Introduction
The contribution of inflammation to wound healing
has attracted considerable attention as a research topic. Wound
healing is a dynamic process and is usually divided into 3 main
phases: Inflammation; tissue formation; and tissue remodeling.
These 3 phases are overlapping. Inflammation is an adaptive
response to tissue stress (1,2). Acute
inflammation is important for wound healing. The infiltrating
immune cells aid the degradation of debris and decrease the number
of invading pathogens. Recruited monocytes can release growth
factors to initiate the formation of granulation tissue (3). However, chronic inflammation can
trigger delayed healing responses. In certain cases, this process
can lead to tissue dysfunction and even carcinogenesis (4). Contrasting behaviors have been observed
in several specific inflammatory factors; for example, interleukin
(IL)-17A can promote enterocyte proliferation and maintain
epithelial barrier integrity in the intestine (5). By contrast, it has also exhibited an
inflammatory role in tumorigenesis (6,7). The
exact role of inflammation in wound healing is yet to be
identified. The mechanisms involved may be used to promote
identification of potential therapeutic targets in tissue repair
and regeneration.
The state between basal homeostatic conditions and
inflammation has been described as para-inflammation (1), which is an adaptive response of the
immune system to low levels of tissue stress (8). The physiological role of
para-inflammation is to maintain homeostasis and restore tissue
functionality (1). This chronic
low-grade inflammation has also been demonstrated to participate in
several pathological conditions. In patients with age-associated
macular degeneration, the by-products of oxidative stress, such as
C-reactive protein and complement, can trigger the
pathophysiological para-inflammatory process (9). This dysregulated para-inflammatory
response contributes to macular damage (8). Notably, para-inflammation serves a more
complex role in tumor development (10), and can repress or promote
tumorigenesis, depending on the activity of the tumor protein
p53(11). However, the association
between para-inflammation and wound healing remains unclear. A more
thorough understanding of para-inflammation has considerable
biological and clinical relevance. It could be hypothesized that a
disruption of para-inflammation may affect cutaneous wound
repair.
Based on the aforementioned data, the present study
aimed to investigate the association between para-inflammation and
tissue repair using a murine cutaneous wound healing model. The
results demonstrated that the expression levels of genes associated
with para-inflammation were significantly altered during the wound
healing process. The analysis performed to identify the
para-inflammation-associated genes in wound tissues revealed
elevated expression levels of solute carrier family 7 member 11
(Slc7a11) in IL-4-induced alternative-activated macrophages.
The inhibition of para-inflammation with sulindac inhibited the
wound healing process. The present study provided novel insights
into the mechanism of wound healing and tissue repair.
Materials and methods
Reagents
HyClone Dulbecco's Modified Eagle Medium (DMEM) was
obtained from GE Healthcare. Fetal bovine serum (FBS) was obtained
from Gibco; Thermo Fisher Scientific, Inc. Murine IL-4 and
interferon-γ (IFNγ) were purchased from PeproTech, Inc. Sulindac
was obtained from Xiya Chemical Co., Ltd. The cystine/glutamate
transporter (xCT) monoclonal antibody was purchased from Abcam
(cat. no. ab175186). GAPDH rabbit antibody was purchased from Cell
Signaling Technology (cat. no. 2118). The horseradish peroxidase
(HRP)-conjugated goat anti-rabbit IgG was purchased from OriGene
Technologies, Inc. (cat. no. ZB 2301).
Cell culture
Bone marrow-derived macrophages (BMDMs) were
cultured as previously described (12), with certain modifications. All animal
procedures were approved by the Sichuan University Institutional
Animal Care and Use Committee. Briefly, the bone marrow was flushed
from the femurs and tibias of 3 C57BL/6 female mice (8-10 weeks
old). The red blood cells were lysed and the mononuclear cells were
maintained in DMEM supplemented with 10% FBS and 100 ng/ml
macrophage colony-stimulating factor for differentiation. Following
7 days of culture, non-adherent cells were removed, and adherent
cells were stimulated by treatment with 20 ng/ml murine IL-4 or
IFNγ for an additional 24 h. Cells were harvested for analysis.
Wound healing model
A total of 22 mice were purchased from Vital River
Laboratories, Co., Ltd. (Beijing, China) and maintained under
specific-pathogen-free conditions between 21 and 27˚C, humidity of
between 40 and 60%, a light/dark cycle of 12 h and ad
libitum access to food and water. The process of murine wound
healing model preparation was performed as previously described
(13,14), with certain modifications. Briefly,
19 female C57BL/6 mice (8-10 week-old) were used to establish an
in vivo wound healing model. Prior to the production of the
wound, the fur on the back of the mice was shaved following
anesthetization. The tissue area was sterilized, the dorsal skin
was stretched at the midline and the tissue was penetrated
generating two full-thickness wounds of 6 mm in diameter on each
side of the midline. For sulindac treatment, the mice were treated
intraperitoneally (i.p.) with 20 mg/kg sulindac for 8 days
consecutively (n=5). The control group (n=5) received a vehicle
solution i.p., which was 5% DMSO, 30% PEG400 and 65% normal saline.
Wound-bearing mice were held carefully during treatment and
examination to avoid secondary trauma. Wound-bearing mice were also
kept in separate cages to avoid secondary trauma. During the wound
healing process, mice were observed for the presence of specific
endpoints, including abnormal bleeding, discharge or severe
infection in the wounds. Images of each wound were captured using a
digital camera (Sonic; Sony Corporation) at the indicated time
points. The degree of wound closure was determined using images as
processed using Adobe Photoshop CS6 (Adobe Systems, Inc.). The
wound area (%) was calculated as follows: (S0-St)/S0x100, where S0
was the original wound area on day 0 and St was the wound area on
the indicated day.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
The wounded tissues were collected at the indicated
times (n=3/group at each time point). Normal skin tissues were used
as controls. Total RNA was extracted from whole tissue or cultured
macrophages using an RNAsimple total RNA extraction kit (Tiangen
Biotech Co., Ltd.). Total RNA was reverse transcribed into cDNA
using reverse transcriptase (Takara Bio, Inc.,) according to the
manufacturer's protocol. qPCR was performed using SYBR®
Premix Ex Taq II (Takara Bio, Inc.,) with specific primer sets. The
PCR assay was performed on a CFX 96 Real-time PCR thermal cycler
(Bio-Rad Laboratories, Inc.). The sequences of the primers used
were as follows: Insulin like growth factor binding protein 4
(Igfbp4) forward, 5'-GGAGCTGTCGGAAATCGAAG-3'; Igfbp4
reverse, 5'-TTGAAGCTGTTGTTGGGATG-3'; lactoperoxidase (Lop)
forward, 5'-TGACCTTGCTCCAGACTGC-3'; Lop reverse,
5'-TTGACCCAGACCTTGACCTC-3'; prostaglandin E synthase (Ptges)
forward, 5'-AGCACACTGCTGGTCATCAA-3'; Ptges reverse,
5'-TCCACATCTGGGTCACTCCT-3'; Slc7a11 forward,
5'-TCTGGTCTGCCTGTGGAGTA-3'; Slc7a11 reverse,
5'-CAAAGGACCAAAGACCTCCA-3'; SRY-box transcription factor 17
(Sox17) forward, 5'-TGAAATATGGCCCACTCACA-3'; Sox17
reverse, 5'-CTGTCTTCCCTGTCTTGGTTG-3'; SRY-box transcription factor
4 (Sox4) forward, 5'-AATTGCACCAACTCCTCAGC-3'; Sox4
reverse, 5'-TCGATTGCAGTTCACGAGAG-3'; TNF receptor superfamily
member 8 (Tnfrsf8) forward, 5'-GAGACTCGGGAAGCCAAGAT-3';
Tnfrsf8 reverse, 5'-GGTGGTCTTGAGTGGTCGAT-3'; toll like
receptor (Tlr) 1 forward, 5'-GGACCTACCCTTGCAAACAA-3';
Tlr1 reverse, 5'-TATCAGGACCCTCAGCTTGG-3'; Tlr2
forward, 5'-GAGCATCCGAATTGCATCA-3'; Tlr2 reverse,
5'-ACAGCGTTTGCTGAAGAGGA-3'; tumor necrosis factor receptor
superfamily member (TNFRSF) 11b forward,
5'-ATGAACAAGTGGCTGTGCTG-3'; TNFRSF11B reverse,
5'-TCACACAGGAGCTGATGACC-3'; TNFRSF19 forward,
5'-CGCTGCCATTCTCTTCCTAC-3'; TNFRSF19 reverse,
5'-TCGATCCTTGAATTCCTGCT-3'; interleukin 1 receptor antagonist
(Il1rn) forward, 5'-TTGTGCCAAGTCTGGAGATG-3'; Il1rn
reverse, 5'-TTCTCAGAGCGGATGAAGGT-3'; 18S rRNA forward,
5'-CGCCGCTAGAGGTGAAATTCT-3'; 18S rRNA and reverse,
5'-CGAACCTCCGACTTTCGTTCT-3'.
Western blot analysis
BMDMs were lysed with RIPA buffer (Beyotime
Institute of Biotechnology) and centrifuged for 10 min at 4˚C, at
12,000 x g to obtain the corresponding lysates. Protein was
quantified using a bicinchoninic protein assay kit (Beyotime
Institute of Biotechnology). The cellular lysates (40 µg protein)
were resolved using a 10% SDS-polyacrylamide gel and transferred
onto a PVDF membrane, which was blocked with 5% skim milk at room
temperature for 1 h. The membrane was incubated with an antibody
against xCT (1:1,000) at 4˚C overnight and subsequently with
HRP-conjugated secondary antibody (1:10,000) at 37˚C for ١ h. The
blots were visualized using the ECL System (Thermo Fisher
Scientific, Inc.). Relative intensity of the indicated blot bands
was quantified using ImageJ software (version 1.8.0-112; National
Institutes of Health).
Statistical analysis
All data are expressed as the mean ± standard
deviation. Statistical analysis was conducted with SPSS 13.0
software (SPSS, Inc.). Statistical comparisons between two groups
were assessed using a Student's t-test. Statistical comparisons
among three groups were analyzed using the one-way analysis of
variance, followed by a Least Significant Difference post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Expression of M2 macrophage-associated
genes in the wound healing process
M2 macrophages have been demonstrated to participate
in tissue repair (15). In addition,
tissue-resident macrophages act as sentinels during homeostasis, in
order to identify and respond to intrinsic and extrinsic stimuli
(10). However, the changes in the
expression pattern of M2-associated genes in wound healing are
unclear. The present study firstly aimed to evaluate the expression
pattern of these genes in murine cutaneous wound tissues. The skin
of the mice was punctured and the wound samples were collected on
days 3 and 8 following wound creation. Normal skin tissues were
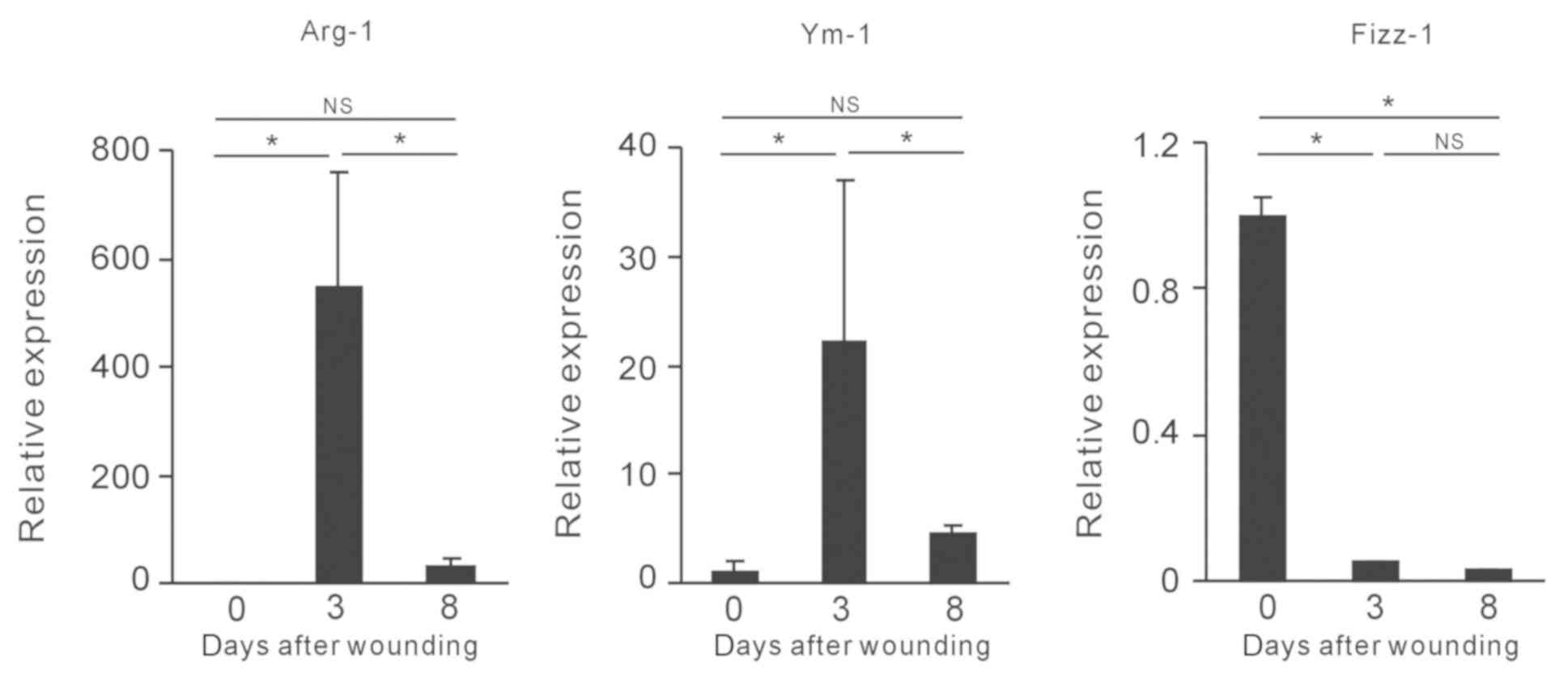
used as controls. The expression levels of well-known markers of M2
macrophages, including arginase-1 (Arg-1), Ym-1, encoded by
chitinase-like protein 3, and found in inflammatory zone (Fizz)-1,
encoded by resistin-like alpha (16,17) were
assessed by RT-qPCR. The highest expression levels of Arg-1 and
Ym-1 were observed on day 3, and were subsequently decreased on day
8 (Fig. 1). No differences in Fizz-1
expression in the wound tissues were observed between days 3 and 8
(Fig. 1). Taken collectively, the
data demonstrated that the expression patterns of the main
M2-associated genes were altered in the wound healing process.
Changes in expression of
para-inflammation-associated genes involved in the wound healing
process
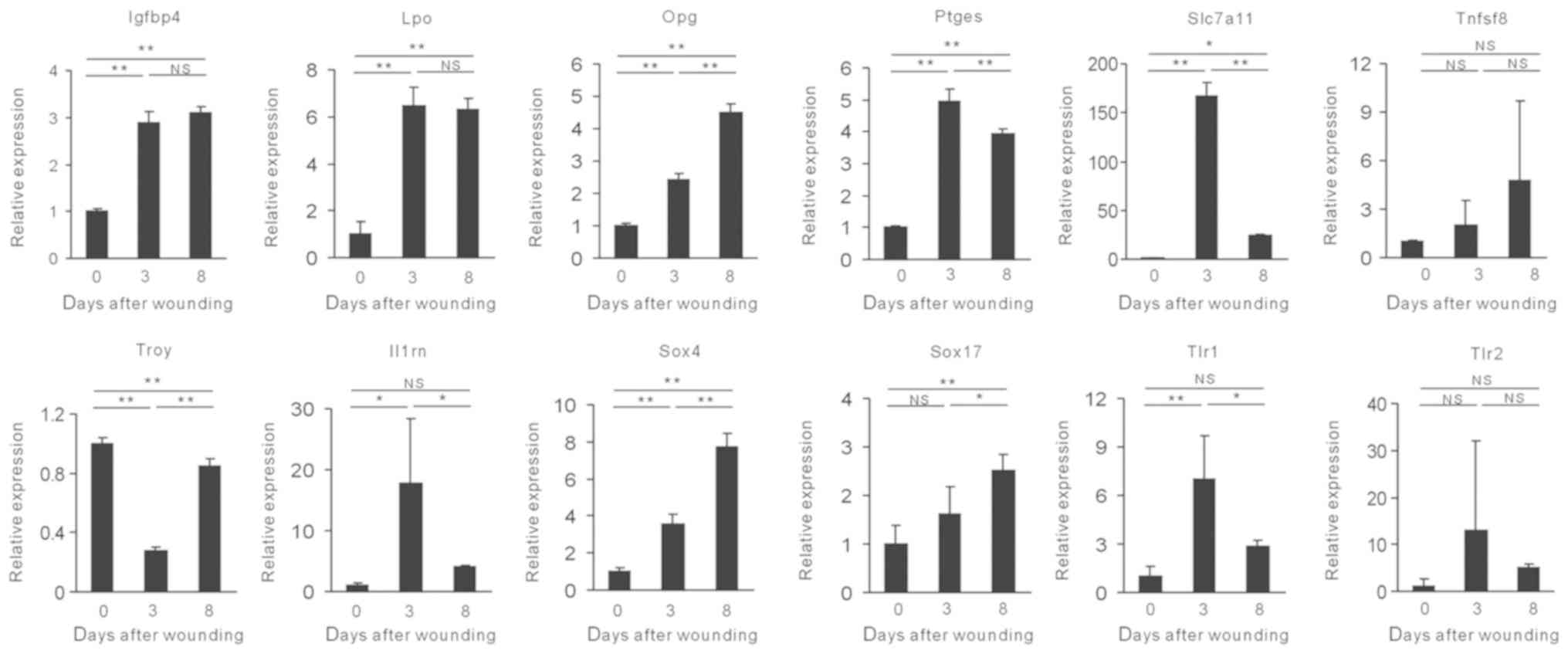
Para-inflammation was identified in a mouse model of
colorectal cancer (11). The
expression levels of the para-inflammation-associated genes were
investigated in wound tissues. The expression levels of these genes
varied according to the different time intervals (Fig. 2). Notably, some of these genes,
including Ptges, Slc7a11, Il1rn, Tlr1
and Tlr2 exhibited the highest expression levels on day 3.
The expression levels of the genes of interest were decreased on
day 8 and were similar to those of the M2-associated markers Arg-1
and Ym-1 (Fig. 2). Notably,
Slc7a11 exhibited the greatest variation during wound
healing (Fig. 2). Taken
collectively, the data suggested that the expression levels of the
genes associated with para-inflammation were significantly altered
in the wound healing process.
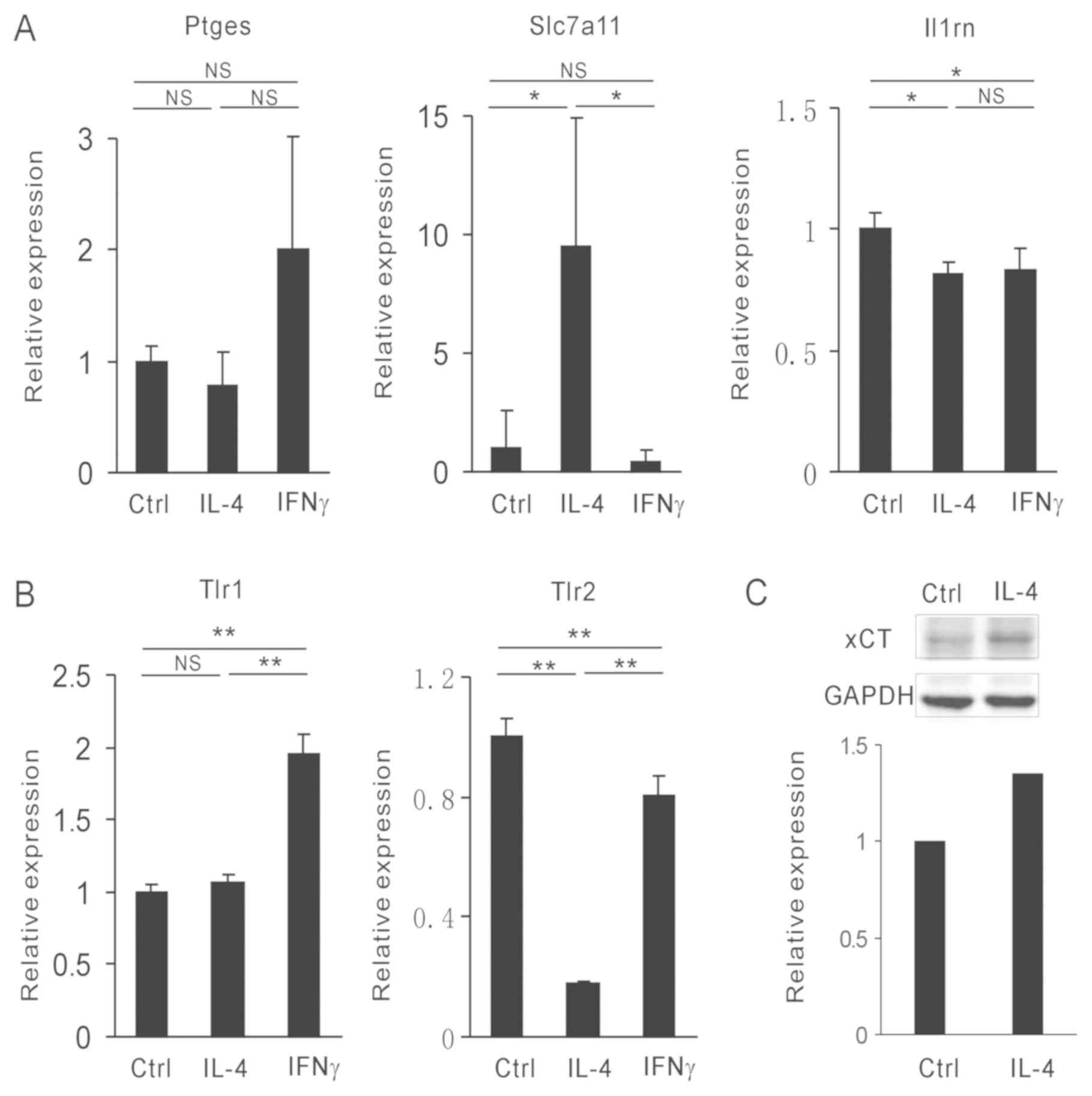
Slc7a11 is highly expressed in M2
macrophages
As the expression levels of specific
para-inflammation genes were similar to those of the M2-associated
genes, subsequent experiments focused on the expression M2
macrophage-associated genes. IFNγ and IL-4 were used to polarize
bone marrow-derived macrophages (BMDMs) into M1 and M2 type
macrophages, respectively. The expression levels of Ptges,
Slc7a11, Il-1rn, Tlr1 and Tlr2 were
analyzed. Notably, only Slc7a11 was increased in the
IL-4-induced M2 macrophages (Fig. 3A
and B). Glutamate transporter xCT is
encoded by Slc7a11 (18). To
confirm this observation, the protein levels of xCT were assessed
by western blot analysis. Following IL-4 stimulation, the levels of
xCT in BMDMs were increased. This results was consistent with that
noted for Slc7a11 mRNA levels (Fig. 3C). Taken collectively, the data
suggested that IL-4 induced Slc7a11 expression in M2
macrophages.
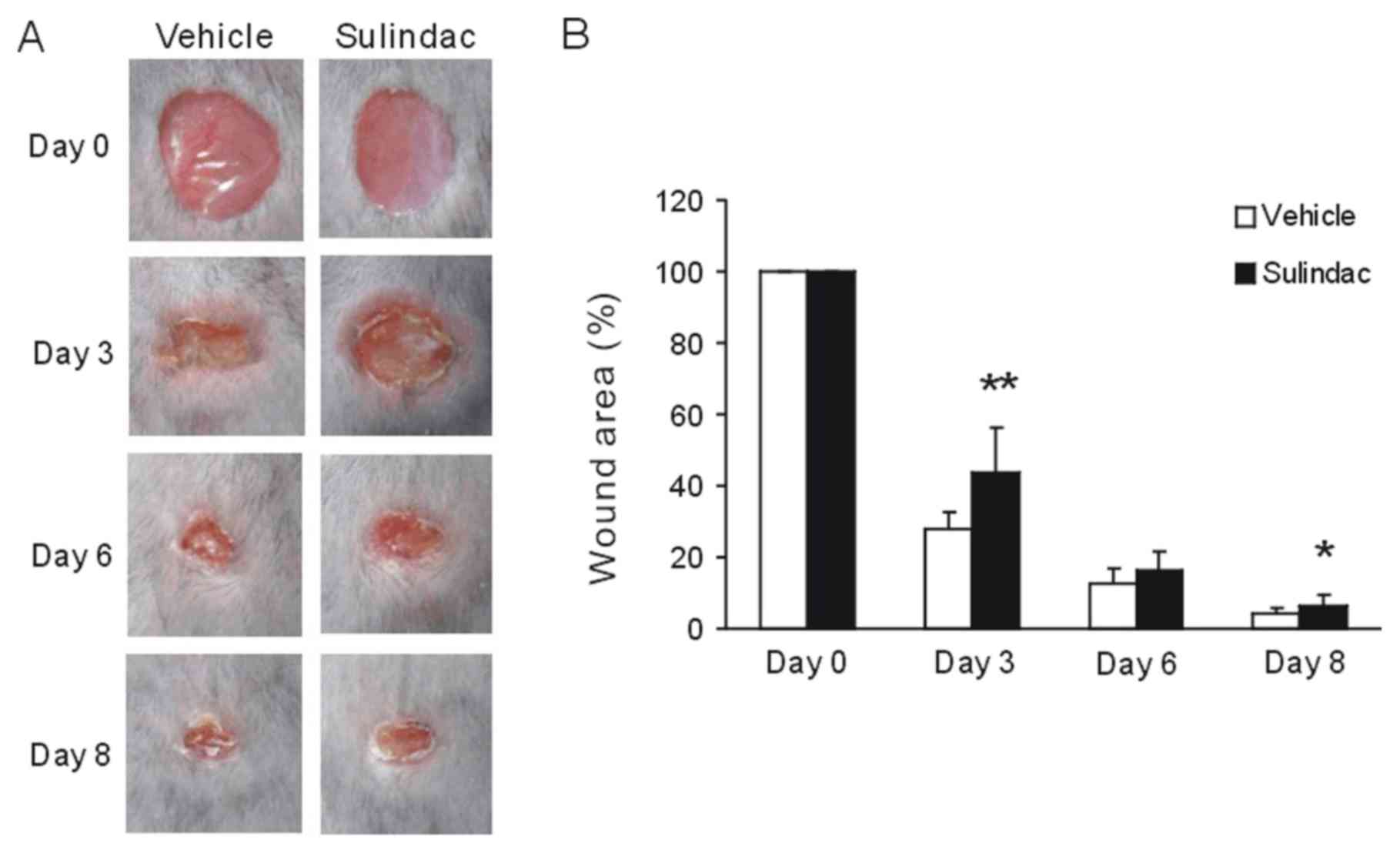
Suppression of para-inflammation by
sulindac inhibits the wound healing process
M2 macrophages have been demonstrated to participate
in the wound healing process (19).
The physiological role of para-inflammation is to reset the
homeostatic threshold of the tissue and restore tissue
functionality (1). The present study
explored whether para-inflammation affected the tissue repair
process in a cutaneous wound healing model. Initially, the wounds
were established in experimental mice and sulindac was subsequently
administered i.p., resulting in inhibition of para-inflammation
(11). The data demonstrated that
the wound areas in sulindac-treated groups were larger compared
with those in the control group; the most notable difference was
observed on day 3 (Fig. 4A and
B). Taken collectively, the data
suggested that the regulation of para-inflammation may inhibit the
wound healing process.
Discussion
The present study demonstrated that the expression
levels of genes associated with para-inflammation were
significantly altered during the wound healing process. Among those
genes, the expression profile of Slc7a11 in the wound
tissues were similar to those associated with M2 macrophages,
demonstrating an important role in tissue repair (15). In addition, the data indicated that
inhibition of para-inflammation gene expression by sulindac
prolonged the wound closure process.
Previous studies have indicated that the
inflammation following tissue damage is a part of the protective
response of the immune system (20).
Accumulating evidence suggests that M2 macrophages exhibit
significant roles in inflammation. Besides, pro-inflammatory M1
macrophages have also been demonstrated to facilitate tissue
repair. Lipopolysaccharide (LPS) is a well-known factor used to
initiate pro-inflammatory responses in macrophages (21). However, LPS has been demonstrated to
activate microglia, which perform neuroprotection against
experimental brain injury (22).
Similarly, zymosan-activated macrophages were confirmed to induce
pro-regenerative and neurotoxic functions (23). Based on these observations,
inflammation is considered to exert pleiotropic roles in the tissue
repair and regeneration process. Para-inflammation is a state
between basal homeostatic conditions and a classic inflammatory
response (1). In a murine cutaneous
wound healing model, several para-inflammation genes were expressed
in the wounded tissues. Following administration of sulindac, a
potent para-inflammation inhibitor, the wound closure rate was
decreased. The results of the present study suggested that
para-inflammation exerted a protective role in the wound healing
process. The therapeutic strategies that induce the
para-inflammation process may be promising for tissue repair and
regeneration.
In the present study, Slc7a11 was expressed
in M2 macrophages. Initially, Slc7a11 expression levels were
similar to those of Arg-1 and Ym-1, all of which were highest on
day 3 and were subsequently decreased on day 8 following wounding.
This result was confirmed using BMDMs. In concordance with these
data, IL-4-induced M2 macrophages exhibited increased levels of
Slc7a11 mRNA expression, as demonstrated by RT-qPCR
analysis, and increased levels of xCT protein, as indicated by
western blot analysis. xCT is a member of a family of amino acid
transporters and is a key player in glutamate/cysteine/glutathione
homeostasis (18). The results were
in concordance with the results from previous studies conducted in
microglia: Although IL-4 suppressed the induction of xCT expression
in the presence of β-amyloid, it increased the expression of the
xCT protein in microglia in the absence of β-amyloid (24).
It is important to note that the present study did
not fully explore the mechanism of para-inflammation in wound
healing promotion. Firstly, the function of Slc7a11 on M2
macrophages was not fully clarified. Certain amino acids were
demonstrated to be essential for the development of M2 macrophages:
Glutamine provided UDP-GlcNac required for the N-linked glycolysis
of macrophages. In the absence of glutamine, the expression levels
of M2-associated genes were decreased in IL-4-induced M2
macrophages (25). As xCT is an
important transporter of glutamate, the function of xCT in the
development of macrophages has to be thoroughly explored. Notably,
the expression levels of xCT were also enhanced in macrophages
stimulated by LPS or by an electrophilic agent (26). High levels of reactive oxygen species
(ROS) induced tissue damage. To prevent the damage caused by ROS,
macrophages utilize a cytosolic redox-buffering system that
consists primary of glutathione (GSH) (27), which can scavenge intracellular ROS
and nitric oxide (NO (28). The
glutamate/cystine transporter system is important for transporting
cystine into cells and exporting glutamate. Cystine is then rapidly
reduced into cysteine, which is the rate-limiting precursor of GSH
(29). A recent study demonstrated
that intracellular cysteine supplied by xCT contributed to NO
production and the decrease of oxidative stress in macrophages. The
ROS levels in xCT-deficient macrophages were increased compared
with those of the wild-type cells (30). xCT deficiency in xCTmu/mu mice causes
sustained inflammation due to the impaired survival of activated
macrophages at the inflammatory site (31). Certain anti-inflammatory reagents,
such as dimethylheptyl-cannabidiol, can upregulate
Slc7a11/xCT expression (32).
Further studies are required to clarify the role of xCT on
macrophage polarization and function, in particular during wound
healing. In addition, whether there is an association between
eicosanoid levels and Slc7a11 expression remains unknown.
Aspirin is a well-known cyclooxygenase (COX) inhibitor that can
inhibit xCT mRNA and protein levels in a dose-dependent manner
(33). The exact interaction between
COX inhibitors and the expression levels of xCT requires further
clarification in future studies.
In summary, the present study demonstrated that
para-inflammation served a protective role in the wound healing
process. The results improve the current understanding of the
contribution of para-inflammation to tissue repair and its
significant potential clinical applications.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81501609, 81772487
and 81500728).
Availability of data and materials
The datasets used and/or analyzed in the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TY and GBS designed the experiments. TY and GPW
wrote the manuscript. GPW and TY performed the experiments. TY, GPW
and GBS analyzed the data. XSJ and ZXC collaborated to design
experiments and analyze the data. All authors read and approved the
final version of this manuscript.
Ethics approval and consent to
participate
All animal procedures were approved by the Sichuan
University Institutional Animal Care and Use Committee (Chengdu,
China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Medzhitov R: Origin and physiological
roles of inflammation. Nature. 454:428–435. 2008.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Medzhitov R: Inflammation 2010: New
adventures of an old flame. Cell. 140:771–776. 2010.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Singer AJ and Clark RA: Cutaneous wound
healing. N Engl J Med. 341:738–746. 1999.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Antonio N, Bonnelykke-Behrndtz ML, Ward
LC, Collin J, Christensen IJ, Steiniche T, Schmidt H, Feng Y and
Martin P: The wound inflammatory response exacerbates growth of
pre-neoplastic cells and progression to cancer. EMBO J.
34:2219–2236. 2015.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Fina D, Sarra M, Fantini MC, Rizzo A,
Caruso R, Caprioli F, Stolfi C, Cardolini I, Dottori M, Boirivant
M, et al: Regulation of gut inflammation and th17 cell response by
interleukin-21. Gastroenterology. 134:1038–1048. 2008.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Grivennikov SI, Wang K, Mucida D, Stewart
CA, Schnabl B, Jauch D, Taniguchi K, Yu GY, Osterreicher CH, Hung
KE, et al: Adenoma-linked barrier defects and microbial products
drive IL-23/IL-17- mediated tumour growth. Nature. 491:254–258.
2012.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Wang K, Kim MK, Di Caro G, Wong J,
Shalapour S, Wan J, Zhang W, Zhong Z, Sanchez-Lopez E, Wu LW, et
al: Interleukin-17 receptor a signaling in transformed enterocytes
promotes early colorectal tumorigenesis. Immunity. 41:1052–1063.
2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Chen M and Xu H: Parainflammation, chronic
inflammation, and age-related macular degeneration. J Leukoc Biol.
98:713–725. 2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Nita M, Grzybowski A, Ascaso FJ and Huerva
V: Age-related macular degeneration in the aspect of chronic
low-grade inflammation (pathophysiological parainflammation).
Mediators Inflamm. 2014(930671)2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Lasry A, Aran D, Butte AJ and Ben-Neriah
Y: Cancer cell-autonomous parainflammation mimics immune cell
infiltration. Cancer Res. 77:3740–3744. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Pribluda A, Elyada E, Wiener Z, Hamza H,
Goldstein RE, Biton M, Burstain I, Morgenstern Y, Brachya G,
Billauer H, et al: A senescence-inflammatory switch from
cancer-inhibitory to cancer-promoting mechanism. Cancer Cell.
24:242–256. 2013.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Okerblom JJ, Schwarz F, Olson J, Fletes W,
Ali SR, Martin PT, Glass CK, Nizet V and Varki A: Loss of CMAH
during human evolution primed the monocyte-macrophage lineage
toward a more inflammatory and phagocytic state. J Immunol.
198:2366–2373. 2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Nyström A, Velati D, Mittapalli VR,
Fritsch A, Kern JS and Bruckner-Tuderman L: Collagen VII plays a
dual role in wound healing. J Clin Invest. 123:3498–3509.
2013.PubMed/NCBI View
Article : Google Scholar
|
|
14
|
Qiang L, Sample A, Liu H, Wu X and He YY:
Epidermal SIRT1 regulates in ammation, cell migration, and wound
healing. Sci Rep. 7(14110)2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wynn TA and Vannella KM: Macrophages in
tissue repair, regeneration, and fibrosis. Immunity. 44:450–462.
2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Nair MG, Gallagher IJ, Taylor MD, Loke P,
Coulson PS, Wilson RA, Maizels RM and Allen JE: Chitinase and Fizz
family members are a generalized feature of nematode infection with
selective up-regulation of Ym1 and Fizz1 by antigen presenting
cells. Infect Immun. 73:385–394. 2005.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Loke P, Nair MG, Parkinson J, Guiliano D,
Blaxter M and Allen JE: IL-4 dependent alternatively-activated
macrophages have a distinctive in vivo gene expression phenotype.
BMC Immunol. 3(7)2002.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Sehm T, Fan Z, Ghoochani A, Rauh M,
Engelhorn T, Minakaki G, Dörfler A, Klucken J, Buchfelder M,
Eyüpoglu IY and Savaskan N: Sulfasalazine impacts on ferroptotic
cell death and alleviates the tumor microenvironment and
glioma-induced brain edema. Oncotarget. 7:36021–36033.
2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Lucas T, Waisman A, Ranjan R, Roes J,
Krieg T, Müller W, Roers A and Eming SA: Differential roles of
macrophages in diverse phases of skin repair. J Immunol.
184:3964–3977. 2010.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Karin M and Clevers H: Reparative
inflammation takes charge of tissue regeneration. Nature.
529:307–315. 2016.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Liu Y, Fang S, Li X, Feng J, Du J, Guo L,
Su Y, Zhou J, Ding G, Bai Y, et al: Aspirin inhibits LPS-induced
macrophage activation via the NF-kB pathway. Sci Rep.
7(11549)2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Chen Z, Jalabi W, Shpargel KB, Farabaugh
KT, Dutta R, Yin X, Kidd GJ, Bergmann CC, Stohlman SA and Trapp BD:
Lipopolysaccharide-induced microglial activation and
neuroprotection against experimental brain injury is independent of
hematogenous TLR4. J Neurosci. 32:11706–11715. 2012.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Gensel JC, Nakamura S, Guan Z, van Rooijen
N, Ankeny DP and Popovich PG: Macrophages promote axon regeneration
with concurrent neurotoxicity. J Neurosci. 29:3956–3968.
2009.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Savchenko VL: Regulation of NADPH oxidase
gene expression with PKA and cytokine IL-4 in neurons and
microglia. Neurotox Res. 23:201–213. 2013.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Jha AK, Huang SC, Sergushichev A,
Lampropoulou V, Ivanova Y, Loginicheva E, Chmielewski K, Stewart
KM, Ashall J, Everts B, et al: Network integration of parallel
metabolic and transcriptional data reveals metabolic modules that
regulate macrophage polarization. Immunity. 42:419–430.
2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Sato H, Tamba M, Ishii T and Bannai S:
Cloning and expression of a plasma membrane cystine/glutamate
exchange transporter composed of two distinct proteins. J Biol
Chem. 274:11455–11458. 1999.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Cai Y, Yang Q, Tang Y, Zhang M, Liu H,
Zhang G, Deng Q, Huang J, Gao Z, Zhou B, et al: Increased
complement C1q level marks active disease in human tuberculosis.
PLoS One. 9(e92340)2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Lewerenz J1, Hewett SJ, Huang Y, Lambros
M, Gout PW, Kalivas PW, Massie A, Smolders I, Methner A, Pergande
M, et al: The cystine/glutamate antiporter system x(c)(-) in health
and disease: From molecular mechanisms to novel therapeutic
opportunities. Antioxid Redox Signal. 18:522–555. 2013.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Cai Y, Yang Q, Liao M, Wang H, Zhang C,
Nambi S, Wang W, Zhang M, Wu J, Deng G, et al: xCT increases
tuberculosis susceptibility by regulating antimicrobial function
and inflammation. Oncotarget. 7:31001–31013. 2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Kobayashi S, Hamashima S, Homma T, Sato M,
Kusumi R, Bannai S, Fujii J and Sato H: Cystine/glutamate
transporter, system xc-, is involved in nitric oxide production in
mouse peritoneal macrophages. Nitric Oxide. 78:32–40.
2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Nabeyama A, Kurita A, Asano K, Miyake Y,
Yasuda T, Miura I, Nishitai G, Arakawa S, Shimizu S, Wakana S, et
al: xCT deficiency accelerates chemically induced tumorigenesis.
Proc Natl Acad Sci USA. 107:6436–6441. 2010.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Juknat A, Kozela E, Kaushansky N,
Mechoulam R and Vogel Z: Anti-inflammatory effects of the
cannabidiol derivative dimethylheptyl-cannabidiol-studies in BV-2
microglia and encephalitogenic T cells. J Basic Clin Physiol
Pharmacol. 27:289–296. 2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Roh JL, Kim EH, Jang H and Shin D: Aspirin
plus sorafenib potentiates cisplatin cytotoxicity in resistant head
and neck cancer cells through xCT inhibition. Free Radic Biol Med.
104:1–9. 2017.PubMed/NCBI View Article : Google Scholar
|