Introduction
Rheumatoid arthritis (RA) is a multi-systemic
inflammatory autoimmune disease that causes joint swelling, pain,
loss of function and stiffness in affected appendages, and
significantly impacts the health and quality of life of patients.
The worldwide prevalence of RA is ~0.24% and it is 2-3 times higher
in females compared with that in males (1-3).
Although the pathogenesis remains to be fully elucidated, it is
well known that inflammatory response-mediated immune dysfunction
is a key factor involved in the pathogenesis (4,5).
High-throughput sequencing is a comprehensive
analysis platform for studying differentially expressed genes in
multiple diseases (6,7). High-throughput sequencing studies have
indicated that various lncRNAs may be associated with a variety of
autoimmune diseases, including RA, systemic lupus erythematosus,
Sjogren's syndrome and multiple sclerosis (8-12).
LncRNAs are also hypothesized to be involved in the differentiation
and activation of CD4+ T cells, which are aberrantly
activated adaptive immune cells in multiple autoimmune diseases
(13-15).
Dysregulated differentiation of CD4+ T cells alter the
balance of interactions between the CD4+ T-cell
subgroups and may be an important factor leading to the development
of RA disease (16). As such, it may
be possible to identify potential therapeutic targets by analyzing
the differential expression of lncRNAs. In the present study, the
lncRNA expression profiles in the peripheral blood and
CD4+ T cells of healthy donors were compared with those
of patients with active RA using high-throughput sequencing.
Further verification was performed using quantitative (q)PCR and
the results supported the potential involvement of specific lncRNA
in CD4+ T-cell differentiation during the development of
RA. These results provide novel insight into the immunological
mechanisms underlying the development of RA.
Materials and methods
Patients and specimens
A total of 19 patients with RA (16 females and 3
males; mean age, 53 years) who visited the Department of
Rheumatology and Immunology of Jiangsu Province Hospital of Chinese
Medicine (Nanjing, China) between March 2016 and June 2016 were
recruited. Patients were recruited if they met the criteria for the
diagnosis of RA set out by the American College of Rheumatology in
1987(17). The patients recruited
were outpatients or hospitalized patients. Peripheral blood was
collected and first used to detect RA-related indicators prior to
administration of glucocorticoids and immunosuppressive agents.
Erythroid sedimentation rate (ESR) was measured using the
Westergren method. An ESR >20 mm/h was considered positive
(18). Rheumatoid factor (RF) and
C-reactive protein (CRP) were measured by immunonephelometry, RF
>15 U/ml and CRP >8 mg/l were considered positive (19,20).
Anti-cyclic citrullinated peptide (anti-CCP) antibody levels were
measured by chemiluminescence microparticle immunoassay, anti-CCP
titers <5.0 RU/ml were judged as negative, and ≥5.0 RU/ml as
positive (21). The Disease Activity
Score 28 (DAS28) were then used to assess RA disease activity. For
each patient in the validation cohort, the disease severity was
assessed according to the DAS28(22). A total of 10 healthy donors (8
females and 2 males; mean age, 50 years) with no history of
autoimmune diseases or administration of immunosuppressive therapy
were used as the control group. The present study was approved by
the Institutional Review Board (IRB) of the Affiliated Hospital of
Nanjing University of Traditional Chinese Medicine (Nanjing, China;
affiliated to Nanjing University of Chinese Medicine; approval no.
2016NL-KS14).
RNA extraction
Total RNA from peripheral blood was extracted using
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. Subsequently, total RNA
was evaluated by 1% agarose gel electrophoresis and observation
under ultraviolet transmitted light, quantified using a NanoDrop
ND-1000 (NanoDrop Technologies; Thermo Fisher Scientific,
Inc.).
High-throughput sequencing
analysis
High-throughput sequencing was performed using an
Arraystar v.3.0 (Arraystar, Inc.). Genome-wide analysis of lncRNA
and mRNA expression profiles in peripheral blood samples from two
patients with RA and two healthy donors was performed using
Arraystar Human lncRNA Microarray version 3.0. In brief, RNA
extraction and evaluation were performed as described above. Sample
labeling (Labeling Kit Agilent p n 5190-0442; Agilent Technologies,
Inc.) and array hybridization was performed according to the
Agilent One-Color Microarray-Based Gene Expression Analysis
protocol (Agilent Technologies, Inc.). Specific experimental
process: mRNA was purified from total RNA using an mRNA isolation
kit after the removal of ribosomal RNA (mRNA-ONLY™ Eukaryotic mRNA
Isolation kit; Epicentre; Illumina, Inc.). Subsequently, each
sample was amplified and transcribed into fluorescent cRNA
(complementary RNA) using a mixture of oligo (dT) and random
primers (Arraystar Flash RNA Labeling kit; Arraystar, Inc.). The
labeled cRNAs were purified using an RNeasy Mini kit (Qiagen). The
concentration and specific activity of the labeled cRNAs (pmol
cyanine 3/µg cRNA) were measured using a NanoDrop-1000 (Thermo
Fisher Scientific, Inc.). A total of 1 µg of each labeled cRNA was
fragmented by adding 5 µl 10X Blocking Agent (Agilent Technologies,
Inc.) and 1 µl of 25X Fragmentation Buffer. The mixture was heated
at 60˚C for 30 min, after which 25 µl 2x GE Hybridization buffer
(Agilent Technologies, Inc.) was added to dilute the labeled cRNA.
A total of 50 µl of hybridization solution was dispensed into a
gasket slide and assembled to obtain the lncRNA microarray slide.
The slides were incubated at 65˚C in an Agilent Hybridization oven
for 17 h. The hybridized arrays were washed, fixed and scanned with
the Agilent DNA Microarray Scanner (cat. no. G2505C; Agilent
Technologies, Inc.).
CD4+ T-cell isolation and
total RNA extraction
CD4+ T cells in the peripheral blood of
each participant were separated by gradient centrifugation at 400 x
g for 30 min at 4˚C and enriched using magnetic beads. In brief, an
equivalent volume of PBS was added to the peripheral blood and the
sample was agitated until it was evenly distributed. Subsequently,
the mixture was added to the lymphocyte separation solution,
followed by centrifugation at 400 x g for 30 min at 4˚C to obtain
peripheral blood mononuclear cells. CD4+ T cells were
isolated using a BD IMag™ cell separation protocol using the
following reagents: Anti-human CD4 particles-dm CD (cat. no.
557767) and IMag buffer (cat. no. 552362; both from BD
Biosciences). Separated CD4+ T cells were frozen in
pre-chilled RNase-free vials for 5 min in liquid nitrogen and
stored at -78˚C prior to RNA extraction.
Reverse transcription (RT)-qPCR
Of the differentially expressed lncRNAs, 21 were
selected (Table SI) according to
the following criteria: i) ≥10-fold up- or downregulation in
expression; ii) the sequence of the lncRNA was in the database
(LncRNADisease; http://cmbi.bjmu.edu.cn/lncrnadisease) and was closely
associated with RA. For RT reactions, Super Script™ III Reverse
Transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.) was
used. qPCR was performed using specific primers, the sequences of
which are provided in Table SII.
The thermocycling conditions were as follows: PCR assay was set at
an initial denaturation step at 95˚C for 10 min, followed by 40
cycles of 95˚C for 10 sec and 60˚C for 1 min. The ViiA 7 Real-time
PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.) was
used for PCR amplification. PCR amplification was performed in
triplicate. The results were quantified using the
2-ΔΔCq method (23).
Flow cytometry and ELISA
Samples were washed with PBS and fixed with
immobilization reagent (BD Biosciences). Type 17 T-helper (Th17)
cells were stained with allophycocyanine mouse anti-human CD4 (cat.
no. 561840; BD Biosciences) for 20 min at 37˚C, phycoerythrin (PE)
mouse anti-human interleukin (IL)-17A (cat. no. 580487; BD
Biosciences) for 30 min at 4˚C, PerCP-Cy™ 5.5 mouse anti-human CD25
(cat. no. 580503; BD Biosciences) for 20 min at 4˚C, PE mouse
anti-human CD127 (cat. no. 561028; BD Biosciences) for for 20 min
at 4˚C. A total of 1x106 of cells were resuspended in 1%
BSA-PBS and the volume of fluorescent antibodies added in to cells
were as listed below: CD4: 1 µl; IL-17A: 2 µl; CD25: 2 µl and
CD127: 2 µl. After incubation with the antibodies, cells were
analyzed using flow cytometry on a BD FACSCalibur (BD
Biosciences).
ELISA kits (cat. nos. m1037279 and m1028599;
Shanghai Enzyme Link) were used to detect the levels of IL-17 and
transforming growth factor (TGF)-β in human serum (1:5 dilution),
according to the manufacturer's instructions.
Volcano plots and hierarchical cluster
analyses
Microarray data were log-transformed and subjected
to quantile normalization. After elimination of unreliable
transcripts, the remaining data of lncRNA and mRNA expression were
further analyzed. The transcripts were distributed according to
statistical significance (y-axis) and magnitude of change (log2
ratio of RA group/normal control group; x-axis). Volcano plots were
used for visualizing genes that were differentially expressed
between the two groups. Hierarchical cluster analysis was performed
to display RNA expression profiles between different samples.
Bioinformatics analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG;
www.genome.ad.jp/kegg) database and Gene
Ontology (GO; www.geneontology.org) analysis were used to explore
the potential role of differentially expressed mRNA in biological
pathways, including biological process, cellular component and
molecular function.
Co-expression network
Co-expression analysis was performed based on the
Pearson correlation coefficient between the coding and non-coding
gene transcripts. Pearson correlation coefficient ≥0.90, P<0.01
and false discovery rate <0.01 were used as thresholds and a
coding-non-coding network was plotted using Cytoscape version 2.8.2
(http://cytoscape.org).
Statistical analysis
Data were analyzed using GraphPad Prism version 5.0
(GraphPad Software, Inc.). A non-parametric Mann-Whitney U-test was
used to compare gene expression between two groups. Spearman rank
correlation analyses were performed and coefficients were
determined to assess the correlation between expression levels of
lncRNAs and clinical features. P<0.05 (bilateral) was considered
to indicate a statistically significant difference.
Results
Differentially expressed lncRNAs and
mRNAs in the peripheral blood of patients with RA
Aberrantly expressed mRNAs and lncRNAs associated
with RA were identified based on the following criteria: ≥2-fold
upregulation or <2-fold downregulation in expression and
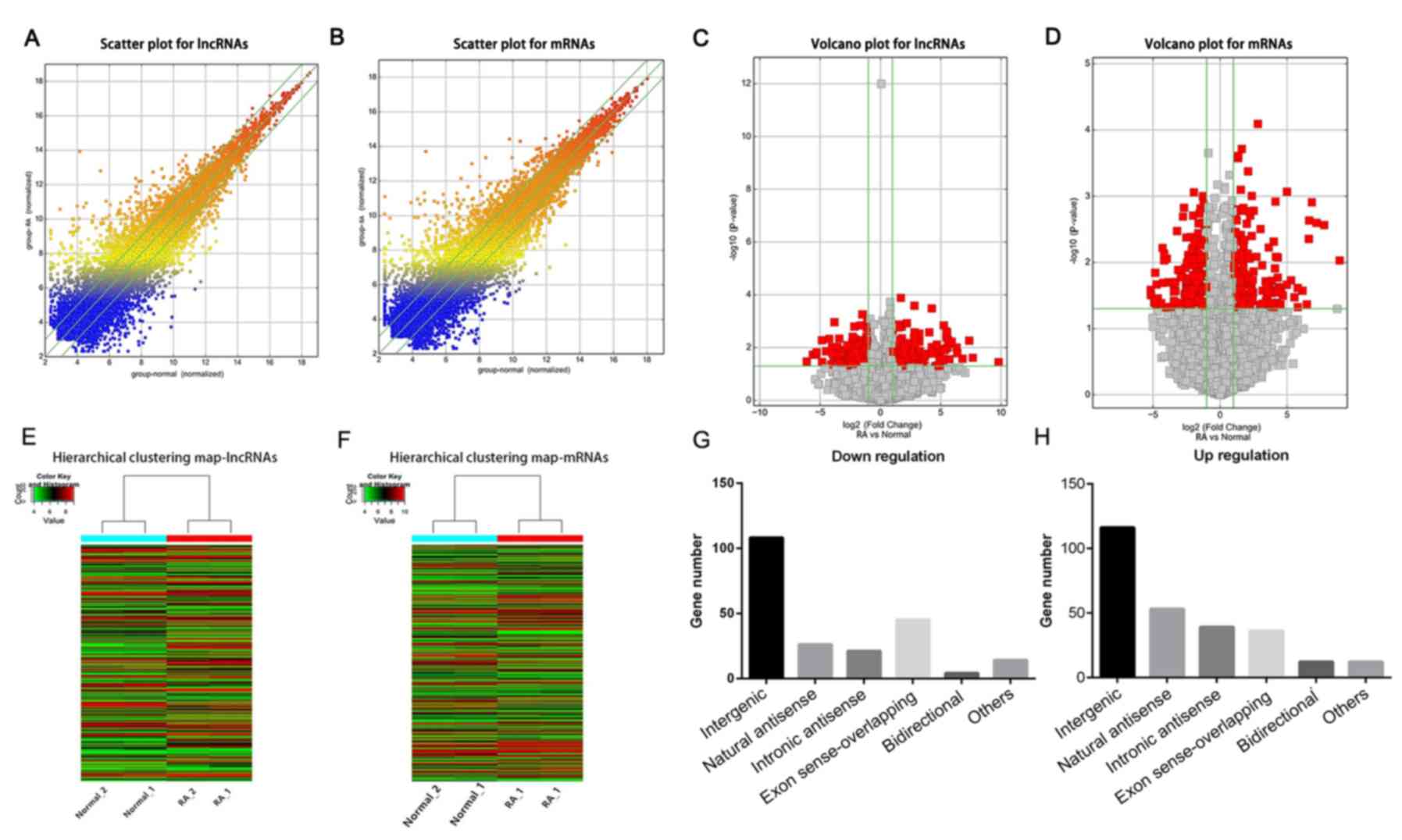
P<0.05. To visualize the differentially expressed lncRNAs and
mRNAs, scatter plots (Fig. 1A and
B) and volcano plots (Fig. 1C and D) were analyzed to further explore the
differences in peripheral blood between patients with active RA and
normal people. A total of 493 differentially expressed lncRNAs and
374 mRNAs were identified in the peripheral blood of patients with
RA, of which 275 lncRNAs and 193 mRNAs were upregulated and the
remaining ones were downregulated. The results of hierarchical
clustering analysis showed distinguishable lncRNA and mRNA
expression profiles between patients with active RA and normal
people. (Fig. 1E and F).
Classification of lncRNAs
Among the 275 significantly upregulated lncRNAs, 116
were associated with intergenic interaction, 53 were natural
antisense, 39 were intronic antisense, 36 were exon sense
overlapping, 12 were bidirectional and 12 were other types of
interactions. Of the 208 significantly downregulated lncRNAs, 108
of these were associated with intergenic interactions, 26 were
natural antisense, 21 were intronic antisense, 45 were exon sense
overlapping, 4 were bidirectional and 14 were other types of
interactions (Fig. 1G and H).
qPCR validation of lncRNA in
peripheral blood
Further identification was performed with qPCR on
peripheral blood plasma of 5 patients and 5 healthy individuals.
Based on these results, 7 of the 11 upregulated lncRNAs and 7 of
the 10 downregulated lncRNAs obtained by chip were confirmed as
differentially expressed (Fig.
2).
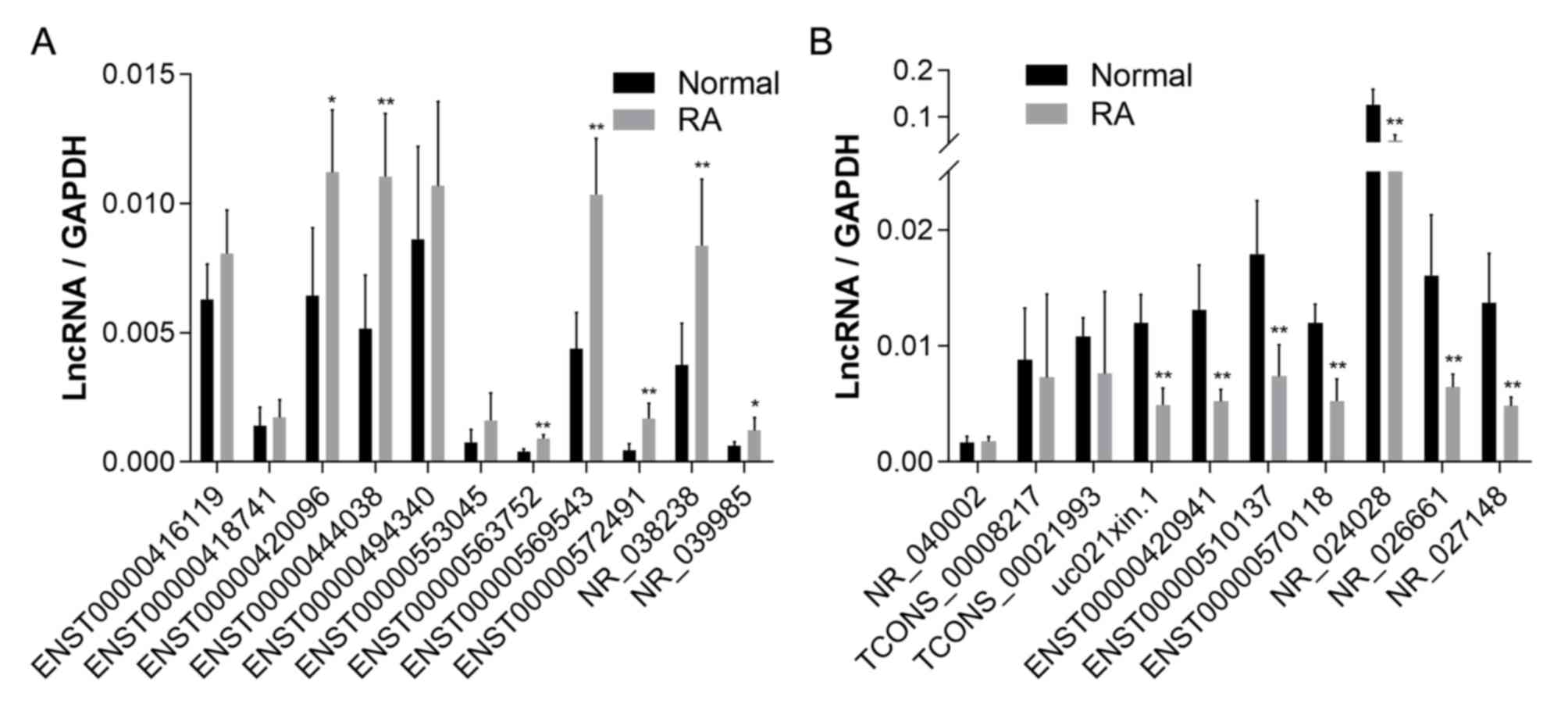 | Figure 2Relative expression of lncRNAs in
peripheral blood determined using quantitative PCR. (A) Expression
of the 11 upregulated lncRNAs (ENST00000416119, ENST00000563752,
ENST00000572491, ENST00000553045, ENST00000494340, ENST00000418741,
ENST00000569543, ENST00000420096, ENST00000444038, NR_038238 and
NR_039985) in the peripheral blood of patients with RA. (B)
Expression of the 10 downregulated lncRNAs (ENST00000510137,
ENST00000570118, NR_024028, TCONS_00021993, uc021xin.1,
TCONS_00008217, ENST00000420941, NR_026661, NR_040002 and
NR_027148) in the peripheral blood of patients with RA.
*P<0.05, **P<0.01,
***P<0.001 vs. normal. ENST, Ensembl transcript;
lncRNA, long non-coding RNA; RA, rheumatoid arthritis. |
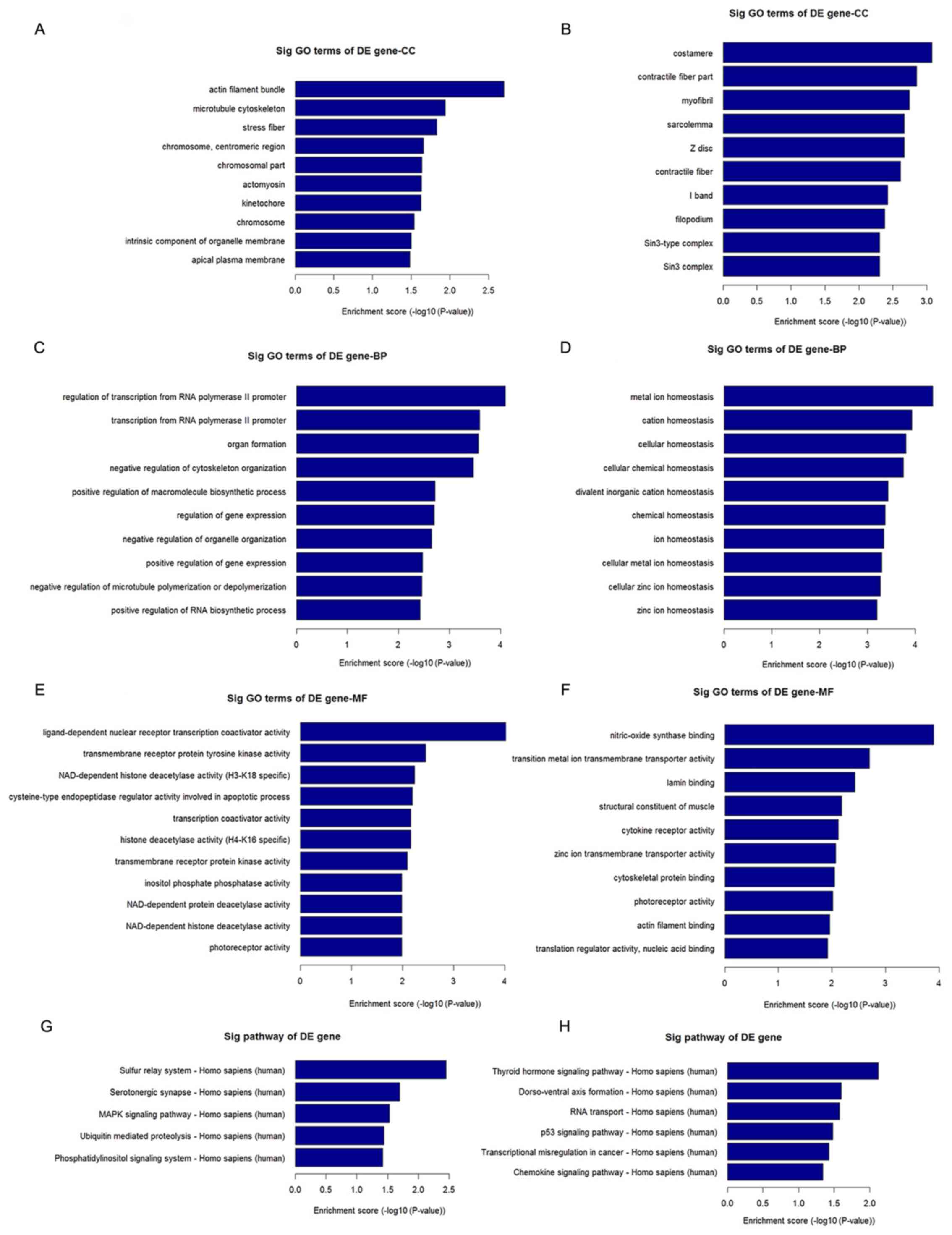
Pathway and GO analyses
As lncRNAs are non-coding RNAs that are not
translated and the majority of them are structurally similar to
mRNA, their function may be reflected in the associated mRNAs.
Pathway and GO enrichment analysis of differentially expressed
mRNAs further demonstrated the regulatory role of lncRNAs (Fig. 3). Pathway analysis indicated that
differentially expressed mRNAs in peripheral blood from patients
with RA were associated with multiple signaling pathways, including
mitogen-activated protein kinase (MAPK) and P53 (Fig. 3G and H). These mRNAs were further characterized
as being involved in the regulation of processes associated with
cell growth and immune regulation (Fig.
3A-F). Of note, the MAPK and P53 signaling pathways are
involved in the differentiation of CD4+ T cells, which
serves an important role in the pathogenesis of RA.
qPCR validation of lncRNA in
CD4+ T cells
To further investigate the alternative expression of
lncRNAs in CD4+ T cells from patients with RA,
peripheral blood was collected from 12 patients with RA and 8
healthy individuals. CD4+ T cells were isolated with
magnetic beads. Based on the qPCR results, one of the less
differentially expressed lncRNAs were excluded for analysis in
CD4+ T cells and finally, a total of 13 lncRNAs (those
previously identified in plasma) were validated. The results
suggested that the differential expression of 7 of the lncRNAs
upregulated in plasma was consistent with that observed in
CD4+ T lymphocytes. Furthermore, two of the lncRNAs
downregulated in plasma were also observed to be downregulated in
CD4+ T lymphocytes (Fig.
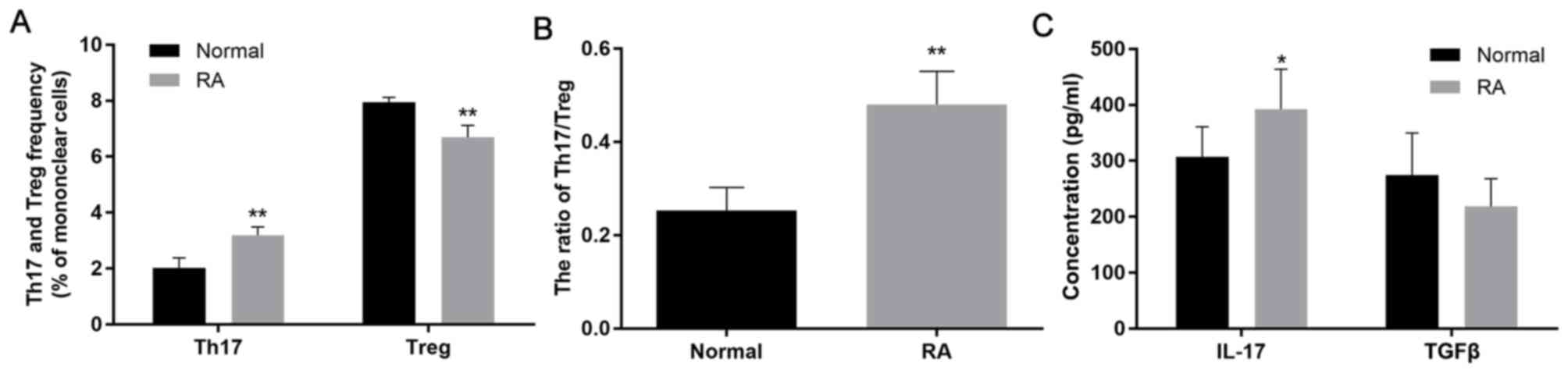
4) ifferentiation of CD4+ T cells in
patients with RA. Differentiation of CD4+ T cells
from patients with RA was observed. The ratio of Th17/T-regulatory
(Treg) cells was upregulated in patients with active RA by flow
cytometry (Fig. 5A and B). IL-17 and TGF-β are important cytokines
that are considered as regulatory sensors in RA (24,25). The
results also suggested an increase in the levels of IL-17 and a
decrease in the release of TGF-β in patients with RA, although
there was no significant difference in TGF-β compared to the normal
control group (Fig. 5C; Table I), demonstrating a difference in the
differentiation of CD4+ T cells.
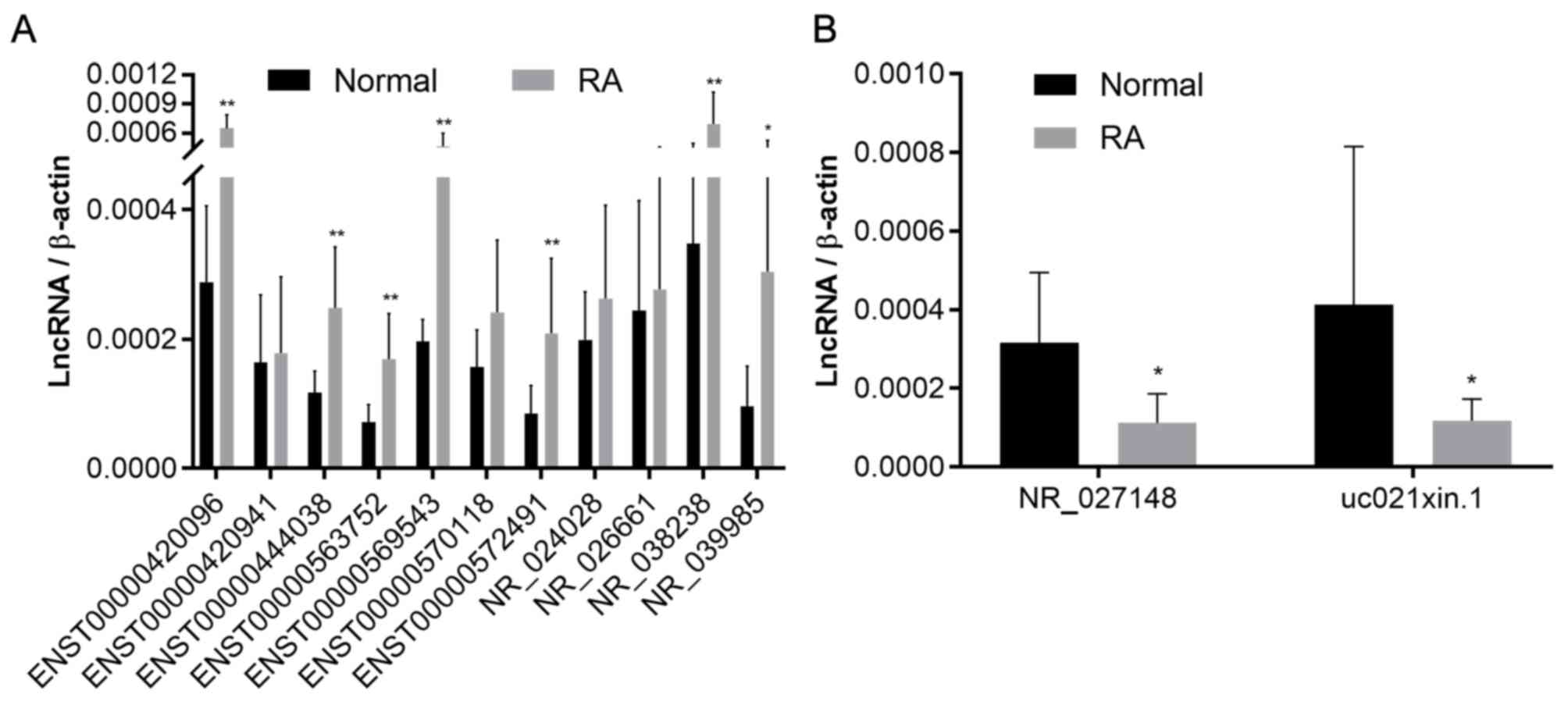 | Figure 4Relative expression of lncRNAs in
CD4+ T cells determined using quantitative PCR. (A)
Expression of ENST00000420096, ENST00000563752, ENST00000572491,
ENST00000569543, ENST00000444038, NR_039985, NR_038238, NR_026661,
NR_024028, ENST00000420941, ENST00000570118 in CD4+ T
cells. (B) Expression of uc021xin.1 and NR_027148 in
CD4+ T cells. *P<0.05,
**P<0.01 vs. normal. ENST, Ensembl transcript; RA,
rheumatoid arthritis; lncRNA, long non-coding RNA. |
 | Table IPlasma levels of IL-17 and TGF-β in
the RA group and the normal control group (pg/ml). |
Table I
Plasma levels of IL-17 and TGF-β in
the RA group and the normal control group (pg/ml).
| Group | n | IL-17 | TGF-β |
|---|
| RA patients | 12 |
392.09±71.69a | 218.47±49.81 |
| Normal | 8 | 307.27±53.87 | 274.28±75.43 |
| P-values | | 0.011 | 0.061 |
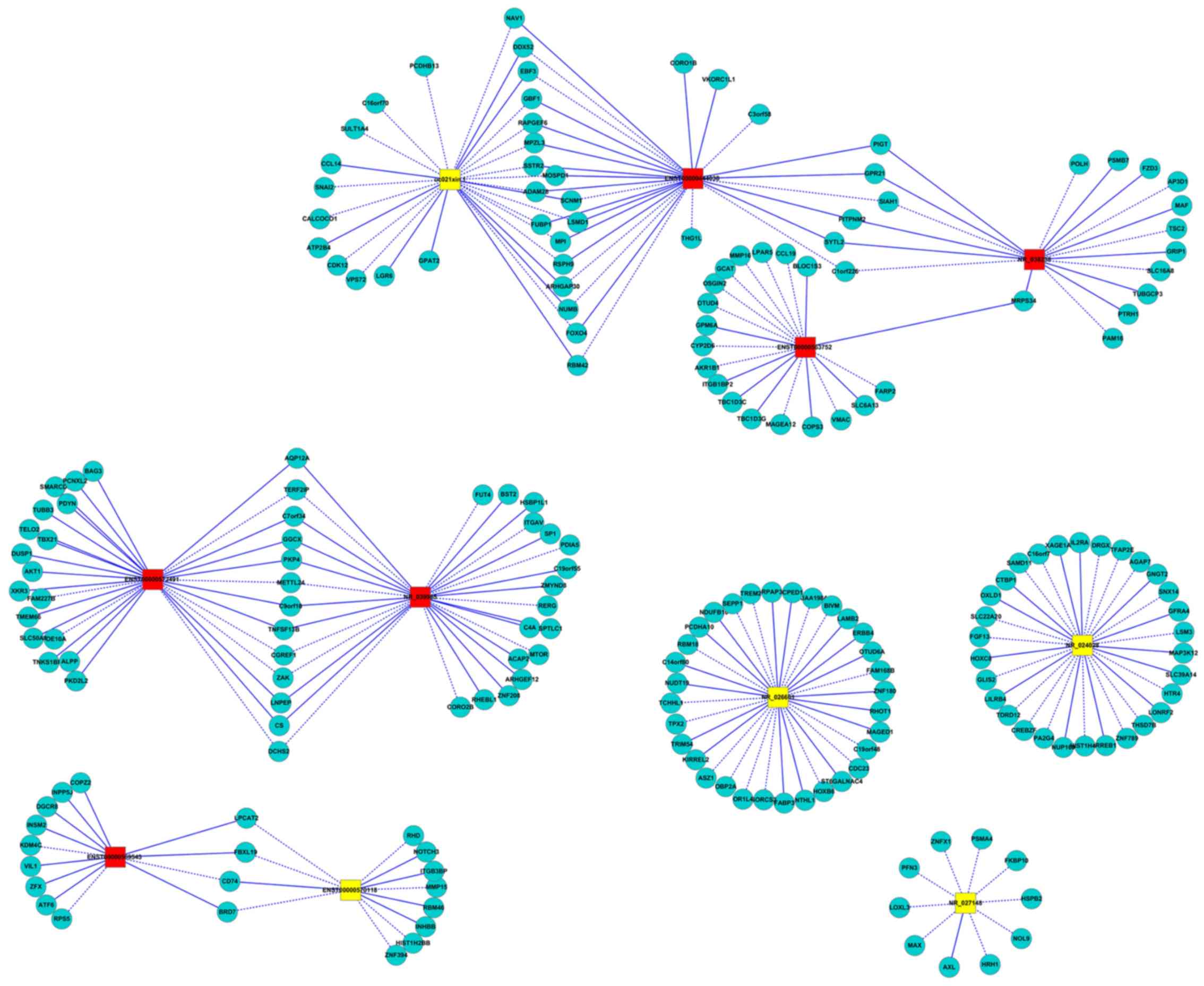
Construction of the coding-non-coding
gene co-expression networks
Functional prediction of lncRNAs was performed based
on the function of the co-expressed mRNAs. A co-expression network
for patients with RA was constructed based on the correlation
analysis between differentially expressed lncRNAs and mRNAs. A
total of 9 central lncRNAs were selected for the co-expression
networks (uc021xin.1, ENST00000444038, NR_038238, NR_039985,
NR_027148, ENST00000563752, ENST00000572491, ENST00000569543,
ENST00000492209 and ENST00000570118), which were verified using
qPCR (Fig. 6). The results indicated
that C-C motif chemokine ligand (CCL)19, CD74 and adaptor-related
protein complex 3 subunit δ1 were correlated with lncRNAs detected
in the network, which were also closely associated with T-cell
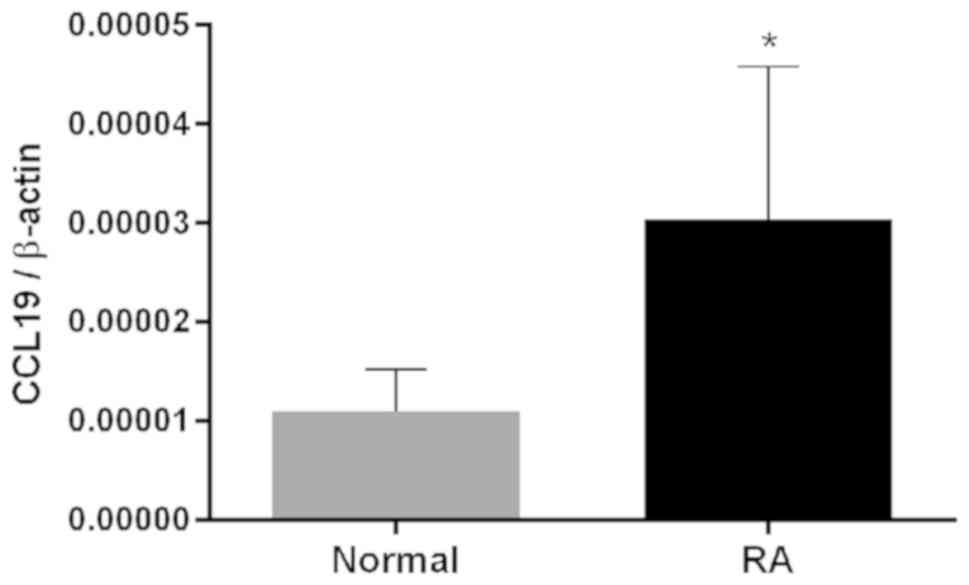
differentiation. RT-qPCR further validated CCL19 as an upregulated
signaling factor in patients with RA (Fig. 7).
Diagnostic value of lncRNA for RA
ROC curves were used to assess the potential
diagnostic value of these 9 lncRNAs in RA. The areas under the ROC
curves of ENST00000420096, ENST00000444038, ENST00000563752,
ENST00000569543, ENST00000572491, NR_027148, NR_038238, NR_039985
and uc021xin.1 were 0.9792, 0.9479, 0.9896, 1.0000, 0.9167, 0.8750,
0.8333, 0.8958 and 0.8438, respectively (Table II, Fig.
8).
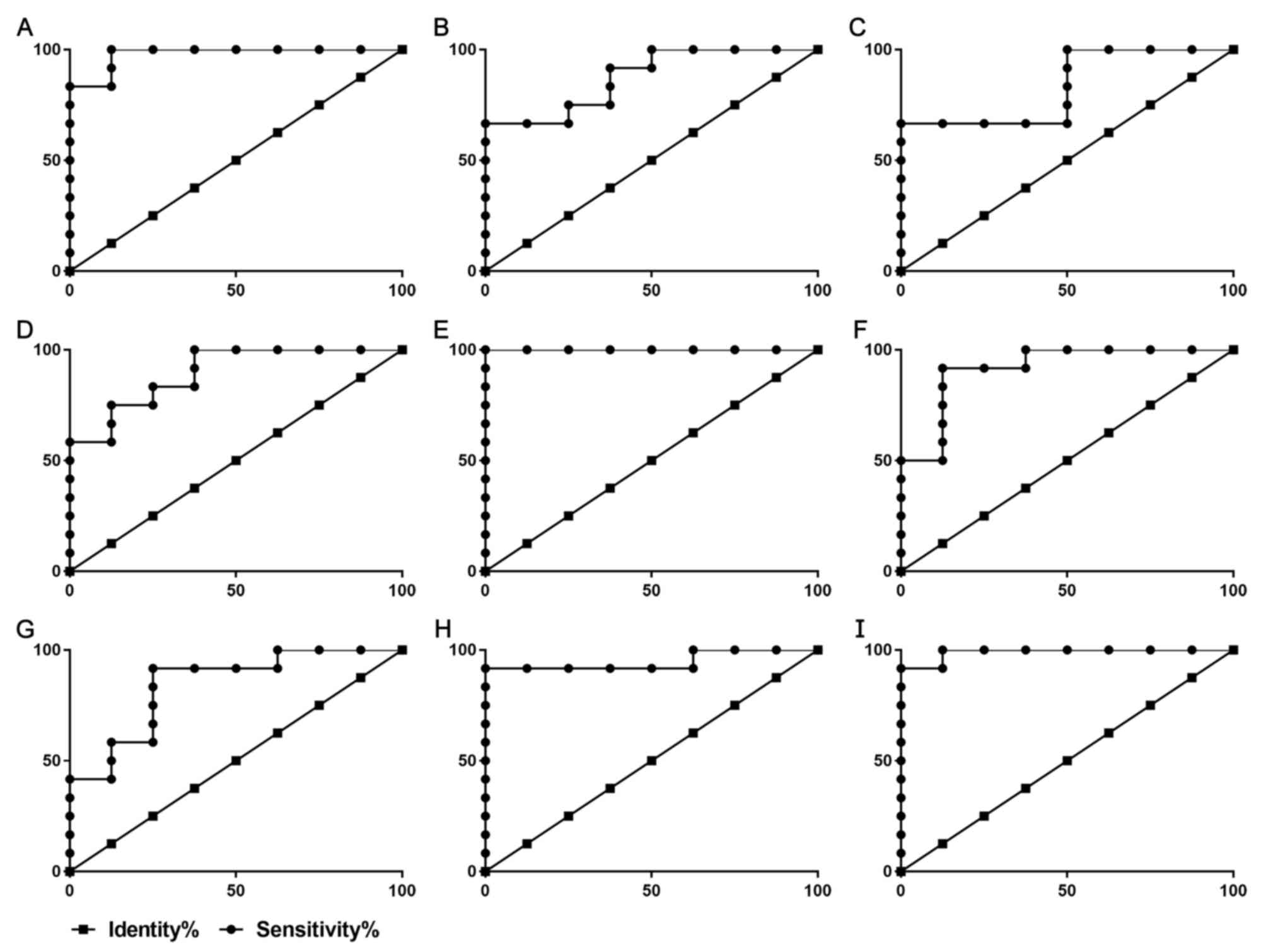 | Figure 8ROC curves for various long
non-coding RNAs to differentiate between healthy individuals and
patients with rheumatoid arthritis. The x-axis displays the
Specificity % and the y-axis displays the Sensitivity %. ROC curves
for patients RA based on the expression of (A) ENST00000420096, (B)
NR_027148, (C) NR_038238, (D) NR_039985, (E) ENST00000569543, (F)
ENST00000572491, (G) uc021xin.1, (H) ENST00000444038, (I)
ENST00000563752. RA, rheumatoid arthritis; ROC, receiver operating
characteristic. |
 | Table IIParameters of the receiver operating
characteristic curve analysis. |
Table II
Parameters of the receiver operating
characteristic curve analysis.
| lncRNA | AUC | Std. Error | 95% confidence
interval | P-value |
|---|
|
ENST00000420096 | 0.9792 | 0.02681 | 0.9266-1.032 | 0.0003903 |
|
ENST00000444038 | 0.9479 | 0.05363 | 0.8428-1.053 | 0.0009147 |
|
ENST00000563752 | 0.9896 | 0.01678 | 0.9567-1.022 | 0.0002906 |
|
ENST00000569543 | 1.000 | 0.0000 | 1.000-1.000 | 0.0002151 |
|
ENST00000572491 | 0.9167 | 0.06784 | 0.7837-1.050 | 0.002039 |
| NR_027148 | 0.8750 | 0.07743 | 0.7232-1.027 | 0.005499 |
| NR_038238 | 0.8333 | 0.09317 | 0.6507-1.016 | 0.01359 |
| NR_039985 | 0.8958 | 0.07068 | 0.7573-1.034 | 0.003386 |
| uc021xin.1 | 0.8438 | 0.09240 | 0.6626-1.025 | 0.01093 |
Correlation analysis between lncRNA
expression and clinical indicators of RA
ESR, CRP, RF and CCP antibodies are conventional
indicators for clinical diagnosis of RA (26). DAS28 is an internationally recognized
method for determining RA activity. Complement 3 (C3) and C4 are
involved in RA autoimmune disorders and inflammatory pathological
processes (27). Pearson correlation
analysis was used to analyze the correlation between target lncRNAs
and ESR, CRP, RF, anti-CCP antibody and DAS28. The correlation
coefficient between ENST00000569543 and C4 was 0.623 (P<0.05)
and the correlation coefficient between ENST00000420096 and
anti-CCP antibody or the DAS 28 score was 0.662 and 0.605,
respectively (P<0.05; Table
III).
 | Table IIICorrelation analysis between lncRNA
and major indicators of RA. |
Table III
Correlation analysis between lncRNA
and major indicators of RA.
| | Correlation
coefficient with major indicators of RA |
|---|
| lncRNA | ESR | RF | CRP | C3 | C4 | CRP | Anti-CCP | DAS 28 |
|---|
|
ENST00000420096 | 0.419 | -0.013 | 0.490 | -0.298 | -0.095 | 0.490 | 0.662a | 0.605a |
|
ENST00000444038 | 0.551 | 0.314 | -0.203 | -0.371 | 0.437 | -0.203 | 0.136 | 0.560 |
|
ENST00000563752 | 0.263 | -0.085 | -0.180 | -0.095 | 0.206 | -0.180 | -0.126 | 0.147 |
|
ENST00000569543 | 0.190 | 0.376 | -0.327 | -0.237 | 0.623a | -0.327 | 0.236 | 0.207 |
|
ENST00000572491 | 0.460 | 0.068 | 0.002 | -0.207 | 0.181 | 0.002 | -0.146 | 0.401 |
| NR_027148 | -0.002 | -0.492 | 0.200 | 0.166 | -0.026 | 0.200 | -0.325 | -0.123 |
| NR_038238 | 0.182 | 0.331 | -0.169 | 0.215 | 0.241 | -0.169 | -0.028 | -0.002 |
| NR_039985 | 0.049 | 0.400 | -0.412 | -0.245 | 0.529 | -0.412 | 0.172 | 0.100 |
| uc021xin.1 | -0.111 | -0.097 | -0.198 | -0.561 | -0.123 | -0.198 | -0.126 | 0.044 |
Discussion
With the development of high-throughput gene
sequencing technology, numerous lncRNAs have been identified and
demonstrated to exert biological functions through interactions
with other cellular macromolecules (28). LncRNAs are expressed in various types
of immune cells and participate in the differentiation and
activation of these cells (29). RA
is an autoimmune disorder, with CD4+ T cells
differentiating into various Th-cell subsets that are thought to be
deeply involved in the pathogenesis of RA (30). However, only few studies have
assessed lncRNAs associated with CD4+ T-cell
differentiation in peripheral blood of patients with active RA
(8,31). In the present study, the expression
of lncRNAs in CD4+ T cells from peripheral blood of
patients with active RA was assessed to provide novel insight into
the pathogenesis of RA.
Based on the microarray data, 493 lncRNAs and 374
differentially expressed mRNAs were identified. LncRNA functional
annotations are still not fully available; however, the majority of
the mRNAs identified are well annotated. Pathway and GO enrichment
analysis of significantly differentially expressed mRNAs revealed
that these mRNAs in the peripheral blood of patients with RA were
primarily involved in the regulation of processes associated with
cell growth and immune regulation. Further analysis demonstrated
that these differentially expressed mRNAs were associated with the
activation of the MAPK and P53 signaling pathways. MAPK
phosphorylation results in the activation of the downstream
inflammatory transcription factor NF-κB, which may control the
activation of the Th17-associated cytokine IL17 and the
Treg-associated cytokine TGF-β (32,33);
this is consistent with the present results. P53 serves an
important role in the differentiation of Treg cells and the
homeostasis of CD4+ T cells (33-35).
The present study suggested that lncRNAs are involved in the
pathogenesis of RA, which is possibly regulated by the
differentiation of CD4+ T lymphocytes.
A co-expression network based on 9 lncRNAs was
constructed. Of these, CCL19 was an overexpressed chemokine in
patients with RA. Various studies have indicated that CCL19 serves
a chemotactic role in B cells, dendritic cells and naïve T cells
via binding to the C-C motif chemokine receptor (CCR)7 (36-38).
CCR7 is an important antigen molecule expressed on the surface of
Treg cells and CCL19 may specifically bind to the receptor CCR7,
which in turn exerts chemotactic activity on Treg cells (39-41).
The results of the present study suggested that CCL19 is
upregulated in patients with active RA, implying that lncRNA may
affect CD4+ T lymphocytes by regulating mRNA-CCL19,
which in turn affects the differentiation of cells. Conversely, the
CCL19/CCR7 signaling pathway is primarily activated through ERK and
p38-associated proteins of the MAPK family (42), which suggests that the activity of
MAPK is closely associated with CCL19/CCR7. These results confirmed
that lncRNA is closely involved in the differentiation of
CD4+ T cells in patients with RA.
Numerous studies have examined the diagnostic value
of various lncRNAs in different types of diseases (43-47).
Studies have investigated the possibility of lncRNAs in fibroblast
synoviocytes as biomarkers for RA (48). The present study explored the
diagnostic value of peripheral blood lncRNAs in RA. ROC curves are
generally used to assess the diagnostic value of markers for a
certain disease (49,50). The association between lncRNAs and
the degree of RA activity was also analyzed. Pearson correlation
analysis suggested that there was a strong correlation between
ENST00000569543 and C4 and between ENST00000420096 with anti-CCP
antibody or DAS. Anti-CCP is a marker that may be used to diagnose
RA (51). DAS is generally
considered the ‘gold standard’ for measuring RA disease activity
(52,53). In the present study, these two
lncRNAs identified in the screen and confirmed by PCR were most
closely associated with RA, which suggested that they may be
potential biomarkers for judging RA disease activity.
In conclusion, a total of 9 lncRNAs were identified
to be involved in the pathogenesis of RA and serve an important
role in the differentiation of CD4+ T cells. ROC curve
analysis indicated that these lncRNAs may also possess diagnostic
value for RA, and ENST00000569543 and ENST00000420096 were most
relevant. These results provide preliminary evidence for the
further study of lncRNAs and their association with RA. Due to the
difficulty in obtaining samples, the sample size was small, which
is a limitation of the present study, also regarding the detection
of IL-17 and TGF-β. Furthermore, the value of lncRNAs in the
diagnosis of RA was preliminarily explored, while horizontal
comparative studies of other autoimmune diseases or osteoarthritis
remain to be performed in the future.
Supplementary Material
Overview of the 21 target long
non-coding RNAs.
List of primers used for real-time
quantitative PCR.
Acknowledgements
The authors thank Director Lu Yan, Department of
Rheumatology, Jiangsu Province Hospital of Chinese Medicine and
Affiliated Hospital of Nanjing University of Chinese Medicine
(Nanjing, China), for her assistance in collecting specimens for
this study.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81573869 and
81673937).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request. All of the data from the microarray trials are stored in
the Gene Expression Omnibus database (dataset no. GSE134087,
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE134087).
Authors' contributions
XZ and LZ conceived and designed the study. ML, KM
and JW performed the experiments. ML and ZF analyzed and
interpreted the experimental data. ML and KM wrote the manuscript.
XZ, LZ and ZF reviewed and edited the manuscript. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
All patients signed informed consent forms and the
study was approved by the IRB of the Affiliated Hospital of Nanjing
University of Traditional Chinese Medicine (Nanjing, China; no.
2016NL-KS14).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Smolen JS, Aletaha D and McInnes IB:
Rheumatoid arthritis. Lancet. 388:2023–2038. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Scott DL, Wolfe F and Huizinga TW:
Rheumatoid arthritis. Lancet. 376:1094–1108. 2010.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Davis JM 3rd: Rheumatoid arthritis: A
severe disease that preventive approaches would greatly benefit.
Clin Ther. 41:1240–1245. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Derksen VFAM, Huizinga TWJ and van der
Woude D: The role of autoantibodies in the pathophysiology of
rheumatoid arthritis. Semin Immunopathol. 39:437–446.
2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Alam J, Jantan I and Bukhari SNA:
Rheumatoid arthritis: Recent advances on its etiology, role of
cytokines and pharmacotherapy. Biomed Pharmacother. 92:615–633.
2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Goldfeder RL, Wall DP, Khoury MJ,
Ioannidis JPA and Ashley EA: Human genome sequencing at the
population scale: A primer on high-throughput DNA sequencing and
analysis. Am J Epidemiol. 186:1000–1009. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Ma Y, Shi N, Li M, Chen F and Niu H:
Applications of next-generation sequencing in systemic autoimmune
diseases. Genomics Proteomics Bioinformatics. 13:242–249.
2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Yuan M, Wang S, Yu L, Qu B, Xu L, Liu L,
Sun H, Li C, Shi Y and Liu H: Long noncoding RNA profiling revealed
differentially expressed lncRNAs associated with disease activity
in PBMCs from patients with rheumatoid arthritis. PLoS One.
12(e0186795)2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Ye H, Wang X, Wang L, Chu X, Hu X, Sun L,
Jiang M, Wang H, Wang Z, Zhao H, et al: Full high-throughput
sequencing analysis of differences in expression profiles of long
noncoding RNAs and their mechanisms of action in systemic lupus
erythematosus. Arthritis Res Ther. 21(70)2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Wang J, Peng H, Tian J, Ma J, Tang X, Rui
K, Tian X, Wang Y, Chen J, Lu L, et al: Upregulation of long
noncoding RNA TMEVPG1 enhances T helper type 1 cell response in
patients with Sjögren syndrome. Immunol Res. 64:489–496.
2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Dolcino M, Tinazzi E, Puccetti A and
Lunardi C: In systemic sclerosis, a unique long non coding RNA
regulates genes and pathways involved in the three main features of
the disease (Vasculopathy, Fibrosis and Autoimmunity) and in
carcinogenesis. J Clin Med. 8(E320)2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Peng QL, Zhang YM, Yang HB, Shu XM, Lu X
and Wang GC: Transcriptomic profiling of long non-coding RNAs in
dermatomyositis by microarray analysis. Sci Rep.
6(32818)2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Mattick JS: The state of long non-coding
RNA biology. Noncoding RNA. 4(E17)2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Panzeri I, Rossetti G, Abrignani S and
Pagani M: Long intergenic non-coding RNAs: Novel drivers of human
lymphocyte differentiation. Front Immunol. 6(175)2015.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Pagani M, Rossetti G, Panzeri I, de Candia
P, Bonnal RJ, Rossi RL, Geginat J and Abrignani S: Role of
microRNAs and long-non-coding RNAs in CD4(+) T-cell
differentiation. Immunol Rev. 253:82–96. 2013.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Kondo Y, Yokosawa M, Kaneko S, Furuyama K,
Segawa S, Tsuboi H, Matsumoto I and Sumida T: Review:
Transcriptional regulation of CD4+ T cell differentiation in
experimentally induced arthritis and rheumatoid arthritis.
Arthritis Rheumatol. 70:653–661. 2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Arnett FC, Edworthy SM, Bloch DA, McShane
DJ, Fries JF, Cooper NS, Healey LA, Kaplan SR, Liang MH and Luthra
HS: The American rheumatism association 1987 revised criteria for
the classification of rheumatoid arthritis. Arthritis Rheum.
31:315–324. 1988.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Jou JM, Lewis SM, Briggs C, Lee SH, De La
Salle B and McFadden S: International Council for Standardization
in Haematology. ICSH review of the measurement of the erythocyte
sedimentation rate. Int J Lab Hematol. 33:125–132. 2011.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Ailus K, Melamies L, Tuomi T, Palosuo T
and Aho K: Measuring rheumatoid factor in nonrheumatoid subjects:
Immunoturbidimetric assay, latex slide test, and enzyme-linked
immunosorbent assay compared. Clin Chem. 37:1766–1769.
1991.PubMed/NCBI
|
|
20
|
Moutachakkir M, Lamrani Hanchi A, Baraou
A, Boukhira A and Chellak S: Immunoanalytical characteristics of
C-reactive protein and high sensitivity C-reactive protein. Ann
Biol Clin (Paris). 75:225–229. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Shen R, Ren X, Jing R, Shen X, Chen J, Ju
S and Yang C: Rheumatoid factor, anti-cyclic citrullinated peptide
antibody, C-reactive protein, and erythrocyte sedimentation rate
for the clinical diagnosis of rheumatoid arthritis. Lab Med.
46:226–229. 2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
McWilliams DF, Kiely PDW, Young A,
Joharatnam N, Wilson D and Walsh DA: Interpretation of DAS28 and
its components in the assessment of inflammatory and
non-inflammatory aspects of rheumatoid arthritis. BMC Rheumatol.
2(8)2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Paradowska A, Maślińiski W,
Grzybowska-Kowalczyk A and Łacki J: The function of interleukin 17
in the pathogenesis of rheumatoid arthritis. Arch Immunol Ther Exp
(Warsz). 55:329–334. 2007.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Astry B, Venkatesha SH and Moudgil KD:
Involvement of the IL-23/IL-17 axis and the Th17/Treg balance in
the pathogenesis and control of autoimmune arthritis. Cytokine.
74:54–61. 2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Sparks JA: Rheumatoid arthritis. Ann
Intern Med. 170:ITC1–ITC16. 2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Okroj M, Heinegård D, Holmdahl R and Blom
AM: Rheumatoid arthritis and the complement system. Ann Med.
39:517–530. 2007.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Guttman M and Rinn JL: Modular regulatory
principles of large non-coding RNAs. Nature. 482:339–346.
2012.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Hur K, Kim SH and Kim JM: Potential
implications of long noncoding RNAs in autoimmune diseases. Immune
Netw. 19(e4)2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Choy E: Understanding the dynamics:
Pathways involved in the pathogenesis of rheumatoid arthritis.
Rheumatology (Oxford). 51 (Suppl 5):v3–v11. 2012.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Luo Q, Xu C, Li X, Zeng L, Ye J, Guo Y,
Huang Z and Li J: Comprehensive analysis of long non-coding RNA and
mRNA expression profiles in rheumatoid arthritis. Exp Ther Med.
14:5965–5973. 2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Liu H, Yao S, Dann SM, Qin H, Elson CO and
Cong Y: ERK differentially regulates Th17- and Treg-cell
development and contributes to the pathogenesis of colitis. Eur J
Immunol. 43:1716–1726. 2013.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zhang Y, Zhang Y, Gu W and Sun B: TH1/TH2
cell differentiation and molecular signals. Adv Exp Med Biol.
841:15–44. 2014.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Jiang D, Lenardo MJ and Zúñiga-Pflücker
JC: p53 prevents maturation to the CD4+CD8+ stage of thymocyte
differentiation in the absence of T cell receptor rearrangement. J
Exp Med. 183:1923–1928. 1996.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Zhang H, Xu CF, Ren C, Wu TT, Dong N and
Yao YM: Novel role of p53 in septic immunosuppression: Involvement
in loss and dysfunction of CD4+ T lymphocytes. Cell Physiol
Biochem. 51:452–469. 2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Asquith DL, Bryce SA and Nibbs RJ:
Targeting cell migration in rheumatoid arthritis. Curr Opin
Rheumatol. 27:204–211. 2015.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Hähnlein JS, Nadafi R, de Jong T,
Ramwadhdoebe TH, Semmelink JF, Maijer KI, Zijlstra IA, Maas M,
Gerlag DM, Geijtenbeek TBH, et al: Impaired lymph node stromal cell
function during the earliest phases of rheumatoid arthritis.
Arthritis Res Ther. 20(35)2018.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Radstake TR, van der Voort R, ten
Brummelhuis M, de Waal Malefijt M, Looman M, Figdor CG, van den
Berg WB, Barrera P and Adema GJ: Increased expression of CCL18,
CCL19, and CCL17 by dendritic cells from patients with rheumatoid
arthritis, and regulation by Fc gamma receptors. Ann Rheum Dis.
64:359–367. 2005.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Förster R, Davalos-Misslitz AC and Rot A:
CCR7 and its ligands: Balancing immunity and tolerance. Nat Rev
Immunol. 8:362–371. 2008.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Hauser MA and Legler DF: Common and biased
signaling pathways of the chemokine receptor CCR7 elicited by its
ligands CCL19 and CCL21 in leukocytes. J Leukoc Biol. 99:869–882.
2016.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Hu Z, Li Y, Van Nieuwenhuijze A, Selden
HJ, Jarrett AM, Sorace AG, Yankeelov TE, Liston A and Ehrlich LIR:
CCR7 modulates the generation of thymic regulatory T cells by
altering the composition of the thymic dendritic cell compartment.
Cell Rep. 21:168–180. 2017.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Shi LJ, Li JH, Cen XM, Yang NP, Yin G and
Xie QB: CCL19/IL-1beta positive feedback loop involved in the
progress of inflammation in rheumatoid arthritis. Sichuan Da Xue
Xue Bao Yi Xue Ban. 46:272–275. 2015.PubMed/NCBI(In Chinese).
|
|
43
|
Viereck J and Thum T: Circulating
noncoding RNAs as biomarkers of cardiovascular disease and injury.
Circ Res. 120:381–399. 2017.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Dang X, Lian L and Wu D: The diagnostic
value and pathogenetic role of lncRNA-ATB in patients with
osteoarthritis. Cell Mol Biol Lett. 23(55)2018.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Fan CN, Ma L and Liu N: Systematic
analysis of lncRNA-miRNA-mRNA competing endogenous RNA network
identifies four-lncRNA signature as a prognostic biomarker for
breast cancer. J Transl Med. 16(264)2018.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Knoll M, Lodish HF and Sun L: Long
non-coding RNAs as regulators of the endocrine system. Nat Rev
Endocrinol. 11:151–160. 2015.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Zhang Y, Xu YZ, Sun N, Liu JH, Chen FF,
Guan XL, Li A, Wang F, Zhao QF, Wang HY, et al: Long noncoding RNA
expression profile in fibroblast-like synoviocytes from patients
with rheumatoid arthritis. Arthritis Res Ther.
18(227)2016.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Linden A: Measuring diagnostic and
predictive accuracy in disease management: An introduction to
receiver operating characteristic (ROC) analysis. J Eval Clin
Pract. 12:132–139. 2006.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Cook NR: Statistical evaluation of
prognostic versus diagnostic models: Beyond the ROC curve. Clin
Chem. 54:17–23. 2008.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Nishimura K, Sugiyama D, Kogata Y, Tsuji
G, Nakazawa T, Kawano S, Saigo K, Morinobu A, Koshiba M, Kuntz KM,
et al: Meta-analysis: Diagnostic accuracy of anti-cyclic
citrullinated peptide antibody and rheumatoid factor for rheumatoid
arthritis. Ann Intern Med. 146:797–808. 2007.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Anderson JK, Zimmerman L, Caplan L and
Michaud K: Measures of rheumatoid arthritis disease activity:
Patient (PtGA) and provider (PrGA) global assessment of disease
activity, disease activity score (DAS) and disease activity score
with 28-joint counts (DAS28), simplified disease activity index
(SDAI), clinical disease activity index (CDAI), patient activity
score (PAS) and patient activity score-II (PASII), routine
assessment of patient index data (RAPID), rheumatoid arthritis
disease activity index (RADAI) and rheumatoid arthritis disease
activity index-5 (RADAI-5), chronic arthritis systemic index
(CASI), patient-based disease activity score with ESR (PDAS1) and
patient-based disease activity score without ESR (PDAS2), and mean
overall index for rheumatoid arthritis (MOI-RA). Arthritis Care Res
(Hoboken). 63 (Suppl 11):S14–S36. 2011.PubMed/NCBI View Article : Google Scholar
|
|
53
|
van Riel PL and Renskers L: The disease
activity score (DAS) and the disease activity score using 28 joint
counts (DAS28) in the management of rheumatoid arthritis. Clin Exp
Rheumatol. 34 (5 Suppl 101):S40–S44. 2016.PubMed/NCBI
|