4A).
For further experiments, siLINC00210#1 was chosen because it
produced the largest knockdown. A549 cells from the siLINC00210#1
group exhibited a significant decrease in cell proliferation, and
in the migratory and invasive ability compared with that in A549
cells from the scramble NC group and the siLINC00210#1 + miR-328-5p
inhibitor group (P<0.05; Fig.
4B-D). However, there was no significant difference in the
proliferation, and migratory and invasive ability of A549 cells
between the scramble NC group and the siLINC00210#1 + miR-328-5p
inhibitor group (Fig. 4B-D). These
results suggest that LINC00210 knockdown may inhibit A549 cell
proliferation, and migratory and invasive ability by promoting
miR-328-5p expression.
Discussion
The present study hypothesized that LINC00210 may be
considered as a novel diagnostic tool and treatment target for
patients with NSCLC. A high LINC00210 expression level was also
found in patients with NSCLC and was associated with poor
prognosis. In addition, the results from the present study revealed
that LINC00210 was primarily expressed in the cytoplasm, and that
knockdown of LINC00210 could inhibit A549 cell proliferation, and
migratory and invasive abilities by promoting miR-328-5p
expression. LINC00210 may therefore serve an oncogenic role in
NSCLC cells by sponging miR-328-5p.
lncRNAs can directly interact with DNA, mRNA or
protein to regulate chromatin modification or structure,
transcription and translation, therefore regulating numerous
physiological and pathological processes, including cell
proliferation or differentiation, stem cell reprogramming,
tumorigenesis or drug resistance (12). lncRNAs are divided into carcinogenic
lncRNAs and tumor suppressor lncRNAs. As with other tumor types,
such as colon cancer and liver cancer (6), the upregulation of carcinogenic lncRNAs
in NSCLC can enhance cell proliferation as well as the migratory
and invasive capacity, and reduce tumor cell apoptosis and tumor
drug sensitivity, including MALAT1, HOTAIR and AFAP1 (13-17).
Recently, numerous carcinogenic lncRNAs have been discovered in
NSCLC, including HOTAIR, MALAT1, colon cancer associated transcript
2, H19 imprinted maternally expressed transcript and AFAP1
antisense RNA 1 (13-17),
whereas SPRY4 intronic transcript 1, maternally expressed 3, GAS6
antisense RNA 1, growth arrest specific 5 and promoter of CDKN1A
antisense DNA damage activated RNA PANDAR have been reported as
tumor suppressor lncRNAs in NSCLC (18-22).
LINC00210 has rarely been investigated in human diseases. A
previous study investigated the effect of LINC00210 in liver
cancer, and reported that LINC00210 is overexpressed in liver
cancer, promoting liver tumor initiating cell self-renewal and
propagation via the activation of the Wnt/β-catenin signaling
pathway (23). Furthermore, another
previous study reported that LINC00210 regulates nasopharyngeal
carcinoma progression via the miR-328-5p/NOTCH3 axis (11). However, the role of LINC00210 in lung
cancer remains unknown. The results from the present study
demonstrate that LINC00210 was significantly upregulated in NSCLC
tissues, and that high LINC00210 expression level was significantly
associated with larger tumor size, advanced TNM stage and lymph
node metastasis in patients with NSCLC. In addition, reducing
LINC00210 expression using siRNA inhibited NSCLC cell
proliferation, and migratory and invasive abilities. As the primary
cause of cancer-associated mortality worldwide, the underlying
mechanisms of NSCLC have not yet been fully elucidated and the
actual therapeutic options remain unsatisfactory. The discovery of
lncRNAs has improved the clinical understanding of NSCLC
tumorigenesis and progression, suggesting that these aforementioned
lncRNAs may be considered as promising biomarkers for the early
diagnosis and treatment of NSCLC. The results from this paper
suggest that LINC00210 may be considered as a novel target for the
diagnosis and treatment of patients with NSCLC.
miRNAs represent a class of endogenous, non-coding
small RNAs found in eukaryotes. They participate in the regulation
of various types of human tumor, such as liver cancer and ovarian
cancer (24,25). In the present study, LINC00210
knockdown inhibited A549 cell proliferation, and migratory and
invasive ability via the promotion of miR-328-5p expression.
miR-328 is a type of microRNA that was been reported to be
associated with the progression of numerous tumors. For example,
Santasusagna et al (25)
reported that miR-328 expression level is reduced in colon cancer
tissues compared with that in adjacent normal tissues, and that
miR-328 can affect colon cancer progression via solute carrier
family 2 member 1/solute carrier family 2 member 1 targeting. In
nasopharyngeal carcinoma, miR-328 is considered as a potential
prognostic and therapeutic marker due to its inhibiting effect on
the epithelial-mesenchymal transition of nasopharyngeal carcinoma
cells (26). Liu et al
(27) demonstrated that low miR-328
expression can predict a poor prognosis in patients with acute
myeloid leukemia. Previous studies also reported that low miR-328
expression level is associated with poor survival in patients with
high-grade glioma, and that miR-328 can impair glioma cell
proliferation and invasive ability. miR-328 was therefore
considered as a favorable prognostic marker in glioma (28,29).
Furthermore, a previous study reported that miR-328 is reduced in
NSCLC, and that miR-328 upregulation can increase NSCLC cell
radiosensitvity via the DNA damage and repair signaling pathway
(30). In the present study,
miR-328-5p expression level was directly reduced following
transfection with LINC00210, which may therefore act as a tumor
suppressor in NSCLC.
In conclusion, the results from the present study
suggest that LINC00210 may be considered as a novel target for the
diagnosis and treatment of NSCLC. In addition, high LINC00210
expression predicted poor prognosis in patients with NSCLC.
Following LINC00210 knockdown, NSCLC cell proliferation, and
migratory and invasive abilities were reduced. Furthermore, the
current study demonstrated that LINC00210 may serve oncogenic role
in NSCLC by sponging miR-328-5p. At present, research on LINC00210
is still at an early stage, and further investigation on LINC00210
is required to determine the underlying mechanism of NSCLC and to
assist with the development of novel treatment options. The present
study did not analyze the effect of LINC00210 on cell cycle;
however this will be performed in future studies.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
ZL and LX contributed to the conception and design
of the present study. NG analyzed and interpreted the results, and
wrote the manuscript. KZ, BG and ZC performed the experiments and
analyzed the data. All authors read and approved the final version
of the manuscript.
Ethics approval and consent to
participate
The present study was approved by The Ethics
Committee of China-Japan Union Hospital of Jilin University.
Written informed consent was provided from all recruited
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Travis WD: The 2015 WHO classification of
lung tumors. Der Pathologe. 35 (Suppl 2)(S188)2014.
|
|
3
|
Meseure D, Drak Alsibai K, Nicolas A,
Bieche I and Morillon A: Long noncoding RNAs as new architects in
cancer epigenetics, prognostic biomarkers, and potential
therapeutic targets. Biomed Res Int. 2015(320214)2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Yao Y, Li J and Wang L: Large intervening
non-coding RNA HOTAIR is an indicator of poor prognosis and a
therapeutic target in human cancers. Int J Mol Sci. 15:18985–18999.
2014.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Liu Z, Sun M, Lu K, Liu J, Zhang M, Wu W,
Wei D, Wang Z and Wang R: The long noncoding RNA HOTAIR contributes
to cisplatin resistance of human lung adenocarcinoma cells via
downregualtion of p21 WAF1/CIP1 expression. PLoS One.
8(e77293)2013.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Cheng N, Cai W, Ren S, Li X, Wang Q, Pan
H, Zhao M, Li J, Zhang Y, Zhao C, et al: Long non-coding
RNAUCA1induces non-T790M acquired resistance to EGFR-TKIs by
activating the AKT/mTOR pathway in EGFR-mutant non-small cell lung
cancer. Oncotarget. 6:23582–23593. 2015..v. PubMed/NCBI View Article : Google Scholar
|
|
7
|
Guo F, Jiao F, Song Z, Li S, Liu B, Yang
H, Zhou Q and Li Z: Regulation of MALAT1 expression by TDP43
controls the migration and invasion of non-small cell lung cancer
cells in vitro. Biochem Biophys Res Commun. 465:293–298.
2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Wu D, Li Y, Zhang H and Hu X: Knockdown of
Lncrna PVT1 enhances radiosensitivity in non-small cell lung cancer
by sponging Mir-195. Cell Physiol Biochem. 42:2453–2466.
2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Zhang S, Li P, Zhao L and Xu L: LINC00210
as a miR-328-5p sponge promotes nasopharyngeal carcinoma
tumorigenesis by activating NOTCH3 pathway. Biosci Rep.
38(BSR20181168)2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Amin MB, Greene FL, Edge SB, Compton CC,
Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR and
Winchester DP: The eighth edition AJCC Cancer Staging Manual:
Continuing to build a bridge from a population-based to a more
‘personalized’ approach to cancer staging. CA Cancer J Clin.
67:93–99. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Geng C, Ziyun W, Dongqing W, Chengxiang Q,
Mingxi L, Xing C, Qipeng Z, Guiying Y and Qinghua C: LncRNADisease:
A database for long-non-coding RNA-associated diseases. Nucleic
Acids Res. 41 (Database Issue):D983–D986. 2013.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Li N, Wang Y, Liu X, Luo P, Jing W, Zhu M
and Tu J: Identification of circulating long noncoding RNA HOTAIR
as a novel biomarker for diagnosis and monitoring of non-small cell
lung cancer. Technol Cancer Res Treat. 16:1060–1066.
2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Li S, Mei Z, Hu HB and Zhang X: The lncRNA
MALAT1 contributes to non-small cell lung cancer development via
modulating miR-124/STAT3 axis. J Cell Physiol. 233:6679–6688.
2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Zhao Z, Wang J, Wang S, Chang H, Zhang T
and Qu J: LncRNA CCAT2 promotes tumorigenesis by over-expressed
Pokemon in non-small cell lung cancer. Biomed Pharmacother.
87:692–697. 2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Huang Z, Lei W, Hu HB, Zhang H and Zhu Y:
H19 promotes non-small-cell lung cancer (NSCLC) development through
STAT3 signaling via sponging miR-17. J Cell Physiol. 233:6768–6776.
2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Deng J, Liang Y, Liu C, He S and Wang S:
The up-regulation of long non-coding RNA AFAP1-AS1 is associated
with the poor prognosis of Patients with NSCLC. Biomed
Pharmacother. 75:8–11. 2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Sun M, Liu XH, Lu KH, Nie FQ, Xia R, Kong
R, Yang JS, Xu TP, Liu YW, Zou YF, et al: EZH2-mediated epigenetic
suppression of long noncoding RNA SPRY4-IT1 promotes NSCLC cell
proliferation and metastasis by affecting the
epithelial-mesenchymal transition. Cell Death Dis.
5(e1298)2014.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu
WQ, Xie WP and Hou YY: Long non-coding RNA MEG3 inhibits NSCLC
cells proliferation and induces apoptosis by affecting p53
expression. BMC Cancer. 13(461)2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Han L, Kong R, Yin DD, Zhang EB, Xu TP and
De W Shu YQ: Low expression of long noncoding RNA GAS6-AS1 predicts
a poor prognosis in patients with NSCLC. Med Oncol.
30(694)2013.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Mei Y, Si J, Wang Y, Huang Z, Zhu H, Feng
S, Wu X and Wu L: Long noncoding RNA GAS5 suppresses tumorigenesis
by inhibiting miR-23a 5 expression in non-small cell lung cancer.
Oncol Res. 25:1027–1037. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Han L, Zhang E, Yin D, Kong R, Xu T, Chen
W, Xia R, Shu Y and De W: Low expression of long noncoding RNA
PANDAR predicts a poor prognosis of non-small cell lung cancer and
affects cell apoptosis by regulating Bcl-2. Cell Death Dis.
6(e1665)2015.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Fu X, Zhu X, Qin F, Zhang Y, Lin J, Ding
Y, Yang Z, Shang Y, Wang L, Zhang Q and Gao Q: Linc00210 drives
Wnt/β-catenin signaling activation and liver tumor progression
through CTNNBIP1-dependent manner. Mol Cancer.
17(73)2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Li J, Li Q, Huang H, Li Y, Li L, Hou W and
You Z: Overexpression of miRNA-221 promotes cell proliferation by
targeting the apoptotic protease activating factor-1 and indicates
a poor prognosis in ovarian cancer. Int J Oncol. 50:1087–1096.
2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Santasusagna S, Moreno I, Navarro A, Muñoz
C, Martinez F, Hernández R, Castellano JJ and Monzo M: miR-328
mediates a metabolic shift in colon cancer cells by targeting
SLC2A1/GLUT1. Clin Transl Oncol. 20:1161–1167. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Lin CH, Chiang MC and Chen YJ:
MicroRNA-328 inhibits migration and epithelial-mesenchymal
transition by targeting CD44 in nasopharyngeal carcinoma cells.
Onco Targets Ther. 11:2375–2385. 2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Liu L, Chen RA, Zhang Y, Fan W, Xiao F and
Yan X: Low expression of circulating microRNA-328 is associated
with poor prognosis in patients with acute myeloid leukemia. Diagn
Pathol. 10(109)2015.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Yuan J, Zheng Z, Zheng Y, Lu X, Xu L and
Lin L: microRNA-328 is a favorable prognostic marker in human
glioma via suppressing invasive and proliferative phenotypes of
malignant cells. Int J Neurosci. 126:145–153. 2016.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wu Z, Sun L, Wang H, Yao J, Jiang C, Xu W
and Yang Z: MiR-328 expression is decreased in high-grade gliomas
and is associated with worse survival in primary glioblastoma. PLoS
One. 7(e47270)2012.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Ma W, Ma CN, Zhou NN, Li XD and Zhang YJ:
Up-regulation of miR-328-3p sensitizes non-small cell lung cancer
to radiotherapy. Sci Rep. 6(31651)2016.PubMed/NCBI View Article : Google Scholar
|