Introduction
Osteoarthritis (OA) affects ~50 million adults in
China, and the currently available treatment strategies only manage
pain (1,2). OA leads to the breakdown and gradual
loss of joint cartilage, triggering debilitating pain and
subsequent disability (3). The
disease mainly affects the aging population, and >37% of
individuals aged 60 years or older are affected by OA (4). At present, arthroscopic surgery and
painkiller therapy are the main treatment options for OA; however,
these treatment strategies result in 36,000,000 ambulatory care
visits and 750,000 hospitalizations annually (5,6), leading
to a high socioeconomic burden (7).
Identifying new treatment strategies for OA is therefore
important.
An increasing number of studies reported that
non-coding RNAs (ncRNAs) are possible mediators of cell
proliferation (8,9). ncRNAs are defined as transcripts of
>200 nucleotides in length with limited or no protein-coding
capacity, which are referred to as long non-coding RNAs (lncRNAs)
(10). lncRNAs serve crucial roles
in multiple diseases (11). For
example, Jiang et al (12)
reported that the lncRNA heart and neural crest derivatives
expressed 2-antisense RNA1 inhibits 5-fluorouracil resistance by
modulating the microRNA (miR)-20a/programmed cell death 4 axis in
colorectal cancer. Furthermore, Li et al (13) reported that the Pvt1 oncogene
regulates chondrocyte apoptosis in OA by acting as a miR-488-3p
sponge. In addition, Xiao et al (14) demonstrated that the lncRNA
MIR4435-2HG is downregulated in OA and can mediate chondrocyte
proliferation and apoptosis. However, the role of miR4435-2HG
during the progression of OA remains unknown. The present study
aimed therefore to investigate the biological function of
MIR4435-2HG in OA in vitro.
Materials and methods
Tissue collection
OA cartilage tissues (taken from patients with OA)
and normal cartilage tissues (n=20 in each group) were collected
from patients at the Second Affiliated Hospital of Inner Mongolia
Medical University between February 2019 and June 2019.
Clinicopathological characteristics were also collected from these
patients who provided written informed consent. Patients were
diagnosed with OA according to The American College of Rheumatology
classification criteria for the diagnosis of OA (15). The present study was approved by the
Institutional Ethical Committee of the Second Affiliated Hospital
of Inner Mongolia Medical University. Patient characteristics are
presented in Table I. Normal tissues
were collected from patients who have suffered from traffic
accidents. These patients provided written informed consent.
Collected tissues were stored at -80˚C until further use.
 | Table IClinical characteristics of patients
with osteoarthritis and healthy controls. |
Table I
Clinical characteristics of patients
with osteoarthritis and healthy controls.
| Clinical
characteristics | Healthy, n | Patients, n | P-value |
|---|
| Age, years | 48±8 | 52±10 | 0.786 |
| Sex | | | |
|
Male | 17 | 18 | 0.821 |
|
Female | 13 | 12 | 0.678 |
| BMI,
kg/m2 | 23.7±2.1 | 24.1±2.5 | 0.788 |
Cell culture
The human chondrocyte cell line CHON-001 and 293T
cells were obtained from the American Type Culture Collection.
Cells were cultured in RPMI-1640 medium (Invitrogen; Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum
(Invitrogen; Thermo Fisher Scientific, Inc.) and 2 mM glutamine
(Sigma-Aldrich; Merck KGaA) andplaced at 37˚C in a humidified
incubator containing 5% CO2. CHON-001 cells were treated
with 10 ng/ml interleukin (IL)-1β (Sigma-Aldrich; Merck KGaA) at
room temperaturefor 24 h to generate an in vitro model of OA
(16). The nuclear factor κB (NF-κB)
inhibitor BAY 11-7085 (10 µM) was purchased from
MedChemExpress.
Cell transfection
293T cells (5x106/well) were transfected
with 1 µg/µl pLVX-IRES-Puro-MIR4435-2HG overexpression (OE) vector
or pLVX-IRES-Puroempty vector (both from Shanghai GenePharma Co.,
Ltd.) using Lipofectamine® 3000 reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. The helper packaging vectors (pLP/VSVG, pLP1 and pLP2)
were obtained from Invitrogen, Thermo Fisher Scientific, Inc., and
used at a concentration of 1 µg/µl. After transfection, cells were
incubated at 32̊C for 48 h. Subsequently, the 293T lentiviral
supernatant containing the retroviral particles, was collected
following centrifugation at 4˚C, 956 x g for 15 min.
For transduction, supernatant was passed through a
45-µm filter (Costar; Corning, Inc.) to obtain viral particles,
which were then added to CHON-001 cells. After transduction for 48
h, cells were selected with puromycin (Sigma-Aldrich; Merck KGaA).
The knockdown efficiency was verified by reverse
transcription-quantitative PCR (RT-qPCR).
RT-qPCR
Total RNA was extracted from tissues or CHON-001
cells using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. Total
RNA was reverse transcribed into cDNA using the PrimeScript RT
reagent kit (Takara Bio, Inc.) according to the manufacturer's
protocol. Subsequently, RT-qPCR was performed using the SYBR premix
Ex Taq II kit (Takara Bio, Inc.) according to the manufacturer's
protocol. All reactions were performed in triplicate as follows: 2
min at 94̊C, followed by 35 cycles at 94̊C for 30 sec and 55̊C for
45 sec. The sequences of the primes used for RT-qPCR were as
follows: MIR4435-2HG forward, 5'-GGAAGTGGTGGCTATGAGTCAG-3' and
reverse, 5'-TGTCAATTTGAAACTTAAAAAGCAG-3'; IL-17A forward,
5'-CTACAACCGATCCACCTCACC-3' and reverse,
5'-AGCCCACGGACACCAGTATC-3'; β-actin forward,
5'-GTCCACCGCAAATGCTTCTA-3' and reverse, 5'-TGCTGTCACCTTCACCGTTC-3';
U6 forward, 5'-CTCGCTTCGGCAGCACAT-3' and reverse
5'-AACGCTTCACGAATTTGCGT-3'. miRNA and mRNA expression levels were
quantified using the 2-ΔΔCq method (17)and normalized to the internal reference
genes β-actin or U6.
Cell Counting Kit-8 (CCK-8) assay
The CCK-8 assay (Beyotime Institute of
Biotechnology) was used to assess cell proliferation according to
the manufacturer's protocol. CHON-001 cells were plated in 96-well
plates at a density of 5x103 cells/well and treated for
0, 24, 48 or 72 h at 37˚C. The cells were cultured with 10 ng/ml
IL-1β alone, MIR4435-2HG OE lentivirus alone, or both (IL-1β +
MIR4435-2HG OE). Subsequently, CHON-001 cells were incubated with
10 µl CCK-8 reagent for 2 h at 37˚C. The absorbance was measured at
a wavelength of 450 nm using a microplate reader (Thermo Fisher
Scientific, Inc.).
Western blotting
Total protein was extracted from cell lines using
RIPA lysis buffer (Beyotime, Institute of Biotechnology). Total
protein was quantified using abicinchoninic acid protein kit
(Thermo Fisher Scientific, Inc.). Protein (40 µg per lane) was
separated by SDS-PAGE on 10% gels and transferred to PVDF membranes
(Thermo Fisher Scientific, Inc.). Subsequently, the membranes were
blocked with 5% skimmed milk in TBST for 1 h at room temperature.
The membranes were incubated overnight at 4̊C with primary
antibodies against collagen II (cat. no. ab239007), matrix
metalloproteinase (MMP)-1 (cat. no. ab118973), MMP13 (cat. no.
ab219620), IL-17A (cat. no. ab136668), p65 (cat. no. ab16502),
inhibitor of κB (IκB; cat. no. ab109330), p-p65 (cat. no. ab6503),
p-IκB (cat. no. ab225650) and β-actin (cat. no. ab179467). All
primary antibodies were purchased from Abcam and diluted at
1:1,000. Following primary incubation, membranes were incubated
with HRP-conjugated secondary antibodies (Abcam; cat. no. ab20272,
1:5,000) for 1 h at room temperature. Protein bands were visualized
using the ECL kit (Thermo Fisher Scientific, Inc.). β-actin was
used as the loading control. The Image-Pro Plus version 6 0
software was used for densitometry analysis.
Immunofluorescence
CHON-001 cells were cultured in 24-well plates
overnight at the density of 5x104 cells/well. Cells were
pre-fixed with 4% paraformaldehyde at room temperature for 10 min
and fixed in pre-cooled methanol at 4˚C for another 10 min.
Subsequently, cells were incubated overnight at 4̊C with a primary
antibody against Ki67 (1:1,000; cat. no. ab270650; Abcam).
Following primary incubation, cells were incubated with a goat
anti-rabbit IgG (HRP-conjugated) antibodies (1:5,000; ab125900,
Abcam) at room temperature for 1 h. Cells were observed using a
CX23 fluorescence microscope (magnification, x200; Olympus
Corporation).
Cell apoptosis analysis
CHON-001 cells were seeded in 6-well plates at a
density of 5x104 cells/well and cultured for 48 h.
CHON-001 cells were centrifuged at 15 x g for 5 min (at 4˚C), and
the pellet was resuspended in 100 µl binding buffer. Subsequently,
5 µl Annexin V-fluorescein isothiocyanate and 5 µl propidium iodide
were added to the cells for 15 min at 4˚C. Samples were analyzed by
flow cytometry (BD Biosciences) and apoptotic cells were analyzed
using the WinMDI software version 2.9 (Invitrogen; Thermo Fisher
Scientific, Inc.).
Downstream target genes
prediction
Downstream target genes of MIR4435-2HG were
predicted by using the publicly available program Starbase 2.0
(http://starbase.sysu.edu.cn/). In
addition, miR-526b-3p targeted gene prediction was performed using
two publicly available programs, TargetScan 7.2 (www.targetscan.org/vert_71/) and miRDB 4.0
(www.mirdb.org). The results obtained from
Starbase were considered to be the downstream target genes of
MIR4435-2HG. The resulting data of TargetScan 7.2 and miRDB 4.0
were selected as the direct gene of miR-526b-3p.
Dual luciferase reporter assay
The partial sequences of MIR4435-2HG and the IL-17A
3'-untranslated region (UTR) containing the putative binding sites
of miR-510-3p were provided by Sangon Biotech Co., Ltd. The
sequences were cloned into the pmirGLO Dual-Luciferase miRNA Target
Expression Vectors (Promega Corporation) to construct wild-type
(WT) reporter MIR4435-2HG and IL-17A vectors. The mutant (MUT)
MIR4435-2HG sequence and IL-17A 3'-UTR containing the putative
binding sites of miR-510-3p were generated using the Q5
Site-Directed Mutagenesis kit (New England Biolabs, Inc.) according
to the manufacturer's protocol. The aforementioned MUT sequences
were cloned into pmirGLO vectors to construct MUT reporter
MIR4435-2HG and IL-17A vectors. The WT or MUT MIR4435-2HG vectors
were transfected into CHON-001 cells together with control
(untransfected cells), 10 nM vector-control (pcDNA-3.1 vector,
Beyotime) or 10 nM miR-510-3p mimic using Lipofectamine®
2000 reagent (Thermo Fisher Scientific, Inc.) for 48 h, according
to the manufacturer's instructions (blank group means untransfected
cells; vector control group means cells transfected with pcDNA-3.1
vector). The sequence of the miR-510-3p mimic was as follows:
5'-AAUCCUUUGUCCCUGGGUGAGA-3'. The sequence of mimic negative
control was as follows: 5'-UCACAACCUCCUAGAAAGAGUAGA-3'. Similarly,
the WT and MUT IL-17A vectors were transfected into 293T cells
together with control, vector-control or miR-510-3p mimics using
Lipofectamine 2000 for 48 h. Relative luciferase activities were
detected using a Dual-Glo Luciferase assay system (Promega
Corporation). The data were normalized to Renilla luciferase
activity.
Statistical analysis
Data were presented as the means ± standard
deviation. Comparison between two groups was analyzed by the
Student's t-test. Multi-group comparison was analyzed using one-way
ANOVA followed by Tukey's post hoc test. Statistical analysis was
conducted using GraphPad Prism version 7 (GraphPad Software, Inc.).
P<0.05 was considered to indicate a statistically significant
difference.
Results
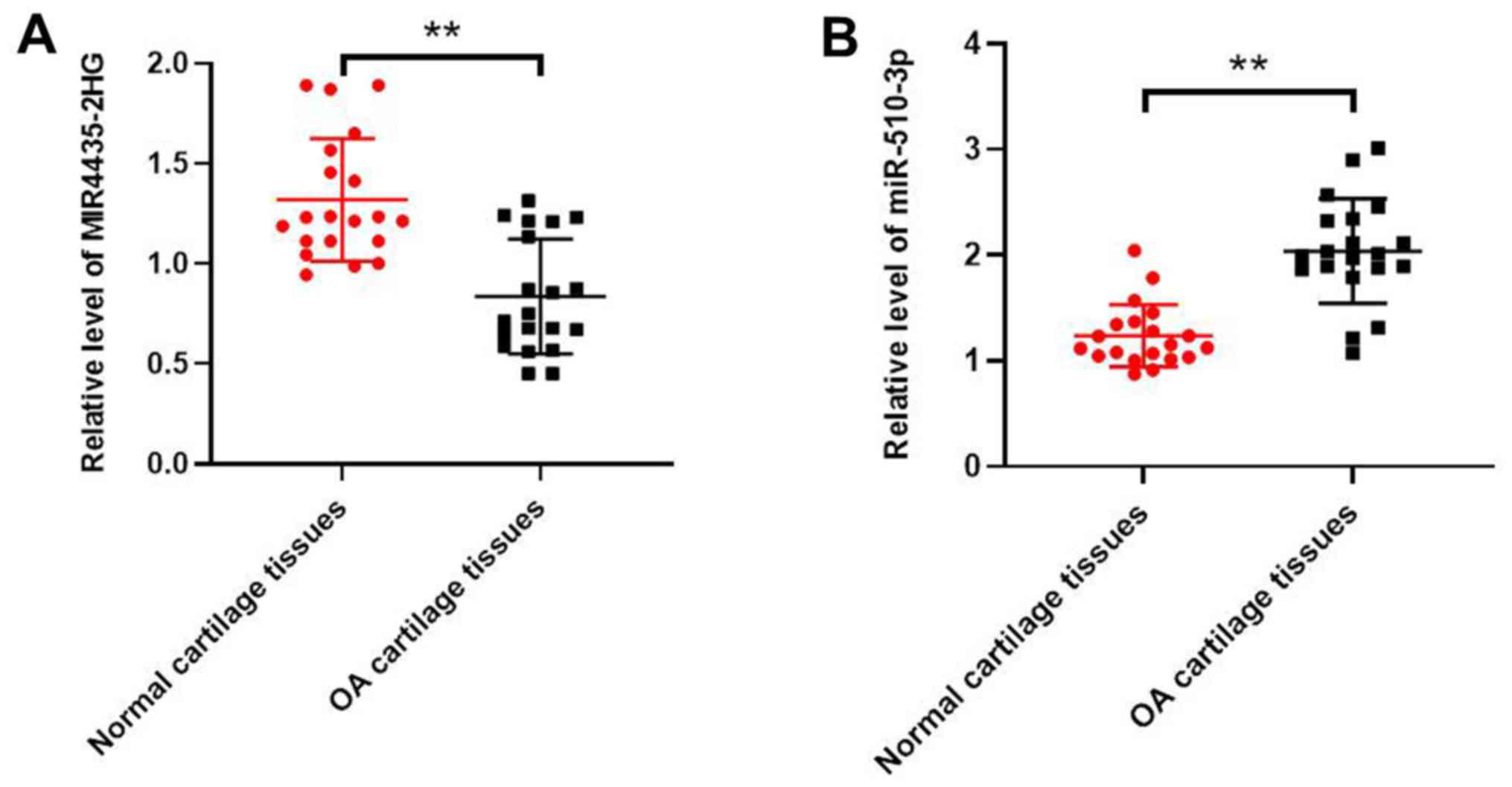
MIR4435-2HG is downregulated
andmiR-510-3p is upregulated in OA tissues
Expression levels of MIR4435-2HG and miR-510-3p were
determined in OA and normal tissues using RT-qPCR. As indicated in
Fig. 1A, MIR4435-2HG expression
level was significantly downregulated in OA tissues compared with
normal tissues. However, the expression level of miR-510-3p was
significantly up regulated in OA tissue compared with normal tissue
(Fig. 1B).
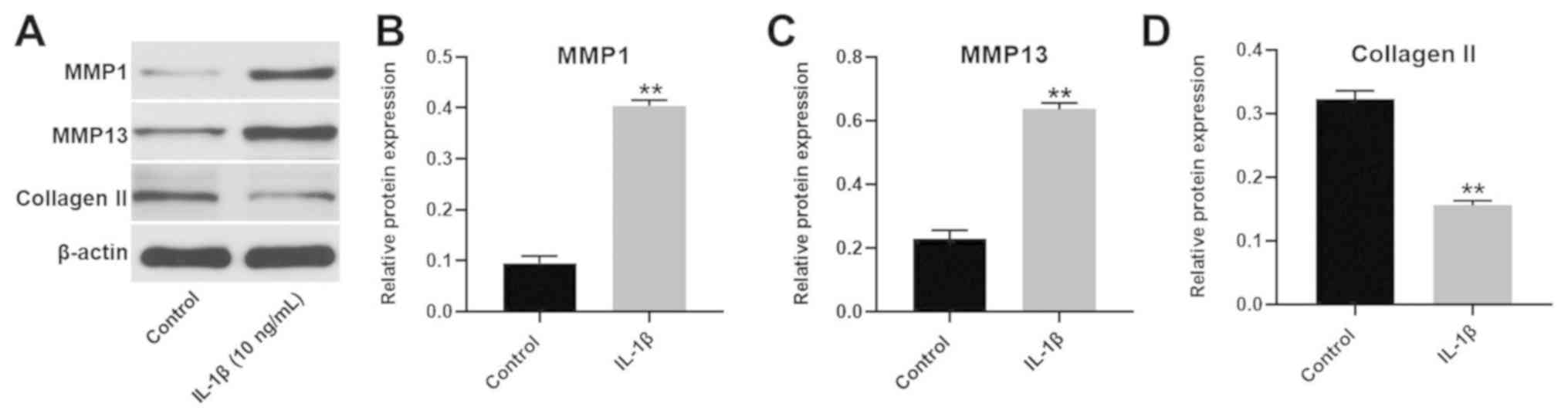
Establishment of an in vitro model of
OA
To establish an in vitro model of OA,
CHON-001 cells were treated with 10 ng/ml IL-1β (Fig. 2). The expression of MMP1 and MMP13 in
CHON-001 cells was significantly increased following IL-1β
treatment. However, the expression of collagen II was significantly
decreased in CHON-001 cells. Since these three proteins are key
markers of OA (18), the results
suggested that the in vitro model of OA had been
successfully established.
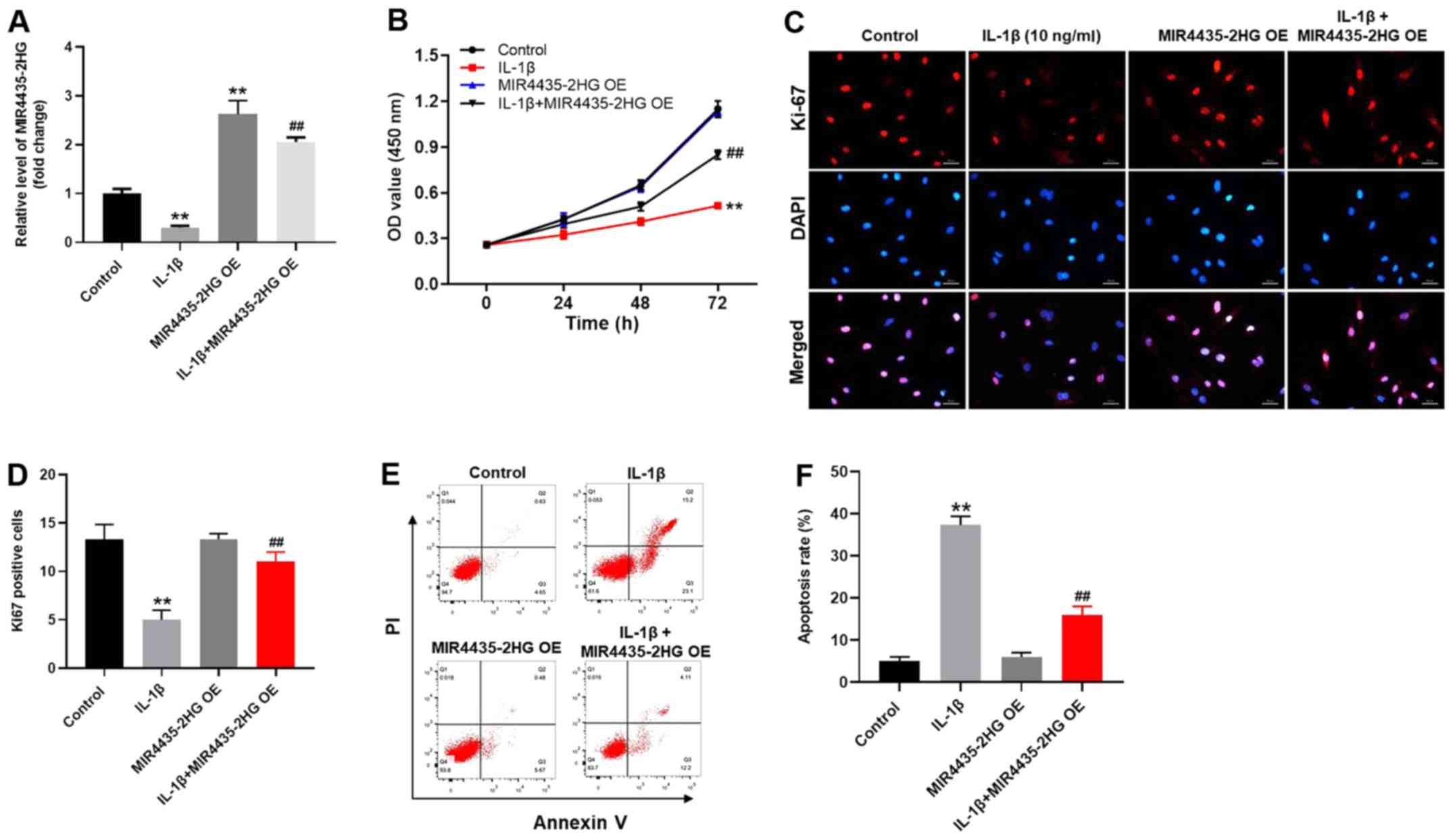
MIR4435-2HG OE significantly promotes
the proliferation of CHON-001 cells treated with IL-1β
To investigate the gene expression, RT-qPCR was
performed (Fig. 3A). The expression
level of MIR4435-2HG in CHON-001 cells was significantly decreased
following IL-1β treatment (Fig. 3A).
However, MIR4435-2HG expression was increased following
transfection with MIR4435-2HG OE, which reversed the inhibitory
effect of IL-1β. MIR4435-2HG may therefore act as an OA suppressor.
In addition, the results from CCK-8 assay demonstrated that
MIR4435-2HG OE significantly increased CHON-001 cell proliferation;
however, IL-1β significantly inhibited the proliferation of
CHON-001 cells (Fig. 3B). Thus,
MIR4435-2HG OE reversed the inhibiting effect of IL-1β on cell
proliferation. Similarly, the Ki-67-positive rate of CHON-001 cells
was significantly decreased by IL-1β, which was reversed by
MIR4435-2HG OE (Fig. 3C and D). Furthermore, IL-1β significantly
increased CHON-001 cell apoptosis; however, MIR4435-2HG OE
inhibited the pro-apoptotic effect of IL-1β treatment on CHON-001
cells (Fig. 3E and F). Taken together, these results indicated
that MIR4435-2HG OE may significantly enhance the proliferation of
CHON-001 cells following treatment with IL-1β.
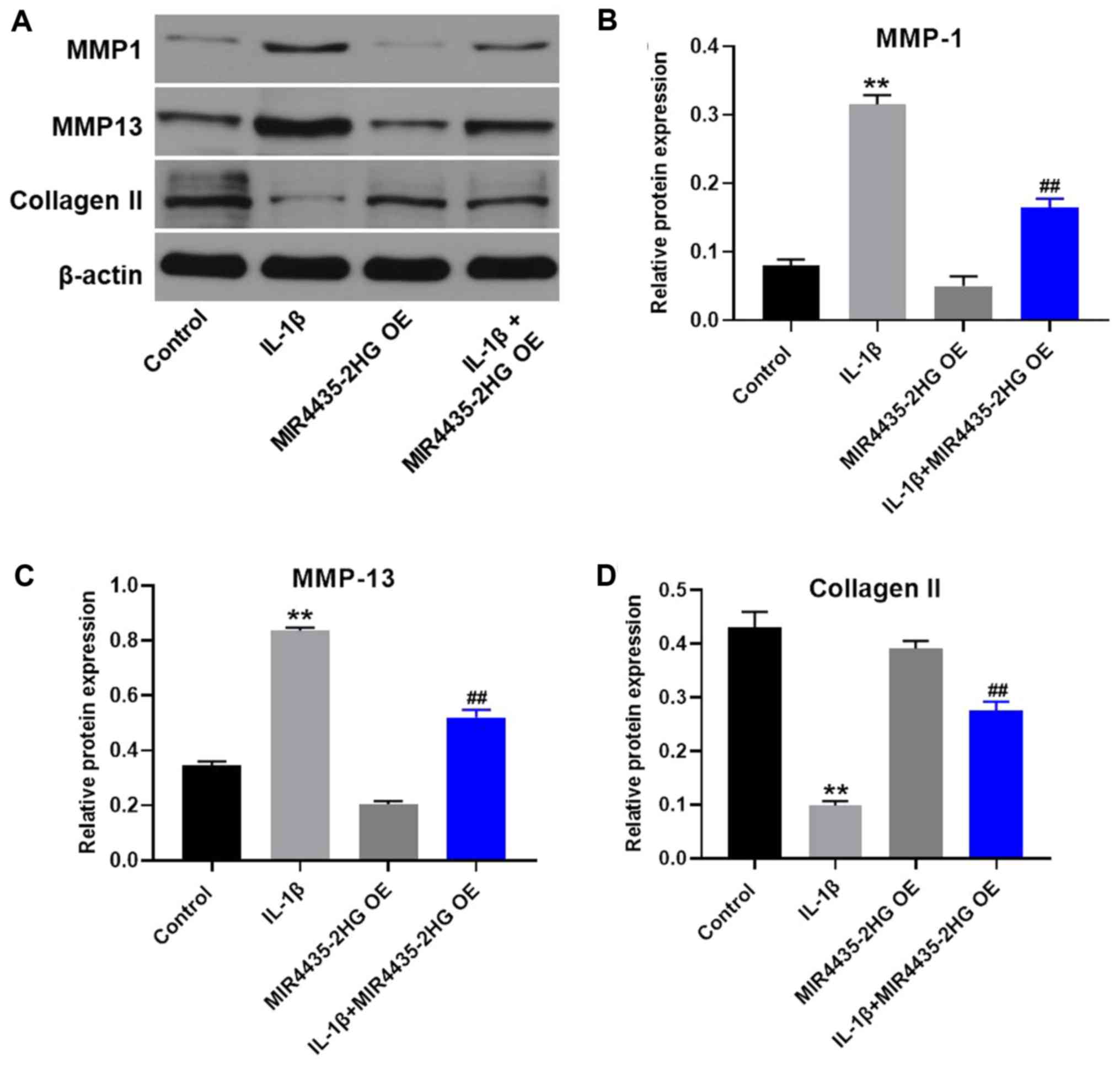
MIR4435-2HG OE downregulates MMP1 and
MMP13 expression and upregulates collagen II expression in
IL-1β-treated CHON-001 cells
Western blotting was used to detect protein
expression in CHON-001 cells. MIR4435-2HG OE significantly
decreased the expression of MMP1 and MMP13 and upregulated the
protein expression of collagen II in IL-1β-treated CHON-001 cells,
compared with IL-1β treatment alone (Fig. 4).
MIR4435-2HG OE significantly
suppresses the progression of OA via the miR-510-3p/IL-17A
axis
To explore the underlying mechanism of MIR4435-2HG
in OA progression in vitro, Starbase, Targetscan, miRDB
predictions were used together with a dual luciferase reporter
assay. According to Starbase predictions, miR-510-3p was identified
as a potential downstream target of miR4435-2HG. Furthermore,
IL-17A was likely to be a potential target of miR-510-3p according
to the Targetscan and miRDB databases (Fig. 5A-D). Since previous studies reported
that IL-17A serves a key role in the progression of OA (19,20),
IL-17A was also considered as a direct target of miR-510-3p in the
present study. The aforementioned results were further verified by
RT-qPCR (Fig. 5E). Expression of
miR-510-3p was not altered by MI miR4435-2HG OE in CHON-001 cells
(Fig. 5F). However, the expression
of IL-17A in CHON-001 cells was significantly decreased in the
presence of miR-510-3p mimics (Fig.
5G). Taken together, these results suggested that MIR4435-2HG
may inhibit OA via the miR-510-3p/IL-17A axis.
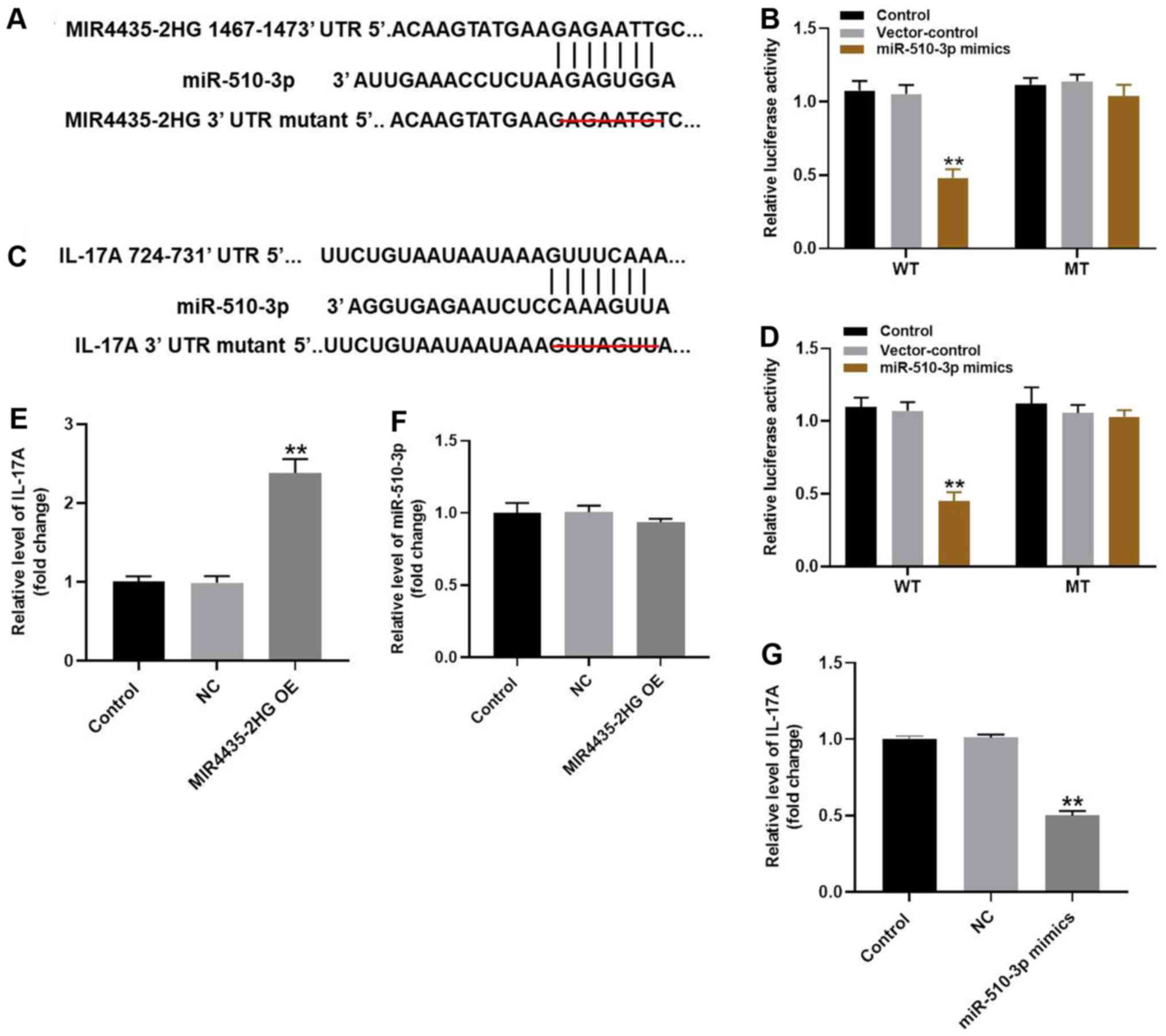 | Figure 5MIR4435-2HG OE significantly
suppressed the progression of OA via the miR-510-3p/IL-17A axis.
(A) Gene structure of MIR4435-2HG at the position of 1467-1473
indicated the predicted target site of miR-510-3p in its 3'UTR,
with a sequence of GACAATC. (B) Luciferase activity was measured in
CHON-001 cells following co-transfection with WT/MT MIR4435-2HG
3'-UTR plasmid and miR-510-3p with the dual luciferase reporter
assay. (C) Gene structure of IL-17A at the position of 724-731
indicated the predicted target site of miR-510-3p in its 3'UTR,
with a sequence of GUUAGUU. (D) Luciferase activity was measured in
CHON-001 cells following co-transfection with WT/MT IL-17A 3'-UTR
plasmid and miR-510-3p with the dual luciferase reporter assay. (E)
Gene expression of IL-17A in CHON-001 cells was detected by
RT-qPCR. (F) Gene expression of miR-510-3p in CHON-001 cells was
detected by RT-qPCR. (G) Gene expression of IL-17A in CHON-001
cells was detected by RT-qPCR. **P<0.01 vs. control.
OE, overexpression; IL, interleukin; miR, microRNA; RT-qPCR,
reverse transcription-quantitative PCR; WT, wildtype; MT, mutant;
UTR, untranslated region; NC (empty vector), negative control. |
MIR4435-2HG OE suppresses OA via
regulation of miR-510-3p/IL-17A axis and NF-κB signaling
pathway
To further investigate the underlying mechanism of
MIR4435-2HG in OA progression, western blotting was performed. The
expression of IL-17A in CHON-001 cells was significantly inhibited
following treatment with IL-1β, and MIR4435-2HG OE partially
reversed IL-1β-mediated effects on IL-17A expression (Fig. 6A and B). MIR4435-2HG OE alone also significantly
enhanced the expression of IL-17A in CHON-001 cells. Furthermore,
IL-1β significantly increased p-p65 and p-IκB expression in
CHON-001 cells. However, IL-1β-induced effects on NF-κB signaling
pathway were significantly suppressed by MIR4435-2HG OE (Fig. 6A, C
and D). In addition, the NF-κB
inhibitor BAY 11-7085 further enhanced the inhibitory effect of
MIR4435-2HG OE on NF-κB signaling pathway (Fig. 6E-G). These results demonstrated that
MIR4435-2HG OE significantly suppressed the progression of OA via
the miR-510-3p/IL-17A axis and by inactivating the NF-κB signaling
pathway.
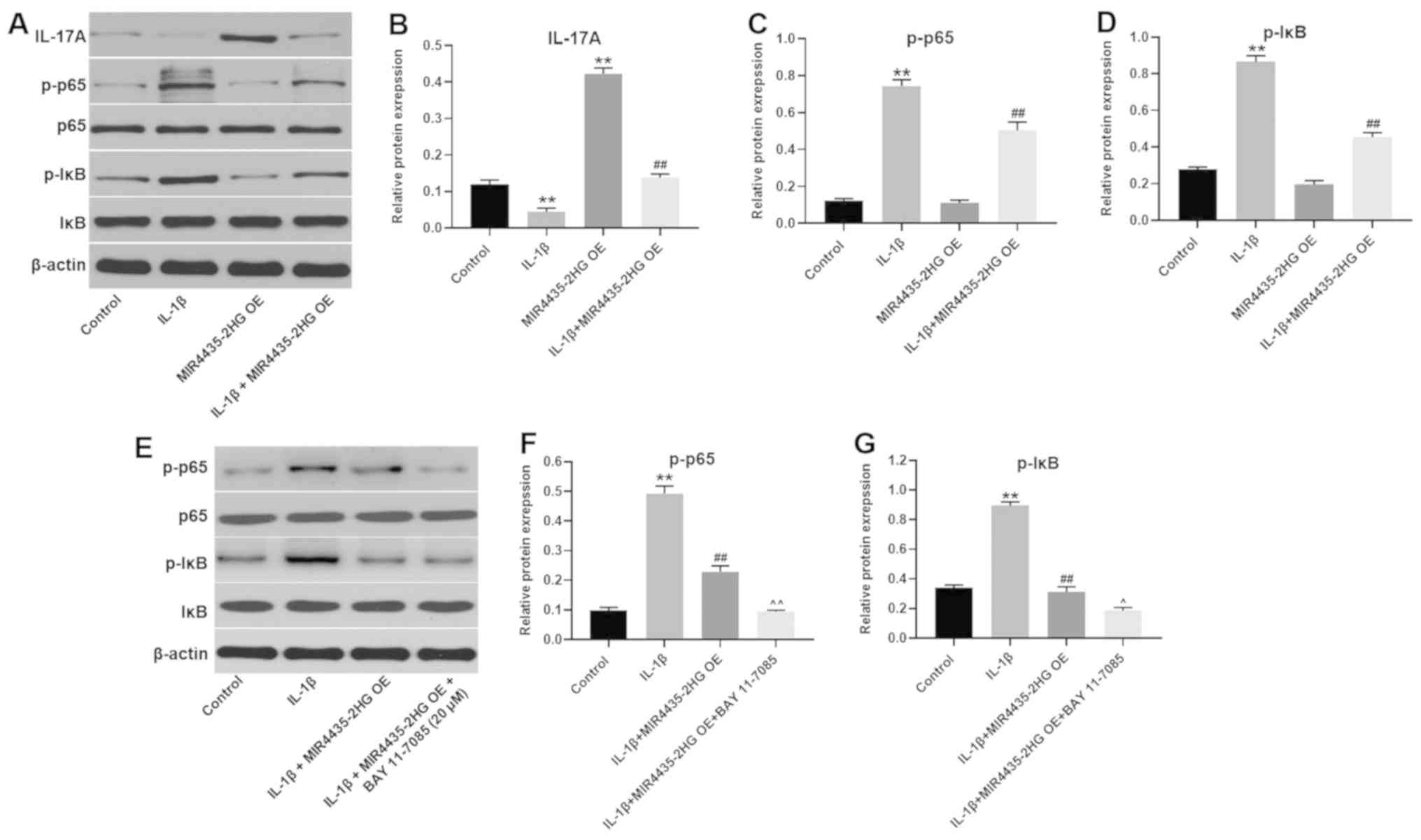 | Figure 6MIR4435-2HG OE suppressed OA
progression via the miR-510-3p/IL-17A axis and inactivation of the
nuclear factor-κB signaling pathway. CHON-001 cells were treated
with IL-1β, MIR4435-2HG OE vector or IL-1β+ MIR4435-2HG OE. Then,
(A) protein expression of IL-17A, p-p65, p65, p-IκB and IκB in
CHON-001 cells was measured by western blotting. (B) Relative
protein expression of IL-17A was normalized to β-actin. (C)
Relative protein expression of p-p65 was normalized to p65. (D)
Relative protein expression of p-IκB was normalized to IκB. (E)
CHON-001 cells were treated with IL-1β, IL-1β+ MIR4435-2HG OE or
IL-1β+ MIR4435-2HG OE + BAY-11-7085. Then, protein expression of
p-p65, p65, p-IκB and IκB in CHON-001 cells were measured by
western blot. (F) Relative protein expression of p-p65 was
normalized to p65. (G) Relative protein expression of p-IκB was
normalized to IκB. **P<0.01 vs. control;
##P<0.01 vs. IL-1β; ^P<0.05,
^^P<0.01 vs. IL-1β+MIR4435-2HG OE. OE,
overexpression; OA, osteoarthritis, miR; microRNA; IL, interleukin;
IκB, inhibitor of κB. |
Discussion
Previous studies suggested that certain lncRNAs are
involved in the pathology of OA (21,22). The
present study demonstrated that MIR4435-2HG was significantly
downregulated in IL-1β-treated CHON-001 cells. Similarly, Wang
et al (16) indicated that
the expression of lncRNA-recombination signal binding protein for
immunoglobulin κ J region (RBPJ, previously known as CBF1)
interacting corepressor displayed an inhibitory role during OA
progression. In addition, a recent study reported that the
inflammatory response plays an important role during the
progression of OA (23). Tan et
al (24) demonstrated that IL-1β
serves a key role in the progression of OA. Furthermore, IL-1β
enhances injury in arthritic cartilage and increases the expression
of matrix-degradation proteins (25). These studies indicated that low
expression of MIR4435-2HG was associated with the development of
OA.
Collagen II, MMP1 and MMP13 are cartilage matrix
components (26,27). Inhibition of collagen II and
upregulation of MMP13 and MMP1 could lead to the progression of OA
(28-30).
In the present study, in vitro experiments were performed to
further explore the biological role of MIR4435-2HG during OA
progression. CHON-001 cell proliferation was inhibited by IL-1β. In
addition, MIR4435-2HGOE significantly decreased the expression of
MMP1 and MMP13 and increased the expression of collagen II in
IL-1β-treated CHON-001 cells. These results were consistent with
previous studies indicating that MIR4435-2HG OE attenuates the
progression of OA by modulating the expression of MMP1, MMP13 and
collagen II.
The mechanism underlying MIR4435-2HG-mediated
inhibition of OA progression in vitro was also investigated.
A dual luciferase reporter assay indicated that miR-510-3p was the
downstream target gene of MIR4435-2HG. miRNAs are highly conserved
ncRNAs that display versatile biological functions (31-33).
Lei et al (34) reported that
the lncRNA small nucleolar RNA host gene 1 could inhibit
IL-1β-induced OA by inhibiting miR-16-5p-mediated p38/MAPK and
NF-κB signaling pathway. In addition, the lncRNA metastasis
associated lung adenocarcinoma transcript 1 promotes the
progression of OA by regulating themiR-150-5p/AKT3 axis (35). However, the function of miR-510-3p in
OA remains unclear. The present study demonstrated that miR-510-3p
may promote OA progression. These findings were similar to previous
studies (36,37), demonstrating that MIR4435-2HG can
inhibit the progression of OA by sponging miR-510-3p.
It has been reported that miRNAs exert their
function by binding to target genes (38,39). To
explore the underlying mechanism of miR-510-3p in the development
of OA, the TargetScan database was used for prediction of
miR-510-3p target genes, and IL-17A was identified as a direct
target of miR-510-3p. IL-17A is an important immune regulator
secreted by Th17 cells (40). It has
been reported that IL-17A servesakey role in the pathogenesis of
autoimmune diseases (41). In
addition, IL-17A deficiency increases susceptibility to multiple
diseases through STAT5 phosphorylation (42). In the present study, a luciferase
reporter assay also identified IL-17A as a direct target gene of
miR-510-3p. In addition, IL-1β treatment significantly decreased
the expression of IL-17A in CHON-001 cells, which suggested that
IL-17A may act as an OA suppressor. Zhang et al (43) reported that IL-17A upregulation could
be involved in inflammatory arthritis by acting as an inhibitor,
which was similar to the results from the present study, suggesting
therefore that IL-17A may be considered as a key regulator during
OA. Taken together, the inhibition of IL-1β-induced CHON-001 cell
damage occurred by upregulating the expression of IL-17A.
NF-κB signaling is a key regulator in multiple
diseases (44,45). NF-κB is an evolutionarily conserved
transcription factor that controls the expression levels of various
cytokines and adhesion molecules, and is involved in responses to
infection, stress and damage (46).
p65 is an important downstream protein of NF-κB (47). Activation of NF-κB results in nuclear
translocation, which is regulated by the targeted phosphorylation
and subsequent degradation of IκB (48). In the present study, MIR4435-2HG OE
inactivated NF-κB signaling pathway, suggesting that NF-κB may
serve as a promoter during OA. Similar to the findings from the
present study, Ren et al (49) reported that IL-17A could activate
NF-κB signaling pathway, further supporting that MIR4435-2HG may
alleviate the progression of OA via IL-17A/NF-κB axis. However, the
present study only focused on the association between miR4435-2HG
and NF-κB signaling pathway. Since IL-6/STAT3 are involved in the
progression of OA (50,51), the effect of MIR4435-2HG on
IL-6/STAT3 signaling should be investigated in future studies.
In conclusion, MIR4435-2HG OE significantly
inhibited the progression of OA in vitro, suggesting that
MIR4435-2HG OE may serve as a novel therapeutic strategy for
OA.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contribution
YL, YY and LD conceived and supervised the current
study. YuqJ and YunJ designed the experiments. YL and LD performed
the experiments. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Ethical Committee of the Second Affiliated Hospital of Inner
Mongolia Medical University. Written informed consent was obtained
from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Thompson RL, Gardner JK, Zhang S and
Reinbolt JA: Lower-limb joint reaction forces and moments during
modified cycling in healthy controls and individuals with knee
osteoarthritis. Clin Biomech (Bristol, Avon). 71:167–175.
2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Brandt KD: Osteoarthritis. Clin Geriatr
Med. 4:279–293. 1988.PubMed/NCBI
|
|
3
|
Mehl J, Imhoff AB and Beitzel K:
Osteoarthritis of the shoulder: Pathogenesis, diagnostics and
conservative treatment options. Orthopade. 47:368–376.
2018.PubMed/NCBI View Article : Google Scholar : (In German).
|
|
4
|
Lukusa A, Malemba JJ, Lebughe P, Akilimali
P and Mbuyi-Muamba JM: Clinical and radiological features of knee
osteoarthritis in patients attending the university hospital of
Kinshasa, Democratic Republic of Congo. Pan Afr Med J.
34(29)2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Andreasson I, Kjellby-Wendt G,
Fagevik-Olsén M, Aurell Y, Ullman M and Karlsson J: Long-term
outcomes of corrective osteotomy for malunited fractures of the
distal radius. J Plast Surg Hand Surg. 54:94–100. 2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Nehrer S, Ljuhar R, Steindl P, Simon R,
Maurer D, Ljuhar D, Bertalan Z, Dima HP, Goetz C and Paixao T:
Automated knee osteoarthritis assessment increases physicians'
agreement rate and accuracy: Data from the osteoarthritis
initiative. Cartilage: Nov 24, 2019 (Epub ahead of print).
|
|
7
|
Gerbrands TA, Pisters MF and Vanwanseele
B: Individual selection of gait retraining strategies is essential
to optimally reduce medial knee load during gait. Clin Biom
(Bristol, Avon). 29:828–834. 2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Hochberg-Laufer H, Neufeld N, Brody Y,
Nadav-Eliyahu S, Ben-Yishay R and Shav-Tal Y: Availability of
splicing factors in the nucleoplasm can regulate the release of
mRNA from the gene after transcription. PLoS Genet.
15(e1008459)2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Qi Y, Ma Y, Peng Z, Wang L, Li L, Tang Y,
He J and Zheng J: Long noncoding RNA PENG upregulates PDZK1
expression by sponging miR-15b to suppress clear cell renal cell
carcinoma cell proliferation. Oncogene: Apr 27, 2020 (Epub ahead of
print).
|
|
10
|
Soares JC, Soares AC, Rodrigues VC,
Melendez ME, Santos AC, Faria EF, Reis RM, Carvalho AL and Oliveira
ON Jr: Detection of the prostate cancer biomarker PCA3 with
electrochemical and impedance-based biosensors. ACS Appl Mater
Interfaces. 11:46645–46650. 2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Tu C, Ren X, He J, Zhang C, Chen R, Wang W
and Li Z: The Value of LncRNA BCAR4 as a prognostic biomarker on
clinical outcomes in human cancers. J Cancer. 10:5992–6002.
2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Jiang Z, Li L, Hou Z, Liu W, Wang H, Zhou
T, Li Y and Chen S: LncRNA HAND2-AS1 inhibits 5-fluorouracil
resistance by modulating miR-20a/PDCD4 axis in colorectal cancer.
Cell Signal. 66(109483)2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Li Y, Li S, Luo Y, Liu Y and Yu N: LncRNA
PVT1 regulates chondrocyte apoptosis in osteoarthritis by acting as
a sponge for miR-488-3p. DNA Cell Biol. 36:571–580. 2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Xiao Y, Bao Y, Tang L and Wang L: LncRNA
MIR4435-2HG is downregulated in osteoarthritis and regulates
chondrocyte cell proliferation and apoptosis. J Orthop Surg Res.
14(247)2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Kloppenburg M, Kroon FP, Blanco FJ,
Doherty M, Dziedzic KS, Greibrokk E, Haugen IK, Herrero-Beaumont G,
Jonsson H, Kjeken I, et al: 2018 update of the EULAR
recommendations for the management of hand osteoarthritis. Ann
Rheumatic Dis. 78:16–24. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Wang X, Fan J, Ding X, Sun Y, Cui Z and
Liu W: Tanshinone I inhibits IL-1β-induced apoptosis, inflammation
and extracellular matrix degradation in chondrocytes CHON-001 cells
and attenuates murine osteoarthritis. Drug Des Devel Ther.
13:3559–3568. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Lin Q and Di YP: Determination and
quantification of bacterial virulent gene expression using
quantitative real-time PCR. Methods Mol Biol. 2102:177–193.
2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
McAlinden A and Im GI: MicroRNAs in
orthopaedic research: Disease associations, potential therapeutic
applications, and perspectives. J Orthop Res. 36:33–51.
2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Lu F, Liu P, Zhang Q, Wang W and Guo W:
Association between the polymorphism of IL-17A and IL-17F gene with
knee osteoarthritis risk: A meta-analysis based on case-control
studies. J Orthop Surg Res. 14(445)2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Kaul NC, Mohapatra SR, Adam I, Tucher C,
Tretter T, Opitz CA, Lorenz HM and Tykocinski LO: Hypoxia decreases
the T helper cell-suppressive capacity of synovial fibroblasts by
downregulating IDO1-mediated tryptophan metabolism. Rheumatology.
59:1148–1158. 2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Lin S, Zhang R, An X, Li Z, Fang C, Pan B,
Chen W, Xu G and Han W: LncRNA HOXA-AS3 confers cisplatin
resistance by interacting with HOXA3 in non-small-cell lung
carcinoma cells. Oncogenesis. 8(60)2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Nanus DE, Wijesinghe SN, Pearson MJ,
Hadjicharalambous MR, Rosser A, Davis ET, Lindsay MA and Jones SW:
Obese osteoarthritis patients exhibit an inflammatory synovial
fibroblast phenotype, which is regulated by the long non coding RNA
MALAT1. Arthritis Rheumatol. 72:1–29. 2019.
|
|
23
|
Mao T, He C, Wu H, Yang B and Li X:
Silencing lncRNA HOTAIR declines synovial inflammation and
synoviocyte proliferation and promotes synoviocyte apoptosis in
osteoarthritis rats by inhibiting Wnt/β-catenin signaling pathway.
Cell Cycle. 18:3189–3205. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Tan C, Zhang J, Chen W, Feng F, Yu C, Lu
X, Lin R, Li Z, Huang Y, Zheng L, et al: Inflammatory cytokines via
up-regulation of aquaporins deteriorated the pathogenesis of early
osteoarthritis. PLoS One. 14(e0220846)2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Kapoor M, Martel-Pelletier J, Lajeunesse
D, Pelletier JP and Fahmi H: Role of proinflammatory cytokines in
the pathophysiology of osteoarthritis. Nat Rev Rheumatol. 7:33–42.
2011.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhang Y, Liu S, Guo W, Hao C, Wang M, Li
X, Zhang X, Chen M, Wang Z, Sui X, et al: Coculture of hWJMSCs and
pACs in oriented scaffold enhances hyaline cartilage regeneration
in vitro. Stem Cells Int. 2019(5130152)2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wang M, Sampson ER, Jin H, Li J, Ke QH, Im
HJ and Chen D: MMP13 is a critical target gene during the
progression of osteoarthritis. Arthritis Res Ther.
15(R5)2013.PubMed/NCBI View
Article : Google Scholar
|
|
28
|
Ji B, Ma Y, Wang H, Fang X and Shi P:
Activation of the P38/CREB/MMP13 axis is associated with
osteoarthritis. Drug Des Devel Ther. 13:2195–2204. 2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zheng W, Feng Z, Lou Y, Chen C, Zhang C,
Tao Z, Li H, Cheng L and Ying X: Silibinin protects against
osteoarthritis through inhibiting the inflammatory response and
cartilage matrix degradation in vitro and in vivo. Oncotarget.
8:99649–99665. 2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Lorenz J, Seebach E, Hackmayer G, Greth C,
Bauer RJ, Kleinschmidt K, Bettenworth D, Böhm M, Grifka J and
Grässel S: Melanocortin 1 receptor-signaling deficiency results in
an articular cartilage phenotype and accelerates pathogenesis of
surgically induced murine osteoarthritis. PLoS One.
9(e105858)2014.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Lin SL and Ying SY: Mechanism and method
for generating tumor-free ips cells using intronic microRNA miR-302
induction. Methods Mol Biol. 1733:265–282. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Eguchi T and Kuboki T: Cellular
reprogramming using defined factors and microRNAs. Stem Cells Int.
2016(7530942)2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Pourrajab F, Vakili Zarch A,
Hekmatimoghaddam S and Zare-Khormizi MR: The master switchers in
the aging of cardiovascular system, reverse senescence by microRNA
signatures; as highly conserved molecules. Prog Biophys Mol Biol.
119:111–128. 2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Lei J, Fu Y, Zhuang Y, Zhang K and Lu D:
LncRNA SNHG1 alleviates IL-1beta-induced osteoarthritis by
inhibiting miR-16-5p-mediated p38 MAPK and NF-kappaB signaling
pathways. Bioscience reports 39, 2019.
|
|
35
|
Zhang Y, Wang F, Chen G, He R and Yang L:
LncRNA MALAT1 promotes osteoarthritis by modulating miR-150-5p/AKT3
axis. Cell Biosci. 9(54)2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Gao H, Peng L, Li C, Ji Q and Li P:
Salidroside alleviates cartilage degeneration through NF-κB pathway
in osteoarthritis rats. Drug Des Devel Ther. 14:1445–1454.
2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Xu C, Sheng S, Dou H, Chen J, Zhou K, Lin
Y and Yang H: α-Bisabolol suppresses the inflammatory response and
ECM catabolism in advanced glycation end products-treated
chondrocytes and attenuates murine osteoarthritis. Int
Immunopharmacol: Apr 22, 2020 (Epub ahead of print).
|
|
38
|
Adlakha YK and Saini N: Brain microRNAs
and insights into biological functions and therapeutic potential of
brain enriched miRNA-128. Mol Cancer. 13(33)2014.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Plé H, Landry P, Benham A, Coarfa C,
Gunaratne PH and Provost P: The repertoire and features of human
platelet microRNAs. PLoS One. 7(e50746)2012.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Wang J, Wang X, Wang L, Sun C, Xie C and
Li Z: MiR-let-7d-3p regulates IL-17 expression through targeting
AKT1/mTOR signaling in CD4(+) T cells. In vitro Cell Dev Biol
Animal. 56:67–74. 2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Zhou X, Chen H, Wei F, Zhao Q, Su Q, Lei
Y, Yin M, Tian X, Liu Z, Yu B, et al: α-mangostin attenuates
pristane-induced lupus nephritis by regulating Th17
differentiation. Int J Rheum Dis. 23:74–83. 2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Me R, Gao N, Dai C and Yu FX: IL-17
Promotes pseudomonas aeruginosa keratitis in C57BL/6 mouse corneas.
J Immunol. 204:169–179. 2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zhang X, Yuan Y, Pan Z, Ma Y, Wu M, Yang
J, Han R, Chen M, Hu X, Liu R, et al: Elevated circulating IL-17
level is associated with inflammatory arthritis and disease
activity: A meta-analysis. Clin Chim Acta. 496:76–83.
2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Ghosh S, May MJ and Kopp EB: NF-kappa B
and Rel proteins: Evolutionarily conserved mediators of immune
responses. Ann Rev Immunol. 16:225–260. 1998.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Puri RV, Yerrathota S, Home T, Idowu JY,
Chakravarthi VP, Ward CJ, Singhal PC, Vanden Heuvel GB, Fields TA
and Sharma M: Notch4 activation aggravates NF-κB mediated
inflammation in HIV-1 associated Nephropathy. Dis Mod Mech. 12:
pii(dmm040642)2019.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Baldwin AS Jr: The NF-kappa B and I kappa
B proteins: New discoveries and insights. Ann Rev Immunol.
14:649–683. 1996.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Schütze S, Wiegmann K, Machleidt T and
Krönke M: TNF-induced activation of NF-kappa B. Immunobiology.
193:193–203. 1995.PubMed/NCBI View Article : Google Scholar
|
|
48
|
TH Lv MM, An XM, Leung WK and Seto WK:
Activation of adenosine A3 receptor inhibits inflammatory cytokine
production in colonic mucosa of patients with ulcerative colitis by
down-regulating the NF-kappaB signaling. J Dig Dis. 21:38–45.
2020.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Ren H, Wang Z, Zhang S, Ma H, Wang Y, Jia
L and Li Y: IL-17A promotes the migration and invasiveness of
colorectal cancer cells through NF-κB-mediated MMP expression.
Oncol Res. 23:249–256. 2016.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Sui C, Zhang L and Hu Y: MicroRNA-let-7a
inhibition inhibits LPS-induced inflammatory injury of chondrocytes
by targeting IL6R. Mol Med Rep. 20:2633–2640. 2019.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Lu W, Ding Z, Liu F, Shan W, Cheng C, Xu
J, He W, Huang W, Ma J and Yin Z: Dopamine delays articular
cartilage degradation in osteoarthritis by negative regulation of
the NF-kappaB and JAK2/STAT3 signaling pathways. Biomed
Pharmacother. 119(109419)2019.PubMed/NCBI View Article : Google Scholar
|