Introduction
Oral cancer is one of the most common malignancies
of the head and neck, with an incidence rate that is increasing
(1,2). Oral and oropharyngeal squamous cell
carcinomas (OSCCs) accounted for 90% of oral cancer incidence in
2012 (3,4). Several therapeutic technologies
including radiotherapy, chemotherapy, traditional surgery and
targeted therapy, have been utilized as major viable treatments for
OSCC. However, patients with OSCC generally have a poor prognosis
and the 5-year survival rate has not improved significantly in the
last 10 years (5). OSCC has a poor
prognosis due to its low response rate to drug-based therapy
(3). Dysregulation of relative
signaling cascades may result in tumorigenesis and cancer
resistance to clinical treatment. Therefore, identifying the
molecular mechanisms of OSCC development and progression is
essential for the development of novel clinical therapies to
improve the survival rate of patients with OSCC.
MicroRNAs (miRNAs or miRs) are a group of non-coding
RNAs, which consist of 20-22 nucleotides (6). miRNAs mediate gene expression by
binding to the 3' untranslated region (UTR) of target mRNAs
(7-9).
miRNAs serve critical roles in the normal development of various
biological processes, including cellular differentiation, cell
proliferation and cell apoptosis (10). Studies have indicated that miR-196a
expression is up-regulated in various types of cancer and its
expression is closely associated with a poor survival rate
(11-14).
Similarly, a number of studies have indicated that miR-196a may
promote the proliferation and cellular invasion of various types of
cancer, including non-small cell lung cancer, gastric cancer and
breast cancer (11-14). Furthermore,
miR-196a has been reported to function by targeting forkhead box O1
(FOXO1) in lung, human liver and cervical cancer cells (15-17).
However, the expression and mechanisms of miR-196a action in OSCCs
are not well established. Previous studies have demonstrated that
the PI3K/Akt pathway is activated in most types of cancer and
suppresses the function of FOXO genes (18-22).
The expression of FOXO1 is upregulated in human OSCCs and is
significantly correlated with the advancement of clinical stages
(23,24). However, the expression profile and
underlying mechanisms of FOXO1 in OSCC remain unknown.
Therefore, the present study aimed to investigate
the role of miR-196a and FOXO1 in OSCC cells and to further explore
its underlying mechanisms of action.
Materials and methods
Cell culture
The human OSCC (SCC9) and primary normal human oral
keratinocyte (HOK) cell lines were obtained from American Type
Culture Collection. Cells were maintained in RPMI 1640 medium
(Invitrogen; Thermo Fisher Scientific, Inc.) containing 10% FBS
(Invitrogen; Thermo Fisher Scientific, Inc.) and 1%
penicillin-streptomycin solution (Sigma-Aldrich; Merck KGaA). Cells
were incubated at 37˚C with 5% CO2.
Plasmids and cell transfection
miR-196a mimic, mimic control, miR-196a inhibitor or
the negative control of miR-196a inhibitor (inhibitor-NC) were
purchased from Guangzhou RiboBio Co., Ltd. Control-small
interfering RNA (siRNA; cat. no. sc-36869) and FOXO1-siRNA (cat.
no. sc-35382) were purchased from Santa Cruz Biotechnology, Inc.
SCC9 cells, which were seeded into six-well plates
(5x104 per well), at the exponential growth phase were
cultured at 37˚C for 24 h in RPMI 1640 medium containing 10% FBS
and then transfected with miR-196a inhibitor, Inhibitor-NC,
control-siRNA, FOXO1-siRNA, or miR-196a inhibitor+ FOXO1-siRNA
using Lipofectamine® 2000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
Cells without any treatment were considered controls. After
incubation at 37˚C for 48 h, transfection efficiency was detected
using RT-qPCR.
Cell viability analysis
An MTT assay was used to detect SCC9 cell
proliferation. 5x103 cells were seeded in 96-well plate
(BD Biosciences) at 37˚C for 24 h. Subsequently, SCC9 cells were
transfected as previously described. Cells were then inhibited at
37˚C for 12, 24 or 48 h. MTT reagent (10 µl; cat. no. C0009;
Beyotime Institute of Biotechnology) was added to each well and
incubated for another 4 h at 37˚C. DMSO (100 µl; Nanjing KeyGen
Biotech Co., Ltd.) was used to dissolve the resulting formazan
crystals. Absorbance at 490 nm was then analyzed using a microplate
reader (Omega Bio-Tek). The experiment was repeated in
triplicate.
Cell apoptosis analysis
SCC9 cells were transfected with miR-196a mimic,
mimic control, miR-196a inhibitor, inhibitor-NC, control-siRNA,
FOXO1-siRNA, or miR-196a inhibitor + FOXO1-siRNA as previously
described. After transfection, cell apoptosis was analyzed using an
Annexin V-FITC/propidium iodide Apoptosis Detection kit (BD
Biosciences) according to the manufacturer's protocol. An FC500
flow cytometer (Beckman Coulter, Inc.) was used to measure the
extent of apoptosis. Each experiment was repeated in triplicate and
CellQuest software version 5.1 (Becton, Dickinson and Company) was
used to analyze the rate of apoptosis.
Transwell migration assay
SCC9 cells were transfected with miR-196a mimic,
mimic control, miR-196a inhibitor, Inhibitor-NC, control-siRNA,
FOXO1-siRNA, or miR-196a inhibitor+ FOXO1-siRNA as previously
described and cultured in serum free RPMI 1640 medium at 37˚C for
48 h. Cells were harvested using trypsin (Gibco; Thermo Fisher
Scientific, Inc.), centrifuged (1,000 x g; 5 min; 4˚C) and
re-suspended in RPMI 1640 medium containing 0.1% FBS. Subsequently,
cells (1x104) were added to the upper chamber of a
transwell apparatus (8 µm pore size; Corning Inc.), while RPMI 1640
culture medium with 10% FBS was added to the lower culture chamber.
Cells were then cultured at 37˚C with 5% CO2 for 48 h.
Cells on the underside of membranes were washed with cold PBS three
times, fixed with 4% paraformaldehyde, at room temperature for 30
min and stained for 10 min at room temperature with 0.1% crystal
violet. The lower membrane cells were counted from 5 random areas
using an inverted light microscope (Olympus IX51; x100
magnification; Olympus Corporation).
Wound-healing assay
To investigate cell migration, wound healing assays
were performed. At 48 h following transfection, transfected SCC9
cells were seeded in 24-well plates (5x105 cells/ml) and
cultured at 37˚C for 24 h, allowing 80% confluency to be reached.
Cells were scratched using a 10 µl pipette tip and the debris were
removed with PBS. After a second incubation in serum-free RPMI 1640
medium at 37˚C for 24 h, cells were washed to remove culture medium
and representative images were captured at x100 magnification using
an inverted light microscope. Wound areas were recorded and
analyzed using Image-Pro Plus 6.0 software (National Institute of
Health). The size of the wound was measured compared with the
original wound following 24 h. Percent of wound (%)=wound width at
24 h/wound width at 0 h x100%. Each experiment was performed in
triplicate.
Reverse transcription-quantitative PCR
(RT-qPCR)
After experimental treatments, total RNA from SCC9
cells and HOK cells was extracted from cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. U6 was used as the
endogenous control for miRNA expression and GAPDH was used as the
endogenous control to normalize mRNA. The PrimeScript™ RT reagent
kit (Takara Bio, Inc.) was used for cDNA syntheses, and the
temperature protocol was as follows: 70˚C for 5 min, 37˚C for 5 min
and 42˚C for 1 h. RT-qPCR was performed using SYBR Premix Ex Taq
(Takara Bio Inc.) according to the manufacturer's protocol. RT-qPCR
was performed as follows: 95˚C for 10 min, followed by 40 cycles of
15 sec at 95˚C, 72˚C for 30 sec and a final dissociation stage
(78˚C for 1.5 min). Relative gene expression was determined using
the 2-ΔΔCq method (25).
All experiments were performed in triplicate.
Western blot assay
After experimental treatments, total protein was
extracted from OSCC cells using RIPA buffer (Auragene Bioscience
Co.). BCA assay kit (Thermo Fisher Scientific, Inc.) was used to
determine the concentration of protein samples. Proteins were
resolved by 10% SDS-PAGE and then transferred onto PVDF membranes.
After blocking with 5% non-fat milk for 1 h at room temperature,
membranes were incubated with the following primary antibodies
overnight at 4˚C: PI3K (cat. no. 4257; 1:1,000; Cell Signaling
Technology, Inc.), FOXO1 (cat. no. 2880; 1:1,000; Cell Signaling
Technology, Inc.), Akt (cat. no. 9272; 1:1,000; Cell Signaling
Technology, Inc.), phosphorylated (p)-Akt (cat. no. 9611; 1:1,000;
Cell Signaling Technology, Inc.), p-PI3K (cat. no. 17366; 1:1,000;
Cell Signaling Technology, Inc.), and GAPDH (cat. no. 5174;
1:1,000; Cell Signaling Technology, Inc.). Subsequently, membranes
were washed three times with PBST and incubated with secondary
antibody (cat. no. 7074; 1:2,000; Cell Signaling Technology, Inc.)
for 2 h at room temperature. Immunoreactive protein bands were
visualized using an ECL detection system (Bio-Rad Laboratories,
Inc.) according to the manufacturer's protocol. Proteins were
quantified using ImageJ version 1.49 software (National Institute
of Health).
Dual luciferase reporter assay
The binding sites between miR-196a and FOXO1 were
investigated using TargetScan bioinformatics software version 7.1
(www.targetscan.org/vert_71). To confirm
direct target binding sites, the wild-type (WT) or mutant (Mut)
FOXO1 genes were inserted into the Dual-luciferase miRNA Target
Expression Vector (Guangzhou RiboBio Co., Ltd.). To point-mutate
the miRNA-196a binding domain on the 3'UTR of FOXO1, the QuikChange
Site-Directed Mutagenesis kit (Agilent Technologies, Inc.) was used
following the manufacturer's protocol. The reporter vectors were
then co-transfected with WT-FOXO1 or MUT-FOXO1 and miR-196a mimic
or mimic control, into SCC9 cells using Lipofectamine®
2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) in
accordance with the manufacturer's protocol. After culturing cells
at 37˚C for an additional 48 h, 100 µl passive lysis buffer
(Promega Corporation) was used to dissociate cells, after which
luciferase activity was assessed using the dual-luciferase assay
system (Promega Corporation) according to the manufacturer's
protocol. All the experiments were performed in triplicate, and
luciferase activity was normalized to Renilla luciferase
activity.
Statistical analysis
All results were expressed as the mean ± standard
deviation. Statistical analysis was performed using Graphpad Prism
6 software (GraphPad Software, Inc.). Comparisons between two
groups were assessed using Student's t-test, and comparisons
between multiple groups were analyzed using one-way ANOVA followed
by Tukey's post-hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-196a expression is increased in
OSCC cells
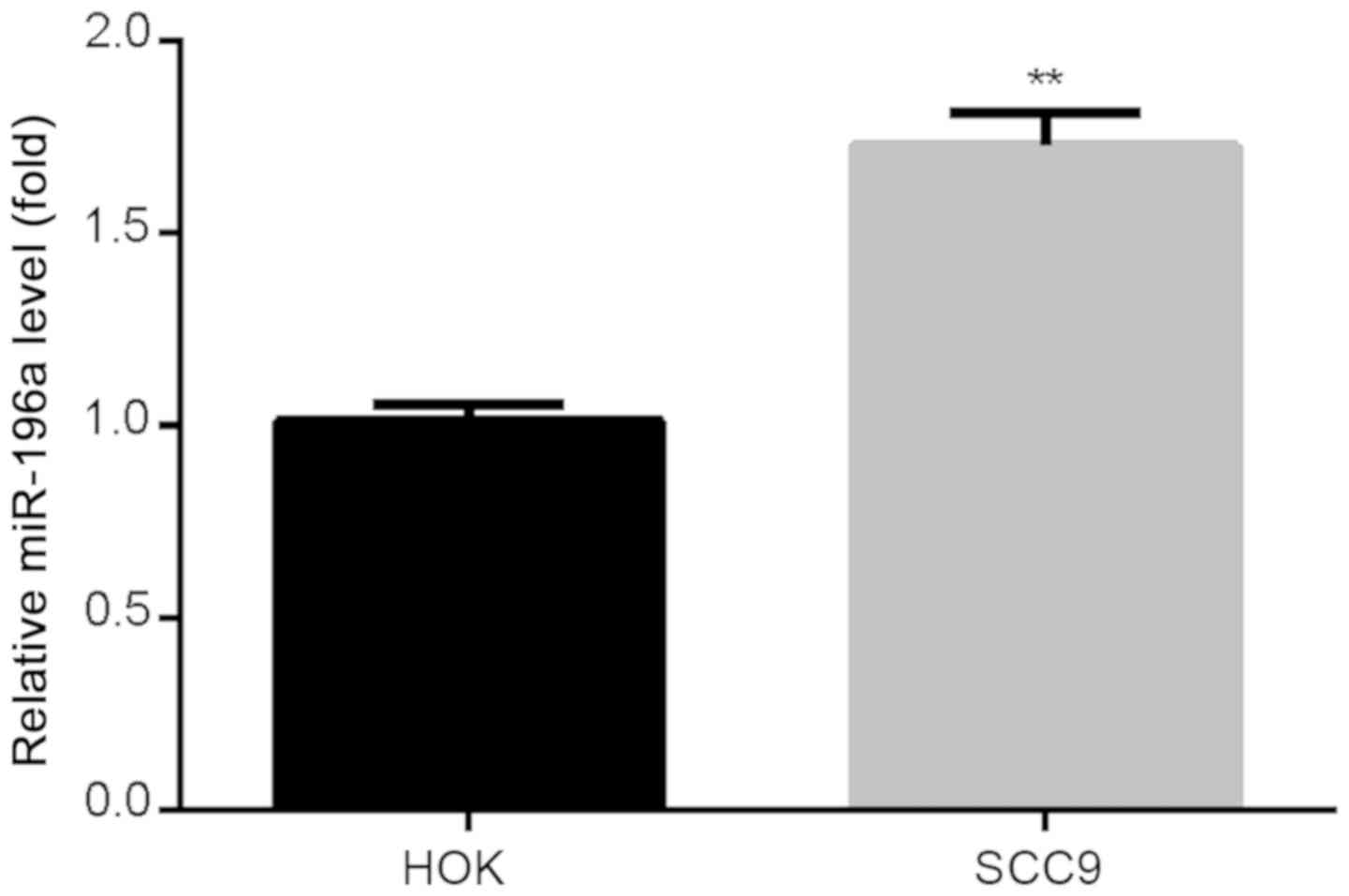
To evaluate the expression of miR-196a in human SCC9
cells and normal HOK cells, RT-qPCR was performed. The results
revealed that the expression of miR-196a was significantly
increased in SCC9 cells compared with HOK cells (Fig. 1).
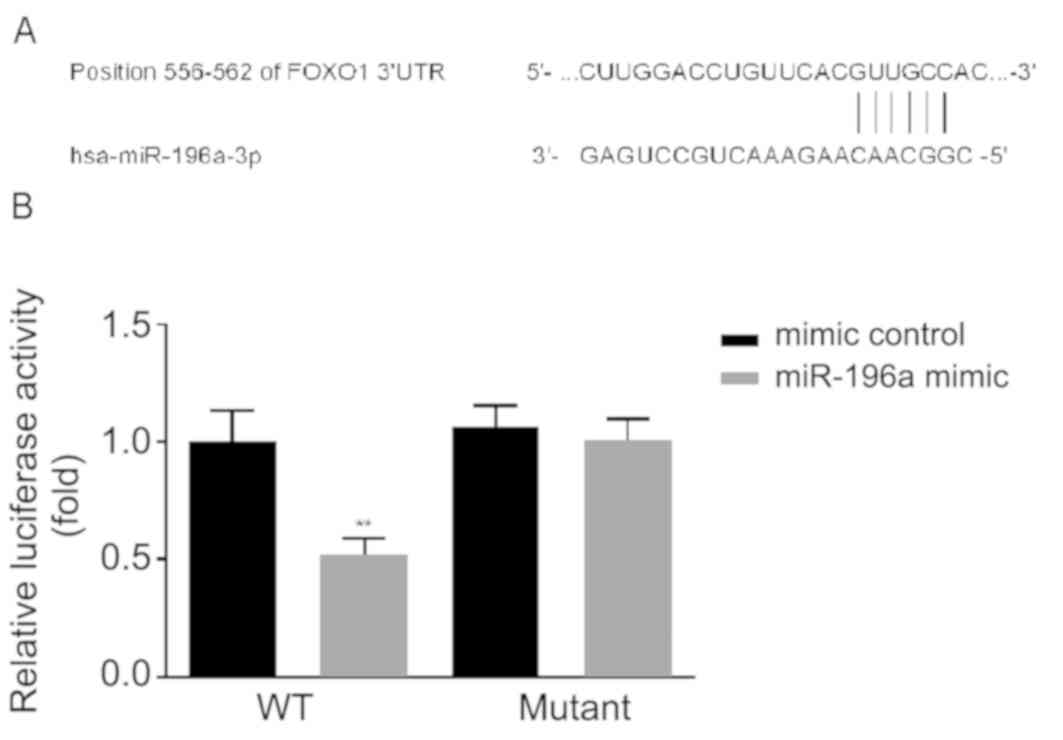
FOXO1 is a target of miR-196a
miR-196a has been reported to function by targeting
FOXO1 in lung, human liver and cervical cancer cells (15-17).
To determine the molecular mechanisms by which miR-196a regulates
the function of OSCC, the associations between FOXO1 and miR-196a
in OSCC were assessed. According to TargetScan bioinformatics
software analysis, binding sites between the 3' UTR of FOXO1 and
miR-196a were identified (Fig. 2A).
Subsequently, dual-luciferase assays were performed. The results
revealed that compared with cells co-transfected with mimic control
and FOXO1-WT, miR-196a significantly suppressed the luciferase
activity of cells co-transfected with miR-196 mimic and FOXO1-WT.
However, no significant changes were observed in cells
co-transfected with miR-196 mimic and FOXO1-MUT (Fig. 2B). The results indicated that FOXO1
is a direct target of miR-196a.
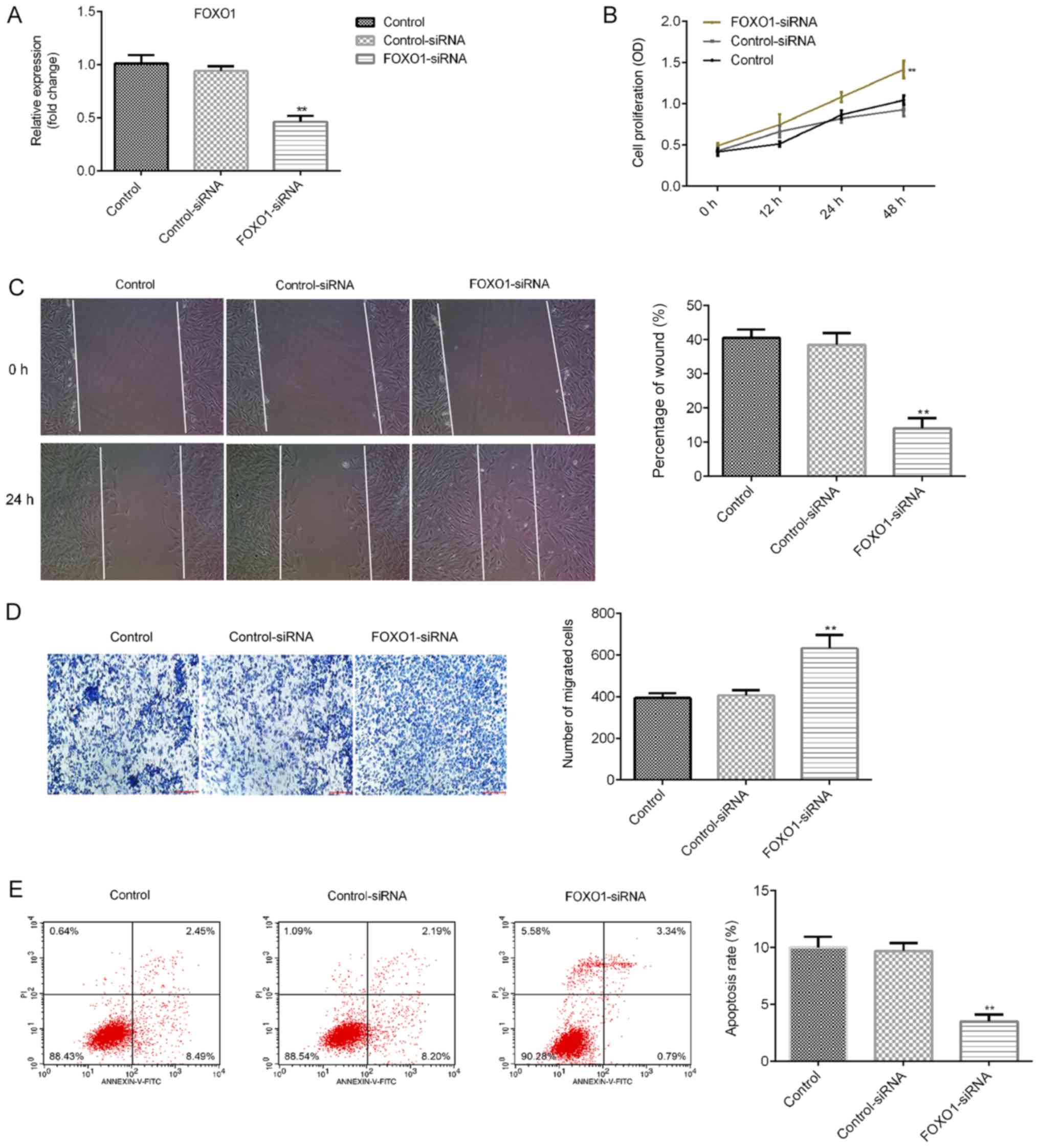
Effect of FOXO1 silencing on OSCC
cells
The effect of FOXO1 silencing on SCC9 cells was
determined. SCC9 cells were transfected with control-siRNA or
FOXO1-siRNA for 48 h, after which the transfection efficiency was
detected via RT-qPCR. As presented in Fig. 3A, when compared with the
control-siRNA group, FOXO1-siRNA significantly reduced the mRNA
levels of FOXO1 in SCC9 cells. Further analysis indicated that
compared with the control-siRNA group, FOXO1-siRNA significantly
increased the proliferation (Fig.
3B) and migration of SCC9 cells (Fig. 3C and D), and inhibited cell apoptosis (Fig. 3E). These results indicated that
miR-196a might affect SCC9 cell proliferation, migration and
apoptosis by regulating FOXO1 expression.
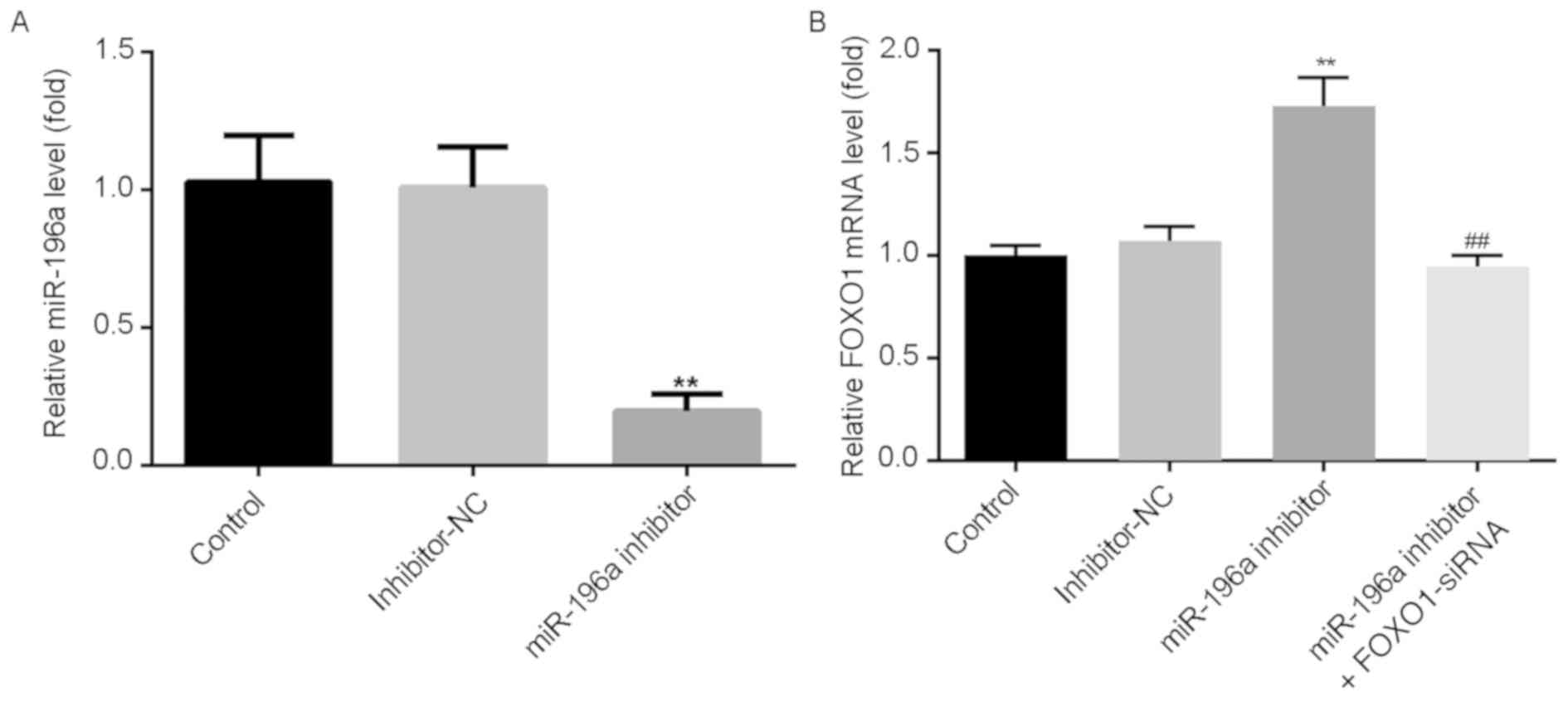
Downregulation of miR-196a resulted in
FOXO1 accumulation and FOXO1 depletion had the opposite effect
Whether miR-196a could reduce the expression of
FOXO1 was investigated by transfecting SCC9 cells with miR-196a
inhibitor, inhibitor-NC or miR-196a inhibitor + FOXO1-siRNA for 48
h. The results revealed that compared with the inhibitor-NC group,
the expression of miR-196a in SCC9 cells was significantly
downregulated following transfection with miR-196a inhibitor
(Fig. 4A). Furthermore, compared to
the inhibitor-NC group, miR-196a inhibitor increased the mRNA
expression of FOXO1 in SCC9 cells, whereas FOXO1-siRNA had the
opposite effect (Fig. 4B). The
results indicated that miR-196a negatively regulated FOXO1
expression in SCC9 cells.
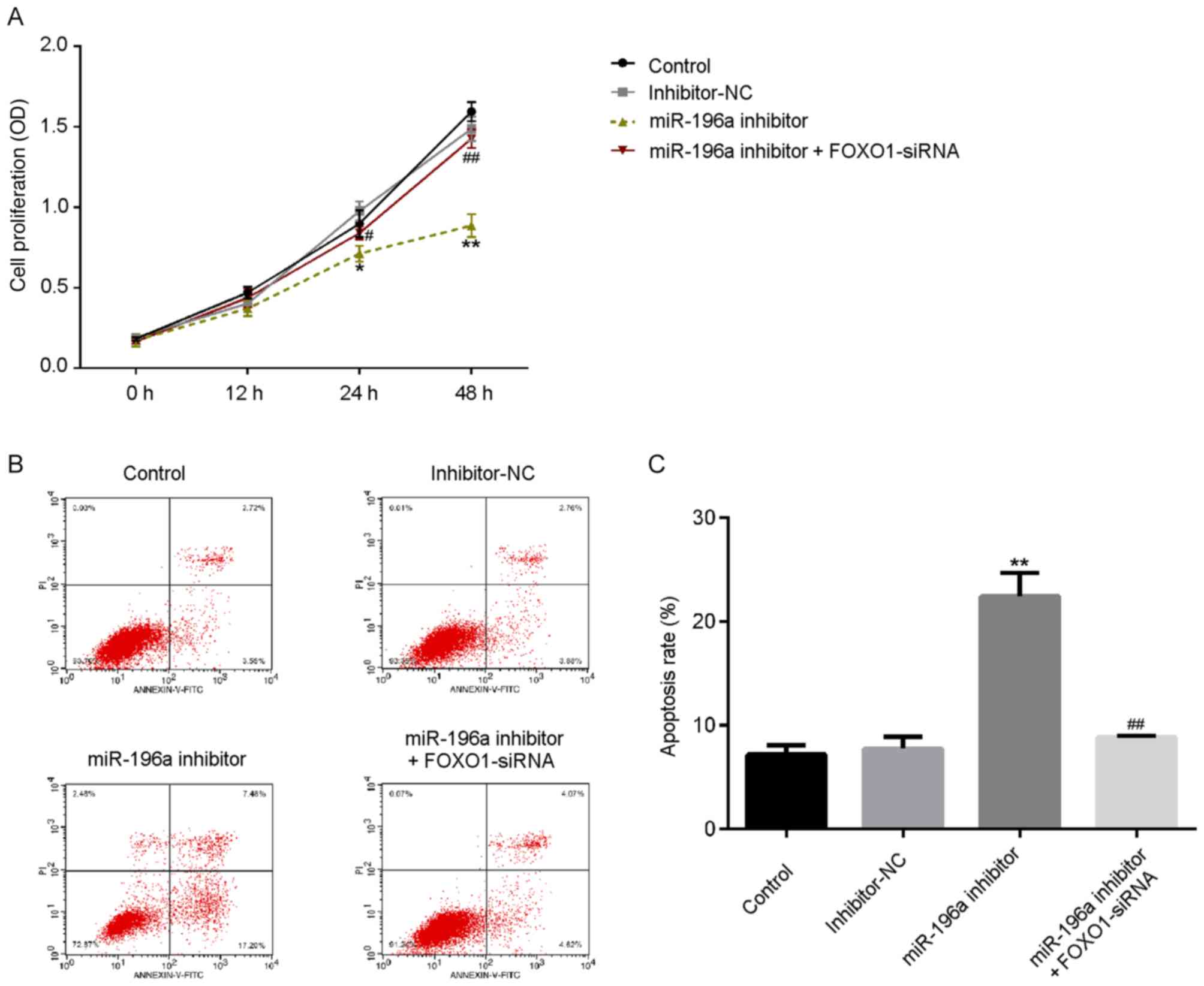
miR-196a inhibitor suppresses cell
proliferation and induces SCC9 cell apoptosis
To investigate the role of miR-196a in OSCC
proliferation, SCC9 cells were transfected with miR-196a inhibitor,
inhibitor-NC or miR-196a inhibitor + FOXO1-siRNA for 48 h. As
presented in Fig. 5A, transfection
of miR-196a inhibitor markedly decreased the proliferation of SCC9
cells, which was subsequently reversed by FOXO1-siRNA.
Additionally, the results revealed that apoptosis rate was
increased in the miR-196a inhibitor group, while this increase was
counteracted by FOXO1-siRNA (Fig. 5B
and C). The results indicated that
miR-196a may serve a vital role in OSCC proliferation and
apoptosis.
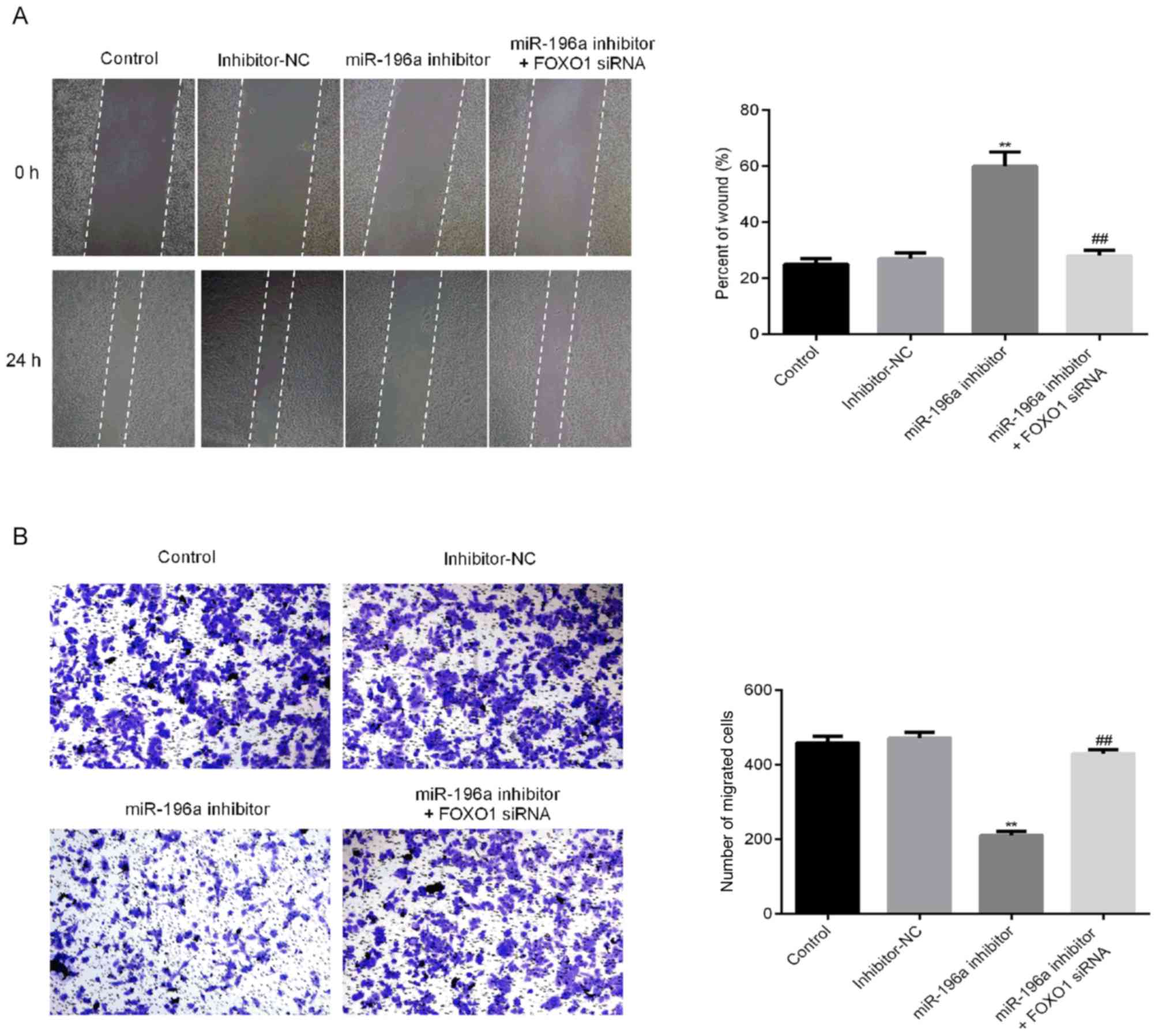
Downregulation of miR-196a inhibited
the migration and invasion capacity of SCC9 cells
To determine the potential regulatory role of
miR-196a in the migration of OSCC cells, wound healing and
transwell invasion assays were performed (Fig. 6A and B). The migration of SCC9 cells in the
miR-196a inhibitor group was significantly decreased, which was
reversed by FOXO1-siRNA treatment. These results indicated that
miR-196a may serve a vital role in the migration of OSCC cells.
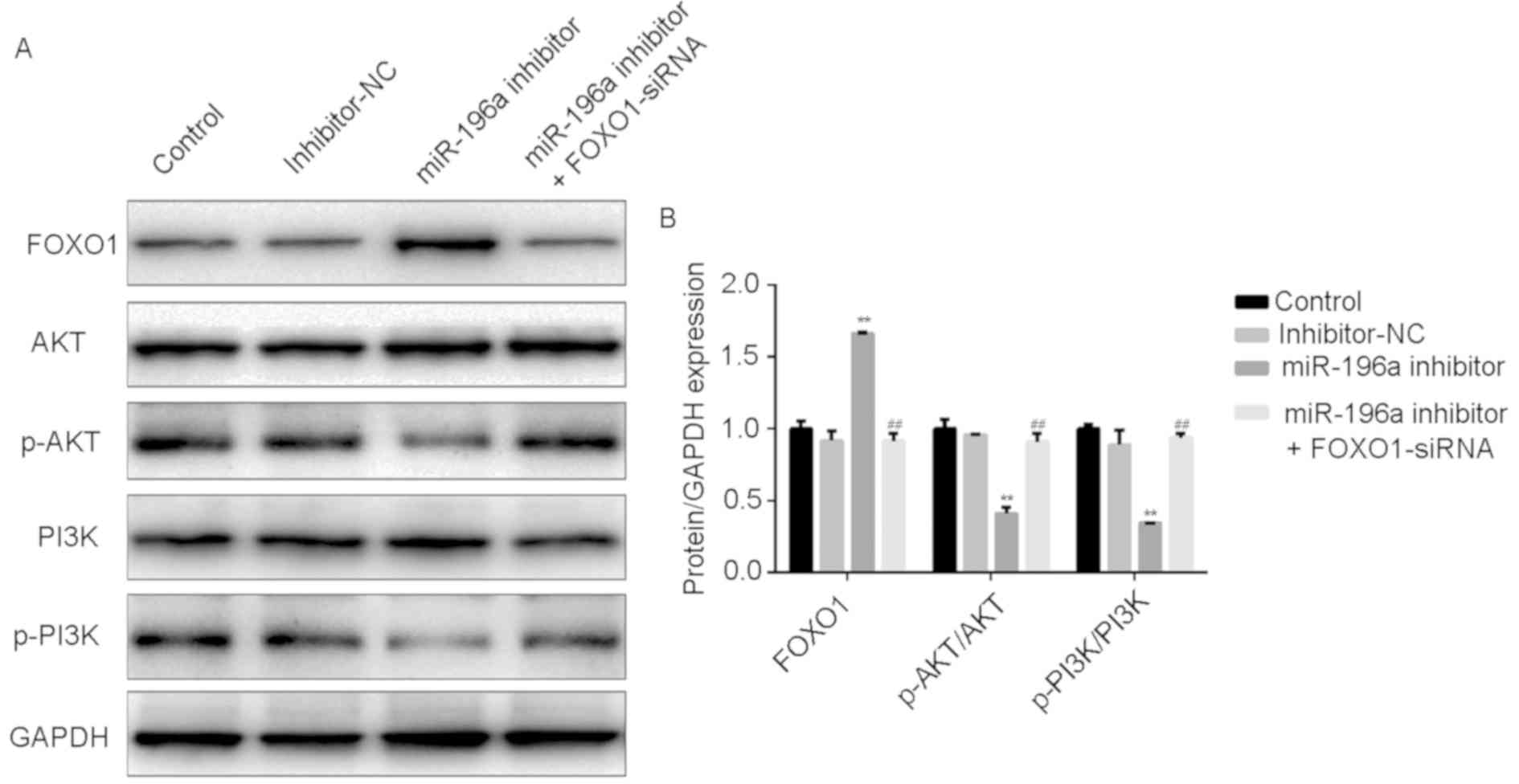
The role of miR-196a in the PI3K/Akt
pathway
To further determine the function of miR-196a in
OSCC cells, western blotting was performed to detect the effect of
miR-196a on the expression of PI3K/Akt pathway proteins. After
transfection, FOXO1 levels and the ratio of
phosphorylated/unphosphorylated PI3K and Akt in SCC9 cells were
measured via western blotting. In SCC9 cells, the downregulation of
miR-196a significantly reduced the protein expression of p-PI3K and
Akt, while the expression of FOXO1 was significantly increased.
Additionally, these effects were reversed by FOXO-siRNA (Fig. 7A and B). These data indicated that miR-196a
modulated the PI3K/Akt signaling pathway by targeting FOXO1 in
OSCC.
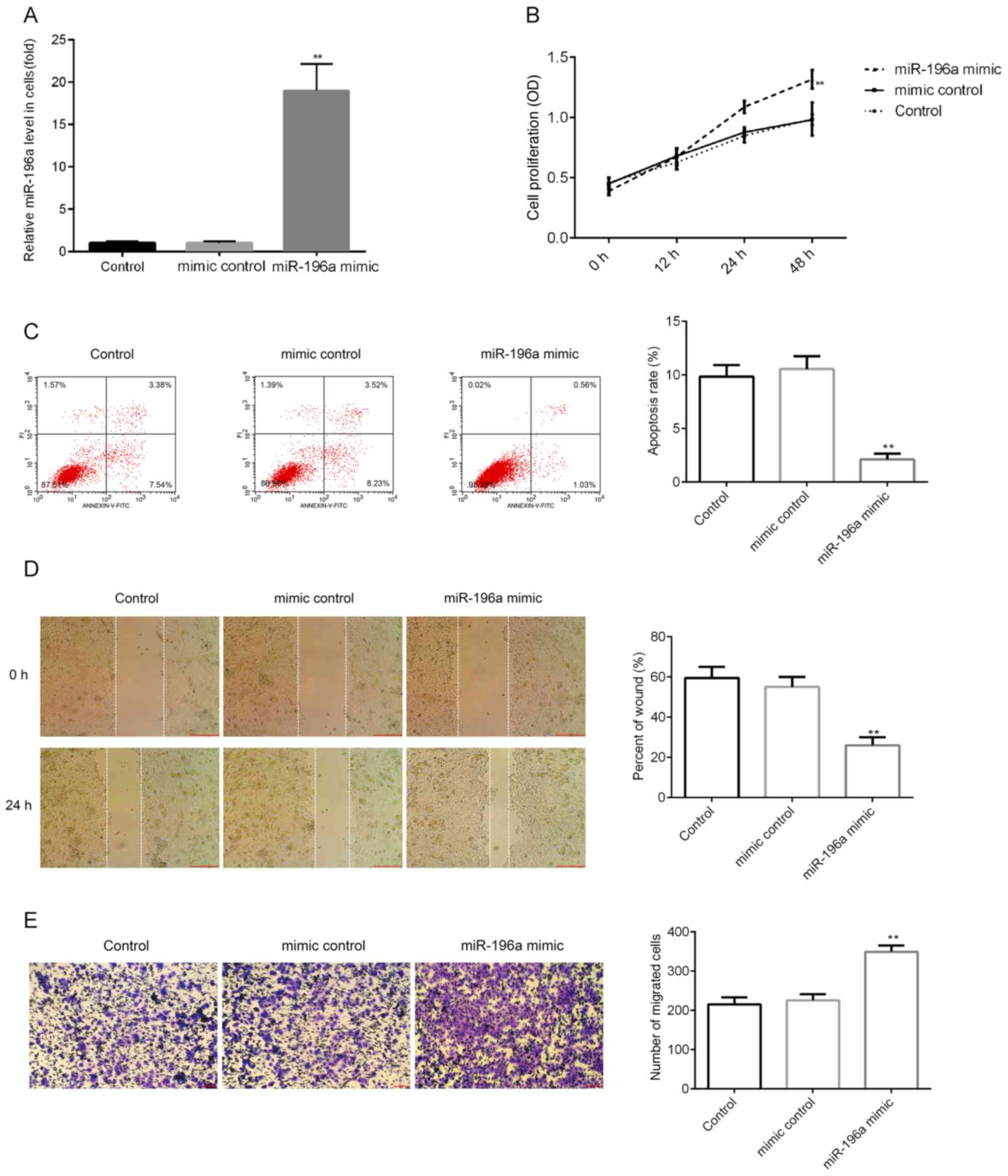
miR-196a mimic promotes cell
proliferation, induces apoptosis and enhances migration in SCC9
cells
The effect of miR-196a upregulation on SCC9 cells
was examined in the current study. SCC9 cells were transfected with
mimic control or miR-196a mimic for 48 h, after which RT-qPCR was
performed to detect transfection efficiency. The results indicated
that miR-196a mimic significantly enhanced miR-196a expression in
SCC9 cells (Fig. 8A). Further
analysis indicated that miR-196a mimic significantly enhanced SCC9
cell proliferation (Fig. 8B),
inhibited cell apoptosis (Fig. 8C)
and enhanced migration (Fig. 8D and
E).
Discussion
Cancer cell invasion and metastasis is arguably the
primary reason of mortality in patients with OSCC (26). Elucidating the underlying molecular
mechanisms of OSCC may therefore allow for the determination of
novel treatment strategies for patients with OSCC. Consistent with
previous research (27), the present
study indicated that miRNA served crucial roles in OSCC cell
invasion and metastasis. miR-196a acted as a negative regulator of
tumor-suppressor genes, promoting cell proliferation and regulating
the expression of several genes including FOXO1, p27, IκBα,
netrin4, mTOR and PI3K/AKT (28,29).
However, the biological function and underlying mechanisms of
miR-196a in OSCC invasion remain unclear.
The present study assessed whether miR-196a had an
effect on the proliferation, apoptosis and migration of OSCC cells
by regulating its target gene, FOXO1. The results of the current
study indicated that the expression of miR-196a was upregulated in
OSCC cells compared with normal HOK cells. miR-196a has been
reported to function by targeting FOXO1 in lung cancer, human liver
cancer and cervical cancer cells (15-17).
In the current study, it was demonstrated that FOXO1 was a direct
target of miR-196a. The FOXO1 gene has been demonstrated to mediate
several biological processes in cancer cells, including cell
proliferation, apoptosis, invasion, migration and angiogenesis
(30). The present study also
investigated the effect of FOXO1 silencing on OSCC cells. It was
determined that FOXO1-siRNA significantly promoted SCC9 cell
proliferation, migration and inhibited SCC9 cell apoptosis. These
results indicated that miR-196a might affect SCC9 cell
proliferation, migration and apoptosis by regulating FOXO1
expression. The results also revealed that miR-196a negatively
mediated the expression of FOXO1 in SCC9 cells. Additionally, the
results demonstrated that downregulation of miR-196a inhibited SCC9
cell growth, induced cell apoptosis and decreased cell migration in
SCC9 cells. The expression of p-PI3K, p-Akt, PI3K, Akt and FOXO1
were subsequently investigated. It was revealed that p-PI3K and
p-Akt were decreased in SCC9 cells following miR-196a
downregulation. Thus, it was hypothesized that the mRNA-196a
inhibitor may affect OSCC cancer cell processes by preventing
PI3K/Akt signaling pathway activation, which may be an important
therapeutic strategy for patients with OSCC. All the previously
described effects of miR-196a inhibitor on SCC9 cells determined in
the present study were reversed following FOXO1 silencing.
Furthermore, it was revealed that miR-196a mimic significantly
enhanced SCC9 cell proliferation and migration, and inhibited cell
apoptosis.
These data indicated that miR-196a was upregulated
in OSCC cells and that the downregulation of miR-196a inhibited
OSCC cell proliferation and migration, and induced OSCC cell
apoptosis due to its regulation of FOXO1. Therefore, the current
resulted indicated a novel therapeutic strategy for OSCC treatment.
miR-196a together with FOXO1 are hypothesized to be a potential
clinical therapy for treating patients with OSCC. However, the
current study is only a preliminary investigation into the role of
miR-196a in OSCCs. To demonstrate a more convincing role for
miR-196a in OSCC, further in-depth studies need to be performed.
For example, the role of miR-196a and FOXO1 in other OSCC cell
lines needs to be reviewed. Furthermore, the role of miR-196a and
FOXO1 in OSCCs should be investigated in vivo.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HS contributed to study design, data collection,
statistical analysis, data interpretation and manuscript
preparation. LL, ML, XW and JZ contributed to data collection and
statistical analysis. SZ contributed to data collection,
statistical analysis and manuscript preparation. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Curado MP and Hashibe M: Recent changes in
the epidemiology of head and neck cancer. Curr Opin Oncol.
21:194–200. 2009.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Warnakulasuriya S: Living with oral
cancer: Epidemiology with particular reference to prevalence and
life-style changes that influence survival. Oral Oncol. 46:407–410.
2010.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Mascolo M, Siano M, Ilardi G, Russo D,
Merolla F, De Rosa G and Staibano S: Epigenetic disregulation in
oral cancer. Int J Mol Sci. 13:2331–2353. 2012.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Sharma A, Mendez E, Yueh B, Lohavanichbutr
P, Houck J, Doody DR, Futran ND, Upton MP, Schwartz SM and Chen C:
Human papillomavirus-positive oral cavity and oropharyngeal cancer
patients do not have better quality-of-life trajectories.
Otolaryngol Head Neck Surg. 146:739–745. 2012.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Reid A, Garrett E, Dibben C and Williamson
L: ‘A confession of ignorance’: Deaths from old age and deciphering
cause-of-death statistics in Scotland, 1855-1949. Hist Fam.
20:320–344. 2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Wang Z and Cai H, Lin L, Tang M and Cai H:
Upregulated expression of microRNA-214 is linked to tumor
progression and adverse prognosis in pediatric osteosarcoma.
Pediatr Blood Cancer. 61:206–210. 2014.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Farh KK, Grimson A, Jan C, Lewis BP,
Johnston WK, Lim LP, Burge CB and Bartel DP: The widespread impact
of mammalian MicroRNAs on mRNA repression and evolution. Science.
310:1817–1821. 2005.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Nohata N, Hanazawa T, Kinoshita T, Okamoto
Y and Seki N: MicroRNAs function as tumor suppressors or oncogenes:
Aberrant expression of microRNAs in head and neck squamous cell
carcinoma. Auris Nasus Larynx. 40:143–149. 2013.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Lamouille S, Subramanyam D, Blelloch R and
Derynck R: Regulation of epithelial-mesenchymal and
mesenchymal-epithelial transitions by microRNAs. Curr Opin Cell
Biol. 25:200–207. 2013.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Ell B and Kang Y: MicroRNAs as regulators
of bone homeostasis and bone metastasis. Bonekey Rep.
3(549)2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Ge J, Chen Z, Li R, Lu T and Xiao G:
Upregulation of microRNA-196a and microRNA-196b cooperatively
correlate with aggressive progression and unfavorable prognosis in
patients with colorectal cancer. Cancer Cell Int.
14(128)2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Liu XH, Lu KH, Wang KM, Sun M, Zhang EB,
Yang JS, Yin DD, Liu ZL, Zhou J, Liu ZJ, et al: MicroRNA-196a
promotes non-small cell lung cancer cell proliferation and invasion
through targeting HOXA5. BMC Cancer. 12(348)2012.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Tsai MM, Wang CS, Tsai CY, Chen CY, Chi
HC, Tseng YH, Chung PJ, Lin YH, Chung IH, Chen CY and Lin KH:
MicroRNA-196a/-196b promote cell metastasis via negative regulation
of radixin in human gastric cancer. Cancer Lett. 351:222–231.
2014.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zhang C, Yao C, Li H, Wang G and He X:
Combined elevation of microRNA-196a and microRNA-196b in sera
predicts unfavorable prognosis in patients with osteosarcomas. Int
J Mol Sci. 15:6544–6555. 2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Guerriero I, D'Angelo D, Pallante P,
Santos M, Scrima M, Malanga D, De Marco C, Ravo M, Weisz A,
Laudanna , et al: Analysis of miRNA profiles identified
miR-196a asa crucial mediator of aberrant PI3K/AKT ignaling in lung
cancer cells. Oncotarget. 8:19172–19191. 2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Yang L, Peng F, Qin J, Zhou H and Wang B:
Downregulation of microRNA-196a inhibits human liver cancer cell
proliferation and invasion by targeting FOXO1. Oncol Rep.
38:2148–2154. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Hou T, Ou J, Zhao X, Huang X, Huang Y and
Zhang Y: MicroRNA-196a promotes cervical cancer proliferation
through the regulation of FOXO1 and p27Kip1. Br J Cancer.
110:1260–1268. 2014.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Guo H, German P, Bai S, Barnes S, Guo W,
Qi X, Lou H, Liang J, Jonasch E, Mills GB and Ding Z: The PI3K/AKT
pathway and renal cell carcinoma. J Genet Genomics. 42:343–353.
2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Gao Y, Ma C, Feng X, Liu Y and Haimiti X:
BF12, a novel benzofuran, exhibits anti-tumor activity by
inhibiting microtubules and the PI3K/Akt/mTOR signaling pathway in
human cervical cancer cells. Chem Biodivers: Jan 17, 2020 (Epub
ahead of print).
|
|
20
|
Sobočan M, Bračič S, Knez J, Takač I and
Haybaeck J: The Communication Between the PI3K/AKT/mTOR pathway and
Y-box binding protein-1 in gynecological cancer. Cancers (Basel).
12(E205)2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
An J, He H, Yao W, Shang Y, Jiang Y and Yu
Z: PI3K/Akt/FoxO pathway mediates glycolytic metabolism in HepG2
cells exposed to triclosan (TCS). Environ Int.
136(105428)2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Nozhat Z, Mohammadi-Yeganeh S, Azizi F,
Zarkesh M and Hedayati M: Effects of metformin on the
PI3K/AKT/FOXO1 pathway in anaplastic thyroid Cancer cell lines.
Daru. 26:93–103. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zheng M, Cao MX, Yu XH, Li L, Wang K, Wang
SS, Wang HF, Tang YJ, Tang YL and Liang XH: STAT3 promotes invasion
and aerobic glycolysis of human oral squamous cell carcinoma via
inhibiting FoxO1. Front Oncol. 9(1175)2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Gao C, Ren C, Liu Z, Zhang L, Tang R and
Li X: GAS5, a FoxO1-actived long noncoding RNA, promotes
propofol-induced oral squamous cell carcinoma apoptosis by
regulating the miR-1297-GSK3β axis. Artif Cells Nanomed Biotechnol.
47:3985–3993. 2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Steeg PS: Tumor metastasis: Mechanistic
insights and clinical challenges. Nat Med. 12:895–904.
2006.PubMed/NCBI View
Article : Google Scholar
|
|
27
|
Sharma M, Sah P, Sharma SS and
Radhakrishnan R: Molecular changes in invasive front of oral
cancer. J Oral Maxillofac Pathol. 17:240–247. 2013.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Huang WC, Chan SH, Jang TH, Chang JW, Ko
YC, Yen TC, Chiang SL, Chiang WF, Shieh TY, Liao CT, et al:
MiRNA-491-5p and GIT1 serve as modulators and biomarkers for oral
squamous cell carcinoma invasion and metastasis. Cancer Res.
74:751–764. 2014.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Nagai H, Hasegawa S, Uchida F, Terabe T,
Ishibashi Kanno N, Kato K, Yamagata K, Sakai S, Kawashiri S, Sato
H, et al: MicroRNA-205-5p suppresses the invasiveness of oral
squamous cell carcinoma by inhibiting TIMP2 expression. Int J
Oncol. 52:841–850. 2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Feng X, Luo Q, Wang H, Zhang H and Chen F:
MicroRNA-22 suppresses cell proliferation, migration and invasion
in oral squamous cell carcinoma by targeting NLRP3. J Cell Physiol.
233:6705–6713. 2018.PubMed/NCBI View Article : Google Scholar
|