Introduction
Lung cancer remains to be the leading cause of
cancer-associated mortality worldwide, where the rate of incidence
continues increasing (1). According
to the GLOBOCAN reports, the incidence of lung cancer stood at 2.1
million occurred worldwide as of 2018, compared with 1.8 million in
2012 (2,3). Non-small cell lung cancer (NSCLC) is
the predominant type of lung cancer, accounting for ~80% of all
reported cases (4). Recently, lung
adenocarcinoma (LUAD) has been reported to be the most predominant
subtype of NSCLC that is experiencing the fastest growth in
incidence (5). Cisplatin is used
extensively as the front-line treatment option for NSCLC and has
been reported to improve the survival outcomes of patients by
impairing the structure and function of DNA in cancer cells
(6). However, its efficacy is
frequently hindered by the development of chemoresistance (7). Tumor cells that are resistant to
chemotherapy exhibit characteristics of malignant behavior,
including high proliferative capability and potent antiapoptotic
ability (8,9). Although studies on cisplatin resistance
have been performed previously (10-12),
the molecular mechanism underlying tumor drug resistance remains
poorly understood.
Over the past number of decades, cancer stem cells
(CSCs) have attracted widespread attention due to their
capabilities of self-renewal and differentiation during cellular
stress or drug resistance (13,14).
CSCs have been reported in several types of human cancer, including
LUAD (15,16). Cluster of differentiation 133 (CD133)
has been previously demonstrated to be a key marker of lung CSCs,
which have the ability to grow indefinitely into tumor spheres in
serum-free medium supplemented with epidermal growth factor (EGF)
and basal fibroblast growth factor (bFGF) (17). Additionally, high expression levels
of pluripotency factors, including SRY-Box 2 (SOX2),
Octamer-binding transcription factor 4 (OCT4), Kruppel like factor
4 (KLF4), NANOG and ATP binding cassette subfamily G member 2
(ABCG2) are associated with enhanced cancer stem cell-like
properties (18,19). Although CSCs represent a small
population of total tumor cells, they serve key roles in tumor
initiation, progression and resistance to radiotherapy and
chemotherapy (20). Previous studies
have reported that CSCs are under the regulation of a number of
signaling pathways, including Wnt/β-catenin, Notch and Hedgehog
signaling pathways (21,22). Therefore, inhibition of CSCs by
targeting the aforementioned signaling pathways may increase the
efficacy of lung cancer therapy.
microRNAs (miRNAs/miRs) are a family of short RNAs
that do not encode proteins. They negatively regulate gene
expression by direct binding to the 3'-untranslated region of their
mRNA targets and participate in the regulation of several
biological processes, including cancer cell proliferation,
apoptosis, sensitivity to chemotherapy and CSCs stemness (23,24).
Previous studies have demonstrated several types of miRNAs to be
either upregulated or downregulated in lung cancer, contributing to
chemoresistance and enhancing stem cell-like properties through
regulation of CSC-associated signaling pathways. Wang et al
(25) reported that miR-181b
overexpression attenuated chemoresistance by regulating cancer stem
cell-like properties and the Notch signaling pathway in NSCLC. In
addition, miR-708-5p has been revealed to suppress stem cell-like
phenotypes in lung cancer by repressing the Wnt/β-catenin signaling
pathway (26). Previous studies have
suggested that upregulated miR-140-3p expression was significantly
associated with reduced cell proliferation, invasion, migration and
sorafenib resistance in a variety of tumors (27-30).
It was also reported previously that miR-140-3p expression was
downregulated in lung squamous cell carcinoma (LUSC) (31), where it has been demonstrated to
function as a tumor suppressor (32). However, the expression profile and
physiological function of miR-140-3p in LUAD remain poorly
understood.
The present study aimed to investigate the effects
of miR-140-3p on cisplatin sensitivity and stem cell-like
properties of LUAD cells and determine the associated molecular
mechanisms that may provide potential therapeutic strategies for
the treatment of LUAD.
Materials and methods
Bioinformatics analysis
The RNA array dataset GSE74190 obtained from the
NCBI/GEO database (https://www.ncbi.nlm.nih.gov/gds/) and RNA seq data
from The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/) were used to analyze
the expression of miR-140-3p in LUAD tissues and adjacent tissues
and over survival rate of patients with LUAD. The median value of
miR-140-3p expression from the TCGA dataset was used to determine
‘low’ and ‘high’ expression.
Cell culture
The human bronchial epithelial cell line BEAS-2B and
lung adenocarcinoma cell lines A549, H1299, H292 and Calu3 were
purchased from the American Type Culture Collection. BEAS-2B cells
were cultured at 37˚C in 5% CO2 in Bronchial Epithelial
Basal Medium (Lonza Group Ltd.) supplemented with 10% FBS (HyClone;
GE Healthcare Life Sciences), whilst the lung adenocarcinoma cell
lines were cultured in RPMI-1640 medium supplemented with 10% FBS
and 1% penicillin/streptomycin (Gibco; Thermo Fisher Scientific,
Inc.) at 37˚C in 5% CO2.
Cell transfection
A549 and Calu3 cells were cultured in six-well
plates (8x105 cells/well) and transfected with plasmids
(pcDNA3.1-ctnnb1 or pcDNA3.1-vector; Shanghai GenePharma Co., Ltd.)
or mimics (miR-140-3p mimics, 5'-UACCACAGGGUAGAACCACGG-3' or
control mimics, 5'-GCAAGAGACAAGCGCUUAGCC-3'; Shanghai GenePharma
Co., Ltd.), using Lipofectamine® 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). Briefly, the plasmids
(2 µg) or mimics (50 nM) were added to 200 µl Opti-MEM medium
(Gibco; Thermo Fisher Scientific, Inc.) in one vial, whilst 4 µl
Lipofectamine® 2000 was diluted in 200 µl Opti-MEM in
another vial. Following incubation for 5 min at room temperature,
the contents of both vials were combined and incubated for a
further 20 min at room temperature before the mixture was added to
the cells. The media in each well was then replaced with fresh
medium 6 h following incubation with the transfection mixture at
37˚C. The transfected cells were harvested 48 h later for
subsequent experimentation.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from the cultured cells
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. Total
RNA was reverse transcribed into cDNA using the Moloney murine
leukemia virus RT kit, with the M-MLV buffer, dNTP and random
primers (all from Promega Corporation). The temperature protocol
for the reverse transcription reaction consisted of cDNA synthesis
at 37˚C for 60 min and termination at 80˚C for 2 min. qPCR was
subsequently performed using the SYBR Green Realtime PCR Master Mix
(Beijing Solarbio Science & Technology Co., Ltd.) in a Bio-Rad
CFX96 system (Bio-Rad Laboratories, Inc.), according to the
manufacturer's protocols. The primer sequences used for qPCR were
designed and synthesized by Guangzhou RiboBio Co., Ltd. (Table I). The following thermocycling
conditions were used for the qPCR: Initial denaturation at 95˚C for
2 min, followed by 40 cycles 94˚C for 20 sec and 60˚C for 30 sec,
and final extension at 72˚C for 30 sec. Relative mRNA or miRNA
expression levels were calculated using the
2-DDCq method (33) and normalized to the internal
reference genes GAPDH and U6, respectively. All experiments were
performed in triplicate.
 | Table IPrimer sequences used for reverse
transcription-quantitative PCR. |
Table I
Primer sequences used for reverse
transcription-quantitative PCR.
| Primer | Forward
(5'-3') | Reverse
(5'-3') |
|---|
| OCT-4 |
GAAGGATGTGGTCCGAGTGT |
GTGAAGTGAGGGCTCCCATA |
| SOX-2 |
ACACCAATCCCATCCACACT |
GCAAACTTCCTGCAAAGCTC |
| KLF-4 |
ACCCACACAGGTGAGAAACC |
ATGTGTAAGGCGAGGTGGTC |
| NANOG |
TTCCTTCCTCCATGGATCTG |
ATCTGCTGGAGGCTGAGGTA |
| ABCG2 |
GTGGCCTTGGCTTGTATGAT |
AACAATTGCTGCTGTGCAAC |
| GAPDH |
AACGGATTTGGTCGTATTG |
GGAAGATGGTGATGGGATT |
| β-catenin |
AAAGCGGCTGTTAGTCACTGG |
CGAGTCATTGCATACTGTCCAT |
| c-Myc |
GGCTCCTGGCAAAAGGTCA |
CTGCGTAGTTGTGCTGATGT |
| Cyclin D1 |
CAATGACCCCGCACGATTTC |
CATGGAGGGCGGATTGGAA |
| miR-140-3p |
CAGTGCTGTACCACAGGGTAGA |
TATCCTTGTTCACGACTCCTTCAC |
| U6 |
CTCGCTTCGGCAGCACATATACT |
ACGCTTCACGAATTTGCGTGTC |
Cell viability assay
After transfection for 48 h, cells were seeded into
96-well plates (4x103 cells/well) and incubated with
RPMI-1640 medium supplemented with 10% FBS and 1%
penicillin/streptomycin at 37˚C in 5% CO2 overnight.
Freshly prepared cisplatin (Jiangsu Haosen Pharmaceutical Group
Co., Ltd.) was added at the indicated concentrations (0, 1, 2, 4, 6
and 8 µg/ml). After treatment for 24 h at 37˚C in 5%
CO2, cells were incubated with 20 µl MTT reagent
(Sigma-Aldrich; Merck KGaA) at 37˚C for 4 h. Following MTT
incubation, the purple formazan crystals were dissolved using
dimethyl sulfoxide (100 µl/well) and cell viability was
subsequently analyzed at a wavelength of 590 nm, using a microplate
reader (Bio-Rad Laboratories, Inc.) All experiments were performed
in triplicate.
Colony formation assay
After transfection for 48 h, cells were seeded into
24-well plates (1x105 cells/well) and incubated with
RPMI-1640 medium at 37˚C in 5% CO2 overnight.
Subsequently, cells were cultured in RPMI-1640 medium supplemented
with cisplatin (5 µg/ml) for 24 h at 37˚C in 5% CO2.
Cell colonies were fixed with pre-cooled 100% methanol at room
temperature for 10 min and stained with 1% crystal violet at room
temperature for 20 min. All experiments were performed in
triplicate.
Caspase-3 activity
Caspase-3 activity was assessed using the Caspase-3
Assay kit according to the manufacturer's protocol (Sigma-Aldrich;
Merck KGaA). Briefly, 1x106 A549 or Calu3 cells were
lysed following treatment with cisplatin (5 µg/ml) for 24 h at 37˚C
in 5% CO2. Assays were performed in 96-well microtiter
plates by incubating 10 µl protein of cell lysate/sample, which was
quantified using the bicinchoninic acid assay kit (Pierce; Thermo
Fisher Scientific, Inc.), in 80 µl reaction buffer (1% NP-40, 20
mmol/l Tris-HCl [pH 7.5], 137 mmol/l NaCl, and 10% glycerol)
supplemented with 10 µl caspase-3 substrate (2 mmol/l Ac-DEVD-pNA).
Lysates were incubated at 37˚C for 4 h and caspase-3 activity was
subsequently analyzed at a wavelength of 405 nm, using a microplate
reader (Bio-Rad Laboratories, Inc.). Caspase-3 activity was
determined by calculating the ratio of optical density at 450 nm of
the cisplatin treated cells to that of untreated cells. All
experiments were performed in triplicate.
Flow cytometric analysis
A549 or Calu3 cells were harvested at a density of
1x106 and washed twice with PBS supplemented with 0.5%
BSA fraction V (Gibco; Thermo Fisher Scientific, Inc.) and 2 mM
EDTA (Sigma-Aldrich; Merck KGaA), prior to incubation with
CD133-phycoerythrin antibody (1:50; cat. no. 372803; BioLegend,
Inc.) for 15 min at room temperature. Cells were washed twice with
PBS and analyzed using a BD FACSCalibur™ flow cytometer (BD
Diagnostics; Becton, Dickinson and Company) and the FlowJo software
(version 10; FlowJo, LLC) (34). All
experiments were performed in triplicate.
Tumor sphere formation assay
Transfected cells were seeded into six-well
ultra-low attachment culture plates (Corning Inc.) at a density of
3x103 cells/well and incubated in DMEM/F12 serum-free
medium supplemented with 20 ng/ml EGF, 20 ng/ml bFGF, 5 µg/ml
insulin, 0.4% BSA and 2% B-27 (Invitrogen; Thermo Fisher
Scientific, Inc.) for 6 days at 37˚C in 5% CO2. The
tumor spheres (diameter >100 µm) were subsequently counted and
images were captured with a light microscope (magnification x200;
Olympus Corporation) (18).
Western blotting
After transfection for 48 h, total protein was
extracted from A549 and Calu3 cells using RIPA lysis buffer
(Beyotime Institute of Biotechnology) at 30 min on ice. For nuclear
protein extraction, A549 and Calu3 cells were isolated in cell
lysis buffer [5 mM PIPES, pH 8.0, 0.85 mM KCl, 0.5% NP40 and 1%
protein inhibitor mixture (Sigma-Aldrich; Merck KGaA)] for 30 min
on ice and centrifuged at 1,000 x g for 20 min at 4˚C. The
supernatant was then removed, where the cell pellet was incubated
-1in nuclear lysis buffer [50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 1%
SDS and 1% protein inhibitor mixture (Sigma-Aldrich; Merck KGaA)]
for 30 min on ice and centrifuged at 10,000 x g for 20 min at 4˚C
to isolate the nuclear protein. Protein was quantified using the
bicinchoninic acid assay kit (Pierce; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol and 40 µg
protein/lane was separated via SDS-PAGE on a 10% gel. The separated
proteins were subsequently transferred onto polyvinylidene fluoride
membranes (EMD Millipore) and blocked with Tris Buffered Saline
with 0.1% Tween 20 supplemented with 5% non-fat dry milk at room
temperature for 1 h. The membranes were incubated with primary
antibodies against β-catenin (1:4,000; cat. no. ab32572; Abcam),
c-Myc (1:1,000; cat. no. 10828-1-AP; ProteinTech Group, Inc.),
cyclin D1 (1:1,000; cat. no. 2978; Cell Signaling Technology,
Inc.), GAPDH (1:5,000; cat. no. 60004-1-Ig; ProteinTech Group,
Inc.), lamin B1 (1:3,000; cat. no. ab16048; Abcam), p53 (cat. no.
2527, 1:1,000, Cell Signaling Technology, Inc.) and phosphorylated
p53 (cat. no. ab1431; 1:1,000; Abcam) overnight at 4˚C. Following
the primary incubation, membranes were incubated with a horseradish
peroxidase-conjugated goat anti-rabbit secondary antibody (1:4,000;
cat. no. ab6721; Abcam) for 1 h at room temperature. Membranes were
then washed three times with PBS supplemented with Tween 20 (15
min/wash). Protein bands were visualized using the enhanced
chemiluminescent substrate kit (Abcam) and Image Lab™ software
(version 2.0; Bio-Rad Laboratories, Inc.). Protein expression
levels were analyzed using the ImageJ software (version 1.41;
National Institutes of Health). GAPDH was used as the internal
control.
Dual-luciferase reporter assay
The transcription factor 7 (TCF) reporter assay
(TOP/FOP) was performed using the Dual-Glo luciferase assay kit
(Promega Corporation), to assess activity of the Wnt/β-catenin
signaling pathway. A total of 5x104 A549 or Calu3 cells
were seeded into 24-well plates and incubated in RPMI-1640 medium
supplemented with 10% FBS and 1% penicillin/streptomycin at 37˚C in
5% CO2 overnight. Subsequently, cells were
co-transfected with 100 ng TOP/FOP flash vector (Promega
Corporation), internal control pRL-TK Renilla luciferase
vector (10 ng) and control mimic or miR-140-3p mimics (50 nM) using
Lipofectamine® 2000. After transfection for 48 h, both
firefly and Renilla luciferase activities were detected in
duplicate/triplicate, according to the manufacturer's protocol
(35). Firefly luciferase activity
was normalized to Renilla luciferase activity.
Statistical analysis
Statistical analysis was performed using SPSS
software (version 19.0; IBM Corp.). Data are presented as the mean
± standard deviation. Log-rank test was used to determine the
statistical significance of Kaplan-Meier overall survival (OS) data
of patients. Student's t-test was used to evaluate the differences
between two groups, whilst one-way ANOVA followed by Bonferroni
post-hoc test was used to measure differences among three groups
and a two-way ANOVA with Bonferroni's correction's was used for
between-subject statistical analyses. All experiments were
performed in triplicate. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-140-3p is downregulated in LUAD
and positively associated with overall survival OS of patients
miR-140-3p expression was assessed in normal lung
tissues and LUAD samples in the GSE74190 dataset downloaded from
the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/gds/?term). The results
demonstrated that miR-140-3p expression was significantly lower in
LUAD samples compared with that in normal lung tissues
(P<0.0001; Fig. 1A), which was
consistent with findings from The Cancer Genome Atlas (TCGA)
database (P<0.0001; Fig. 1B).
Survival analysis of LUAD data from TCGA database demonstrated that
patients with low miR-140-3p expression levels exhibited
significantly lower OS rate compared with those with high
miR-140-3p expression levels (P=0.023; Fig. 1C). RT-qPCR analysis subsequently
indicated that miR-140-3p expression was significantly lower in
LUAD cell lines (A549, H292, H1299 and Calu3) compared with human
normal bronchial epithelial BEAS-2B cells, particularly in A549 and
Calu3 cells (P<0.01; Fig. 1D).
These two cell lines (A549 and Calu3) were therefore selected for
further experimentation. Taken together, these results suggest that
miR-140-3p expression was downregulated in LUAD and positively
associated with the OS of patients.
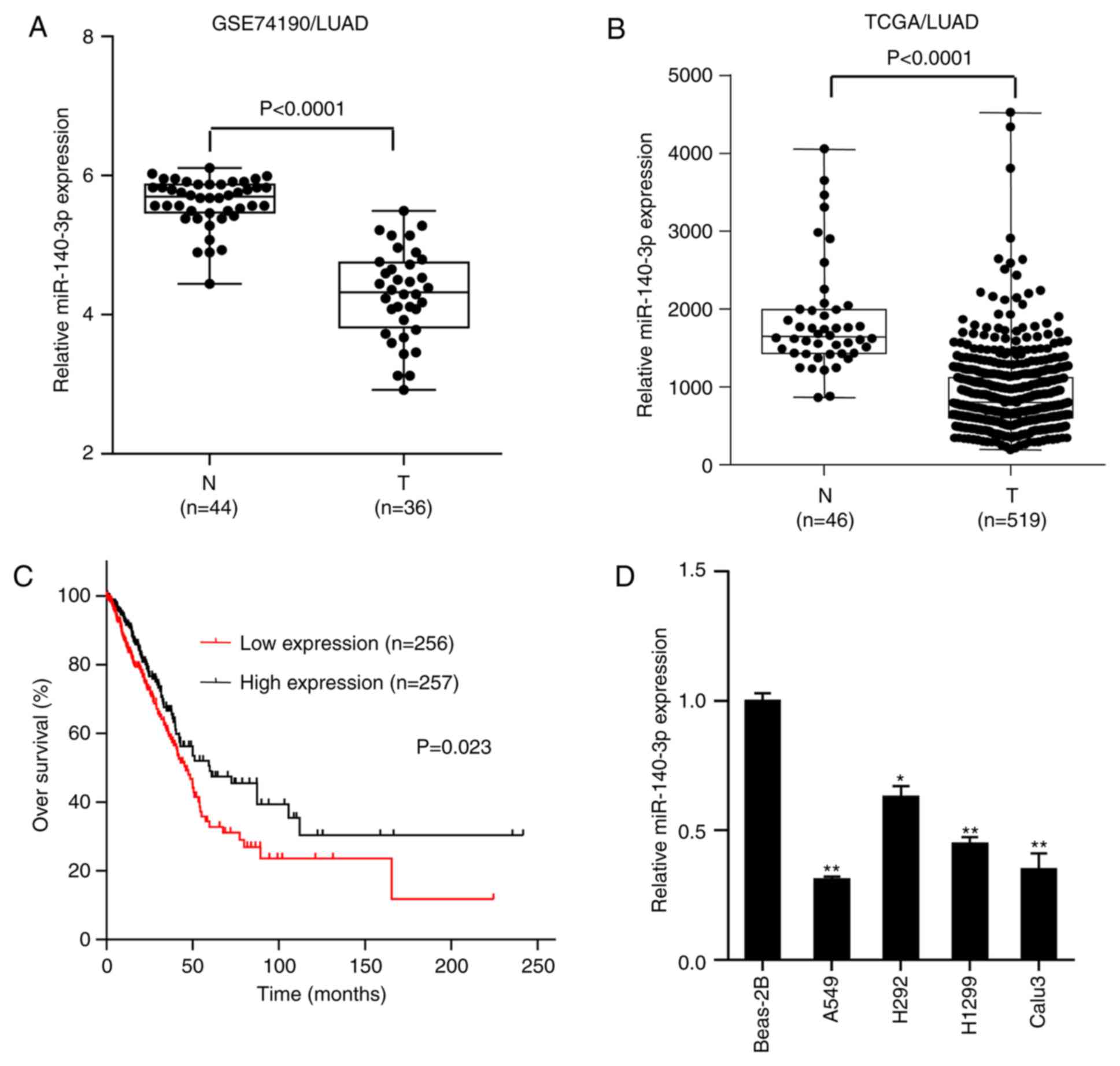 | Figure 1miR-140-3p expression is
downregulated in LUAD, which is in turn positively associated with
OS of patients. Comparison of miR-140-3p expression between LUAD
samples and normal lung tissues in data obtained from the (A)
GSE74190 dataset from Gene Expression Omnibus and (B) TCGA
database. (C) Kaplan-Meier OS analysis of patients with LUAD,
downloaded from TCGA database. (D) Reverse
transcription-quantitative PCR analysis of miR-140-3p expression in
the lung epithelial cell line BEAS-2B and four LUAD cell lines
A549, H292, H1299 and Calu3. Data are presented as the mean ±
standard deviation from three independent experiments.
*P<0.05 and **P<0.01 vs. BEAS-2B. miR,
microRNA; LUAD, lung adenocarcinoma; OS, overall survival; TCGA,
The Cancer Genome Atlas; N, normal tissue; T, tumor tissue. |
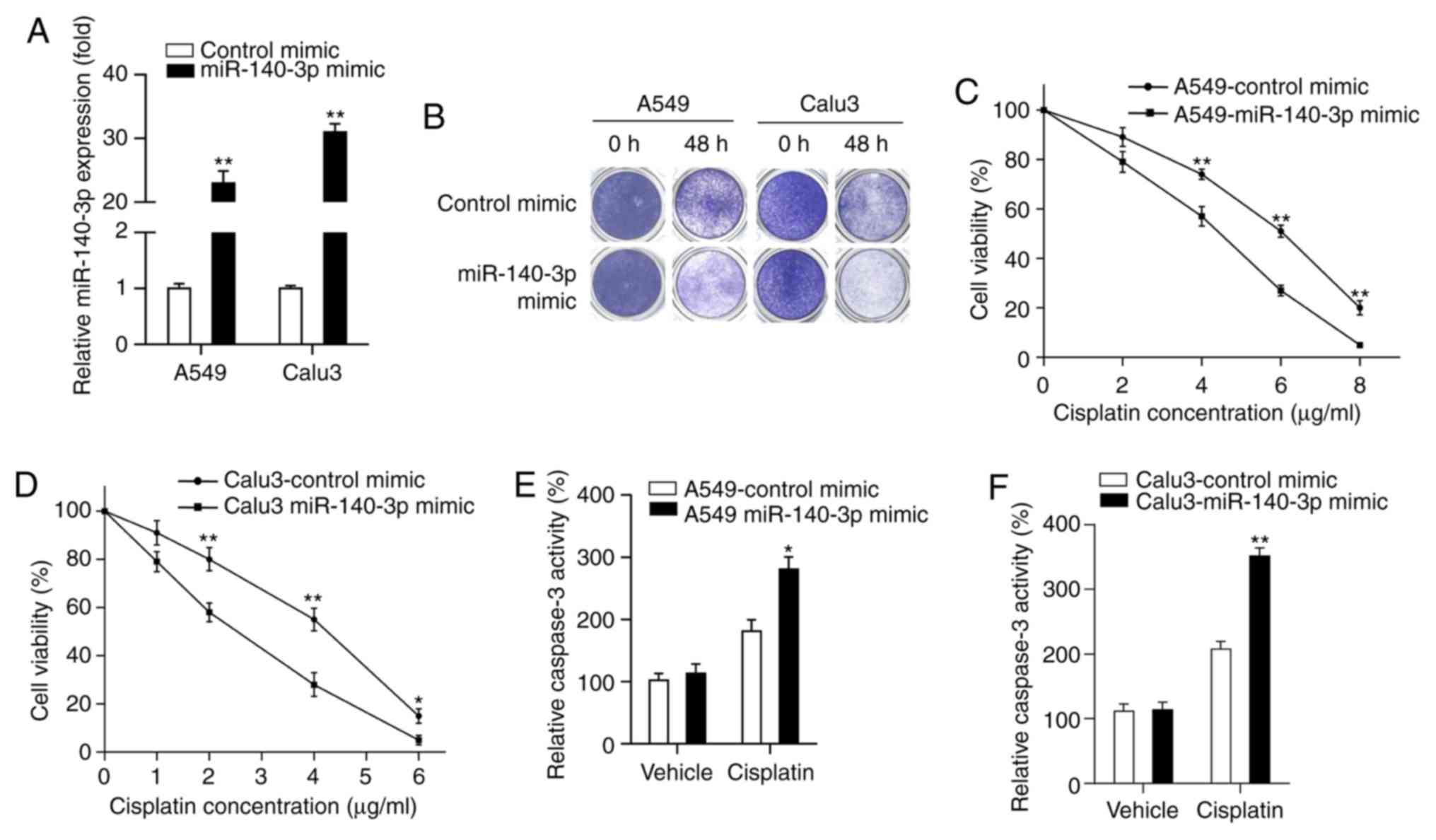
miR-140-3p enhances the sensitivity of
LUAD cells to cisplatin
miR-140-3p has been reported to serve as a key tumor
suppressor in several types of malignancies, where patients with
lower miR-140-3p expression levels had poor prognoses (28-31,33).
Therefore, the present study hypothesized that upregulated
miR-140-3p expression may be beneficial for the treatment of
patients with LUAD. Cisplatin is frequently used as the first-line
treatment for NSCLC, which has been reported to improve survival
outcomes (6). The effect of
miR-140-3p on LUAD cisplatin sensitivity was investigated in the
present study. miR-140-3p mimics or control mimics were first
transfected into A549 and Calu3 cells, where miR-140-3p expression
was significantly higher in the two cell lines transfected with the
miR-140-3p mimic compared with those transfected with the control
mimic (P<0.01; Fig. 2A). Results
from colony formation assay demonstrated that fewer cells survived
in the group overexpressing the miR-140-3p mimic treated with
cisplatin compared with cells transfected with the control mimic
(Fig. 2B). MTT assay indicated that
cell viability was significantly decreased in cells transfected
with the miR-140-3p mimic treated with different concentrations
(A549 cells, 4, 6 and 8 µg/ml; Calu3 cells, 2, 4 and 6 µg/ml) of
cisplatin compared with those transfected with the control mimic
(P<0.05 and P<0.01; Fig. 2C
and D). Subsequently, the effect of
miR-140-3p on cell apoptosis was measured by assessing caspase-3
activity, following treatment with cisplatin. Caspase-3 activity
was found to be significantly increased following miR-140-3p
overexpression compared with that in the control mimics group after
treatment with cisplatin (P<0.05 and P<0.01; Fig. 2E and F). Collectively, these results suggest that
miR-140-3p enhanced sensitivity of LUAD cells to cisplatin.
miR-140-3p decreases stem cell-like
properties of LUAD cells
The presence of CSCs is considered a predominant
cause of chemoresistance (36).
Therefore, the present study investigated the effects of miR-140-3p
on the stem cell-like properties in LUAD cells. Flow cytometry
analysis of the CSC marker CD133 demonstrated that miR-140-3p
overexpression significantly reduced the percentage of
CD133+ cells compared with the control mimic group
(P<0.01; Fig. 3A and B). Tumor sphere formation assay indicated
that cells transfected with the miR-140-3p mimic formed fewer and
smaller spheres compared with cells transfected with the control
mimic (P<0.05; Fig. 3C). RT-qPCR
analysis also demonstrated significantly reduced expression levels
of genes associated with stemness (SOX2, OCT4, KLF4, NANOG and
ABCG2) in cells overexpressing the miR-140-3p mimic compared cells
transfected with the control mimic (P<0.01; Fig. 3D and E). Taken together, these results suggest
that miR-140-3p overexpression can attenuate stem cell-like
properties of LUAD cells.
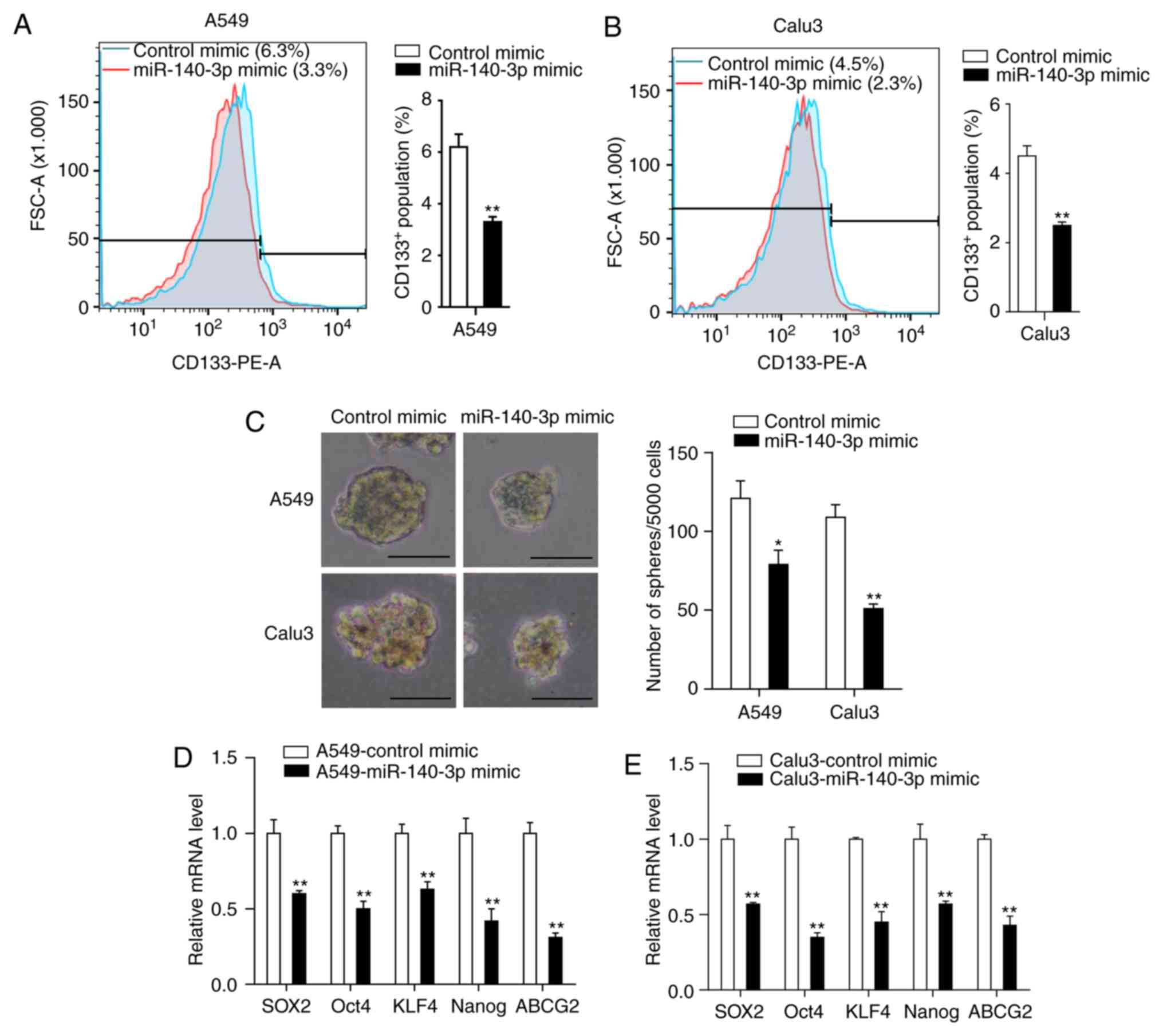 | Figure 3miR-140-3p attenuates the stem
cell-like properties of LUAD cells. The percentage of
CD133+ cells in (A) A549 and (B) Calu3 cells was
analyzed by flow cytometry. (C) Representative images and the
number of spheres with diameter >100 µm formed of A549 and Calu3
cells following transfection with miR-140-3p or the control mimic.
mRNA expression levels of SOX2, OCT4, KLF4, NANOG and ABCG2 in (D)
A549 and (E) Calu3 cells following transfection with miR-140-3p or
the control mimic. Data are represented as the mean ± standard
deviation from three independent experiments. *P<0.05
and **P<0.01 vs. control mimic. Scale bar, 100 µm.
miR, microRNA; LUAD, lung adenocarcinoma; CD133, cluster of
differentiation 133; SOX2, SRY-Box 2; OCT4, Octamer-binding
transcription factor 4; KLF4, Kruppel like factor 4; ABCG2, ATP
binding cassette subfamily G member 2; PE, phycoerythrin; FSC,
Forward versus side scatter. |
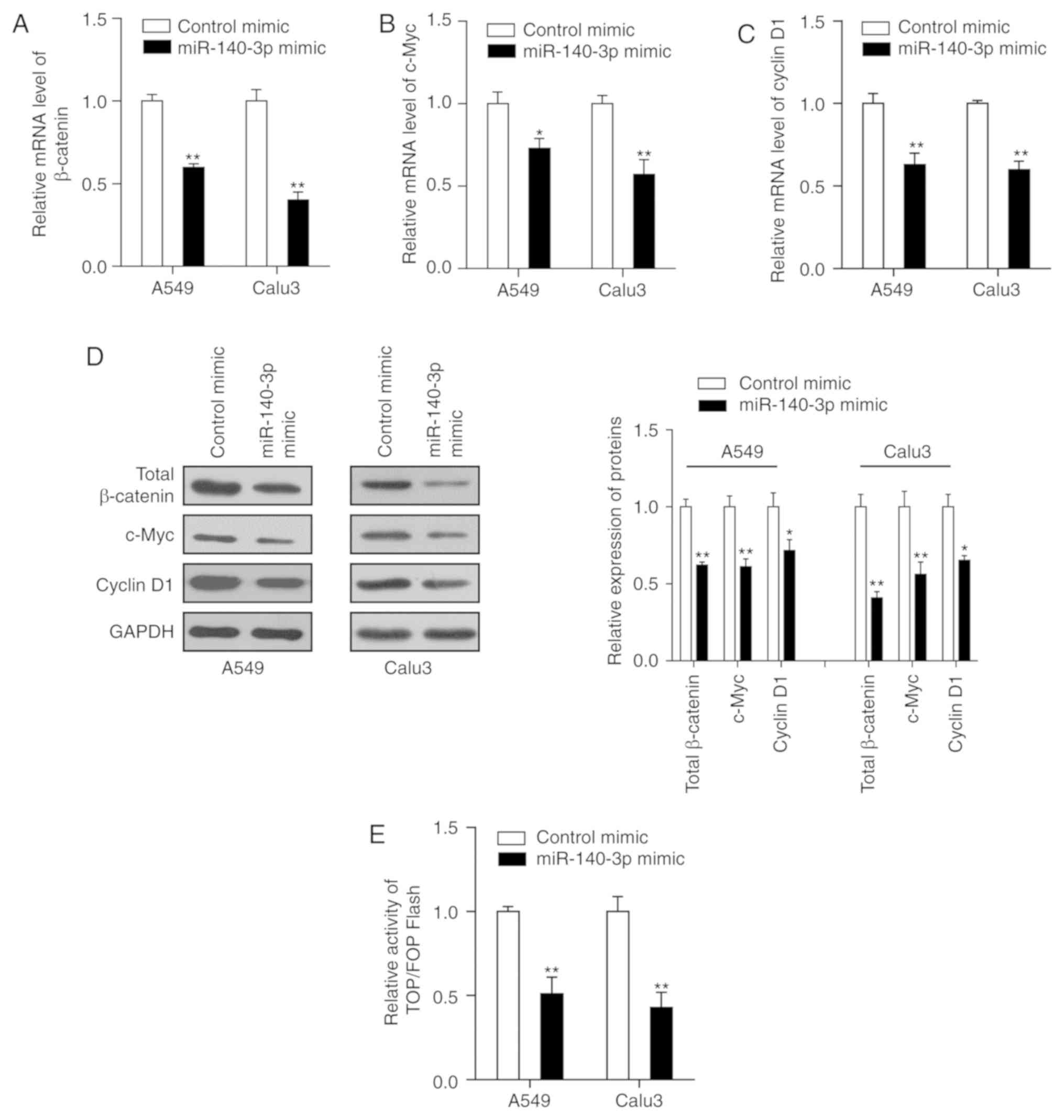
miR-140-3p attenuates the stem
cell-like properties and cisplatin resistance in LUAD cells by
inhibiting the Wnt/β-catenin signaling
The Wnt/β-catenin signaling pathway is speculated to
serve a key role in the self-renewal capacity of CSCs and is
activated in several types of human malignancies including LUAD
(37). Therefore, the present study
hypothesized that upregulated miR-140-3p expression may repress the
Wnt/β-catenin signaling, thereby attenuating the stem cell-like
properties and cisplatin resistance in LUAD. The results
demonstrated that transfection of both LUAD cell lines with the
miR-140-3p mimic significantly reduced the mRNA and protein
expression levels of β-catenin, c-Myc and cyclin D1 compared with
cells transfected with control mimics (P<0.05; Fig. 4A-D). In addition, the dual-luciferase
assay suggested that β-catenin/TCF transcriptional activity was
significantly reduced in cells co-transfected with miR-140-3p mimic
compared with cells transfected with control mimic (P<0.01;
Fig. 4E).
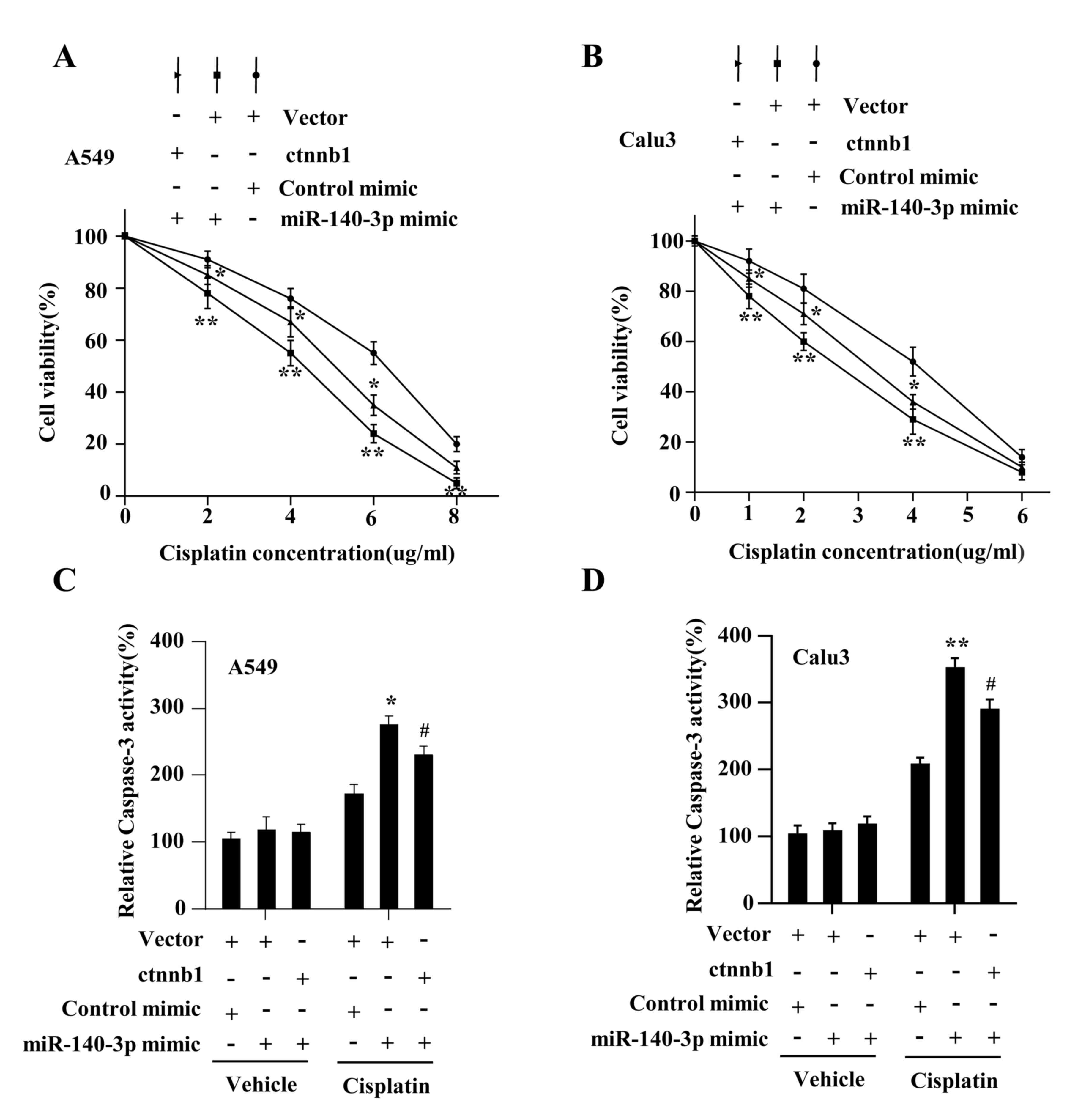
The present study investigated whether Wnt/β-catenin
signaling mediated the miR-140-3p-attenuated stem cell-like
properties and cisplatin resistance. The results demonstrated that
β-catenin expression on both protein and mRNA levels were
significantly increased in A549 and Calu3 cells following
transfection with plasmids overexpressing β-catenin compared with
cells transfected with the vector plasmids (P<0.01; Fig. 5A and B). The levels of total and nuclear
β-catenin were found to be restored by co-transfecting the
β-catenin plasmid with the miR-140-3p mimic in LUAD cells
(P<0.05; Fig. 5C).
Co-transfecting LUAD cells with the β-catenin plasmid also
significantly reversed the inhibitory effects of miR-140-3p mimics
on β-catenin/TCF transcriptional activity (P<0.05; Fig. 5D), CD133+ expression
(P<0.05; Fig. 5E), sphere
formation ability (P<0.05; Fig.
5F). Co-transfection with the β-catenin plasmid also reversed
the effects of miR-140-3p on cell viability and antiapoptotic
ability, in response to cisplatin treatment (P<0.05; Fig. 6A-D). In addition, the status of p53
in the assessed LUAD cell lines were investigated further but no
significant difference was found between cells transfected with the
miR-140-3p mimic and those transfected with the control mimic at
protein level (data not shown), suggesting that these phenotypic
changes mediated by miR-140-3p are p53-independent. Taken together,
these results suggest that miR-140-3p attenuated stem cell-like
properties and cisplatin resistance in LUAD by repressing the
Wnt/β-catenin signaling.
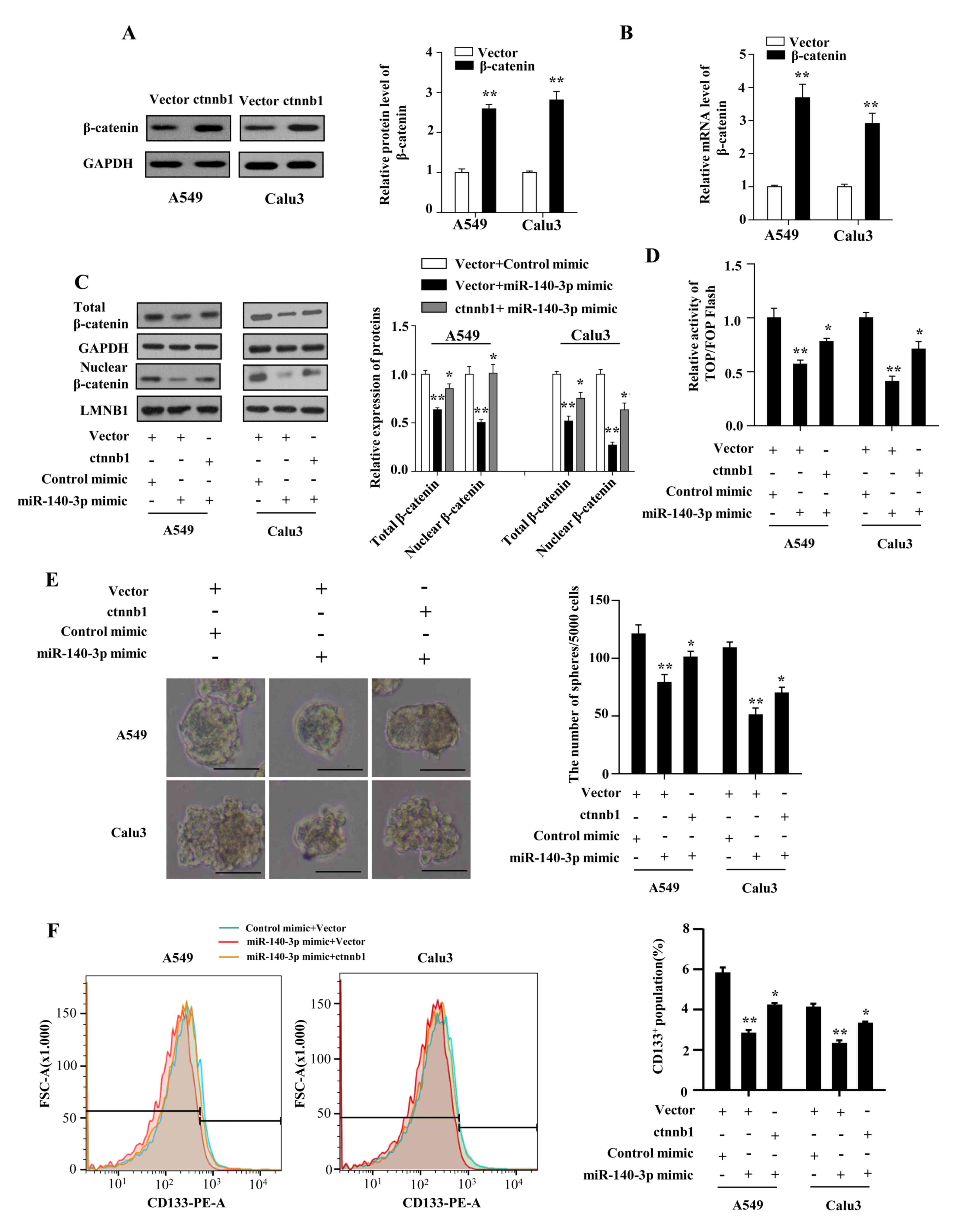 | Figure 5miR-140-3p attenuates the stem
cell-like properties in LUAD by suppressing Wnt/β-catenin
signaling. (A) Protein and (B) mRNA expression levels of β-catenin
in A549 and Calu3 cells following transfection with either the
empty vector or vector expressing β-catenin. **P<0.01
vs. vector. (C) Protein levels of total and nuclear β-catenin,
*P<0.05 vs. vector + miR-140-3p mimic;
**P<0.01 vs. vector + control mimic. (D) TOP/FOP
flash activity, *P<0.05 vs. vector + miR-140-3p
mimic, **P<0.01 vs. vector + control mimic. (E)
number of spheres with diameter >100 µm formed by A549 and Calu3
cells. Scale bar, 100 µm. *P<0.05 vs. vector +
miR-140-3p mimic, **P<0.01 vs. vector + control
mimic. (F) percentage of CD133+ cells analyzed using
flow cytometry following co-transfection with either the empty
vector or vector expressing β-catenin and miR-140-3p mimic or the
control mimic. *P<0.05 vs. vector + miR-140-3p mimic,
**P<0.01 vs. vector + control mimic. Data are
represented the mean ± standard deviation from three independent
experiments. miR, microRNA; LUAD, lung adenocarcinoma; CD133,
cluster of differentiation 133; ctnnb1, β-catenin gene. |
Discussion
Increasing evidence suggests that miRNAs serve key
roles in regulating NSCLC progression, therapeutic resistance and
stem cell-like properties, all of which constitute an obstacle to
the successful treatment of NSCLC (25,38,39). The
present study investigated the significance of miR-140-3p
expression in LUAD. Based on publicly available data from the GEO
and TCGA databases, the results demonstrated that miR-140-3p
expression was significantly lower in LUAD samples compared with
normal lung tissues, which was found to be positively associated
with patient survival, suggesting that miR-140-3p may function as
an effective prognostic marker for patients with LUAD.
Although cisplatin is applied extensively for
treating LUAD (40), chemotherapy
resistance continues to be a major challenge. Therefore, increasing
the cisplatin sensitivity of LUAD cells can serve as a useful
therapeutic strategy. Huang et al (32) previously reported that miR-140-3p
served as a tumor suppressor in LUSC, whilst Li et al
(30) demonstrated that miR-140-3p
enhanced the sensitivity of hepatocellular carcinoma cells to
sorafenib. Therefore, the present study hypothesized that
miR-140-3p may regulate the development of cisplatin sensitivity in
LUAD. The combined results of the colony formation, MTT and
caspase-3 assays demonstrated that upregulation of miR-140-3p
expression enhanced the sensitivity of LUAD cells to cisplatin.
Accumulating evidence supports the hypothesis that CSCs contribute
to one of the major mechanisms of tumor cell chemoresistance
(41,42). Therefore, the stem cell-like
properties of LUAD cells were also investigated in the present
study. The results indicated that upregulating miR-140-3p
expression reduced the CD133+ cell population,
attenuated tumor sphere formation and decreased the expression of
pluripotency factors in LUAD cells. Taken together, these results
suggest that miR-140-3p may serve a key role in regulating
sensitivity to cisplatin and the stem cell-like properties of LUAD
cells, thereby acting as a therapeutic target in LUAD.
Over the past few decades, key pathways involved in
maintaining cancer stem cell-like properties, including the
Wnt/β-catenin, Notch and Hedgehog signaling pathways, have garnered
widespread attention (43,44). In particular, the Wnt/β-catenin
signaling pathway has become prominent in studies involving NSCLC,
since targeting this signaling pathway has been demonstrated to
improve anti-CSC-based treatment efficacy (18). Therefore, the present study
investigated the effects of miR-140-3p on activation of the
Wnt/β-catenin signaling pathway. It was found that the upregulation
of miR-140-3p repressed Wnt/β-catenin signaling activation, whilst
reactivation of Wnt/β-catenin signaling by β-catenin overexpression
partially restored cisplatin resistance and cancer stem cell-like
properties of LUAD cells. In addition, considering that the tumor
suppressor p53 appears to be at the center of tumor-associated
events, including stemness and treatment resistance (45,46), the
present study further investigated the status of p53 in the
assessed LUAD cell lines, though no significant difference was
observed between cells transfected with the miR-140-3p mimic and
control mimic at protein level (data not shown), suggesting that
these phenotypic changes mediated by miR-140-3p are
p53-independent. Taken together, results of the present study
indicate that miR-140-3p/Wnt/β-catenin signaling had a notable
effect on cancer stem cell-like properties, which may serve a novel
regulatory role in the development of cisplatin resistance in
patients with LUAD.
The present study had certain limitations. Despite
concluding that miR-140-3p expression was downregulated in LUAD, it
lacked sufficient data to prove the abnormal expression of
miR-140-3p in cisplatin-resistant patient tissues or cell lines.
Despite demonstrating that miR-140-3p was positively associated
with OS of patients with LUAD, whether miR-140-3p affects the
prognosis of patients following treatment with cisplatin requires
further investigation.
To the best of our knowledge, the present study was
the first to demonstrate that miR-140-3p enhanced cisplatin
sensitivity and attenuated stem cell-like properties, by repressing
Wnt/β-catenin signaling in LUAD cells. Taken together, results of
the present study suggest that miR-140-3p may serve a role as an
effective treatment target, whereby targeting miR-140-3p may
increase the efficacy of cisplatin in patients with LUAD.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Key R & D
Plan of Lianyungang High-tech Zone (grant no. ZD201928).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SW and DW designed the experiments and wrote the
manuscript. SW and HW performed the experiments. YP and XY
contributed to data analysis. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Travis WD, Brambilla E, Nicholson AG,
Yatabe Y, Austin JHM, Beasley MB, Chirieac LR, Dacic S, Duhig E,
Flieder DB, et al: The 2015 world health organization
classification of lung tumors: Impact of genetic, clinical and
radiologic advances since the 2004 classification. J Thorac Oncol.
10:1243–1260. 2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386.
2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Han L, Xu G, Xu C, Liu B and Liu D:
Potential prognostic biomarkers identified by DNA methylation
profiling analysis for patients with lung adenocarcinoma. Oncol
Lett. 15:3552–3557. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Fennell DA, Summers Y, Cadranel J, Benepal
T, Christoph DC, Lal R, Das M, Maxwell F, Visseren-Grul C and Ferry
D: Cisplatin in the modern era: The backbone of first-line
chemotherapy for non-small cell lung cancer. Cancer Treat Rev.
44:42–50. 2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Chang A: Chemotherapy, chemoresistance and
the changing treatment landscape for NSCLC. Lung Cancer. 71:3–10.
2011.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zarogoulidis P, Petanidis S, Kioseoglou E,
Domvri K, Anestakis D and Zarogoulidis K: MiR-205 and miR-218
expression is associated with carboplatin chemoresistance and
regulation of apoptosis via Mcl-1 and survivin in lung cancer
cells. Cell Signal. 27:1576–1588. 2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Tian H, Liu S, Zhang J, Zhang S, Cheng L,
Li C, Zhang X, Dail L, Fan P, Dai L, et al: Enhancement of
cisplatin sensitivity in lung cancer xenografts by
liposome-mediated delivery of the plasmid expressing small hairpin
RNA targeting survivin. J Biomed Nanotechnol. 8:633–641.
2012.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhang Y, Yuan Y, Li Y, Zhang P, Chen P and
Sun S: An inverse interaction between HOXA11 and HOXA11-AS is
associated with cisplatin resistance in lung adenocarcinoma.
Epigenetics. 14:949–960. 2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Li J, Wang Y, Song Y, Fu Z and Yu W:
MiR-27a regulates cisplatin resistance and metastasis by targeting
RKIP in human lung adenocarcinoma cells. Mol Cancer.
13(193)2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zhan J, Wang P, Li S, Song J, He H, Wang
Y, Liu Z, Wang F, Bai H, Fang W, et al: HOXB13 networking with
ABCG1/EZH2/Slug mediates metastasis and confers resistance to
cisplatin in lung adenocarcinoma patients. Theranostics.
9:2084–2099. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Clarke MF, Dick JE, Dirks PB, Eaves CJ,
Jamieson CH, Jones DL, Visvader J, Weissman IL and Wahl GM: Cancer
stem cells-perspectives on current status and future directions:
AACR workshop on cancer stem cells. Cancer Res. 66:9339–9344.
2006.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Visvader JE and Lindeman GJ: Cancer stem
cells in solid tumours: Accumulating evidence and unresolved
questions. Nat Rev Cancer. 8:755–768. 2008.PubMed/NCBI View
Article : Google Scholar
|
|
15
|
Lathia J, Liu H and Matei D: The clinical
impact of cancer stem cells. Oncologist. 17(0517)2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Bora-Singhal N, Mohankumar D, Saha B,
Colin CM, Lee JY, Martin MW, Zheng X, Coppola D and Chellappan S:
Novel HDAC11 inhibitors suppress lung adenocarcinoma stem cell
self-renewal and overcome drug resistance by suppressing Sox2. Sci
Rep. 10(4722)2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Eramo A, Lotti F, Sette G, Pilozzi E,
Biffoni M, Di Virgilio A, Conticello C, Ruco L, Peschle C and De
Maria R: Identification and expansion of the tumorigenic lung
cancer stem cell population. Cell Death Differ. 15:504–514.
2008.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Lu CH, Yeh DW, Lai CY, Liu YL, Huang LR,
Lee AY, Jin SC and Chuang TH: USP17 mediates macrophage-promoted
inflammation and stemness in lung cancer cells by regulating
TRAF2/TRAF3 complex formation. Oncogene. 37:6327–6340.
2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Chiou SH, Wang ML, Chou YT, Chen CJ, Hong
CF, Hsieh WJ, Chang HT, Chen YS, Lin TW, Hsu HS and Wu CW:
Coexpression of oct4 and nanog enhances malignancy in lung
adenocarcinoma by inducing cancer stem cell-like properties and
epithelial-mesenchymal transdifferentiation. Cancer Res.
70:10433–10444. 2010.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Donnenberg VS and Donnenberg AD: Multiple
drug resistance in cancer revisited: The cancer stem cell
hypothesis. J Clin Pharmacol. 45:872–877. 2005.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Takebe N, Miele L, Harris PJ, Jeong W,
Bando H, Kahn M, Yang SX and Ivy SP: Targeting notch, hedgehog, and
Wnt pathways in cancer stem cells: Clinical update. Nat Rev Clin
Oncol. 12:445–464. 2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Chatterjee S and Sil PC: Targeting the
crosstalks of Wnt pathway with hedgehog and notch for cancer
therapy. Pharmacol Res. 142:251–261. 2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Park EY, Chang E, Lee EJ, Lee HW, Kang HG,
Chun KH, Woo YM, Kong HK, Ko JY, Suzuki H, et al: Targeting of
miR34a-NOTCH1 axis reduced breast cancer stemness and
chemoresistance. Cancer Res. 74:7573–7582. 2014.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Bitarte N, Bandres E, Boni V, Zarate R,
Rodriguez J, Gonzalez-Huarriz M, Lopez I, Javier Sola J, Alonso MM,
Fortes P and Garcia-Foncillas J: MicroRNA-451 is involved in the
self-renewal, tumorigenicity, and chemoresistance of colorectal
cancer stem cells. Stem Cells. 29:1661–1671. 2011.PubMed/NCBI View
Article : Google Scholar
|
|
25
|
Wang X, Meng Q, Qiao W, Ma R, Ju W, Hu J,
Lu H, Cui J, Jin Z, Zhao Y and Wang Y: MiR-181b/Notch2 overcome
chemoresistance by regulating cancer stem cell-like properties in
NSCLC. Stem Cell Res Ther. 9(327)2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Liu T, Wu X, Chen T, Luo Z and Hu X:
Downregulation of DNMT3A by miR-708-5p inhibits lung cancer stem
cell-like phenotypes through repressing Wnt/β-catenin signaling.
Clin Cancer Res. 24:1748–1760. 2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Sun T, Song Y, Yu H and Luo X:
Identification of lncRNA TRPM2-AS/miR-140-3p/PYCR1 axis's
proliferates and anti-apoptotic effect on breast cancer using
co-expression network analysis. Cancer Biol Ther. 20:760–773.
2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Zhang QY, Men CJ and Ding XW: Upregulation
of microRNA-140-3p inhibits epithelial-mesenchymal transition,
invasion, and metastasis of hepatocellular carcinoma through
inactivation of the MAPK signaling pathway by targeting GRN. J Cell
Biochem. 120:14885–14898. 2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zhou Y, Wang B, Wang Y, Chen G, Lian Q and
Wang H: MiR-140-3p inhibits breast cancer proliferation and
migration by directly regulating the expression of tripartite motif
28. Oncol Lett. 17:3835–3841. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Li J, Zhao J, Wang H, Li X, Liu A, Qin Q
and Li B: MicroRNA-140-3p enhances the sensitivity of
hepatocellular carcinoma cells to sorafenib by targeting
pregnenolone X receptor. Onco Targets Ther. 11:5885–5894.
2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Tan X, Qin W, Zhang L, Hang J, Li B, Zhang
C, Wan J, Zhou F, Shao K, Sun Y, et al: A 5-microRNA signature for
lung squamous cell carcinoma diagnosis and hsa-miR-31 for
prognosis. Clin Cancer Res. 17:6802–6811. 2011.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Huang H, Wang Y, Li Q, Fei X, Ma H and Hu
R: MiR-140-3p functions as a tumor suppressor in squamous cell lung
cancer by regulating BRD9. Cancer Lett. 446:81–89. 2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Cao S, Wang Z, Gao X, He W, Cai Y, Chen H
and Xu R: FOXC1 induces cancer stem cell-like properties through
upregulation of beta-catenin in NSCLC. J Exp Clin Cancer Res.
37(220)2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Cui Y, Ma W, Lei F, Li Q, Su Y, Lin X, Lin
C, Zhang X, Ye L, Wu S, et al: Prostate tumour overexpressed-1
promotes tumourigenicity in human breast cancer via activation of
Wnt/β-catenin signalling. J Pathol. 239:297–308. 2016.PubMed/NCBI View Article : Google Scholar
|
|
36
|
MacDonagh L, Gray SG, Breen E, Cuffe S,
Finn SP, O'Byrne KJ and Barr MP: Lung cancer stem cells: The root
of resistance. Cancer Lett. 372:147–156. 2016.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Jiang N, Zou C, Zhu Y, Luo Y, Chen L, Lei
Y, Tang K, Sun Y, Zhang W, Li S, et al: HIF-1a-regulated miR-1275
maintains stem cell-like phenotypes and promotes the progression of
LUAD by simultaneously activating Wnt/β-catenin and notch
signaling. Theranostics. 10:2553–2570. 2020.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Li T, Ding ZL, Zheng YL and Wang W:
MiR-484 promotes non-small-cell lung cancer (NSCLC) progression
through inhibiting apaf-1 associated with the suppression of
apoptosis. Biomed Pharmacother. 96:153–164. 2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Ma Z, Cai H, Zhang Y, Chang L and Cui Y:
MiR-129-5p inhibits non-small cell lung cancer cell stemness and
chemoresistance through targeting DLK1. Biochem Biophys Res Commun.
490:309–316. 2017.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Zhang Y, Du H, Li Y, Yuan Y, Chen B and
Sun S: Elevated TRIM23 expression predicts cisplatin resistance in
lung adenocarcinoma. Cancer Sci. 111:637–646. 2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Chen MJ, Wu DW, Wang YC, Chen CY and Lee
H: PAK1 confers chemoresistance and poor outcome in non-small cell
lung cancer via beta-catenin-mediated stemness. Sci Rep.
6(34933)2016.
|
|
42
|
MacDonagh L, Gray SG, Breen E, Cuffe S,
Finn SP, O'Byrne KJ and Barr MP: BBI608 inhibits cancer stemness
and reverses cisplatin resistance in NSCLC. Cancer Lett.
428:117–126. 2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Lathia JD and Liu H: Overview of cancer
stem cells and stemness for community oncologists. Targeted Oncol.
12:387–399. 2017.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Najafi M, Farhood B and Mortezaee K:
Cancer stem cells (CSCs) in cancer progression and therapy. J Cell
Physiol. 234:8381–8395. 2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Fu X, Wu S, Li B, Xu Y and Liu J:
Functions of p53 in pluripotent stem cells. Protein Cell. 11:71–78.
2020.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Cao X, Hou J, An Q, Assaraf YG and Wang X:
Towards the overcoming of anticancer drug resistance mediated by
p53 mutations. Drug Resist Updat. 49(100671)2019.PubMed/NCBI View Article : Google Scholar
|