Introduction
Allergic rhinitis (AR) is a type of immunoglobulin
(Ig)E-mediated type I allergic disease that affects the nasal
mucosa. It is clinically characterized by itching of the nasal
passages, sneezing, nasal hypersecretion and nasal mucosal
swelling, and 60-70% of patients frequently present with eye
itching, reddish eye and/or lacrimation (1). Extensive attention has been paid to the
treatment of AR, including antihistamines, glucocorticoids,
decongestants and immunotherapy (2);
however, favorable therapeutic effects have not been fully achieved
due to the diverse pathogenic factors and complex pathogenesis of
AR. AR pathogenesis has been the subject of numerous studies
(3). In particular, mast cells
(MCs), which are the major effector cells of AR, have become an
important research topic (4,5).
The RNA interference (RNAi) technique has become a
common and effective tool for the study of gene function. This
technique typically involves introducing a chemically synthesized
small interfering RNA (siRNA) or short hairpin RNA (shRNA) into
cells in order to interfere with a target gene, and specifically
downregulate its expression (6). C-C
chemokine receptor type 3 (CCR3) is a single strand
G-protein-coupled receptor containing seven hydrophobic
transmembrane domains, which was originally suggested to be
specifically expressed on the surfaces of eosinophils (EOSs)
(7). Previous studies conducted by
the present research team demonstrated that CCR3 downregulation by
RNAi markedly alleviated nasal cavity symptoms, significantly
reduced CCR3 mRNA expression in the peripheral blood, bone marrow
and nasal lavage fluid, and notably decreased EOS infiltration in a
mouse model of AR. In addition, degranulated proteins in the EOSs
of peripheral blood, bone marrow and nasal lavage fluid were
significantly decreased in the AR model mice by CCR3 RNAi, and
in vitro experiments revealed that CCR3 downregulation in
EOSs promoted apoptosis and inhibited proliferation (8,9).
However, other studies have reported that CCR3 is also expressed on
the surfaces of Th2 cells (10), MCs
(11-13)
and basophils (14). Ochi et
al (13) demonstrated that human
MC progenitors express four chemokine receptors, namely CXCR2,
CCR3, CXCR4 and CCR5; however, only CCR3 is maintained until MC
maturation. Brightling et al (11) reported that MCs migrate to CCR3, and
that the application of a CCR3 inhibition with a specific blocking
antibody could significantly reduce the migration of MCs to CCR3.
Similarly, Miyazaki et al (15) demonstrated that a CCR3 blockade by
mAb or specific CCR3 antagonist was able to reduce the amount of
histamine and β hexosaminidase secreted following MC-activated
degranulation.
In the present study, CCR3 lentiviral vector
plasmids were constructed and transfected into mouse MCs. The
efficacy of the transfection was determined by assessing CCR3 mRNA
and protein expression in the MCs. Furthermore, the effects of
CCR3-shRNA transfection on MC proliferation, apoptosis and
chemotaxis were evaluated, in order to provide a theoretical
foundation for the further investigation of AR pathogenesis.
Materials and methods
Animals
Male Balb/c mice (n=5; weight 20±2 g; 4-6 weeks old)
were purchased from the Laboratory Animal Science Center of
Nanchang University. All animals were housed in cages with free
access to food and water and were acclimated for 1 week at a
controlled temperature of 24˚C and relative humidity of 55-65%,
under a 12-h light/dark cycle (lights on at 7:00 a.m.) prior to
experimental surgery. All efforts were made to minimize suffering.
Animal procedures were conducted according to the Guidelines for
Care and Use of Laboratory Animals and were approved by the Animal
Care and Use Committee of The Second Affiliated Hospital of
Nanchang University.
Reagents
Mouse interleukin (IL)-3 and mouse stem cell factor
(SCF) were purchased from Promega Corporation. TRIzol reagent (cat.
no. CW0580S), Ultrapure RNA extraction kit (cat. no. CW0581M),
HiFiScript cDNA synthesis kit (cat. no. CW2569M) and UltraSYBR
Mixture (cat. no. CW0957M) were all purchased from Beijing CoWin
Biotech Co., Ltd. Annexin V-fluorescein isothiocyanate
(FITC)/propidium iodide (PI) Apoptosis kit (cat. no. AP101-100-kit)
was purchased from Hanzhou Multi Sciences (Lianke) Biotech Co.,
Ltd. RIPA Lysis Buffer (cat. no. C1053) was purchased from Applygen
Technologies, Inc. SuperSignal® West Pico
chemiluminescent substrate (cat. no. RJ239676) was obtained from
Thermo Fisher Scientific, Inc. Polyvinylidene difluoride (PVDF)
membranes (cat. no. IPVH0001) were purchased from Merck KGaA. Mouse
monoclonal primary antibody against GAPDH (1:2,000; cat. no. TA-08)
and horseradish peroxidase (HRP)-conjugated goat anti-mouse and
anti-rabbit IgG secondary antibodies (1:2,000, cat. nos. ZB-2305
and cat. no. ZB-2301, respectively) were purchased from Beijing
Zhongshan Jinqiao Biotechnology Co., Ltd. Rabbit monoclonal primary
antibody against CCR3 (1:500; cat. no. ab32512) was purchased from
Abcam. PE-CD117 (cat. no. 555714) and FITC-FCεRI α (cat. no.
553376) were purchased from Becton Dickinson. Mouse histamine ELISA
kit (cat. no. CEA927Ge) was purchased from USCN Life Sciences, Inc.
and mouse β-hexosaminidase ELISA kit (cat. no. SBJ-M0352) was
purchased from SBJBio.
Culture of mouse bone marrow-derived
MCs
BALB/c mice were sacrificed by cervical dislocation.
Femurs and tibias were isolated, immersed in 75% ethanol for 5 min
and rinsed with PBS. The ends of the femurs and tibias were cut
off. Bone marrow was flushed out from the bones using RPMI-1640 and
was collected on a plate. A single-cell suspension of bone marrow
was subsequently prepared via filtering the bone marrow with a
100-mesh sieve. Cells were collected following centrifugation at
188.9 x g at 4˚C for 5 min and were washed twice with PBS.
Subsequently, cells were cultured in RPMI-1640 supplemented with
10% FBS (Gibco; Thermo Fisher Scientific, Inc.)., 10 µg/ml
streptomycin, 100 IU/ml penicillin, 50 µmol/l non-essential amino
acids, 10 ng/ml IL-3 and 10 ng/ml SCF and were placed at 37˚C in a
humidified incubator containing 5% CO2.
Toluidine blue staining
The medium were changed every two days, and the mast
cells were detected using Toluidine blue staining. Following 4
weeks of culture, 0.5 ml cell suspension was collected with a
Pasteur tube, added dropwise onto an autoclaved cover glass covered
with polylysine and then air-dried. Cells were stained with
toluidine blue for 15 min at room temperature, washed with water,
followed by acetone differentiation, gradual ethanol (95, 85 and
75%) dehydration, xylene hyalinization and neutral resin mounting
at room temperature. Slides were imaged using a microscope (CX41;
Olympus Corporation; magnification, x200).
Flow cytometry for MC
identification
Cells cultured for 4 weeks were collected by
horizontal centrifugation (1889 x g; 5 min) at 4˚C, washed twice
and resuspended in PBS, resuspended, and the cell concentration was
adjusted to 1x106/ml. Cell suspension (100 µl) was put
into two tubes, and incubated with 0.5 µl PE-CD117 and 0.2 µl
FITC-FCεRI α, the specific MCs markers (16), in the dark at 4˚C for 30 min. A
volume of 1 ml PBS containing 2.5% FBS was added into each tube for
30 sec, cells were washed twice in PBS and centrifuged at 188.9 x g
for 1 min at 4˚C, and then the supernatant was discarded.
Eventually, cells were resuspended in 0.5 ml PBS containing 2.5%
FBS. Cells were analyzed by flow cytometry (NovoCyte 2060R) with
NovoExpress_1.2.5_Setup_Cn_170605.20914 software provided by ACEA
Biosciences Inc.
Construction of CCR3-shRNA lentivirus
vector
The CCR3 gene-specific mRNA sequence was obtained
from the Genebank database (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Subsequently, targeting sequences interfering with the CCR3 gene
were searched for using the siRNA online design tool from Ambion;
Thermo Fisher Scientific, Inc. (https://www.thermofisher.com/uk/en/home/brands/invitrogen/ambion.html).
Three CCR3 target sequences were designed by General Biosystems and
screened using Blast (http://blast.ncbi.nlm.nih.gov/Blast.cgi; Table I).
 | Table IPrimer sequences of the
CCR3-shRNAs. |
Table I
Primer sequences of the
CCR3-shRNAs.
| Primer name | Direction | Primer sequences
(5'-3') |
|---|
| CCR3-shRNA1 | Forward |
GATCCGGCAGCATTGCCTGAATTTATCTTCCTGTCAGAATAAATTCAGGCAATGC |
| | | TGCCTTTTTG |
| | Reverse |
AATTCAAAAAGGCAGCATTGCCTGAATTTATTCTGACAGGAAGATAAATTCAGG |
| | | CAATGCTGCCG |
| CCR3-shRNA2 | Forward |
GATCCGCAGCATTGCCTGAATTTATCCTTCCTGTCAGAGATAAATTCAGGCAATG |
| | | CTGCTTTTTG |
| | Reverse |
AATTCAAAAAGCAGCATTGCCTGAATTTATCTCTGACAGGAAGGATAAATTCAG |
| | | GCAATGCTGCG |
| CCR3-shRNA3 | Forward |
GATCCGCTCTTCCTCTCCTCATTATGCTTCCTGTCAGACATAATGAGGAGAGGAA |
| | | GAGCTTTTTG |
| | Reverse |
ATTCAAAAAGCTCTTCCTCTCCTCATTATGTCTGACAGGAAGCATAATGAGGAG |
| | | AGGAAGAGCG |
Cell transfection
MC cells were seeded in a 24-well plate at a density
of 1x105/well and grown at 37˚C for 18-24 h prior to
lentivirus transfection. On the second day, the medium was replaced
by 2 ml fresh medium containing 6 µg/ml polybrene and 10 µl virus
suspension, and cells were incubated at 37˚C for 4 h. After 4 h
incubation at 37˚C with the vectors, 2 ml fresh medium was added to
dilute the polybrene and the cells were further cultured for 72, 96
and 144 h, and virus-containing medium was replaced by the fresh
medium in the blank control group, vector control group and
CCR3-shRNA group.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA from each group of cells was isolated
using TRIzol (cat. no. CW0580S; CoWin Biosciences) according to the
manufacturer's instructions. cDNA was synthesized from 1 µg total
RNA using an HiFiScript cDNA synthesis kit (cat. no. CW2569M; CoWin
Biosciences) following the manufacturer's instructions. The thermal
conditions of reverse transcription were as follows: 37˚C for 15
min and 85˚C for 5 sec. qPCR analysis was then performed to
evaluate CCR3 mRNA expression using UltraSYBR mixture (cat. no.
CW0957M; CoWin Biosciences). The primers were designed as follows:
CCR3, forward 5'-CGCTATCCAGAGGGTGAAG-3' and reverse
5'-AGCAGTGGGTGTAGGCAAT-3' (predicted amplicon length, 328 bp); and
GAPDH, forward 5'-AAGAAGGTGGTGAAGCAGG-3' and reverse
5'-GAAGGTGGAAGAGTGGGAGT-3' (predicted amplicon length, 111 bp). The
qPCR cycles were performed as follows: 94˚C for 10 min, followed by
40 cycles of 95˚C for 15 sec, 58˚C for 30 sec and 72˚C for 30 sec,
and a final extension at 72˚C for 10 min. The relative expression
level of CCR3 was normalized to endogenous control GAPDH and was
expressed as 2-ΔΔCT (17).
Western blotting
Cells in each group were lysed using RIPA buffer on
the ice for 30 min and the protein concentration was estimated
using a bicinchoninic acid assay kit. Proteins (60 µg) were boiled
at 95˚C for 5 min in 500 µl NuPAGE 4X LDS sample buffer
(Invitrogen; Thermo Fisher Scientific, Inc.) containing 5%
β-mercaptoethanol. Subsequently, proteins (60 µg/lane) were
separated using 10% NuPAGE Bis-Tris precast gels (Nanjing KeyGen
Biotech Co., Ltd) and transferred onto PVDF membranes. Membranes
were washed with TBS supplemented with 0.1% Tween 20 (TBST),
blocked with 3% bovine serum albumin (Applygen Technologies, Inc.)
in TBST for 2 h at room temperature, and incubated at 4˚C overnight
with anti-CCR3 and anti-GAPDH primary antibodies. Following three
washes of 10 min with TBST, membranes were incubated with secondary
antibodies for 2 h at room temperature. After three washes of 5 min
with TBST, the signal on the membrane was detected using
SuperSignal® West Pico chemiluminescent substrate (cat.
no. RJ239676; Thermo Fisher Scientific, Inc.) and imaged with a
ChemiDoc system (Bio-Rad Laboratories, Inc.). The data were
analyzed via densitometry using ImageJ software 1.8.0 (National
Institutes of Health) and normalized to expression of the internal
control GAPDH.
Detection of MC proliferation with
Cell Counting kit-8 (CCK-8) assay
Cells in each group were collected following 96 h
transfection, washed twice with PBS, and seeded in a 96-well plate
at a density of 2.5x105/ml. Then, 100 µl culture medium
(RPMI-1640 supplemented with 10% FBS) was added to each well (in
pentaplicate for each group) and cells were cultured at 37˚C for 0,
12, 48, 72 and 96 h. CCK8 reagent (20 µl; Nanjing KeyGen Biotech
Co., Ltd) was then added to each well and cells were incubated at
37˚C for 4 h. Absorbance at 450 nm was then measured using a
microplate reader.
Flow cytometry for detection of MC
apoptosis
Cells were collected following 96 h transfection,
and the cell concentration was adjusted to 2.0x105/ml.
Cell suspension from each group was centrifuged (698.8 x g, 3 min)
at 4˚C and the supernatant was discarded. PBS (300 µl) was then
added to each tube and the mixture was gently mixed. Subsequently,
5 µl Annexin V-FITC and 5 µl PI were added to each tube and cells
were incubated in the dark at room temperature for 10 min. Cells
were then transferred into special centrifuge tube for flow
apoptosis detection, and apoptosis was eventually detected using a
flow cytometer (NovoCyte 2060R) and analyzed with
NovoExpress_1.2.5_Setup_Cn_170605.20914 software provided by ACEA
Biosciences Inc.
Transwell assay for detection of MC
chemotaxis
Cells were collected following 96 h transfection,
centrifuged (111.8 x g; 5 min) at room temperature and washed twice
with PBS. Cells in each group were resuspended in medium containing
10% FBS and the concentration was adjusted to
2.0x105/ml. Medium containing 10% FBS (800 µl in total)
and 1 nM chemokines leukotriene B4 (LTB4; cat. no. 71160-24-2;
Hubei Jusheng Technology Co. Ltd.) (12) were added to each well of a 24-well
plate to evaluate the chemotaxis of MCs. Chambers (pore size 8 µm;
BD Biosciences) were placed inside the wells, 200 µl cell
suspension from each group was added into the chambers and cells
were cultured at 37˚C for 36 h. MCs were the suspended cells, which
did not adhere to the filter membrane. Flow cytometry was
subsequently used to detect the number MCs that had migrated into
the lower chamber. Subsequently, the chambers were removed and the
medium contained in the 24-well plate was collected and centrifuged
at 1,889.4 x g at room temperature for 5 min before discarding the
supernatant. Gr-1-FITC (5 µl) (BD Biosciences) and C-kit PE (5 µl)
(BD Biosciences) were then added into each tube and the mixture was
gently mixed and incubated in the dark for 10 min at room
temperature to detect chemotaxis (18). The single-cell suspension obtained
was then transferred into the special flow cytometry centrifuge
tube and analyzed by using a flow cytometer (NovoCyte 2060R) and
analyzed with NovoExpress_1.2.5_Setup_Cn_170605.20914 software
(ACEA Biosciences Inc.). C-kit positive cells were considered to
represent the number of MCs cells that had migrated.
Detection of MC degranulation by
ELISA
MC degranulation was evaluated using
β-hexosaminidase and Histamine detection. Cells were collected
following 96 h transfection, centrifuged at 4˚C (111.8 x g; 5 min)
and washed twice with PBS. Cells were resuspended in medium
containing 10% FBS and the concentration was adjusted to
5.0x106/ml and seeded into 24-well plate (in
sextaplicate for each group). Subsequently, 10 µg/ml anti-DNP IgE
(19) was added into each well for 1
h at 37˚C to promote the MCs activation and degranulation. Then, 40
ng/ml HSA-DNP was added for 1 h at 37˚C to simulate the
sensitization process and trigger allergic reactions. Subsequently,
plates were placed on ice for 10 min in order to terminate the
degranulation reaction, and centrifuged at 4˚C (1,889.4 x g; 5 min)
to collect the supernatants from each well. Then, 50 µl of 1 mM
4-nitrophenyl-N-acetyl β-D-glucoside (dissolved into 100 mM citric
acid-sodium citrate, pH 4.4) was added to each supernatant, and the
mixture was transferred into a 96-well plate and incubated for 1 h
at 37˚C. Sodium carbonate buffer solution (200 µl) was then added
to terminate the reaction. Absorbance was read at 405 nm using a
microplate reader, and the amount of β-hexosaminidase and Histamine
was expressed as its proportion of total enzymes in the
unstimulated cells (20).
Statistical analysis
All data were analyzed using SPSS 19.0 software (IBM
Corp.). Data are expressed as the means ± standard deviation.
Multiple group comparison was performed using one-way ANOVA
followed by Tukey's post hoc test. Each experiment was repeated
three times independently. P<0.05 was considered to indicate a
statistically significant difference.
Results
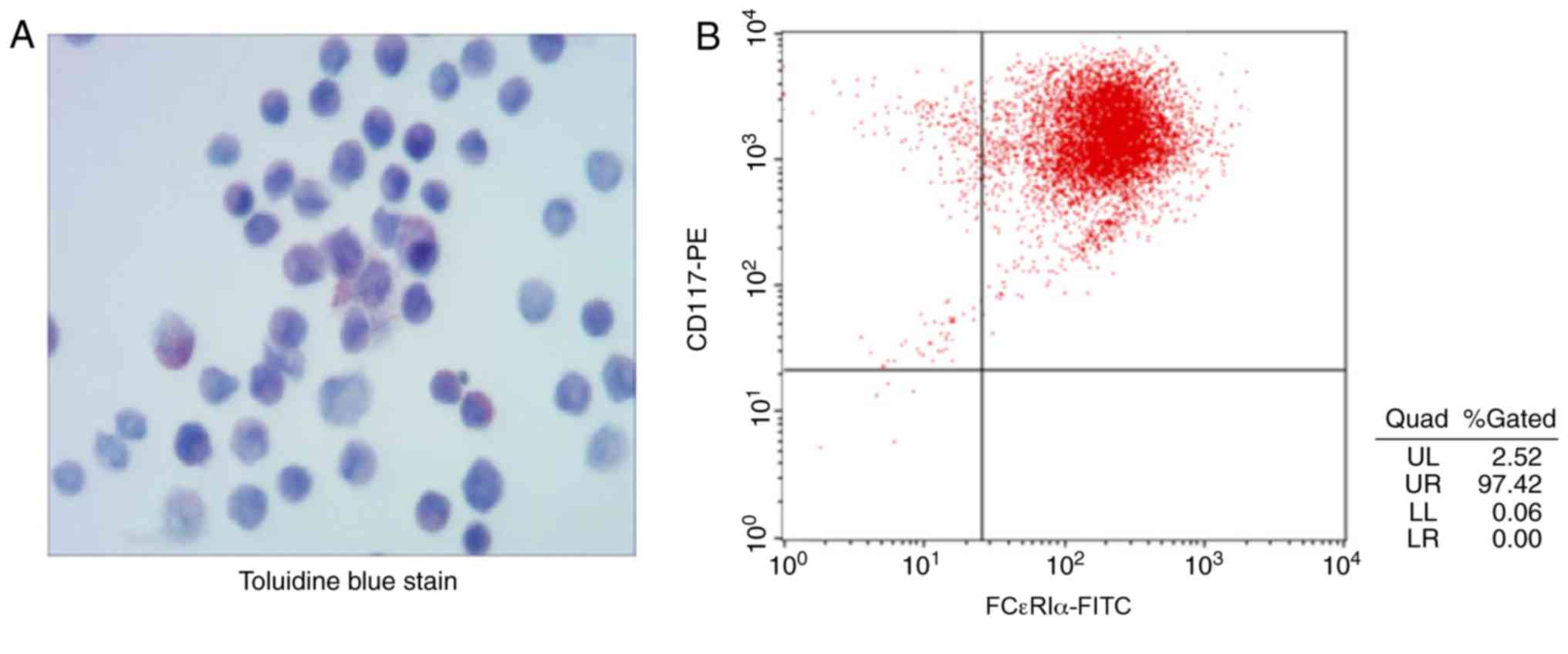
Identification of MCs
MCs were identified through toluidine blue staining
following 4 weeks of culture of the primary MCs, since there are
substances in the cytoplasm of MCs, such as heparin and histamine,
that create a metachromatic purple stain when they are treated with
toluidine blue (21) (Fig. 1A). The results in Fig. 1B demonstrated that 97.42% of the
cells were positively stained for the cell surface antigens CD117
and FCεRI α. These results indicate that MCs were successfully
induced.
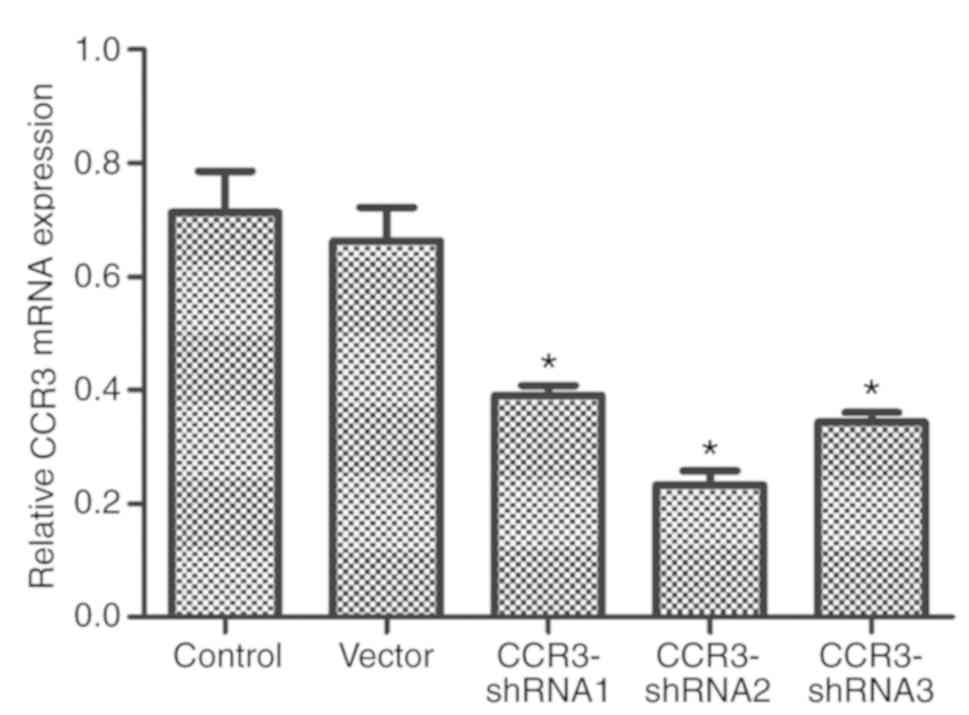
Screening of CCR3-shRNA effects
As presented in Fig.
2, the three CCR3-shRNAs significantly reduced CCR3 mRNA
expression compared with that in the control cells. In addition,
CCR3-shRNA2 appeared to have the strongest CCR3 downregulating
effect and was therefore selected for subsequent experiments.
Statistical analysis was not performed statistical analysis between
the three groups. The CCR3-shRNA2 downregulated CCR3 expression the
most as shown in Fig. 2 and was thus
selected for the following experiments.
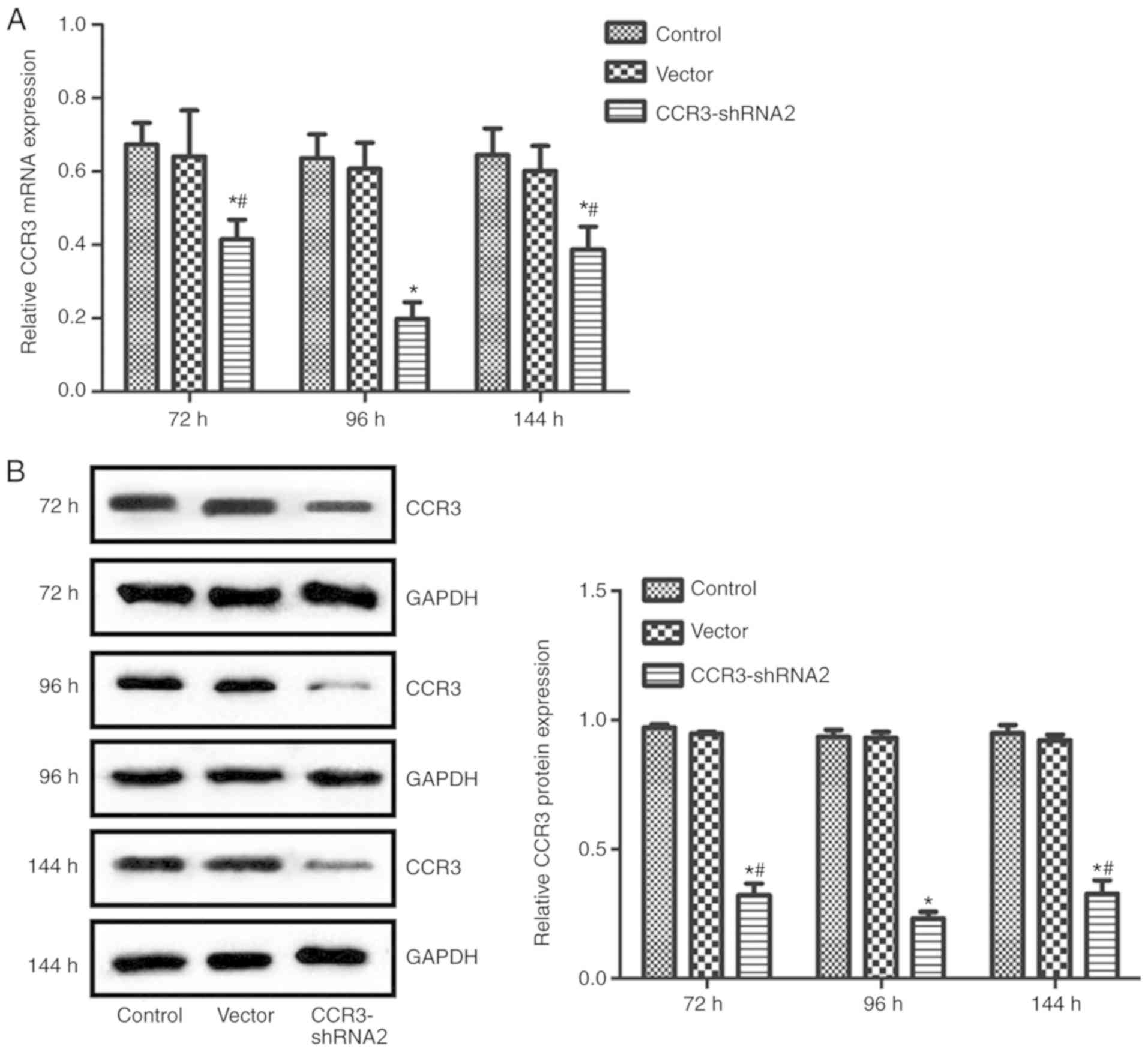
CCR3-shRNA2 transfection efficiency
and determination of the optimum transfection time
CCR3 expression at the mRNA and protein levels were
detected at 72, 96 and 144 h post transfection with CCR3-shRNA2.
The results in Fig. 3 suggest that
CCR3 mRNA and protein expression levels in cells transfected with
CCR3-shRNA2 were significantly reduced at each time point compared
with the control group. In addition, the CCR3 expression in the
CCR3-shRNA2 group at 96 h was significantly lower compared with
that at 72 and 144 h (P<0.05). The transfection time of 96 h was
therefore selected for subsequent experiments.
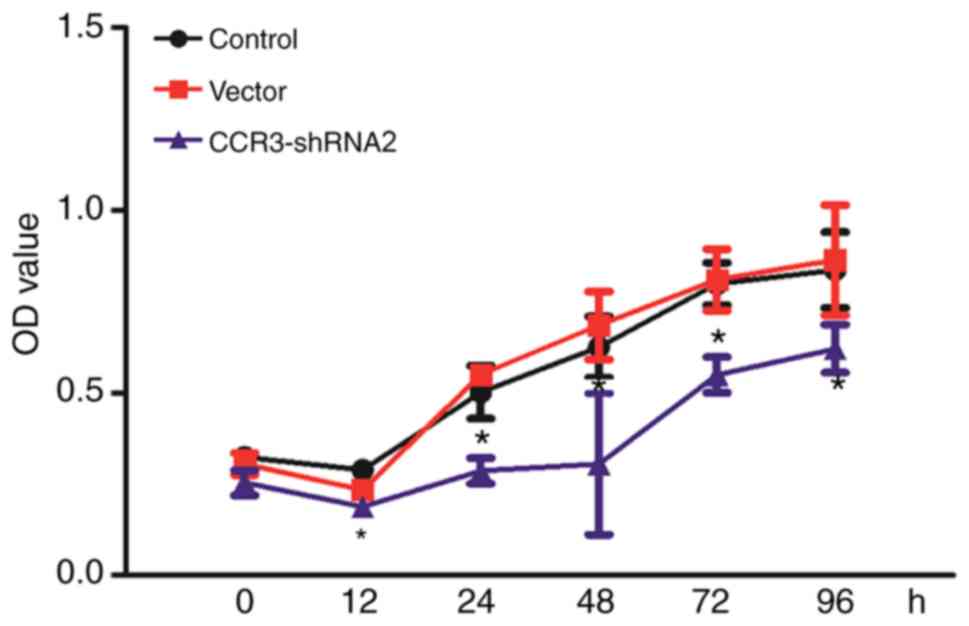
CCR3-shRNA2 transfection decreases MC
proliferation
CCK-8 assay was used to determine the effect of
CCR3-shRNA2 transfection on proliferation. As presented in Fig. 4, cells in each group proliferated in
a time dependent manner. However, compared with the control group,
CCR3-shRNA transfection significantly reduced the cell
proliferation at each time point after 12 h (P<0.05). These
findings demonstrated that CCR3 downregulation decreased MC cell
proliferation.
CCR3-shRNA2 transfection promotes MC
apoptosis
Following 96 h of cell transfection with
CCR3-shRNA2, apoptosis was assessed by flow cytometry using
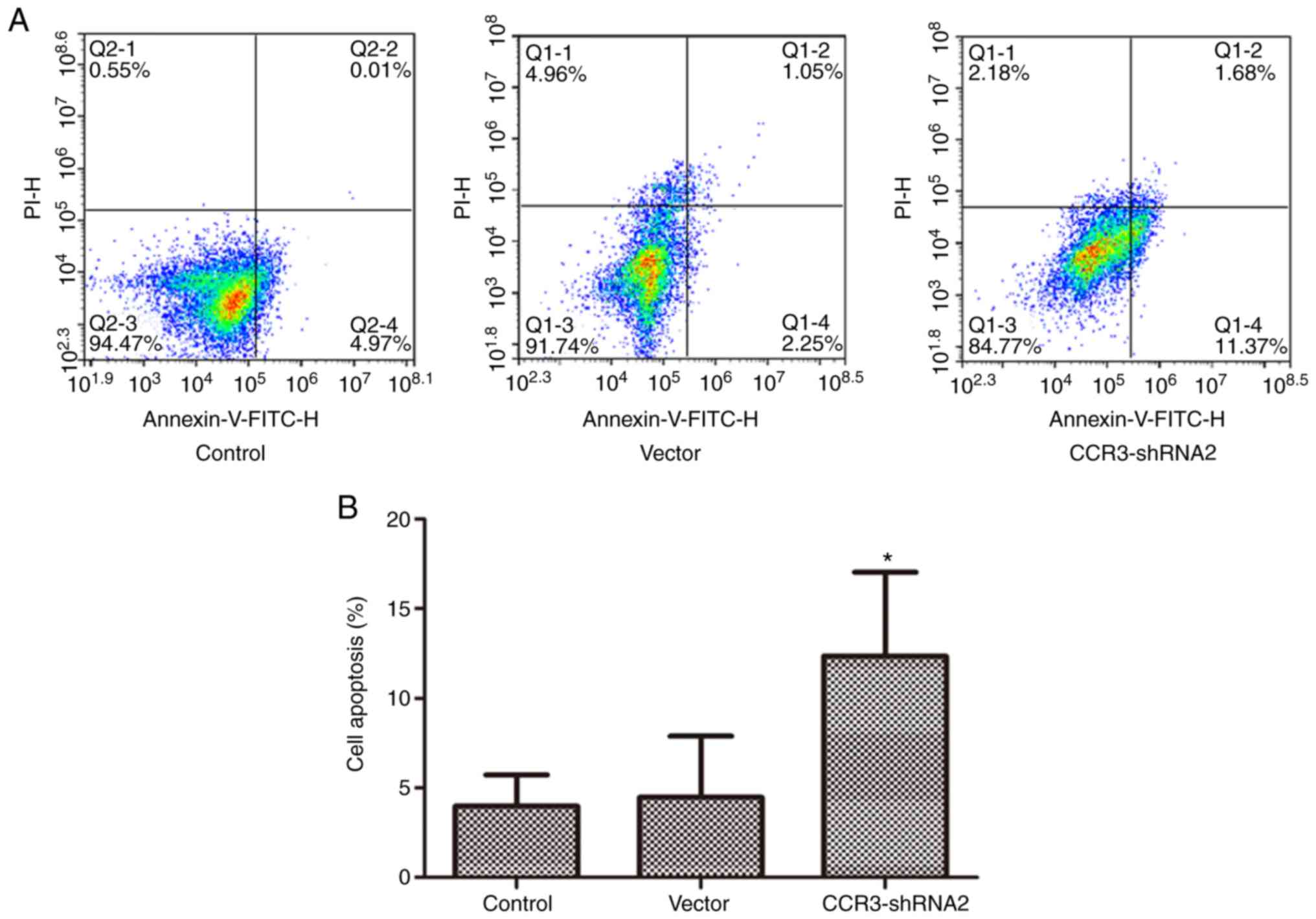
AnnexinV-FITC/PI staining. As presented in Fig. 5, apoptosis in the CCR3-shRNA2 group
was significantly increased compared with that in the control group
(P<0.05). This result indicates that CCR3 downregulation
promoted apoptosis in MCs.
CCR3-shRNA2 transfection decreases MC
chemotaxis
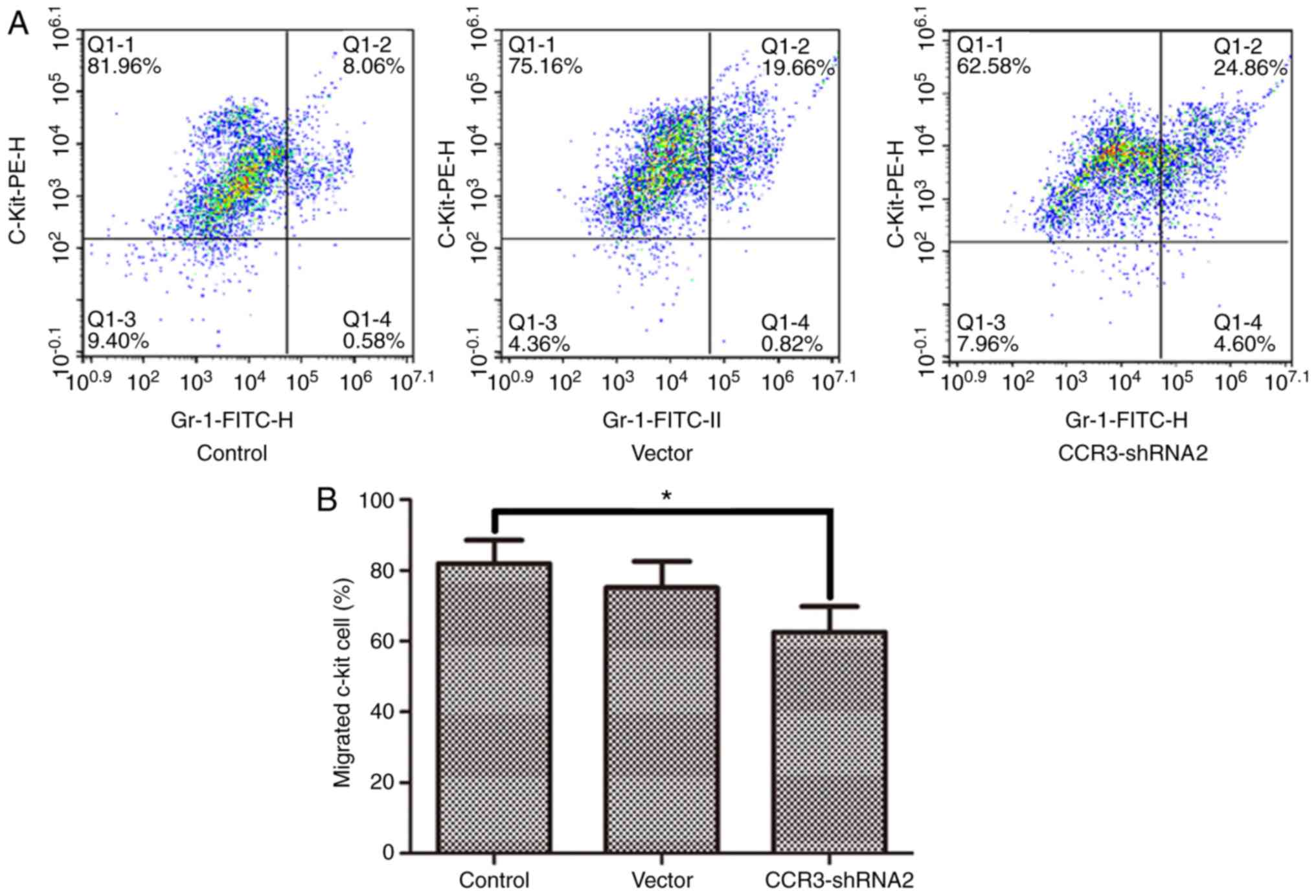
The migration of MCs towards the chemokine LTB4 was
detected using a Transwell assay. The results in Fig. 6 demonstrate that MC chemotaxis was
significantly reduced following CCR3-shRNA2 transfection
(P<0.05).
CCR3-shRNA2 transfection decreases the
degranulation of MCs
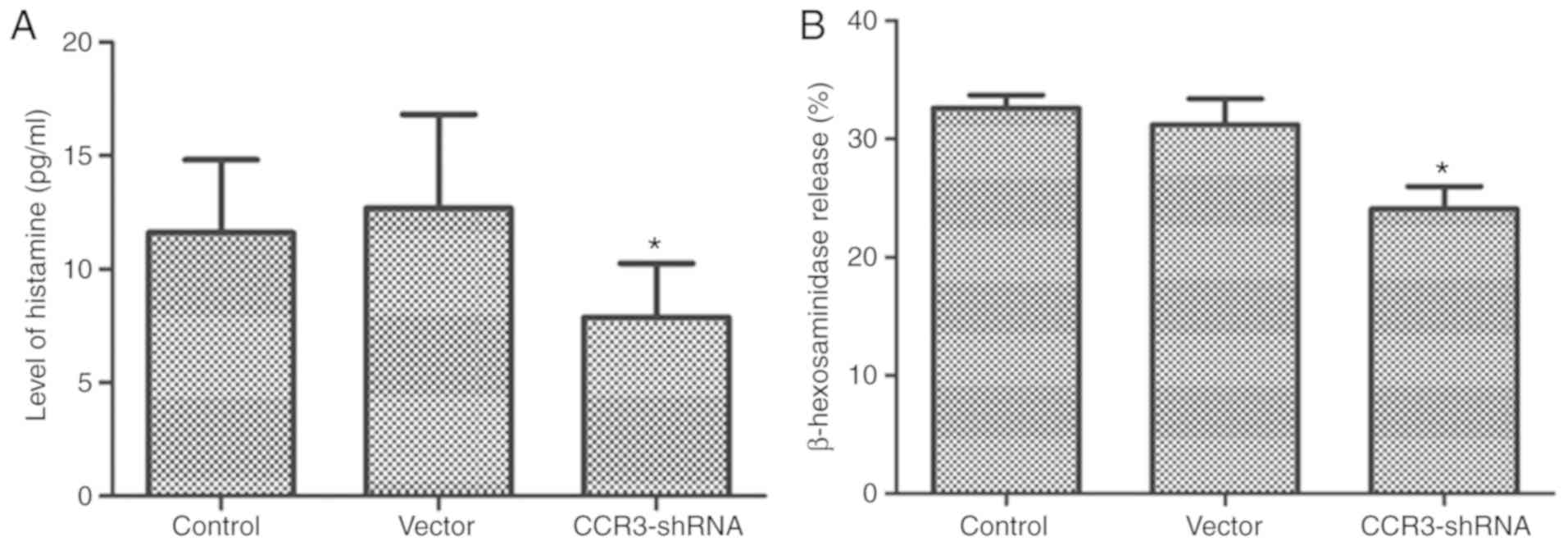
Histamine and β-hexosaminidase are granules
synthesized by MCs. They are important markers of MC degranulation
(22). ELISAs were performed to
detect the histamine and β-hexosaminidase release rate during MC
degranulation. The results in Fig. 7
demonstrate that the release of histamine and β-hexosaminidase into
the supernatant by CCR3-shRNA2 transfected MCs was significantly
reduced compared with that of control cells (P<0.05).
Discussion
MCs were first isolated by Ehrlich et al from
blood samples and connective tissue in 1878(23). Since then, MCs have been considered
as the major effector cells in allergic diseases, including asthma,
atopic dermatitis and AR (5,24). The binding of antigen-specific IgE to
FCεRI to sensitize MCs has been convincingly linked to the
pathophysiology of anaphylaxis and other acute allergic reactions
(25). MCs originate from bone
marrow-derived pluripotent hematopoietic cells and circulate as
immature precursors. However, after entering tissues, they develop
into functional MCs via multiple signals that prompt their
proliferation and differentiation (26,27). SCF
and IL-3 are the most important stimulating factors for MC
maturation. IL-3 is also called the polyclonal colony stimulating
factor, and is the most potent cytokine able to promote early-stage
mouse MC differentiation (28). SCF
maintains MC survival and promotes MC proliferation, and its
concentration will directly affect the number of MCs in the
circulation and tissues, In addition, SCF can assist IL-3-mediated
MC differentiation and serves a key role in late stage MC
maturation (29). At present, SCF
and IL-3 are frequently used to induce the differentiation of mouse
bone marrow-derived MCs; however, the concentration of each factor
used varies between studies. In the present study, the
concentration of 10 ng/ml was used for both factors, medium was
replaced every 7 days, and cells were identified 4 weeks after the
induction of differentiation through toluidine blue staining and
flow cytometry. The flow cytometry results demonstrated that
>97% cells were positively stained for CC117 and FCεRI,
confirming that the purity of MCs obtained was high. With regards
to the role of MCs in AR, Lin et al (30) investigated a mouse mode of AR and the
results from toluidine blue staining of the nasal mucosa suggested
that MC infiltration was significantly increased; furthermore, the
concentrations of specific IgE and histamine in peripheral blood
and IL-4, IL-9 and IL-17 in nasal lavage fluid were significantly
increased compared with those in normal controls. In addition,
following downregulation of potassium calcium-activated channel
subfamily N member 4 (KCa3.1) expression using a lentivirus vector
plasmid, the symptoms of AR in the AR model mice were markedly
ameliorated, MC infiltration into the nasal mucosa was reduced, and
the concentrations of specific IgE and histamine in peripheral
blood and of IL-4, IL-9 and IL-17 in nasal lavage fluid were
significantly decreased. Furthermore, in vitro experiments
demonstrated that KCA3.1 downregulation in the MC cell line P815
significantly downregulated MC degranulation and IL-6 and IL-8
release (30). Shao et al
(31) used the traditional Chinese
medicine Shenqi in AR mice and the MC cell line RBL-2H3, and
reported that this treatment could regulate MC degranulation and
treat AR. Zhang et al (32)
described similar results using the traditional Chinese medicine
curcumin. These findings suggest that MC proliferation, local
infiltration and degranulation serve key roles in the pathogenesis
of AR. Reducing MC proliferation, infiltration and degranulation,
and promoting MC apoptosis might therefore be crucial in the
treatment of AR.
Previous studies conducted by the present research
team demonstrated that CCR3 downregulation can promote eosinophil
apoptosis, and suppress the proliferation and degranulation of
eosinophils, which could therefore be used to treat AR (8,9).
However, the role of the CCR3 gene in MCs proliferation or
apoptosis remains controversial (12,33).
Collington et al (12)
compared bone marrow-derived MCs from CCR3 knockout and wild-type
mice, and reported that CCR3 knockout mice shared a similar MC
phenotype with wild-type mice, and that both MC types had similar
migratory capacity towards the chemokines LTB4 and SCF, suggesting
that the CCR3 gene has no influence on mouse MC chemotaxis.
Furthermore, Brightling et al (11) demonstrated that MCs can migrate
towards CCR3 chemokine, and that the use of CCR3 inhibitor can
markedly decrease the migratory capacity of MCs towards this
chemokine. Miyazaki et al (15) reported that the histamine and
β-hexosaminidase release following MC degranulation in CCR3 gene
knockout mice was significantly decreased, suggesting that the role
of CCR3 gene in MC degranulation remains unclear. The present study
demonstrated that MC proliferation was significantly decreased
following CCR3 downregulation in mouse-derived MCs compared with
the control group. In addition, the migratory capacity of
CRR3-shRNA transfected MCs towards the chemokine LTB4 was also
significantly reduced compared with the control group, indicating
that CCR3 interference could reduce MC proliferation and
chemotaxis. HSA-DNP and anti-DNP IgE were used to activate MCs
proliferation, in order to assess MC degranulation. The results
demonstrated that the levels of histamine and β-hexosaminidase
release following MC degranulation were significantly decreased in
the CCR3-shRNA transfected MC group compared with the control
group, suggesting that CCR3 interference may suppress MC
degranulation. Furthermore, the results of flow cytometry and CCK-8
assays demonstrated that CCR3 interference may promote MC apoptosis
and reduce MC proliferation.
This study exhibits some limitations. Firstly, as
the study was only restricted to the in vitro detection of
MC function, the results should be further verified using in
vivo experiments. Secondly, the study only determined the
effect of MC phenotype and CCR3 function on MC chemotaxis and
degranulation, and the mechanisms involved were not evaluated.
Future investigation will therefore focus on the underlying
mechanisms of CCR3.
In conclusion, CCR3 interference may promote MC
apoptosis and reduce MC proliferation, chemotaxis and
degranulation, thereby alleviating the MC-mediated allergic
inflammatory reaction observed in AR. To the best of our knowledge,
the present study was the first to investigate the effect of CCR3
interference on MCs and to explore the roles of CCR3 and MCs in the
pathogenesis of AR. The findings from this study may provide a
theoretical basis for the use of CCR3 as a potential target in the
treatment of AR and lay a favorable foundation for further
investigation.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81560171) and the
Talent Team Project of Jiangxi Province (grant no.
20161BCB24010).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
HP isolated the cells from mouse, cultured cells and
performed cell biology experiments. BL performed plasmid
construction. XZ designed this study and was a major contributor in
writing the manuscript. YL performed plasmid transfection and
identification. YJ analyzed the data. SW contributed to the design
of the study and revised the manuscript. All authors read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
All of the animal procedures were conducted in
accordance with the Guidelines for Care and Use of Laboratory
Animals, and were approved by the Animal Care and Use Committee of
The Second Affiliated Hospital of Nanchang University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bousquet J, Schunemann HJ, Hellings PW,
Arnavielhe S, Bachert C, Bedbrook A, Bergmann KC, Bosnic-Anticevich
S, Brozek J, Calderon M, et al: MACVIA clinical decision algorithm
in adolescents and adults with allergic rhinitis. J Allergy Clin
Immunol. 138:367–374.e2. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Utomo BSR, Hatta M, Sirait RH, Pratiwi S
and Massi MN: The role of cytokine interleukin-2, transcription
factor of FoxP3 in the immunological regulation of allergic
rhinitis. Int J Otolaryngol Head Neck Surg. 7:7–19. 2017.
|
|
3
|
Eifan AO and Durham SR: Pathogenesis of
rhinitis. Clin Exp Allergy. 46:1139–1151. 2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Im YS, Lee B, Kim EY, Min JH, Song DU, Lim
JM, Eom JW, Cho HJ, Sohn Y and Jung HS: Antiallergic effect of
gami-hyunggyeyeongyotang on ovalbumin-induced allergic rhinitis in
mouse and human mast cells. J Chin Med Assoc. 79:185–194.
2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Modena BD, Dazy K and White AA: Emerging
concepts: Mast cell involvement in allergic diseases. Transl Res.
174:98–121. 2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Ma J, Song Y, Wu B, Jiang M, Li K, Zhu C
and Wen F: Production of transgenic rice new germplasm with strong
resistance against two isolations of Rice stripe virus by RNA
interference. Transgenic Res. 20:1367–1377. 2011.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Liu Y, Zhu X and Zhang H: Effects of
chemokine receptor 3 gene silencing by RNA interference on
eosinophils. Exp Ther Med. 13:215–221. 2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zhu XH, Liao B, Liu K and Liu YH: Effect
of RNA interference therapy on the mice eosinophils CCR3 gene and
granule protein in the murine model of allergic rhinitis. Asian Pac
J Trop Med. 7:226–230. 2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Zhu XH, Liao B, Xu Y, Liu K, Huang Y,
Huang QL and Liu YH: Downregulation of mouse CCR3 by lentiviral
shRNA inhibits proliferation and induces apoptosis of mouse
eosinophils. Mol Med Rep. 15:696–702. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Andalib A, Doulabi H, Maracy MR, Rezaei A
and Hasheminia SJ: CCR3, CCR4, CCR5, and CXCR3 expression in
peripheral blood CD4+ lymphocytes in gastric cancer patients. Adv
Biomed Res. 2(31)2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Brightling CE, Kaur D, Berger P, Morgan
AJ, Wardlaw AJ and Bradding P: Differential expression of CCR3 and
CXCR3 by human lung and bone marrow-derived mast cells:
Implications for tissue mast cell migration. J Leukoc Biol.
77:759–766. 2005.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Collington SJ, Westwick J, Williams TJ and
Weller CL: The function of CCR3 on mouse bone marrow-derived mast
cells in vitro. Immunology. 129:115–124. 2010.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ochi H, Hirani WM, Yuan Q, Friend DS,
Austen KF and Boyce JA: T helper cell type 2 cytokine-mediated
comitogenic responses and CCR3 expression during differentiation of
human mast cells in vitro. J Exp Med. 190:267–280. 1999.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Khanolkar A, Burden SJ, Hansen B, Wilson
AR, Philipps GJ and Hill HR: Evaluation of CCR3 as a basophil
activation marker. Am J Clin Pathol. 140:293–300. 2013.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Miyazaki D, Nakamura T, Ohbayashi M, Kuo
CH, Komatsu N, Yakura K, Tominaga T, Inoue Y, Higashi H, Murata M,
et al: Ablation of type I hypersensitivity in experimental allergic
conjunctivitis by eotaxin-1/CCR3 blockade. Int Immunol. 21:187–201.
2009.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Zhang WN, Wu K, Zhou HM, Peng QZ, He WT,
Gao Y, Lin XG, Fang ZM and Chen ZH: Cultivation and identification
of bone marrow mast cells in mouse. J Intern Intensive Med.
15:42–44. 2009.
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Vink E, Willemien V, Michiel V, Wilko S,
Evert-Jan V, Blankestijn P, Yao Y, Harrison J, Davis G and Sammut
I: Novel invasive strategies in antihypertensive treatment-Renal
sympathetic denervation, baroreflex stimulation. Nephrol Dial
Transplant. 27 (Suppl 2)(ii38)2012.
|
|
19
|
Varga JM, Kalchschmid G, Klein GF and
Fritsch P: Mechanism of allergic cross-reactions-I. Multispecific
binding of ligands to a mouse monoclonal anti-DNP IgE antibody. Mol
Immunol. 28:641–654. 1991.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Bojarová P, Bruthans J and Křen V:
β-N-Acetylhexosaminidases-the wizards of glycosylation. Appl
Microbiol Biotechnol. 103:7869–7881. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Dahal BK, Kosanovic D, Messinger J,
Fischer Y, Hoffmann K, Antel J, Husen B, Hanke N, Mayet S and
Ghofrani HA: Role of mast cells and chymase in pulmonary vascular
remodeling. Eur Respir J. 38(1524)2011.
|
|
22
|
Zudaire E, Martínez A, Garayoa M, Pío R,
Kaur G, Woolhiser MR, Metcalfe DD, Hook WA, Siraganian RP and Guise
TA: Adrenomedullin is a cross-talk molecule that regulates tumor
and mast cell function during human carcinogenesis. Am J Pathol.
168:280–291. 2006.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Fisher ER: Tissue mast cells. J Am Med
Assoc. 173:171–173. 1960.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Krystel-Whittemore M, Dileepan KN and Wood
JG: Mast cell: A multi-functional master cell. Front Immunol.
6:1–12. 2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Galli SJ and Tsai M: IgE and mast cells in
allergic disease. Nat Med. 18(693)2012.PubMed/NCBI View
Article : Google Scholar
|
|
26
|
Ribatti D: The development of human mast
cells. An historical reappraisal. Exp Cell Res. 342:210–215.
2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Schmetzer O, Valentin P, Church MK, Maurer
M and Siebenhaar F: Murine and human mast cell progenitors. Eur J
Pharmacol. 778:2–10. 2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Desai A, Jung MY, Olivera A, Gilfillan AM,
Prussin C, Kirshenbaum AS, Beaven MA and Metcalfe DD: IL-6 promotes
an increase in human mast cell numbers and reactivity through
suppression of suppressor of cytokine signaling 3. J Allergy Clin
Immunol. 137:1863–1871.e6. 2016.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Bérard F, Ferrier Le Bouëdec MC, Bouillet
L, Reguiai Z, Barbaud A, Cambazard F, Milpied B, Pelvet B, Kasujee
I and Gharbi H: Omalizumab in patients with chronic spontaneous
urticaria nonresponsive to H1-antihistamine treatment: Results of
the phase IV open-label SUNRISE study. Br J Dermatol. 180:56–66.
2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Lin H, Zheng CQ, Li J, Yang C and Hu L:
Lentiviral shRNA against KCa3.1 inhibits allergic response in
allergic rhinitis and suppresses mast cell activity via PI3K/AKT
signaling pathway. Sci Rep. 5(13127)2015.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Shao YY, Zhou YM, Hu M, Li JZ, Chen CJ,
Wang YJ, Shi XY, Wang WJ and Zhang TT: The Anti-allergic rhinitis
effect of traditional Chinese medicine of shenqi by regulating mast
cell degranulation and Th1/Th2 cytokine balance. Molecules. 22(pii:
E504)2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Zhang N, Li H, Jia JH and He MQ:
Anti-inflammatory effect of curcumin on mast cell-mediated allergic
responses in ovalbumin-induced allergic rhinitis mouse. Cell
Immunol. 298:88–95. 2015.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Gangwar RS, Landolina N, Arpinati L and
Levi-Schaffer F: Mast cell and eosinophil surface receptors as
targets for anti-allergic therapy. Pharmacol Ther. 170:37–63.
2017.PubMed/NCBI View Article : Google Scholar
|