Introduction
Ovarian cancer (OVC) is a deadly gynecologic malignancy (1). In 2015, 1.2 million women suffered from OVC worldwide, and 16,1100 patients succumbed to OVC (2). Extensive efforts have been made to improve treatment; cisplatin [also known as platinum diamminodichloride (DDP)]-based therapy can cause DNA impairment and induce apoptosis of OVC cells and has become the standard treatment for OVC (3). However, the majority of patients eventually develop DDP-resistant OVC with a 5-year survival rate of ~40% (1). Previous studies have reported that resistance to DDP treatment may be induced by the dysregulation of certain oncogenes and tumor suppressors in OVC cells (4-7). Therefore, it is necessary to explore the underlying mechanisms and develop novel strategies to overcome OVC resistance to DDP.
MicroRNAs (miRNAs) are evolutionarily conserved small non-protein coding RNAs consisting of ~22 nucleotides (8). miRNAs regulate the expression of their target genes post-transcriptionally by binding to their target mRNA sequences (9). miRNAs function as the regulators of various cellular processes, including cell proliferation, survival, differentiation and motility, which suggests that miRNA-based therapies mat be a promising strategy for disease treatment (10,11). Previous studies have demonstrated that miRNAs serve an important role in mediating the sensitivity of cancer cells to chemotherapeutic agents and that the dysregulation of miRNA may lead to the development of chemoresistance (12,13). For example, miR-363 reversed the resistance to the combined treatment with doxorubicin, DDP and fluorouracil in gastric cancer by negatively regulating F-box and WD repeat domain-containing 7(14); miR-539 decreased the chemoresistance to DDP in non-small cell lung cancer cells by directly targeting doublecortin-like kinase 1(15); miRNA-125a-5p increased the chemosensitivity of esophageal squamous cell carcinoma cells to DDP by inhibiting the STAT3 signaling pathway (16). In OVC, miR-335-5p reduced DDP resistance by targeting the 3'-UTR of Bcl2-like protein 2 (BCL2L2), an antiapoptotic protein of the Bcl-2 family (3); miR-205-5p promoted DDP resistance by directly regulating the PTEN/AKT pathway (17). The evidence above indicated that miRNAs may be associated with DDP resistance.
Due to the heterogeneity of OVC, differential miRNA expression and molecular mechanisms may contribute to OVC development (18). Numerous studies have reported that miR-654-3p acts as a tumor suppressor in breast (19), papillary thyroid (20) and non-small cell lung (21) cancer. In addition, miR-654-3p serves an oncogenic role in oral squamous cell carcinoma by targeting GRB2-related protein to facilitate proliferation, chemoresistance and metastasis through the Ras/mitogen-activated protein kinase signaling pathway (22). In OVC, low miR-654-3p levels have been detected in OVC cells or their exosomes compared with normal cells (23). However, whether miR-654-3p modulates DDP resistance in OVC and the underlying mechanisms still remain underexplored. The present study aimed to investigate the role and mechanism of miR-654-3p in DDP resistance in OVC.
Materials and methods
Microarray assay
The schematic diagram of all experimental procedures is provided in Fig. S1. The miRNA expression profile in the GSE58469 dataset and the mRNA expression profile in the GSE58470 dataset (24) were downloaded from the GEO (Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo) database. Data from three DDP-sensitive and three DDP-resistant IGROV-1 cell samples were available. Subsequently, differentially expressed genes (DEGs) were identified using limma package (version 3.22.7) in R language (version 3.4.1) (25). To compare the profile differences in DDP-sensitive and resistant IGROV-1 cell lines, the input values were log transformed fold-change (FC) of mRNA or miRNA expression levels. The criteria of P-value <0.05 and log2|FC|>1 were used to screen the differentially expressed miRNAs and mRNAs. The pheatmap package (version 2.0; https://davetang.org/muse/2018/05/15/making-a-heatmap-in-r-with-the-pheatmap-package/) in R language was used to generate heatmaps. The log2FC values of differentially expressed mRNAs obtained from the Limma differential analysis were used for Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment Gseaplot analysis (version 3.0) (26). The enriched signaling pathways are presented using dot plots.
Clinical specimens
A total of 20 OVC and 20 adjacent healthy ovarian tissue specimens were obtained at the time of surgery from female patients diagnosed with OVC by pathologic histology or a hysteroscopy at the Center for Reproductive Medicine, Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University between September 2014 and February 2017. Adjacent tissues were collected 2-3 cm from the tumor site. This study was approved by the Ethics Committees of Shanghai Jiao Tong University, and written informed consent was obtained from all patients. None of the patients received such preoperative therapies as radiation, chemotherapy, or immunotherapy. The average age of the patients was 52 years (range, 36-68). Tumor stages were as follows: i) stage I, 5 patient; ii) stage II, 12 patients; iii) stage III, 2 patients; and iv) stage IV, 1 patients.
Cell line and cell culture
The immortalized ovarian epithelial cell line (IOSE386) was purchased from the American Type Culture Collection (ATCC) and cultured in RPMI-1640 medium (Corning, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.). IGROV-1 and 293T cell lines were purchased from ATCC and were cultured in standard DMEM (HyClone; GE Healthcare Life Sciences) supplemented with 10% FBS, 100 U/ml penicillin and 100 mg/ml streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.). All cell lines were maintained in a 37˚C incubator with humidified atmosphere containing 95% air and 5% CO2. DDP-resistant IGROV-1 subline was established by continuous exposure to DDP. Briefly, IGROV-1 cells were exposed to incremental increases of DDP concentration; the initial DDP dose was 1.52 µM (IC80 of the sensitive cell line) for 48 h once a week. Once the cells had recovered and entered the exponential growth phase, the DDP concentration was gradually increased to 15.20 µM. After 6 months of DDP treatment for 6 cycles, the resulting resistant line IGROV-1/DDP was subcultured and treated monthly with 15.20 µM DDP to maintain the chemoresistance. IGROV-1/DDP cells were cultured without DDP for ~14 days prior to subsequent experiments.
Cell transfection
IGROV-1/DDP cells were transfected with si-QPRT (50 nM), negative control siRNA (si-NC; 50 nM), pcDNA3.1 empty plasmid (150 nM), pcDNA3.1-QPRT (150 nM), microRNA mimic (miR-mimic; 30 nM), microRNA inhibitor (miR-inhibitor; 30 nM), scrambled miR control (miR-NC; 30 nM), or co-transfected with si-QPRT (50 nM) and miR-inhibitor (30 nM) using Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.) for 48 h at 37˚C in a humidified incubator with 5% CO2. pcDNA-3.1 was purchased from Clontech Laboratories, Inc. pcDNA3.1-QPRT were synthesized by GenePharma Technology Co., Ltd, and the miR-654-3p mimic and inhibitor were purchased from Guangzhou RiboBio Co., Ltd. The sequences of si-QPRT sense, 5'-CUACUUGUGUUAUCUGUAAAU-3' and antisense, 5'-UUACAGAUAACACAAGUAGUU-3') and si-NC sense, 5'-UUCUCCGAACGUGUCACGUTT-3' and antisense, 5'-ACGUGACACGUUCGGAGAATT-3') were designed by DSIR (http://biodev.extra.cea.fr/DSIR/DSIR.html), a tool for siRNA (19 or 21 nt) and shRNA target design based on the publication by Vert et al (27) and synthesized by GenePharma Technology Co., Ltd. Except for the IGROV-1 cells, which were cultured without DDP, the untransfected IGROV-1/DDP and transfected IGROV-1/DDP cells were maintained in media containing 8 µM DDP.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA from cells (IGROV-1, IGROV-1/DDP or various transfected-IGROV-1/DDP cells) were extracted using TRIzol® reagent (Ambion; Thermo Fisher Scientific, Inc.), and total RNA was quantified using ND-2000 spectrophotometer (Thermo Fisher Scientific, Inc.). Then 1 µg of total RNA was used to synthesize cDNA using the ReverTra Ace qPCR RT Kit (Toyobo Life Science) according to the manufacturer's instructions. RT-qPCR was performed using a SYBR® RT-PCR kit (Toyobo Life Science) on the ABI 7500 Fast real-time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The primers used for RT-qPCR analysis are presented in Table I. For the detection of miR-654-3p, total miRNA was extracted from IGROV-1 cells using miRVana miRNA Isolation kit (Ambion; Thermo Fisher Scientific, Inc.) according to the standard procedure (28). The extracted miRNA was converted to cDNA using a TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) The RT reaction was incubated as follows: 30 min at 16˚C, 30 min at 42˚C, 5 min at 85˚C. RT-qPCR analysis was performed using the TaqMan 2X Universal PCR Master mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) on the ABI 7500 Fast real-time PCR system. The qPCR thermocycling conditions were as follows: 95˚C for 10 min, followed by 40 cycles of denaturation at 95˚C for 15 sec and annealing/elongation at 60˚C for 1 min. The primers used were obtained from Thermo Fisher Scientific, Inc. (miR-654-3p and U6, cat. no. 4427975). The relative expression levels of mRNA or miRNAs were normalized to those of the internal controls GAPDH or U6 using the 2-ΔΔCq method (29).
 |
Table I
mRNA primers used for reverse transcription-quantitative PCR.
|
Table I
mRNA primers used for reverse transcription-quantitative PCR.
| Gene name |
Gene bank ID |
Primer sequences (5'→3') |
Product size, bp |
| QPRT |
NM_014298.4 |
F:TGTTGAAGGATAACCATGTGGTG |
106 |
| |
|
R:CTGCTGCATTCCACTTCCA |
|
| GAPDH (human) |
NM_002046.6 |
F:TGCACCACCAACTGCTTAGC |
87 |
| |
|
R:GGCATGGACTGTGGTCATGAG |
|
Western blotting
IGROV-1 cells were lysed in RIPA buffer (Beyotime Institute of Biotechnology) containing fresh proteinase inhibitor 1X phenylmethylsulfonyl fluoride (Beyotime Institute of Biotechnology). The protein concentration was measured using a BCA kit (Beyotime Institute of Biotechnology), and equal amounts of protein were separated by SDS-PAGE (10%). Proteins were transferred to a PVDF membrane (EMD Millipore) and incubated overnight with primary antibodies against QRPT (1:1,000; Abcam; cat. no. ab171944), cleaved-caspase3 (1:1,000; Abcam; cat. no. ab32042), PI3K (1:1,000; Abcam; cat. no. ab154598), phosphorylated (p-)PI3K (1:1,000; Affinity Biologicals, Inc.; cat. no. AF3242), AKT (1:1,000; Abcam; cat. no. ab179463), p-AKT (1:1,000; Abcam; cat. no. ab215873), permeability glycoprotein (P-gp; 1:1,000; Abcam; cat. no. ab216656) and GAPDH (1:5,000; Cell Signaling Technology, Inc.; cat. no. 5174). Subsequently, the membrane was incubated with the horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (1:5,000; cat. no. ab205718; Abcam) at room temperature for 1 h, and the signals were detected using an ECL kit (Invitrogen; Thermo Fisher Scientific, Inc.; cat. no. WP20005). Protein expression levels were analyzed with Image-Pro Plus software 6.0 (Media Cybernetics, Inc.). All experiments were performed in triplicate.
MTT assay
MTT assay was performed to detect cell viability. Briefly, cells were transfected with miRNA mimic, inhibitor, si-QPRT or the corresponding negative controls following DDP treatments. At the indicated time points (24, 48, 72 and 96 h) post-treatment, the medium was replaced with fresh medium containing 200 µg/ml MTT and incubated at 37˚C for 4 h. The purple formazan crystals were dissolved with DMSO. Absorbance was measured at 570 nm using a microplate reader (Biotek Instruments, Inc.), and cell viability was calculated based on the absorbance of the control cells.
BrdU staining
BrdU staining was performed to assess the proliferation of IGROV-1 and IGROV-1/DDP cells. IGROV-1/DDP cells were transfected with si-QPRT, si-NC, miR-mimic, miR-inhibitor, miR-NC, or si-QPRT + miR-inhibitor for 24 h prior to 24-h DDP treatment. Subsequently, cells were then fixed in cold ethanol and acetic acid mixture (2:1) for 10 min at room temperature and washed using PBS (pH 7.4) supplemented with 1% Triton X-100. Then, fixed cells were incubated with 4 N HCl to denature DNA for 15 min at 37˚C and 0.1 M sodium borate (pH 8.5) was used for neutralizing. The cells were washed with 0.1% NP-40 (Beyotime Institute of Biotechnology) in PBS for three times and blocked for 30 min at room temperature. Finally, cells were incubated with 10 µM BrdU (Sigma-Aldrich; Merck KGaA) for 6 h. For immunodetection, cells were incubated with goat anti-BrdU antibody (1:1,000; cat. no. B2531; Sigma-Aldrich; Merck KGaA) for 2 h at room temperature. Cells were washed with PBS and incubated with a fluorescein-conjugated anti-human immunoglobulin antibody (Thermo Fisher Scientific, Inc.) for 1 h at room temperature. Following washing with PBS, cells were stained with DAPI (Sigma-Aldrich; Merck KGaA). Immunostaining images were captured using Axiovert 200 fluorescence microscope (Carl Zeiss AG; magnification, x100).
Hoechst 33258 staining
The morphological features of apoptotic IGROV-1/DDP cells were detected by Hoechst 33258 staining (Beyotime Institute of Biotechnology). Following 48-h transfection, the cells (IGROV-1 and IGROV-1/DDP) were washed with PBS twice and fixed with 4% paraformaldehyde for 30 min at room temperature. The cells were washed with PBS and stained with Hoechst 33258 (5 µg/ml) for 20 min at 37˚C in the dark. Subsequently, cells were washed with PBS and images of the apoptotic cells exhibiting Hoechst 33258 fluorescence were captured by a fluorescence microscope (Nikon Corporation). The percentage of apoptotic cells was semi-quantified based on the Image-Pro Plus software 6.0 (Media Cybernetics, Inc.).
Wound-healing assay
For the wound-healing assay in ovarian cancer cell lines, transfected IGROV-1/DDP cells were seeded in 96-well plates (5x103 cells/well), and artificial wounds were made using a 200 µl pipette tip. The ability of the cells to migrate was evaluated by comparing the distance between the two edges in the images captured at the beginning of the experiment (0 h) with those at 24 h. Hoechst 33258 was used to stain the cell nucleus, and phase-contrast images were captured using the LeicaDMI300B (Leica) at x40 magnification.
Luciferase assay
The predicted binding sites of QPRT and miR-654-3p were predicted by TargetScan (version 7.2; www.targetscan.org). Then the human QPRT 3'-untranslated region (UTR) containing the predicted miR-654-3p binding sites was cloned into a psi-CHECK2 vector to obtain the QPRT 3'-UTR-luciferase construct. The QPRT 3'-UTR-mutant luciferase construct was obtained from the QPRT 3'-UTR construct by mutating the complementary sequence of the miR-654-3p seed region (5'-CAGACAU-3' to 5'-AUUACGA-3'). The constructs were co-transfected with synthesized miR-654-3p miRNA mimics or mimic NC (Shanghai GenePharma Co., Ltd.) into 293T cells using Lipofectamine® 3000 (Life Technologies). At 24 h, the luciferase assay was performed using the Dual Luciferase Reporter Assay System (Promega Corporation), and the mean luciferase activity for each miRNA was calculated relative to the mean for the negative control.
Statistical analysis
The data are expressed as the mean ± SD. Statistical comparisons between two groups were performed using Student's t-test. Multiple groups were compared using one-way ANOVA, followed by Tukey's post hoc test. GraphPad Prism 6.0 (GraphPad Software, Inc.) was used for all analyses, and P<0.05 was considered to indicate a statistically significant difference.
Results
Differential expression of miRNAs and mRNAs in the DDP-resistant OVC cell subline
To identify potential regulators of DDP resistance in OVC cells, the microarray profiles from GEO datasets GSE58469 and GSE58470 were obtained. Hierarchical clustering analysis separated the samples into two groups, IGROV-1 (cisplatin-sensitive; IGROV-1_1, IGROV-1_2 and IGROV-1_3) and IGROV-1/OHP (cisplatin-resistant; IGROV-1/OHP_1, IGROV-1/OHP_2 and IGROV-1/OHP_3). The top 10 miRNAs and mRNAs with the highest FC in the expression between the two groups were used to generate the heatmaps (Fig. 1A and B). The results indicated that a set of miRNAs and mRNAs were frequently aberrantly expressed in DDP-resistant cells. miR-654-3p was downregulated (Fig. 1A) and QPRT mRNA was upregulated (Fig. 1B) in DDP-resistant OVC cells compared with their respective controls.
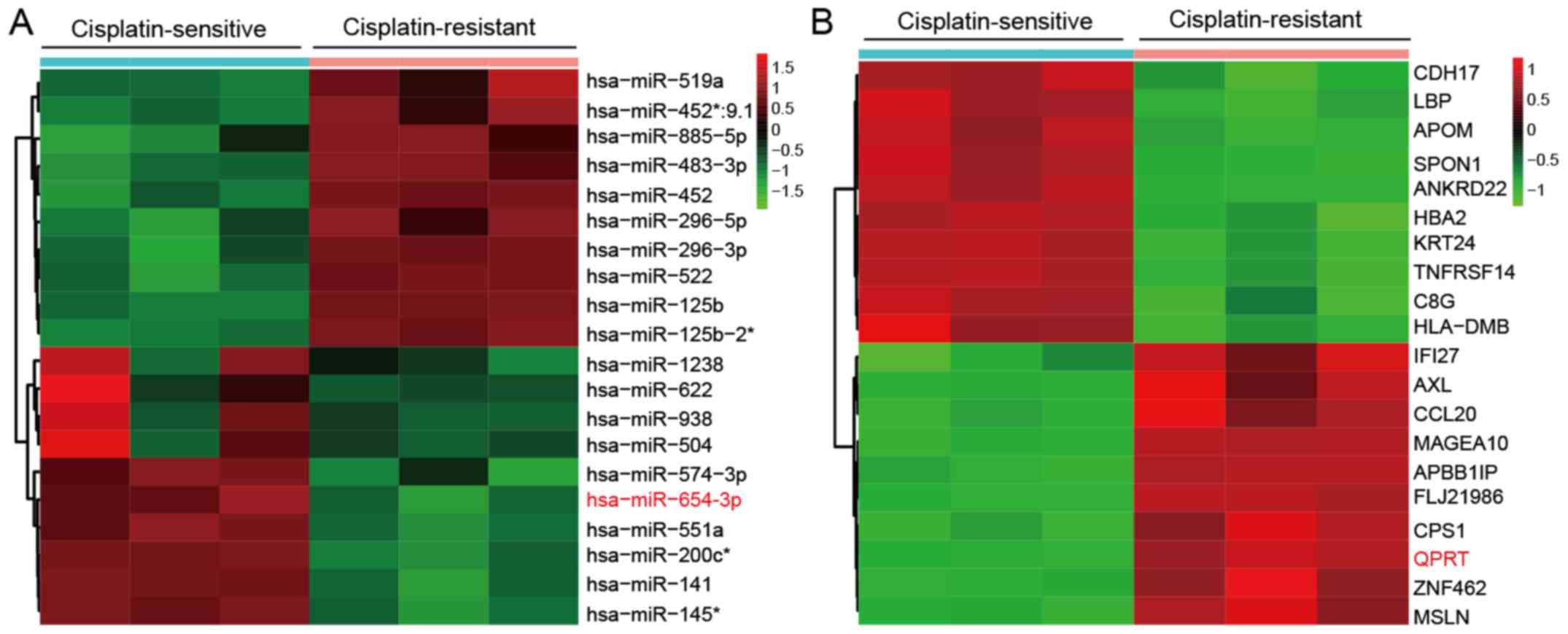 |
Figure 1
Differential expression of miRNAs and mRNAs in cisplatin-sensitive and -resistant IGROV-1 cell lines. (A) Heatmap of the top 10 upregulated (red) and downregulated (green) microRNAs from cisplatin-sensitive and resistant IGROV-1 cell lines detected by microarray analysis. (B) Heatmap of selected upregulated (red) and downregulated (green) mRNAs from the microarray analysis. miR-654-3p and QPRT are highlighted. n=3 for each condition. miR, microRNA; QPRT, quinolinate phosphoribosyl transferase.
|
KEGG analysis of differentially expressed mRNAs
To further explore the signaling pathway involved in DDP resistance of OVC cells, the differentially expressed mRNAs were subjected to KEGG analysis. The results revealed that the differentially expressed genes were enriched in several pathways, including ‘PI3K-Akt signaling pathway’ (Fig. 2A). In addition, Gseaplot analysis revealed that the running enrichment score of the PI3K-AKT signaling pathway in DDP-resistant IGROV-1 cell lines was >0, which indicated that the PI3K-AKT signaling pathway was activated in these cells (Fig. 2B).
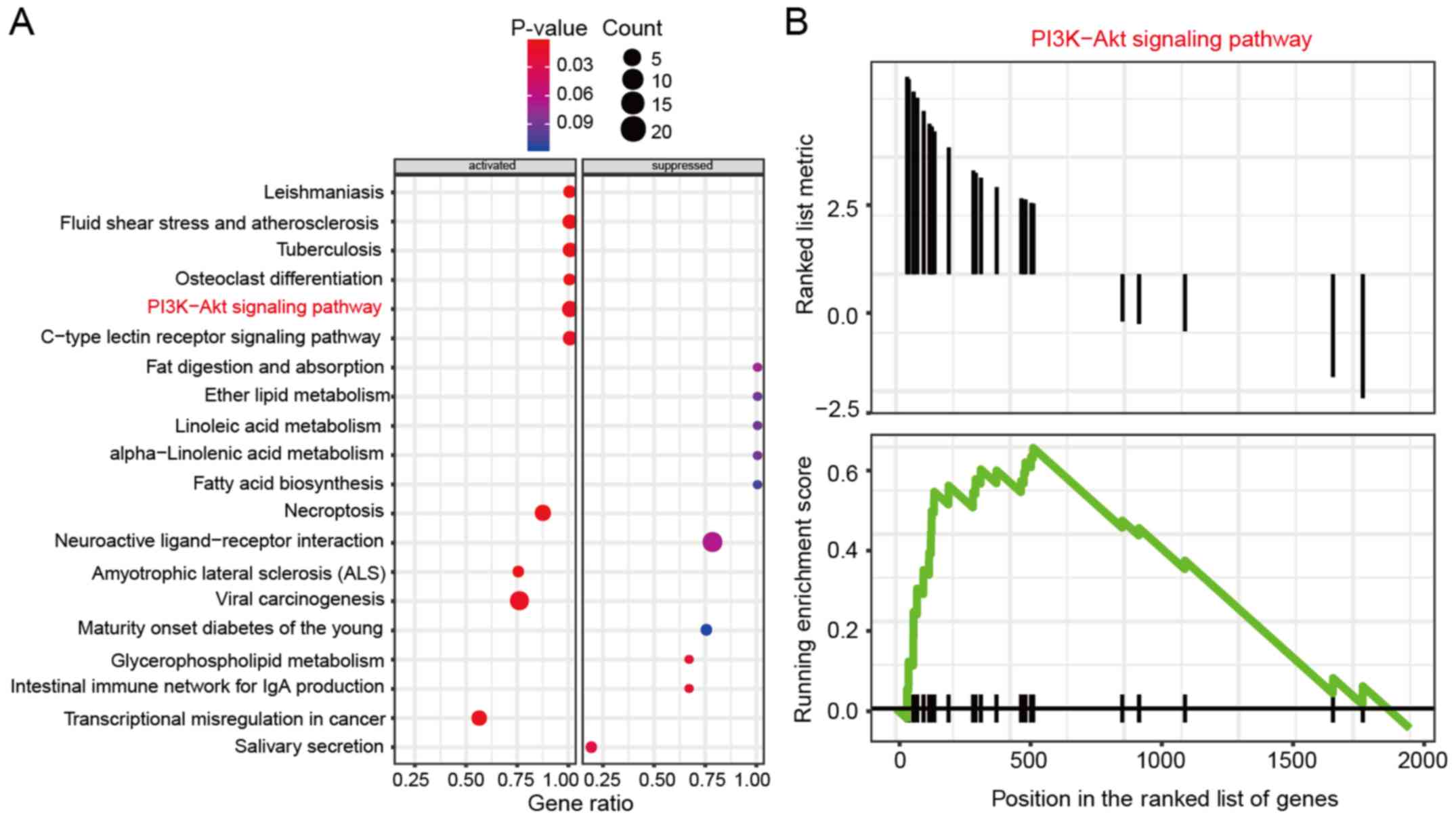 |
Figure 2
PI3K-AKT signaling pathway is activated in cisplatin-resistant IGROV-1 cell lines. (A) KEGG enrichment analysis demonstrated that the PI3K-AKT signaling pathway was activated in cisplatin-resistant IGROV-1 cell lines. (B) Gseaplot analysis revealed that the running enrichment score of PI3K-AKT signaling pathway in cisplatin-resistant IGROV-1 cell lines was >0, indicating that PI3K-AKT signaling pathway was activated.
|
miR-654-3p is downregulated in OVC tissues and cell lines
To explore the role of miR-654-3p in OVC, the expression levels of miR-654-3p in OVC and healthy tissue samples, as well as IGROV-1 and IOSE386 cells, were determined by RT-qPCR. miR-654-3p expression levels were downregulated in OVC tumor samples and IGROV-1 OVC cells compared with healthy samples and IOSE386normal human ovarian cells, respectively (Fig. 3A and B). To confirm the successful establishment of DDP-resistant IGROV-1 cell line, the IC50 of the sensitive IGROV-1 and resistant IGROV-1/DDP OVC cells were calculated; the results demonstrated that the IC50 was 7.13±0.82 µM for IGROV-1 and 23.67±0.96 µM for IGROV-1/DDP, which indicated that the IGROV-1/DDP cell line was indeed DDP-resistant (Fig. 3C). In addition, the protein expression levels of P-gp, which is responsible for DDP resistance of OVC, were detected in IGROV-1 and IGROV-1/DDP cells by western blot analysis. As presented in Fig. 3D and E, P-gp protein expression was notably increased in IGROV-1/DDP cells compared with that in IGROV-1 cells, which further demonstrated the successful establishment of a DDP-resistant cell line. To confirm the results obtained from the microarray analysis, a thorough analysis of miRNA expression in IGROV-1 and IGROV-1/DDP was performed by RT-qPCR. The results demonstrated that IGROV-1/DDP cells exhibited significantly lower miR-654-3p levels compared with IGROV-1 cells (Fig. 3F).
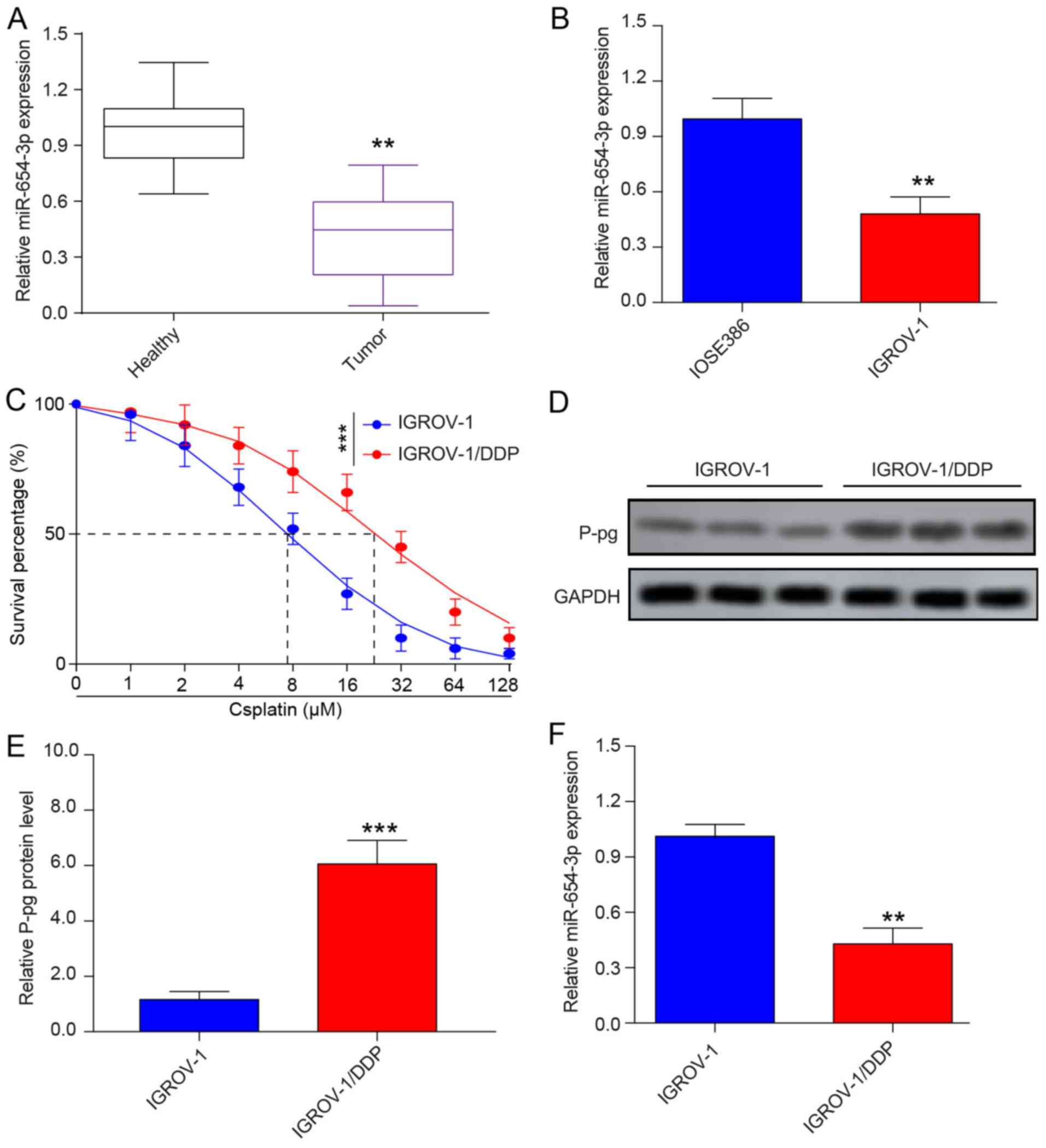 |
Figure 3
miR-654-3p is downregulated in ovarian cancer tissues and cell lines. (A) The relative miR-654-3p expression was determined by RT-qPCR in healthy tissues (n=20) and ovarian cancer tissues (n=20). (B) miR-654-3p expression levels in the ovarian cancer cell line IGROV-1 and in the normal human ovarian epithelial cell line IOSE386 were detected by RT-qPCR. (C) MTT assay of DDP-sensitive IGROV-1 and DDP-resistant IGROV-1 cells. (D and E) Western blotting was performed to examine the protein level of P-gp in IGROV-1 and IGROV-1/DDP cells. (F) RT-qPCR analysis of miR-654-3p expression levels in IGROV-1 and IGROV-1/DDP cells. **P<0.01 and ***P<0.001 vs. control or IGROV-1. miR, microRNA; RT-qPCR, reverse transcription-quantitative PCR; DDP, cisplatin; P-gp, permeability glycoprotein; IGROV-1/DDP, DDP-resistant IGROV-1 cells.
|
Overexpression of miR-654-3p enhances the DDP sensitivity of IGROV-1/DDP cells
As miR-654-3p expression is significantly downregulated in tumor tissues and cell lines of OVC, it may function as a tumor suppressor. Therefore, gain-of-function experiments were preformed. IGROV-1/DDP cells were transfected with miR-654-3p mimic and miR-NC. An increase in mature miR-654-3p was detected in the miR-654-3p mimic-transfected cells compared with miR-NC-transfected cells; no notable changes were observed in miR-654-3p expression levels following transfection with miR-NC compared with untransfected IGROV-1/DDP cells (Fig. 4A). MTT assay was performed to investigate the viability of IGROV-1/DDP cells transfected with miR-654-3p mimic. Overexpression of miR-654-3p increased the toxicity of 8 µM DDP in IGROV-1/DDP cells compared with the miR-NC-transfected group (Fig. 4B). In addition, the BrdU cell proliferation assay was performed following miR-654-3p mimic transfection; fewer cells were labeled with BrdU in the miR-654-3p overexpression group compared with cells transfected with miR-NC (Fig. 4C and D).
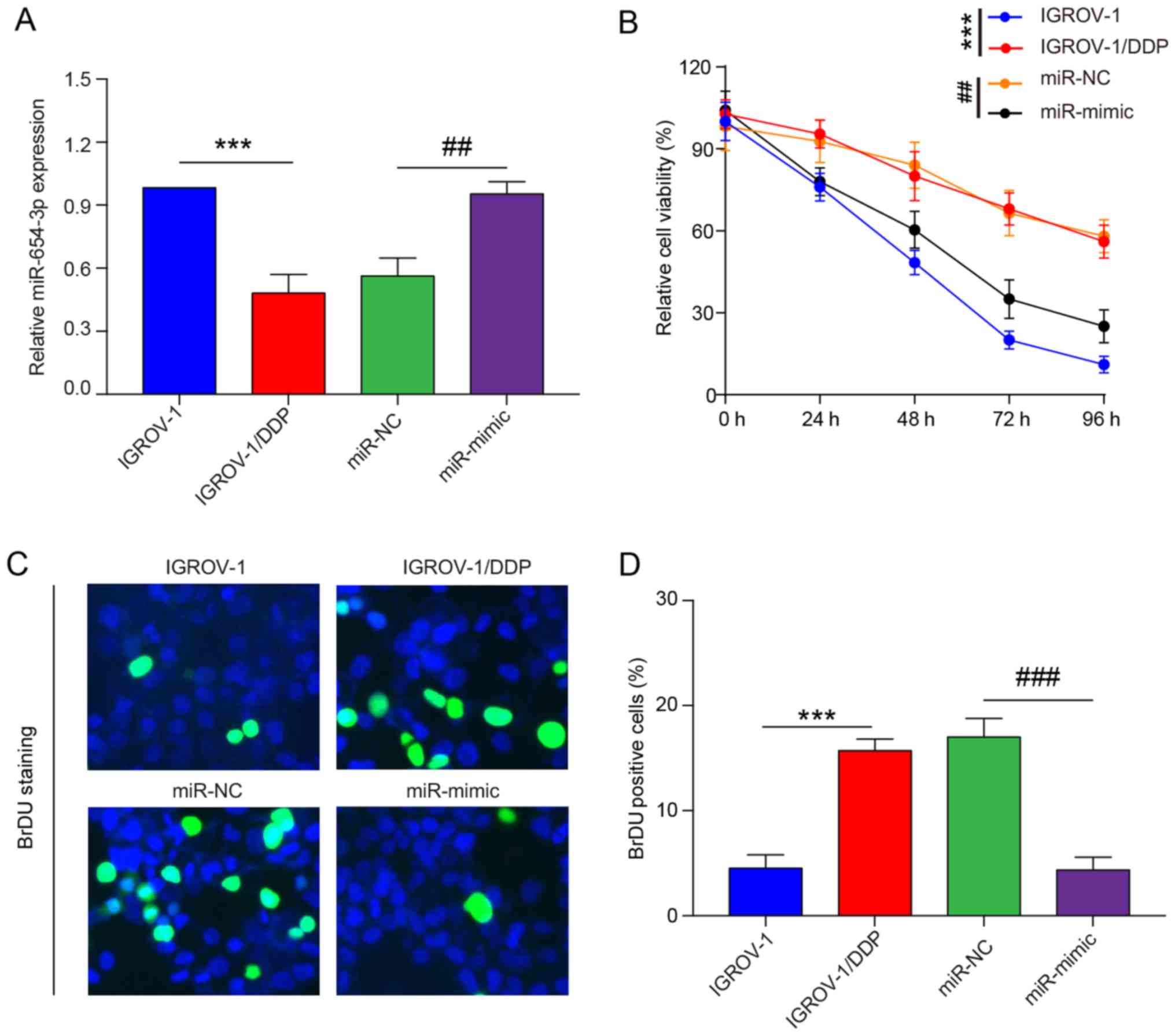 |
Figure 4
Restoration of miR-654-3p sensitizes ovarian cancer cells to cisplatin in vitro. IGROV-1/DDP cells were seeded and transfected with miR-NC and miR-mimic respectively. (A) At 48 h post-transfection, reverse transcription-quantitative PCR was performed to determine the miR-654-3p expression levels. (B) At 24 h post-transfection, cells were treated with 8 µM cisplatin. The effects of miR-654-3p on cell viability were determined at different time points (0, 24, 48, 72 and 96 h) by MTT assay. (C) At 24 h post-transfection, cells were treated with 8 µM cisplatin for 48 h, and DNA biosynthesis was detected by BrdU labelling (green). DAPI was used to stain cell nuclei (blue). Scale bar, 100 µm. (D) BrdU-positive cells were counted and subjected to statistical analysis. ***P<0.001 vs. IGROV-1; ##P<0.01 and ###P<0.001 vs. miR-NC. IGROV-1/DDP, cisplatin-resistant IGROV-1 cells; miR, microRNA; NC, negative control.
|
To determine whether apoptosis contributed to the cell proliferation inhibition by miR-654-3p, Hoechst 33258 staining was performed. As presented in Fig. 5A and B, higher levels of nucleus cleavage and chromatin condensation were observed in the miR-654-3p mimic group, and overexpression of miR-654-3p greatly increased the number of total apoptotic cells in IGROV-1/DDP compared with the miR-NC group. Wound-healing assay was performed to determine whether miR-654-3p exhibited inhibitory effects on IGROV-1/DDP cell migration; the results demonstrated that overexpression of miR-654-3p efficiently inhibited IGROV-1/DDP migration (Fig. 5C and D). In addition, the expression levels of apoptosis-related protein cleaved-caspase3 were analyzed by western blotting. Following 8 µM DDP treatment for 48 h, the expression levels of cleaved-caspase3 were higher in the miR-654-3p mimic group compared with those in the miR-NC group, which indicated that the overexpression of miR-654-3p may promote apoptosis in IGROV-1/DDP cells (Fig. 5E and F). The expression levels of p-PI3K and p-AKT were also analyzed in these samples, as the PI3K/AKT signalling pathway promotes DDP resistance in OVC (30,31). The phosphorylation levels of PI3K and AKT were high in the untransfected and miR-NC-transfected cells, which was reversed by miR-654-3p overexpression (Fig. 5E and F).
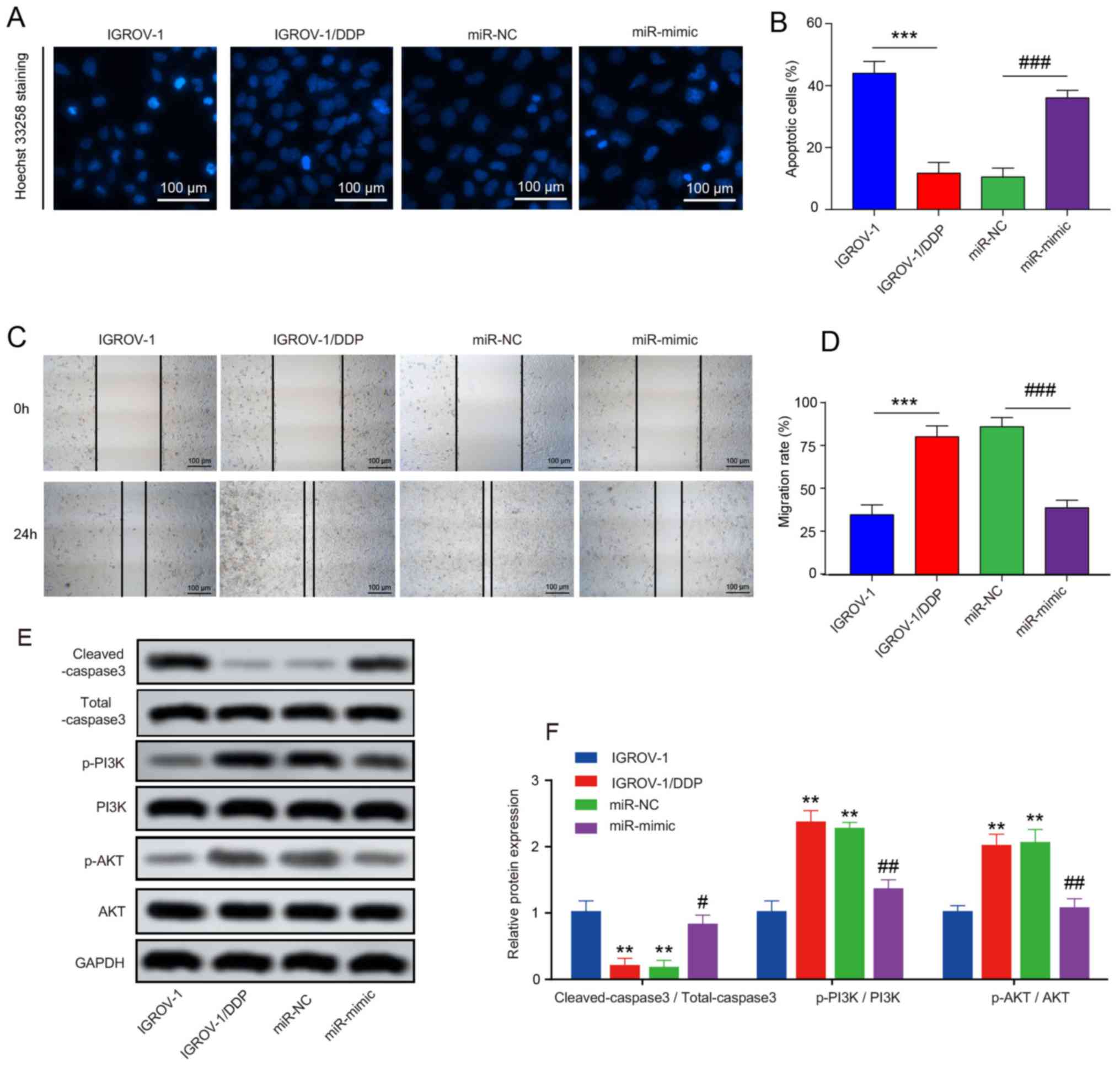 |
Figure 5
Effects of miR-654-3p upregulation on cisplatin-induced apoptosis and migration of OVC cells. (A) Hoechst-positive cells were observed under a fluorescence microscope. Normal cells displayed intact nuclei and adqulis chromatin. Shrunken cells with condensed or fragmented nuclei were considered to exhibit typical morphological features of apoptosis. Scale bar, 100 µm. (B) The percentage of apoptotic cells is expressed as the means ± SD of 3 independent experiments. (C) Wound-healing assay was performed. At 48 h, OVC cells were stained by Hoechst 33258 solution. Scale bar, 100 µm. (D) The migration rate was compared between IGROV-1/DDP cells transfected with miR-NC and miR-mimic. (E) Western blotting was used to detect the protein expression levels of cleaved-caspase3, Total caspase3, PI3K, p-PI3K, AKT and p-AKT. (F) Quantification of the western blotting results. **P<0.01 and ***P<0.001 vs. IGROV-1; #P<0.05, ##P<0.01 and ###P<0.001 vs. miR-NC. OVC, ovarian cancer; IGROV-1/DDP, cisplatin-resistant IGROV-1 cells; miR, microRNA; NC, negative control.
|
QPRT is a direct target of miR-654-3p
To further investigate the molecular mechanisms of miR-654-3p, miR-654-3p target gene prediction was performed using Targetscan. QPRT was selected for further analysis due to its antiapoptotic properties (32). A miR-654-3p binding site in QPRT 3'-UTR was identified (Fig. 6A) and further validated using a luciferase assay. The results domenstrated that co-transfection of the pGL3 reporter plasmid with miR-654-3p mimic exhibited a ~45% reduction in luciferase activity compared with miR-NC. In addition, the inhibition of luciferase activity by miR-654-3p was reversed when the QPRT 3'-UTR was mutated (Fig. 6B).
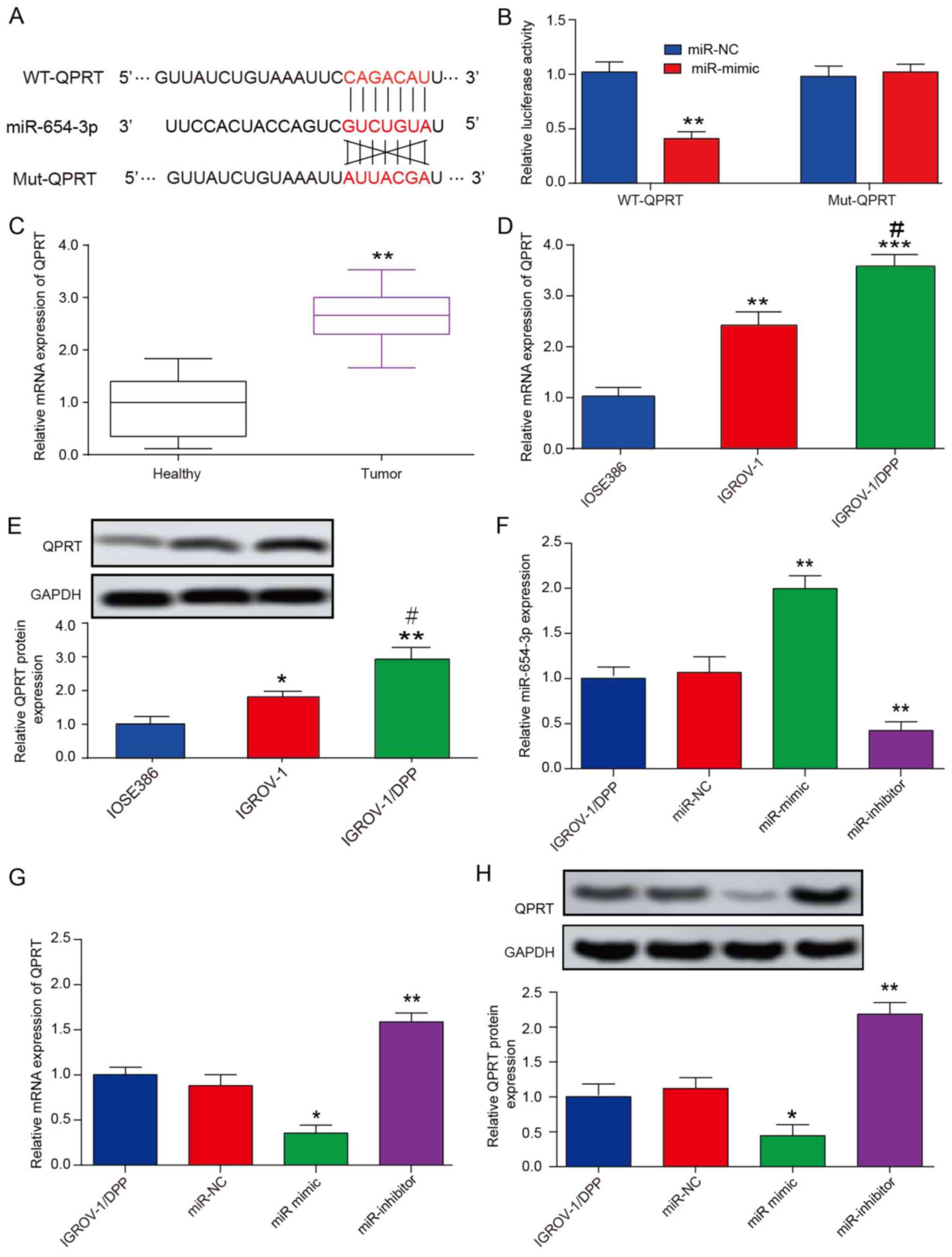 |
Figure 6
miR-654-3p directly targets QPRT. (A) Predicted binding sites of miR-654-3p to QPRT 3'-UTR. (B) Luciferase activity of 293T cells transfected with WT or Mut 3'-UTR of QPRT plus miR-654-3p mimic and scramble control (miR-NC). **P<0.01 vs. miR-NC group. (C) RT-qPCR analysis of the QPRT mRNA levels in healthy (n=20) and ovarian cancer (n=20) tissues. **P<0.01 vs. Healthy group. (D) RT-qPCR analysis of the expression levels of QPRT in IOSE386, IGROV-1 and IGROV-1/DDP cell lines. **P<0.01 and ***P<0.001 vs. IOSE386 group; #P<0.05 vs. IGROV-1 group. (E) Western blotting detected that QPRT protein expression was significantly increased in IGROV-1/DDP cells compared with that in IGROV-1. *P<0.05 and **P<0.01 vs. IOSE386 group; #P<0.05 vs. IGROV-1 group. (F) RT-qPCR was used to determine miR-654-3p expression levels in cells transfected with miR-NC, miR-654-3p mimic and miR-654-3p inhibitor. **P<0.01 vs. miR-NC group. (G) RT-qPCR and (H) western blotting were used to detect the mRNA and protein expression levels of QPRT following 48-h transfection. *P<0.05, **P<0.01 vs. miR-NC group. Data are presented as the mean ± SD. QPRT, quinolinate phosphoribosyl transferase; UTR, untranslated region; WT, wild-type; Mut, seed sequence-mutated; IGROV-1/DDP, cisplatin-resistant IGROV-1 cells; miR, microRNA; NC, negative control; RT-qPCR, reverse transcription-quantitative PCR.
|
Western blotting and RT-qPCR results demonstrated that QPRT was upregulated in OVC samples and IGROV-1 cells compared with adjacent healthy tissues and IOSE386 human normal ovarian cells, respectively (Fig. 6C and D). In addition, QPRT expression was also upregulated in IGROV-1/DDP cells compared with IGROV-1 cells at the mRNA and protein levels (Fig. 6D and E). The expression levels of miR-654-3p and QPRT following transfection with the miR-654-3p mimic or inhibitor in IGROV-1/DDP cells were measured by qRT-PCR (Fig. 6F-H). Of note, miR-654-3p overexpression reduced QPRT mRNA and protein expression levels in IGROV-1/DDP, whereas downregulation of miR-654-3p was associated with the upregulation QPRT (Fig. 6G and H). These results indicated that miR-654-3p may target QPRT by transcriptional and translational regulation.
Downregulation of miR-654-3p enhances DDP resistance in QPRT-knockdown OVC cells
To investigate the functional involvement of QPRT in miR-654-3p-mediated DDP resistance, si-QPRT, miR-654-3p inhibitor and si-QPRT + miR-654-5p inhibitor were transfected into IGROV-1/DDP cells. Following transfection, the mRNA and protein levels of QPRT were signifcantly increased in the si-QPRT + miR-inhibitor group compared with those in the si-QPRT group (Fig. 7A and F). The results of the MTT assay demonstrated that following DDP treatment, the viability of IGROV-1/DDP cells transfected with si-QPRT was decreased compared with that of the si-NC group, whereas cells co-transfected with miR-654-5p inhibitor and si-QPRT exhibited a moderate change in the cell viability compared with si-QPRT (Fig. 7B). Consistently with the results of cell viability analysis, the proportion of BrdU-positive cells at 24 h in the si-QPRT group was decreased compared with the si-NC group, but signifcantly increased in the si-QPRT + miR-654-5p inhibitor group compared with that in the si-QPRT group (Figs. 7C and S2A), which indicated that miR-654-5p inhibitor may have rescued the suppressive effect of si-QPRT on IGROV-1/DDP cell proliferation. Subsequently, Hoechst 33528 dye staining was performed to detect cell apoptosis. The percentage of Hoechst-positive cells was reduced from 34.27±2.42% in the si-QPRT group to 7.85±2.66% in the si-QPRT + miR-654-5p inhibitor group (Figs. 7D and S2B). The increased protein expression level of cleaved caspase-3 induced by si-QPRT was also reduced in the si-QPRT + miR-654-5p inhibitor group (Figs. 7F and S2C) In addition, knockdown of QPRT signifcantly suppressed the cell migratory capacity, but an increased migration rate was observed in the si-QPRT + miR-654-5p inhibitor group compared with the si-QPRT group (Fig. 7E). In addition, miR-654-3p- and QPRT-induced PI3K and AKT phosphorylation was evaluated by western blot analysis; the levels of p-PI3K and p-AKT were lower in cells transfected with si-QPRT following DDP treatment compared with those in cells transfected with si-NC. By contrast, the PI3K and AKT phosphorylation levels were partially reversed in the si-QPRT + miR-654-5p inhibitor group (Fig. 7F). In addition, QPRT overexpression increased the levels of p-PI3K, p-AKT and P-gp and inhibited the protein expression of cleaved-caspase3 (Fig. 8A and B). Taken together, these results revealed that miR-654-3p may enhances the DDP sensitivity in OVC by suppressing QPRT.
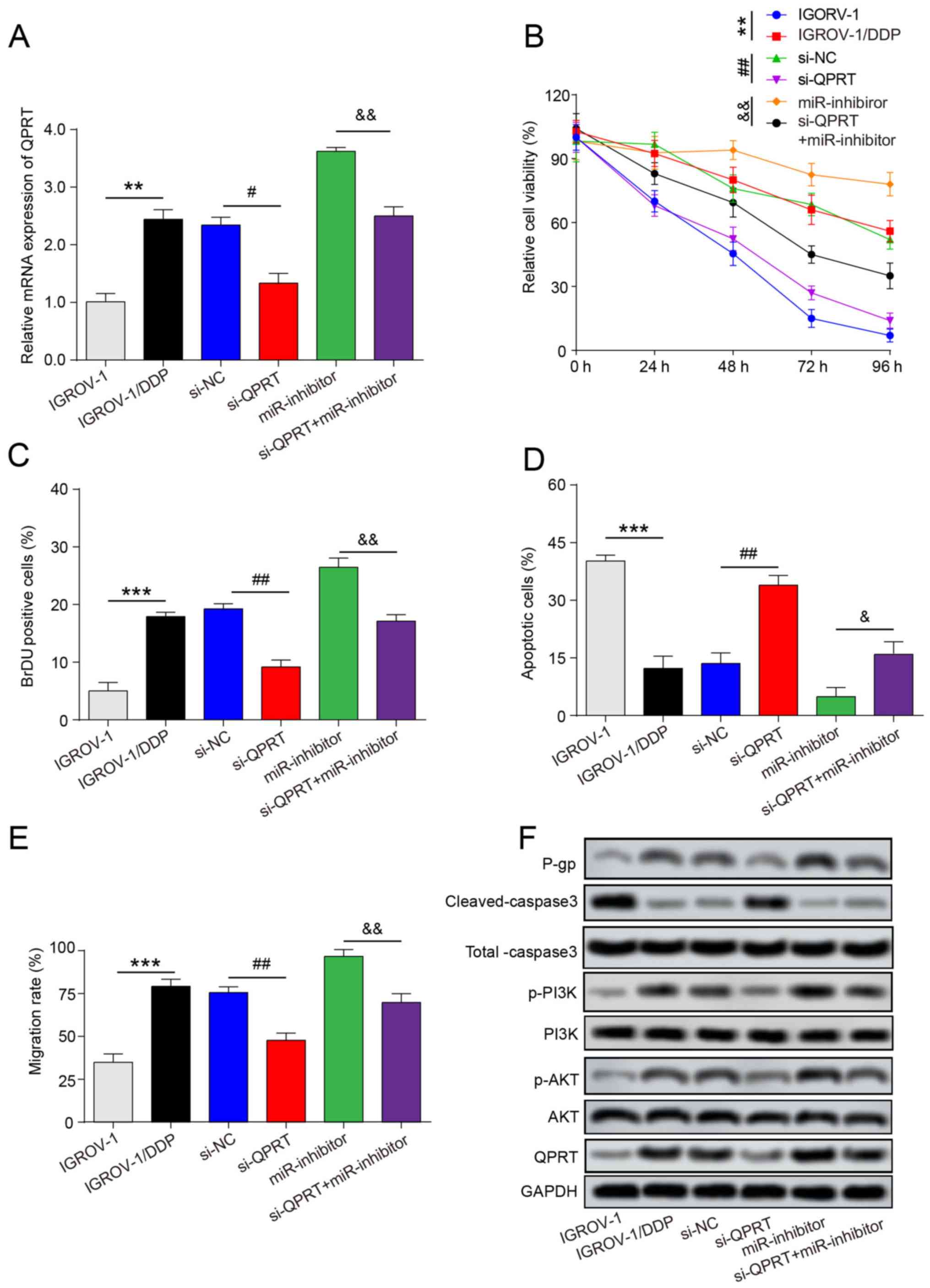 |
Figure 7
QPRT is involved in miR-654-3p suppressed cisplatin resistance of IGROV-1/DDP cells. IGROV-1/DDP cells were transfected with si-NC, si-QPRT, miR-inhibitor and si-QPRT + miR-inhibitor. (A) Post-transfection, reverse transcription-quantitative PCR was used to detect the mRNA expression of QPRT. (B) Following transfection and cisplatin treatment, MTT assay was used to examine cell viability. (C) Following transfection and cisplatin treatment, DNA biosynthesis was detected by BrdU labelling, and BrdU-positive cells were counted and subjected to statistical analysis. (D) Hoechst 33258 staining was performed, and the percentage of apoptotic cells is expressed as the means ± SD of 3 independent experiments. (E) Cell migration was detected by wound-healing assay. (F) Western blotting was used to detect the protein expression levels of P-gp, Total caspase3, cleaved-caspase3, PI3K, p-PI3K, AKT, p-AKT and QPRT. **P<0.01 and ***P<0.001 vs. IGROV-1; #P<0.05 and ##P<0.01 vs. si-NC; &P<0.05 and &&P<0.01 vs. miR-inhibitor. QPRT, quinolinate phosphoribosyl transferase; IGROV-1/DDP, cisplatin-resistant IGROV-1 cells; miR, microRNA; si-NC, negative control small interfering RNA; si-QPRT, small interfering RNA targeting QPRT; P-gp, permeability glycoprotein.
|
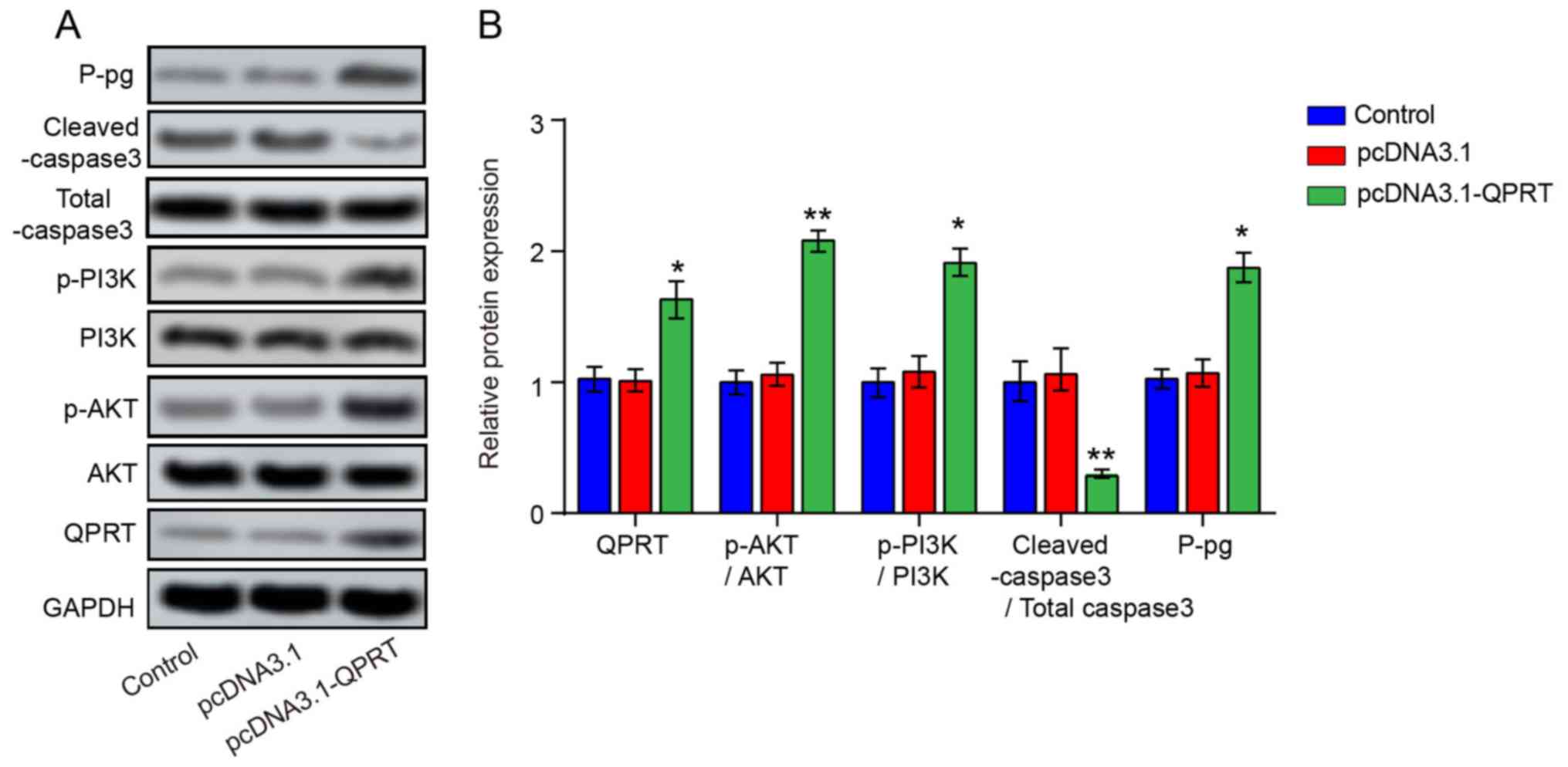 |
Figure 8
QPRT overexpression promotes P-gp protein expression and activates the PI3K/AKT signaling pathway of IGROV-1/DDP cells. (A) QPRT overexpression promoted PI3K, p-AKT and P-gp expression and inhibited cleaved-caspase3 expression. (B) Quantification of QPRT, PI3K, p-AKT, P-gp, Total caspase3 and cleaved-caspase3 protein expression. *P<0.05 and **P<0.01 vs. pcDNA3.1. QPRT, quinolinate phosphoribosyl transferase; P-gp, permeability glycoprotein; IGROV-1/DDP, cisplatin-resistant IGROV-1 cells.
|
Discussion
DDP is a first-line chemotherapeutic agent used in the treatment of OVC. However, ~25% of patients develop recurrence and metastasis within 6 months after chemotherapy due to DDP resistance, which is a major challenge in OVC treatment (1). Although previous studies have demonstrated that aberrant expression of miRNAs is associated with DDP resistance (33,34), the regulatory mechanism of miR-654-3p underlying OVC chemoresistance remains unknown. The results of the present study demonstrated that downregulation of miR-654-3p may be involved in DDP resistance in OVC. Ectopic expression of miR-654-3p significantly inhibited the proliferation, survival and migration of OVC cells and increased the sensitivity of IGROV-1/DDP cells to DDP by targeting QPRT. In addition to QPRT, miR-654-3p also mediated the PI3K/AKT signaling pathway, which is associated with DDP resistance of patients with OVC. The results of the present study provide a novel option for therapeutic intervention of DDP resistance.
Multiple studies have demonstrated that miRNA dysregulation is involved in DDP resistance in OVC. let-7e was significantly downregulated in DDP-resistant OVC cells, whereas overexpression of let-7e enhanced the sensitivity of DDP-resistant OVC cells to DDP (35). Zhang et al (36) reported low expression levels of miR-1294 in DDP-resistant tissues and cells, revealing the role of miR-1294 in DDP resistance. Vera et al (37) demonstrated that DNA methylation of miR-7 participated in the OVC DDP resistance through MAF bZIP transcription factor G (MAFG); Yu et al (33) reported the mediation of DDP resistance in OVC by miR-21 by targeting PTEN. Additionally, Li et al (38) demonstrated that miR-146a-5p facilitated OVC DDP resistance by suppressing several antiapoptotic genes, including baculoviral IAP repeat containing (BIRC), BIRC2, BCL2L2 and X-linked inhibitor of apoptosis. Overexpression of miR-139-5p can reverse DDP resistance in OVC by targeting C-Jun (39). However, the expression levels of miR-654-3p in OVC samples have not been studied previously. The results of the present study demonstrated that miR-654-3p levels were significantly downregulated in DDP-resistant ovarian cancer cells. In addition, overexpression of miR-654-3p increased the DDP sensitivity of DDP-resistant OVC cells in vitro.
miRNAs normally function as negative regulators of gene expression by binding to the 3'-UTRs of their target mRNAs (40). In the present study, TargetScan was used to identify and characterize the targets of miR-654-3p. Based on the top10 differentially overexpressed genes identified by the microarray assay, QPRT, which is a key enzyme for de novo synthesis of nicotinamide adenine dinucleotide (NAD+) (41), was selected. The results of further analysis revealed a negative association between QPRT and miR-654-3p levels in OVC cells. Luciferase reporter assay and other in vitro assays based on ectopic modulation of miR-654-3p expression in cultured cells demonstrated that miR-654-3p directly targeted QPRT. QPRT has previously been reported to be involved in several tumors, including glioma and leukemia. QPRT has been demonstrated to enhance the resistance of glioma cells to oxidative stress and radiation by activating the NAD+ salvage pathway (42). In addition, antiapoptotic QPRT upregulation in leukemic K562 cells enhanced their resistance to imatinib (32). However, a comprehensive study on the function and mechanism of QPRT is still lacking and has not been reported in OVC. The results of the present study demonstrated that miR-654-3p downregulated the expression of QPRT. siRNA knockdown of QPRT decreased the viability, proliferation and migratory capacity of IGROV-1/DDP cells. In addition, QPRT-knockout cells exhibited higher levels of apoptosis and were more susceptible to DDP treatment compared with the control group. The co-application of si-QPRT and miR-654-3p mimic independently reduced DDP resistance. The effects of si-QPRT on cellular function were reversed by the miR-654-3p inhibitor. Thus, the results of the present study suggest that the suppressive effects of miR-654-3p on the survival and chemoresistance of OVC cells may have occurred through the inhibition of QPRT expression.
QPRT protein has been reported to bind to the active caspase3, which decreases the caspase3 activity in the presence or absence of apoptotic inducers (43). siRNA knockdown of QPRT increases the likelihood of spontaneous cell death (43). Consistently, in the present study, the expression level of cleaved-caspase3 was increased following miR-654-3p transfection in IGROV-1/DDP cells, and QPRT-deficient cells exhibited higher cleaved-caspase3 expression compared with the control group. In addition, miR-654-3p and QPRT regulated PI3K/AKT signaling. Recently, PI3K/AKT signaling has been demonstrated to be involved in the pathogenesis and DDP chemoresistance in OVC (30,31,44). The results of the present study demonstrated that overexpression of miR-654-3p could inhibit the activation of PI3K/AKT pathways, and targeting the miR-654-3p/QPRT/PI3K/AKT axis may be significant to overcome the problem of DDP resistance.
A previous study has suggested that a reduction in QPRT, which is the only enzyme catabolizing quinolinic acid (QUIN), leads to an accumulation of QUIN, which in turn decreases NAD+ production (35). NAD+ is a cofactor for enzymes such as the DNA repair protein poly (ADP-ribose) polymerase (PARP). Therefore, reduced NAD+ production can inhibit PARP activity (32,36). In addition, Teng et al (45) have reported that PARP2 knockdown by siRNA resulted in decreased expression of PI3K, AKT and p70S6K mRNA and protein in cervical cancer in vitro. However, whether the PI3K-AKT pathway is the direct effector of QPRT, or QPRT achieves PI3K-AKT activation through regulating other molecules such as PARP needed further investigation. P-gp is an ATP-dependent drug efflux pump for xenobiotic compounds with broad substrate specificity; it is responsible for decreased drug accumulation in multidrug-resistant cells and often mediates the development of resistance to anticancer drugs (46). The results of the present study demonstrated that enhanced miRNA-654-3p inhibited P-gp by targeting QPRT, which may contribute to the sensitivity of IGROV-1/DPP cells to DPP.
In summary, the present study demonstrated that downregulation of miR-654-3p contributed to DDP resistance in IGROV-1 cells. Restoration miR-654-3p through ectopic expression significantly sensitized IGROV-1/DPP cells to DDP treatment in vitro. The underlying mechanisms may be regulated through QPRT mRNA. In addition to QPRT, miR-654-3p also inactivated the PI3K/AKT signaling. Taken together, these results suggested that miR-654-3p may be a biomarker for evaluating responsiveness to DDP treatment and a potential therapeutic target in OVC.
Supplementary Material
Figure S1. Schematic diagram of the experimental procedure. QPRT, quinolinate phosphoribosyl transferase; miR, microRNA; IGROV.1/DDP, cisplatin.resistant IGROV.1 cells.
Figure S2. (A) Representative images of BrDU staining. (B) Hoechest 33258 staining. (C) Wound healing assay after different transfection. Scale bar, 100 μm.
Acknowledgements
Not applicable.
Funding
The present study was supported in part by the Shanghai Commission of Science and Technology (grant no. 17DZ2271100).
Authors' contributions
YCN substantially contributed to the conception and design of the study. JT and XFS acquired, analyzed and interpreted the data. YN drafted the manuscript. TZ designed the research, performed some experiments and revised the manuscript critically for important intellectual content and acquired funding. All authors provided final approval of the version to be published.
Availabilty of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
All procedures performed in studies involving human participants approved by the Ethics Committee of The School of Medicine, Shanghai Jiao Tong University (Shanghai, China). Informed consent was obtained from all individual participants included in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
1
|
Cao L, Wan Q, Li F and Tang CE: miR-363 inhibits cisplatin chemoresistance of epithelial ovarian cancer by regulating snail-induced epithelial-mesenchymal transition. BMB Rep. 51:456–461. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
GBD 2015 LRI Collaborators;. Troeger C, Forouzanfar M, Rao PC, Khalil I, Brown A, Swartz S, Fullman N, Mosser J, Thompson RL, et al: Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory tract infections in 195 countries: A systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect Dis. 17:1133–1161. 2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Liu R, Guo H and Lu S: miR-335-5p restores cisplatin sensitivity in ovarian cancer cells through targeting BCL2L2. Cancer Med. 7:4598–4609. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Feng X, Liu N, Deng S, Zhang D, Wang K and Lu M: miR-199a modulates cisplatin resistance in ovarian cancer by targeting Hif1alpha. Onco Targets Ther. 10:5899–5906. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Liu Y, Han S, Li Y, Liu Y, Zhang D, Li Y and Zhang J: MicroRNA-20a contributes to cisplatin-resistance and migration of OVCAR3 ovarian cancer cell line. Oncol Lett. 14:1780–1786. 2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Teng Y, Zuo X, Hou M, Zhang Y, Li C, Luo W and Li X: A double-negative feedback interaction between microRNA-29b and DNMT3A/3B contributes to ovarian cancer progression. Cell Physiol Biochem. 39:2341–2352. 2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Zou J, Yin F, Wang Q, Zhang W and Li L: Analysis of microarray-identified genes and microRNAs associated with drug resistance in ovarian cancer. Int J Clin Exp Pathol. 8:6847–6858. 2015.PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 116:281–297. 2004.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Inoue K, Deng Z, Chen Y, Giannopoulou E, Xu R, Gong S, Greenblatt MB, Mangala LS, Lopez-Berestein G, Kirsch DG, et al: Bone protection by inhibition of microRNA-182. Nat Commun. 9(4108)2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Hayes J, Peruzzi PP and Lawler S: MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol Med. 20:460–469. 2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Olive V, Minella AC and He L: Outside the coding genome, mammalian microRNAs confer structural and functional complexity. Sci Signal. 8(re2)2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Ju J: Implications of miRNAs in colorectal cancer chemoresistance. Int Drug Discov. 2011(pii: 2063)2011.PubMed/NCBI
|
|
13
|
Kutanzi KR, Yurchenko OV, Beland FA, Checkhun VF and Pogribny IP: MicroRNA-mediated drug resistance in breast cancer. Clin Epigenetics. 2:171–185. 2011.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Siddik ZH: Cisplatin Mode of cytotoxic action and molecular basis of resistance. Oncogene. 22:7265–7279. 2003.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Deng H, Qianqian G, Ting J and Aimin Y: miR-539 enhances chemosensitivity to cisplatin in non-small cell lung cancer by targeting DCLK1. Biomed Pharmacother. 106:1072–1081. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Zhao Y, Ma K, Yang S, Zhang X, Wang F, Zhang X, Liu H and Fan Q: MicroRNA-125a-5p enhances the sensitivity of esophageal squamous cell carcinoma cells to cisplatin by suppressing the activation of the STAT3 signaling pathway. Int J Oncol. 53:644–658. 2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Shi X, Xiao L, Mao X, He J, Ding Y, Huang J, Peng C and Xu Z: miR-205-5p mediated downregulation of PTEN contributes to cisplatin resistance in C13K human ovarian cancer cells. Front Genet. 9(555)2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Deb B, Uddin A and Chakraborty S: miRNAs and ovarian cancer: An overview. J Cell Physiol. 233:3846–3854. 2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Tan YY, Xu XY, Wang JF, Zhang CW and Zhang SC: miR-654-5p attenuates breast cancer progression by targeting EPSTI1. Am J Cancer Res. 6:522–532. 2016.PubMed/NCBI
|
|
20
|
Geraldo MV, Nakaya HI and Kimura ET: Down-regulation of 14q32-encoded miRNAs and tumor suppressor role for miR-654-3p in papillary thyroid cancer. Oncotarget. 8:9597–9607. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Xu C, Zheng Y, Lian D, Ye S, Yang J and Zeng Z: Analysis of microRNA expression profile identifies novel biomarkers for non-small cell lung cancer. Tumori. 101:104–110. 2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Lu M, Wang C, Chen W, Mao C and Wang J: miR-654-5p targets GRAP to promote proliferation, metastasis, and chemoresistance of oral squamous cell carcinoma through Ras/MAPK signaling. DNA Cell Biol. 37:381–388. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zhang S, Zhang X, Fu X, Li W, Xing S and Yang Y: Identification of common differentially-expressed miRNAs in ovarian cancer cells and their exosomes compared with normal ovarian surface epithelial cell cells. Oncol Lett. 16:2391–2401. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Arrighetti N, Cossa G, De Cecco L, Stucchi S, Carenini N, Corna E, Gandellini P, Zaffaroni N, Perego P and Gatti L: PKC-alpha modulation by miR-483-3p in platinum-resistant ovarian carcinoma cells. Toxicol Appl Pharmacol. 310:9–19. 2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Smyth GK: Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 3(Article3)2004.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Yao J, Zhang X, Li J, Zhao D, Gao B, Zhou H, Gao S and Zhang L: Silencing TRIP13 inhibits cell growth and metastasis of hepatocellular carcinoma by activating of TGF-β1/smad3. Cancer Cell Int. 18(208)2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Vert JP, Foveau N, Lajaunie C and Vandenbrouck Y: An accurate and interpretable model for siRNA efficacy prediction. BMC Bioinformatics. 7(520)2006.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Wang Z and Zhu F: MicroRNA-100 is involved in shrimp immune response to white spot syndrome virus (WSSV) and Vibrio alginolyticus infection. Sci Rep. 7(42334)2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Amini-Farsani Z, Sangtarash MH, Shamsara M and Teimori H: miR-221/222 promote chemoresistance to cisplatin in ovarian cancer cells by targeting PTEN/PI3K/AKT signaling pathway. Cytotechnology. 70:203–213. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Bugide S, Gonugunta VK, Penugurti V, Malisetty VL, Vadlamudi RK and Manavathi B: HPIP promotes epithelial-mesenchymal transition and cisplatin resistance in ovarian cancer cells through PI3K/AKT pathway activation. Cell Oncol (Dordr). 40:133–144. 2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Ullmark T, Montano G, Jarvstrat L, Jernmark Nilsson H, Håkansson E, Drott K, Nilsson B, Vidovic K and Gullberg U: Anti-apoptotic quinolinate phosphoribosyltransferase (QPRT) is a target gene of Wilms' tumor gene 1 (WT1) protein in leukemic cells. Biochem Biophys Res Commun. 482:802–807. 2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Yu X, Zheng H, Chan MT and Wu WK: Modulation of chemoresponsiveness to platinum-based agents by microRNAs in cancer. Am J Cancer Res. 7:1769–1778. 2017.PubMed/NCBI
|
|
34
|
Samuel P, Pink RC, Brooks SA and Carter DR: miRNAs and ovarian cancer: A miRiad of mechanisms to induce cisplatin drug resistance. Expert Rev Anticancer Ther. 16:57–70. 2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Cai J, Yang C, Yang Q, Ding H, Jia J, Guo J, Wang J and Wang Z: Deregulation of let-7e in epithelial ovarian cancer promotes the development of resistance to cisplatin. Oncogenesis. 2(e75)2013.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Zhang Y, Huang S, Guo Y and Li L: miR-1294 confers cisplatin resistance in ovarian Cancer cells by targeting IGF1R. Biomed Pharmacother. 106:1357–1363. 2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Vera O, Jimenez J, Pernia O, Rodriguez-Antolin C, Rodriguez C, Sanchez Cabo F, Soto J, Rosas R, Lopez-Magallon S, Esteban Rodriguez I, et al: DNA methylation of miR-7 is a mechanism involved in platinum response through MAFG overexpression in cancer cells. Theranostics. 7:4118–4134. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Li X, Jin Y, Mu Z, Chen W and Jiang S: MicroRNA-146a-5p enhances cisplatininduced apoptosis in ovarian cancer cells by targeting multiple anti-apoptotic genes. Int J Oncol. 51:327–335. 2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Jiang Y, Jiang J, Jia H, Qiao Z and Zhang J: Recovery of miR-139-5p in ovarian cancer reverses cisplatin resistance by targeting C-Jun. Cell Physiol Biochem. 51:129–141. 2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Calin GA and Croce CM: MicroRNA signatures in human cancers. Nat Rev Cancer. 6:857–866. 2006.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Poyan Mehr A, Tran MT, Ralto KM, Leaf DE, Washco V, Messmer J, Lerner A, Kher A, Kim SH, Khoury CC, et al: De novo NAD+ biosynthetic impairment in acute kidney injury in humans. Nat Med. 24:1351–1359. 2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Sahm F, Oezen I, Opitz CA, Radlwimmer B, von Deimling A, Ahrendt T, Adams S, Bode HB, Guillemin GJ, Wick W and Platten M: The endogenous tryptophan metabolite and NAD+ precursor quinolinic acid confers resistance of gliomas to oxidative stress. Cancer Res. 73:3225–3234. 2013.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Ishidoh K, Kamemura N, Imagawa T, Oda M, Sakurai J and Katunuma N: Quinolinate phosphoribosyl transferase, a key enzyme in de novo NAD(+) synthesis, suppresses spontaneous cell death by inhibiting overproduction of active-caspase-3. Biochim Biophys Acta. 1803:527–533. 2010.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Liu M, Bamodu OA, Huang WC, Zucha MA, Lin YK, Wu ATH, Huang CC, Lee WH, Yuan CC, Hsiao M, et al: 4-Acetylantroquinonol B suppresses autophagic flux and improves cisplatin sensitivity in highly aggressive epithelial cancer through the PI3K/Akt/mTOR/p70S6K signaling pathway. Toxicol Appl Pharmacol. 325:48–60. 2017.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Teng P, Jiao Y, Hao M and Tang X: microRNA-383 suppresses the P13K-AKT-MTOR signaling pathway to inhibit development of cervical cancer via downregulating PARP2. J Cell Biochem. 117:5243–5252. 2018.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Zhang H, Gong J, Zhang H and Kong D: Induction of apoptosis and reversal of permeability glycoprotein-mediated multidrug resistance of MCF-7/ADM by ginsenoside Rh2. Int J Clin Exp Pathol. 8:4444–4456. 2015.PubMed/NCBI
|






















